Abstract
Background & objectives:
Streptococcus pneumoniae (pneumococcus) is a highly invasive extracellular pathogen that causes diseases such as pneumonia, otitis media and meningitis. This study was undertaken to determine the serotype diversity and penicillin susceptibility of S. pneumoniae isolated from paediatric patients in a tertiary teaching hospital in Malaysia.
Methods:
A total of 125 clinical isolates collected from January 2013 to May 2015 were serotyped using seven sequential multiplex polymerase chain reactions. The susceptibility of these isolates to penicillin was also investigated.
Results:
Serotypes detected among the isolates were serotypes 3, 6A/B, 6C, 11/A/D/F, 15A/F, 19A, 19F, 23A, 23F, 34. Serotypes 19F and 6A/B were the most prevalent serotypes detected. Most of the S. pneumoniae were isolated from nasopharyngeal samples of children below five years of age. Majority of the isolates were penicillin susceptible. Only 5.6 per cent of the isolates were non-susceptible to penicillin, mostly of serotype 19F.
Interpretation & conclusions:
Our study revealed the distribution of various serotypes in S. pneumoniae isolates obtained from children in a teaching hospital at Kuala Lumpur, Malaysia and decreasing rates of penicillin resistance among them. The shifts in serotypes and susceptibility to penicillin from time to time have been observed. Continuous monitoring and surveillance are pivotal for better infection control and management of pneumococcal infections among children.
Keywords: Capsular typing, pneumococcus, pneumonia, sequential multiplex PCR, serotyping, Streptococcus pneumoniae
Streptococcus pneumoniae is the most common cause of bacterial pneumonia in children contributing to the high percentage of morbidity and mortality worldwide. Pneumococcal pneumonia is highly associated with high mortality rate of children below five years old, particularly in the developing countries1. Increasing incidence of antibiotic resistance in S. pneumoniae strains worldwide is imposing challenges for the effective treatment of S. pneumoniae infections2. Penicillin has been used as the primary antibiotic to treat S. pneumoniae infections. However, the widespread use of penicillin has instigated the emergence of penicillin-resistant S. pneumoniae strains, driving the needs for alternative treatment of the diseases2. Since prevalence and distribution of penicillin-resistant strains differ geographically3, surveillance and continual monitoring of penicillin susceptibility among the local S. pneumoniae strains is important.
S. pneumoniae can be classified into >90 distinct serotypes based on its reaction with type-specific antisera against the capsular polysaccharides. S. pneumoniae serotypes vary in invasiveness, virulence and antibiotic resistance. Differential ability of the serotypes to cause invasive diseases is dependent on the chemical structure of the capsular polysaccharides and rarely the thickness of the capsules1. Serotypes that are associated with invasive pneumococcal diseases (IPD) among children, such as 19A, 3, 7F, 23F, 11, 6A, 6B, 14, 8, 18C and 19F have been described previously4,5,6. Among these common serotypes in children, serotypes 19A, 3, 7F, 23F, 14 and 8 were also present in adults5,6.
Precise and accurate serotyping is essential in the epidemiological study of S. pneumoniae and its serotypes. Several serotyping methods have been introduced to deduce the serotypes of S. pneumoniae including Quellung test which is the gold standard technique7. Unfortunately, this test is time-consuming and requires interpretation expertise. To overcome these problems, latex agglutination tests and counterimmunoelectrophoresis were introduced8,9. Later on, PCR-based assays became preferable over the conventional methods. PCR-based serotyping assays are designed to identify serotype-specific target sequences within the pneumococcal capsular gene cluster to deduce the S. pneumoniae serotypes10.
Capsular polysaccharides as the major virulence factor of S. pneumoniae are also used as immunogens in pneumococcal vaccines. S. pneumoniae infections are preventable by vaccination. In Malaysia, the pneumococcal vaccines are not included in the National Childhood Immunisation programme11. However, continuous surveillance on the diversity, prevalence and penicillin susceptibility of the S. pneumoniae serotypes are vital to enhance the existing vaccination and treatment protocols5. Therefore, this study was aimed to report on the diversity, occurrence and penicillin susceptibility of S. pneumoniae serotypes among paediatric patients in a tertiary care teaching hospital in Malaysia.
Material & Methods
The study was approved by the Medical Ethics Committee of University Malaya Medical Center (UMMC) (MECID: 20146-336), Kuala Lumpur, Malaysia. Electronic medical records were reviewed to obtain patients’ characteristics (age, gender, ethnicity and source of isolates) and penicillin susceptibility data (penicillin-susceptible, intermediate or resistant).
Bacterial isolates: One hundred and twenty five S. pneumoniae isolates were collected from Medical Microbiology Laboratory, UMMC. The isolates were previously isolated from respiratory tract clinical specimens from all paediatric patients (age ranged from one month to 12 yr) admitted to UMMC from January 2013 to May 2015. The source of the isolates included nasopharyngeal specimens (NPS), sputum and bronchoalveolar lavage (BAL, mini-BAL). Clinical specimens were cultured and subjected to identification using VITEK 2 system (bioMérieux, Inc., Hazelwood, Mo., USA). The confirmed S. pneumoniae isolates were cryopreserved. To revive the isolates, 100 μl of the stock was suspended in 400 μl of brain heart infusion broth and incubated overnight at 37°C with 5 per cent CO2. One loopful of the overnight culture was streaked on sheep blood agar and further incubated under the same conditions.
Genomic DNA extraction: The extraction of DNA was performed by boiling lysis of bacteria as described by Yap et al12 with slight modification. Briefly, countable α-haemolytic S. pneumoniae colonies were picked from blood agar plate, and washed with phosphate buffered saline. The cells were centrifuged for 10 min at 11,337× g. The supernatant was eliminated and the pellet was re-suspended in 100 μl of ultrapure water, subjected to boiling at 99°C in a water bath for 10 min, cooled on ice for five minutes and stored at −20°C until further use.
PCR confirmation of Streptococcus pneumoniae and capsular typing: PCR was performed to reconfirm the identity of the revived culture prior to capsular typing of S. pneumoniae isolates. Primers targeting transketolase (RecP), a housekeeping gene of S. pneumoniae as described by Enright and Spratt13 were used for PCR confirmation of S. pneumoniae. PCR was performed in a final reaction volume of 25 μl comprising 0.5 U of Taq polymerase (Promega, Madison, USA), 1.5 mM MgCl2, 1× buffer, 0.2 mM of deoxynucleotide triphosphates (dNTPs), 0.4 mM of primers RecP-f and RecP-r and 100 ng of genomic DNA. PCR amplifications were carried out in a Veriti® Thermal Cycler (Applied Biosystems, USA) using the following cycling conditions: 94°C for five min and 30 cycles of 94°C for 15 sec, 50°C for 30 sec and 72°C for 45 sec. Following amplification, all reactions were extended at 72°C for 10 min.
Capsular typing was carried out using 30 serotype specific primer pairs14,15,16,17. Primer sequences and PCR protocols of the capsular typing assay were obtained from the previous study18. Briefly, primers with varying concentrations were grouped into seven multiplex reactions to allow differentiation of PCR products based on respective band sizes. Each reaction comprised five to six primer pairs targeting respective serotypes including a primer targeting the CpSA gene, a conserved region located at the cps locus that served as an internal control. PCR reactions were performed in a 25 μl reaction mixture comprising 0.5 U of Taq polymerase, 1.5 mM MgCl2, 1× buffer, 0.2 mM of dNTPs and 100 ng/μl of genomic DNA. PCR amplifications were carried out in a Veriti® Thermal Cycler using the following cycling conditions: 94°C for four min and 30 cycles of 94°C for 45 sec, 54°C for 45 sec and 65°C for two min and 30 sec without extension.
Four microliters of PCR products obtained from both molecular identification and capsular typing assays were analyzed on one per cent LE agarose gel electrophoresis (Promega, Madison, USA) and stained with SYBR® Safe DNA Gel Stain (Invitrogen Corporation, USA). A100 bp DNA ladder was included in all gels. The gels with resulting bands were viewed under an ultraviolet transilluminator.
Statistical analysis: Statistical analysis was performed using IBM SPSS Modeller v.16 software (IBM Corp, Armonk, New York, USA). Random Forest analysis was used to study the relationship between serotype frequency and patient demographics.
Results
The patients belonged to four ethnic groups (Malay, Chinese, Indian and others) and categorized into two major age categories (<5 yr old, n=115 and >5 yr old, n=10). Majority (92.0%) of the patients were below five years while only 8.0 per cent were above five years. Most (n=113, 90.4%) of the isolates were isolated from NPS. The remaining isolates were isolated from sputum (n=6, 4.8%), nasal swabs (n=1, 0.8%), BAL (n=2, 1.6%) and mBAL (n=3, 2.4%).
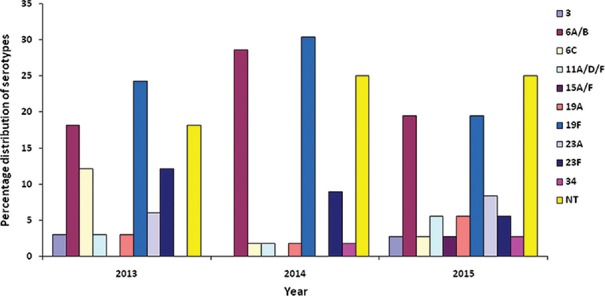
RecP PCR-screening and multiplex capsular typing: Prior to serotyping, identity of all the isolates was re-confirmed by PCR targeting RecP gene and all were found to be RecP positive. Based on multiplex PCR serotyping, 10 serotypes representing eight different serogroups were determined among the 125 isolates (Figure). Only 96 isolates (76.8%) were serotyped; 29 isolates (23.2%) were categorized as non-typeable (NT) by the multiplex PCR used in the study. All isolates were CpsA positive including the NT isolates. Majority of our isolates belonged to serotype 19F (25.6%), followed by serotypes 6A/B (23.2%), 23F (8.8%), 6C (4.8%), 23A (4.0%), 11A/D/F (3.2%), 19A (3. 2%), 34 (1.6%), 3 (1.6%) and 15A/F (0.8%).
Figure.
Serotype diversity of Streptococcus pneumoniae isolates isolated from paediatric patients (January 2013 to May 2015).
Thirty three S. pneumoniae isolated in 2013 were represented by eight serotypes (3, 6A/B, 6C, 11A/D/F, 19A, 19F, 23A and 23F). In 2014, the 56 S. pneumoniae isolates comprised of seven serotypes (6A/B, 6C, 11A/D/F, 19A, 19F, 23F and 34). In 2015 between January and May, 10 serotypes (3, 6A/B, 6C, 11A/D/F, 15A/F, 19A, 19F, 23A, 23F and 34) were identified among the 36 S. pneumoniae isolates. The diversity and percentage of distribution of serotypes were different each year. However, serotypes 19F and 6A/B were consistently prevalent between January 2013 and May 2015.
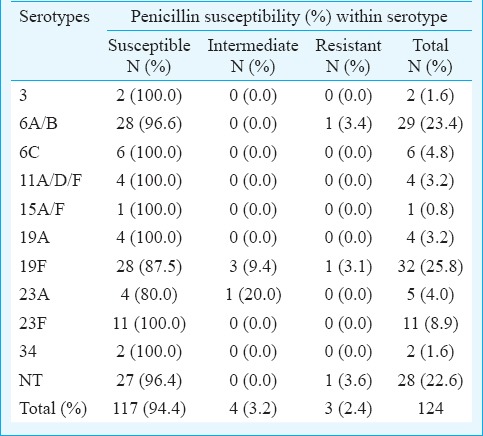
Penicillin susceptibility: Penicillin susceptibility patterns of the 124 S. pneumoniae isolates were gathered from the electronic records and tabulated with respect to the serotypes (Table I). One isolate was excluded from the penicillin susceptibility analysis due to the absence of required information on the system. S. pneumoniae isolates were categorized based on their susceptibility to penicillin namely penicillin-susceptible, penicillin intermediate and penicillin-resistant. Of the 124 S. pneumoniae isolates, 94.4 per cent were penicillin-susceptible, 5.6 per cent were penicillin intermediate and 2.4 per cent were penicillin-resistant. With respect to the serotype distribution, 3.4 per cent isolates of serotype 6A/B were penicillin resistant. Further, 9.4 and 3.1 per cent isolates of serotype 19F that were penicillin intermediate and penicillin-resistant, respectively. Separately, 20.0 per cent isolates of serotype 23A were penicillin intermediate while 3.6 per cent of NT isolates were penicillin resistant. Rest of the isolates were susceptible to penicillin.
Table I.
Penicillin susceptibility with respect to serotype distribution of Streptococcus pneumoniae isolates collected from January 2013 to May 2015
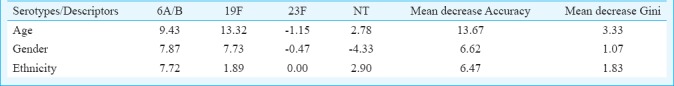
For statistical analysis, serotypes distributed with more than 10 cases (serotype 6A/B, 19F, 23F and NT) were selected for Random Forest analysis. Based on the model built using three predictors namely ethnicity, age and gender, the overall error rate of the model was high at 56.3 per cent. However, good prediction rate of 83.0 per cent was achieved for serotype 19F (Table II). The high prediction rate was mainly contributed by the strong correlation between serotype 19F and age (Table III).
Table II.
Confusion matrix generated by SPSS modeller for serotypes 6A/B, 19F, 23F and NT
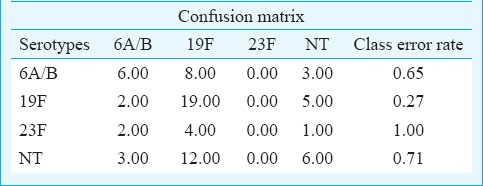
Table III.
Rank of variable importance to the model generated by SPSS modeller for serotypes 6A/B, 19F, 23F and NT based on three descriptors (age, ethnicity and gender)
Discussion
Pneumonia is the major cause of morbidity and mortality in children. S. pneumoniae is the major causative agent of pneumonia thus imposing a serious need in understanding the epidemiology of the bacterium. There are over 90 serotypes of S. pneumoniae identified to date. These serotypes are known to cause distinct clinical presentations of pneumococcal diseases19. Accurate serotyping of the bacterium is vital to study the serotype distribution and potential in causing pneumococcal diseases. The seven sequential multiplex PCRreactions for S. pneumoniae serotyping were previously developed by Pai et al10. It was later adapted and modified in other studies to suit their objectives. For instance, the sequential multiplex PCR reactions have been modified to target only IPD-associated serotypes20. The multiplex PCR has also been used to determine serotypes associated with pneumococcal carriage as well as IPD in some studies21. In our study, we adapted the multiplex PCR assay described by Jourdain et al18 as this was a modified assay to specifically detect pneumococcal carriage associated serotypes. It is generally perceived that pneumococcal carriage precedes infections such as pneumonia and IPD. Hence, the baseline data on the prevalence and diversity of the serotypes associated with pneumococcal carriage and infections will be useful for further evaluation of pneumococcal vaccines in reducing pneumococcal diseases.
Our results suggested that serotypes 19F and 6A/B were the most prevalent serotypes. The findings were in agreement with the previous reports on the prevalence of different serotypes of S. pneumoniae around the world. For instance, serotype 19F and 6B were reported as two of the most common serotypes among the 171 S. pneumoniae isolates collected from children in China from 2006 to 2008, where 19F and 6B serotypes were commonly detected from children below five years old while serotype 19F was the predominant serotype among children above five years of age22. In another study, serotypes 6A, 6B and 19F were reported as the common S. pneumoniae serotypes among 22 different serotypes detected in 367 Lithuanian children under six with serotype 6B (15.8%) as the most common serotypes detected, followed by 19F (13.9%) and 6A (9.3%) as the sixth most common serotype23. In addition, serotypes 19F and 6B were also found to be the most common S. pneumoniae serotypes detected among 835 Moscowian children below six years old24. In contrast, Rijal et al25 reported low distribution of serotypes 19F (2.1%) and serotype 6B (2.1%) among 60 Nepalese children. Serotypes 1 (27.7%), 5 (19.1%) and 4 (8.5%) were more common serotypes reported in the study with distribution above 5 per cent. However, serotypes 1 and 4 were not detected and serotype 5 was not targeted by the sequential multiplex PCR reactions used in our study.
In Malaysia, the most common serotypes (1, 2, 4, 6A/B, 7A/F, 7C, 9V/A, 9N/L, 11A, 14, 15/BC, 17F, 18A/B/C, 19A, 19F, 20, 23F, 23A, 23B) of S. pneumoniae have been identified previously26,27,28,29,30. In 2011, serotypes 6A/6B, 11A, 15B/C, 19A, 19F, 23A and 23F were detected among 41 Malaysian isolates by Shakrin et al30. Sixteen isolates (39.0%) were of serotype 6A/6B, followed by 22.0 per cent isolates of serotype 23F. Only 12.2 per cent isolates of serotype 19F were detected30. On the other hand, serotypes of 151 S. pneumoniae isolates from both children and adult patients in UMMC from 1999 to 2007 have been reported29. The strains were serotyped into different serogroups including serotypes 1, 3, 6A/B, 7C/B/40, 7F/A, 9V/A 10A, 11A/D, 12F/A, 14, 15A, 15B/C, 16F, 18C/B/A/F, 19A, 19F, 23F, 34, 35B and 35F/47F. The most prevalent serotypes among children under five were serotypes 19F (46.8%), 23F (6.4%), 1 (6.4%) and 14 (6.4%)29. Our study results concurred with these findings29,30 as serotypes 19F and 6A/B contributed >18.0 per cent annually throughout the study. It is also noteworthy that more than six per cent of the isolates reported by Le et al29 belonged to serotypes 1 and 14. However, these two serotypes were not detected in our study and indicated a possibility of a shift in serotype distribution trends.
Most of our isolates (94.4%) were penicillin susceptible. Among the seven non-susceptible S. pneumoniae isolates, four were of serotype 19F. Although our study revealed a lower percentage of penicillin non-susceptible S. pneumoniae isolates compared to Le et al29, serotype 19F remained as the significant contributor to the rate of penicillin non-susceptible S. pneumoniae isolates in both studies. Desa et al31 reported 22.9 per cent of their S. pneumoniae isolates as penicillin resistant, and 18.4 per cent as penicillin intermediate.
In conclusion, our study showed the distribution of various serotypes in S. pneumoniae isolates from children in a teaching hospital at Malaysia with decreasing incidence of penicillin-resistant isolates. However, the association of serogroups distribution and vaccination plan was not analyzed due to the absence of clinical data and patient's vaccination records.
Footnotes
Financial support & sponsorship: This study was supported by University of Malaya Research Grant (UMRG: RP026B-14HTM) and University of Malaya High Impact Research (grant number: H-50001- 00-A000025).
Conflicts of Interest: None.
References
- 1.AlonsoDeVelasco E, Verheul AF, Verhoef J, Snippe H. Streptococcus pneumoniae: Virulence factors, pathogenesis, and vaccines. Microbiol Rev. 1995;59:591–603. doi: 10.1128/mr.59.4.591-603.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dagan R. Antibiotic resistance and the potential impact of pneumococcal conjugate vaccines. Commun Dis Intell Q Rep. 2003;27(Suppl):S134–42. [PubMed] [Google Scholar]
- 3.Hackel M, Lascols C, Bouchillon S, Hilton B, Morgenstern D, Purdy J, et al. Serotype prevalence and antibiotic resistance in Streptococcus pneumoniae clinical isolates among global populations. Vaccine. 2013;31:4881–7. doi: 10.1016/j.vaccine.2013.07.054. [DOI] [PubMed] [Google Scholar]
- 4.Syrjänen RK, Kilpi TM, Kaijalainen TH, Herva EE, Takala AK. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J Infect Dis. 2001;184:451–9. doi: 10.1086/322048. [DOI] [PubMed] [Google Scholar]
- 5.Jauneikaite E, Mary Carnon Jefferies J, William Vere Churton N, Tzer Pin Lin R, Lloyd Hibberd M, Charles Clarke S, et al. Genetic diversity of Streptococcus pneumoniae causing meningitis and sepsis in Singapore during the first year of PCV7 implementation. Emerg Microbes Infect. 2014;3:e39. doi: 10.1038/emi.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imöhl M, Reinert RR, Ocklenburg C, van der Linden M. Association of serotypes of Streptococcus pneumoniae with age in invasive pneumococcal disease. J Clin Microbiol. 2010;48:1291–6. doi: 10.1128/JCM.01937-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Habib M, Porter BD, Satzke C. Capsular serotyping of Streptococcus pneumoniae using the quellung reaction. J Vis Exp. 2014;(84):e51208. doi: 10.3791/51208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trollfors B, Burman L, Dannetun E, Llompart J, Norrby R. Serotyping of Streptococcus pneumoniae strains by coagglutination and counterimmunoelectrophoresis. J Clin Microbiol. 1983;18:978–80. doi: 10.1128/jcm.18.4.978-980.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slotved HC, Kaltoft M, Skovsted IC, Kerrn MB, Espersen F. Simple, rapid latex agglutination test for serotyping of pneumococci (Pneumotest-latex) J Clin Microbiol. 2004;42:2518–22. doi: 10.1128/JCM.42.6.2518-2522.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol. 2006;44:124–31. doi: 10.1128/JCM.44.1.124-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darus N, Akmal S, Redzuan A. Putrajaya, Malaysia: Malaysian Health Technology Assessment Section (MaHTAS); 2013. [accessed on November 12, 2015]. Ministry of Health Malaysia. Pneumococcal conjugate vaccine for children below five years old. Available from: http://www.moh.gov.my/ [Google Scholar]
- 12.Yap PSX, Ahmad Kamar A, Chong CW, Yap IKS, Thong KL, Choo YM, et al. Intestinal carriage of multidrug-resistant gram-negative bacteria in preterm-infants during hospitalization in neonatal Intensive Care Unit (NICU) Pathog Glob Health. 2016;110:238–46. doi: 10.1080/20477724.2016.1229884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: Identification of clones associated with serious invasive disease. Microbiology. 1998;144(Pt 11):3049–60. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho Mda G, Pimenta FC, Gertz RE, Jr, Joshi HH, Trujillo AA, Keys LE. PCR-based quantitation and clonal diversity of the current prevalent invasive serogroup 6 pneumococcal serotype, 6C, in the United States in 1999 and 2006 to 2007. J Clin Microbiol. 2009;47:554–9. doi: 10.1128/JCM.01919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.da Gloria Carvalho M, Pimenta FC, Jackson D, Roundtree A, Ahmad Y, Millar EV, et al. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J Clin Microbiol. 2010;48:1611–8. doi: 10.1128/JCM.02243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dias CA, Teixeira LM, Carvalho Mda G, Beall B. Sequential multiplex PCR for determining capsular serotypes of pneumococci recovered from Brazilian children. J Med Microbiol. 2007;56:1185–8. doi: 10.1099/jmm.0.47347-0. [DOI] [PubMed] [Google Scholar]
- 17.Saha SK, Darmstadt GL, Baqui AH, Hossain B, Islam M, Foster D, et al. Identification of serotype in culture negative pneumococcal meningitis using sequential multiplex PCR: Implication for surveillance and vaccine design. PLoS One. 2008;3:e3576. doi: 10.1371/journal.pone.0003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jourdain S, Drèze PA, Vandeven J, Verhaegen J, Van Melderen L, Smeesters PR, et al. Sequential multiplex PCR assay for determining capsular serotypes of colonizing S.pneumoniae. BMC Infect Dis. 2011;11:100. doi: 10.1186/1471-2334-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song JY, Nahm MH, Moseley MA. Clinical implications of pneumococcal serotypes: Invasive disease potential, clinical presentations, and antibiotic resistance. J Korean Med Sci. 2013;28:4–15. doi: 10.3346/jkms.2013.28.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azzari C, Moriondo M, Indolfi G, Massai C, Becciolini L, de Martino M, et al. Molecular detection methods and serotyping performed directly on clinical samples improve diagnostic sensitivity and reveal increased incidence of invasive disease by Streptococcus pneumoniae in Italian children. J Med Microbiol. 2008;57:1205–12. doi: 10.1099/jmm.0.2008/000935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miernyk K, Debyle C, Harker-Jones M, Hummel KB, Hennessy T, Wenger J, et al. Serotyping of Streptococcus pneumoniae isolates from nasopharyngeal samples: Use of an algorithm combining microbiologic, serologic, and sequential multiplex PCR techniques. J Clin Microbiol. 2011;49:3209–14. doi: 10.1128/JCM.00610-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue L, Yao K, Xie G, Zheng Y, Wang C, Shang Y, et al. Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae isolates that cause invasive disease among Chinese children. Clin Infect Dis. 2010;50:741–4. doi: 10.1086/650534. [DOI] [PubMed] [Google Scholar]
- 23.Usonis V, Stacevičienė I, Petraitienė S, Vaičiūnienė D, Alasevičius T, Kirslienė J, et al. Streptococcus pneumoniae nasopharyngeal colonisation in children aged under six years with acute respiratory tract infection in Lithuania, February 2012 to March 2013. Euro Surveill. 2015;20:34–41. doi: 10.2807/1560-7917.es2015.20.13.21079. [DOI] [PubMed] [Google Scholar]
- 24.Mayanskiy N, Alyabieva N, Ponomarenko O, Lazareva A, Katosova L, Ivanenko A, et al. Serotypes and antibiotic resistance of non-invasive Streptococcus pneumoniae circulating in pediatric hospitals in Moscow, Russia. Int J Infect Dis. 2014;20:58–62. doi: 10.1016/j.ijid.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Rijal B, Tandukar S, Adhikari R, Tuladhar NR, Sharma PR, Pokharel BM, et al. Antimicrobial susceptibility pattern and serotyping of Streptococcus pneumoniae isolated from Kanti children hospital in Nepal. Kathmandu Univ Med J (KUMJ) 2010;8:164–8. doi: 10.3126/kumj.v8i2.3551. [DOI] [PubMed] [Google Scholar]
- 26.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: Global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 27.Yasin RM, Zin NM, Hussin A, Nawi SH, Hanapiah SM, Wahab ZA, et al. Current trend of pneumococcal serotypes distribution and antibiotic susceptibility pattern in Malaysian hospitals. Vaccine. 2011;29:5688–93. doi: 10.1016/j.vaccine.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Jauneikaite E, Jefferies JM, Hibberd ML, Clarke SC. Prevalence of Streptococcus pneumoniae serotypes causing invasive and non-invasive disease in South East Asia: A review. Vaccine. 2012;30:3503–14. doi: 10.1016/j.vaccine.2012.03.066. [DOI] [PubMed] [Google Scholar]
- 29.Le CF, Palanisamy NK, Mohd Yusof MY, Sekaran SD. Capsular serotype and antibiotic resistance of Streptococcus pneumoniae isolates in Malaysia. PLoS One. 2011;6:e19547. doi: 10.1371/journal.pone.0019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shakrin NN, Balasubramaniam SD, Yusof HA, Mastuki MF, Masri SN, Taib NM, et al. Evaluation of PCR-based approach for serotype determination of Streptococcus pneumoniae. Trop Biomed. 2013;30:338–44. [PubMed] [Google Scholar]
- 31.Desa MNM, Lin TK, Yasin RM, Parasakthi N. Penicillin susceptibility and molecular characteristics of clinical isolates of Streptococcus pneumoniae at the university of Malaya medical center, Kuala Lumpur, Malaysia. Int J Infect Dis. 2003;7:190–7. doi: 10.1016/s1201-9712(03)90051-8. [DOI] [PubMed] [Google Scholar]