Abstract
Background
Non-small cell lung cancer (NSCLC) accounts for about 85% of all types of lung cancer. Methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) is involved in DNA methylation, and DNA methylation is related to tumorigenesis. The role of MTHFD1 in NSCLC was examined in our study.
Material/Methods
The correlation between the expression of MTHFD1 and the clinicopathological features of patients diagnosed with lung cancer was investigated using the chi-square test. The viability and apoptosis of NCI-H1299 cells was respectively detected using cell counting kit-8 and flow cytometry assays. The expression levels of MTHFD1, apoptosis-related factors and DNA methyltransferase-related factors were assessed by quantitative real-time PCR (qRT-PCR) and western blot assays.
Results
We found that MTHFD1 expression in the tumor tissues and cells was higher than that of adjacent normal tissues and cells. The survival time of patients with high MTHFD1 expression was shorter than those with low MTHFD1 expression. The expression level of MTHFD1 was related to tumor size, TNM stage, histologic grade, and metastasis, but not linked to gender and age. Besides, si-MTHFD1 significantly decreased the viability of cells in a time-dependent manner, and increased cell apoptosis. When cells were transfected with MTHFD1-siRNA, the levels of surviving and B-cell lymphoma-2 (Bcl-2) were attenuated, while p53 and Bcl-2 associated X protein (Bax) levels were enhanced. Moreover, si-MTHFD1 markedly downregulated the expression levels of DNA methyltransferase 1 (DNMT1), DNMT3a, and DNMT3b.
Conclusions
Collectively, our results proved that MTHFD1 silencing obviously reduced the proliferation and enhanced the apoptosis of NSCLC via suppressing DNA methylation.
MeSH Keywords: Apoptosis; Carcinoma, Non-Small-Cell Lung; DNA Methylation; Methylenetetrahydrofolate Dehydrogenase (NADP)
Background
Lung cancer is the highest incidence and mortality of malignant tumors worldwide, and 1.5 million people all over the world die from lung cancer every year [1]. Non-small cell lung cancer (NSCLC) accounts for about 85% of all types of lung cancers, and more than 70% of these have advanced diagnosis, and have lost the chance of operative treatment. The 5-year survival rate is less than l5%. The high mortality rate of lung cancer is related to the lack of early detection methods and poor treatment measures [2–5]. In order to improve survival rate, we must achieve early diagnosis and surgical treatment. Although a lot of research and exploration have been carried out on the formation and development mechanism of lung cancer, the exact mechanism is not yet clear.
DNA methylation is one of the gene epigenetic modifications that may exist in all higher organisms [6–8]. In mammalian body, DNA methylation refers to a reaction of methylation of fifth carbon atoms of cytosine nucleotides (C) to 5 methyl cytosine (5′-mC) under the catalysis of DNA methyltransferase (DNMTs), s-adenosyl methionine as a methyl donor. Methyl cytosine can be converted to thymidine (T) by deamination, resulting in the mutation of C-T and the inactivation of some genes. This process regulates gene expression [9,10]. DNA methylation often occurs in the CG dinucleotide dense zone, which is called the CpG island [11,12]. When the CpG sequence of some tumor suppressor genes CpG island is hypermethylation, it can increase the degree of chromosome helix and silencing and deletion of tumor suppressor gene, thereby inducing tumor growth [13,14]. Increasing evidence illustrates that DNA methylation is involved in the occurrence of tumor, and it is an important biomarker to measure various tumors [15–17].
The methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) enzyme takes part in DNA methylation, and regulates the tumoral suppressor genes, thereby affecting the behavior of tumors [18]. The MTHFD1 1958G>A variants, are involved in association with defect of nerve canal and embryonic development, which also is related to various cancers [19–22]. Concretely, the genotypes of MTHFD1, including execu1958AA and 1958GA, have a protective effect on the tumor via regulation of DNA methylation [23]. Meanwhile, it has been reported that MTHFD1 variant is linked to the risk of lung cancer [24]. Nevertheless, the roles of MTHFD1 on proliferation and apoptosis of NSCLC are still unclear.
Herein, we explored the expression levels of MTHFD1 in the LC tissues and cells, and the relationship between MTHFD1 expression and the pathological features and the 5-year survival rate of lung cancer patients. Moreover, we analyzed the roles of MTHFD1 proliferation and apoptosis of NSCLC, and further tested whether the molecular mechanism mediates DNA methylation.
Material and Methods
Tissue source
The 35 samples of lung cancer tissues and adjacent normal breast tissues were obtained from lung cancer patients who were in The First Affiliated Hospital of Zhejiang Chinese Medical University from May 2015 to April 2017. Patient informed consents were acquired, and all patients agreed that tissues could be applied to our research. The Ethics Committee also approved this study.
Cell culture
Human normal lung epithelial cell line (BEAS-2B) and NSCLC cell lines (A549, NCI-H1299, NCI-H1650, and NCI-H460) were provided from Shanghai Gefan Biotechnology Co., Ltd. Dulbecco’s Modified Eagle Medium (DMEM; Haibo, Qingdao, Shandong, China) including 10% fetal bovine serum (FBS; Bena, Beijing, China) and a mixture of penicillin and streptomycin (Yuanmu, Shanghai, China) was used to culture BEAS-2B cells. Roswell Park Memorial Institute 1640 (RPMI-1640; Haibo, Qingdao, Shandong, China) medium supplemented with 10% FBS was used to culture A549, NCI-H1299, NCI-H1650, and NCI-H460 cells. All cells were maintained in an incubator with 95% humidified and 5% CO2 (WJ-185I; Santn, Shanghai, China) at 37°C.
RNA interference and transfection
The human MTHFD1-target siRNA and unspecific scrambled siRNA vectors were synthesized by BioVector (Beijing, China). Then, NCI-H1299 cells were transfected with plasmids by Hieff Trans™ Lipofectamine Reagent (Yeasen, Shanghai, China) for 48 hours.
Quantitative real-time PCR (qRT-PCR)
First, the tissues and cells of total RNA were harvested and lysed by RNA extraction kit (Promega, Beijing, China). Second, 1 μg of RNA was used to synthesize cDNA by SuperScript® VILO™ cDNA Synthesis kit (Thermo, Shanghai, China) following the manufacturer’s instructions. Third, SYBR® Premix Ex Taq™ II kit (Bao biologic engineering, Dalian, Liaoning, China) was used to amply cDNA (Takara, Beijing, China) according to the protocols of the manufacturer. GAPDH was considered as the internal control. The sequences of primers were listed in the Table 1. The expression level of each gene is quantitatively compared through the 2−ΔΔCT formula.
Table 1.
Sequences of the primers.
| Primer name | Sequence (5′-3′) | Product size (bp) |
|---|---|---|
| MTHFD1-Forward | AAAGAGAGGGCGAGCTTCAT | |
| MTHFD1-Reverse | CCTCTTCAGACAGCAGACCA | 250 |
| p53-Forward | GCCCCTCCTCAGCATCTTAT | |
| p53-Reverse | AAAGCTGTTCCGTCCCAGTA | 246 |
| survivin-Forward | TTCTCAAGGACCACCGCATC | |
| survivin-Reverse | GTTTCCTTTGCATGGGGTCG | 206 |
| Bax-Forward | AACATGGAGCTGCAGAGGAT | |
| Bax-Reverse | CCAATGTCCAGCCCATGATG | 208 |
| Bcl-2-Forward | TTCTTTGAGTTCGGTGGGGT | |
| Bcl-2-Reverse | CTTCAGAGACAGCCAGGAGA | 207 |
| DNMT1-Forward | ACTGGCTTTGATGGAGGTGA | |
| DNMT1-Reverse | ACCGTGGTCTCGATCTTGTT | 206 |
| DNMT3a-Forward | TCTCCAAGTCCCCATCCATG | |
| DNMT3a-Reverse | CAGCCATTTTCCACTGCTCT | 193 |
| DNMT3b-Forward | TTGCTGTTGGAACCGTGAAG | |
| DNMT3b-Reverse | TCCTTGGGGCGTGAGTAATT | 226 |
| GAPDH-Forward | CCATCTTCCAGGAGCGAGAT | |
| GAPDH-Reverse | TGCTGATGATCTTGAGGCTG | 222 |
Western blotting
The tissues and cells of total proteins were harvested and lysed with high efficiency RIPA lysate (Solarbio, Beijing, China). The contents of proteins were analyzed using protein analyzer (Imagin 200, MD Pacific, Tianjing, China). Each protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein was bound to PVDF membranes (Tengxiang; Wenzhou, Zhejiang, China). Then, membranes were transferred to TBST containing 5% skimmed milk powder for 1.5 hours. The corresponding primary antibody was used to incubate the membranes at 4°C refrigerator for 24 hours. (anti-MTHFD1, Abcam, ab18708, dilution: 1: 1000; anti-p53, R&D, IC13551G, dilution: 1: 800; anti-survivin, R&D, IC886G, dilution: 1: 500; anti-Bcl-2 associated X protein (Bax), Abcam, ab32503, dilution: 1: 1000; anti-B-cell lymphoma-2 (Bcl-2), Abcam, ab32370, dilution: 1: 800; anti-DNA methyltransferase 1 (DNMT1), CST, 5032, dilution: 1: 800; anti-DNMT3a, CST, 2160, dilution: 1: 700; anti-DNMT3b, CST, 67259, dilution: 1: 800; anti-GAPDH, R&D, 2275-PC-100, dilution: 1: 1000). Subsequently, membranes were incubated with the secondary antibodies (donkey anti-rabbit IgG, R&D, NL004, 1: 5000; mouse anti-rabbit IgG, CST, 93702, 1: 6000; goat anti-mouse IgG, Abcam, ab6785, 1: 8000) at 37°C for 1 hour. The membranes were exposed by Bio-RAD GelDoc XR+ (V140130, Vedeng, Suzhou, China).
Cell counting kit-8 assay
The viability of NCI-H1299 cells was detected by CCK-8 (Solarbio, Beijing, China) following the manufacturer instructions. First, cells were incubated in 96-well plates (2×103 cell/well) in incubator for 24 hours. Second, cells were respectively exposed to 0.1% PBS (control), unspecific scrambled siRNA vector (empty vector), MTHFD1-target siRNA (MTHFD1-siRNA), and 5-Aza-dc (positive control) for 12, 24, and 48 hours. Third, CCK-8 reagent was added to cells for 4 hours. Absorbance at 450 nm was assessed by SpectraMax iD3 multifunctional enzyme labeling instrument (Molecular Devices, Shanghai, China).
Flow cytometry assay
The apoptosis of NCI-H1299 cells was tested using Annexin V-FITC/PI apoptosis detection kit (BestBio, Shanghai, China) as the standardized method. The cells were incubated in 6-well plates (5×104 cell/well) in incubator for 24 hours. Then, the cells were treated as before. Subsequently, the cells were digested by 0.25% trypsin for 2 minutes (Kete, Jiangsu, China). The supernatant was removed, and the cells were resuspended using incubation buffer. The Annexin V-FITC and propidium iodide (PI) were dripped into cells in the dark at room temperature for 25 minutes. BD FACS Aria III flow cytometry (H143461, Huan Xi, Shanghai, China) was used to analyze the cell apoptosis.
Statistical analysis
The samples were analyzed in triplicate, and each experiment was conducted at least 3 times. All experimental data was presented as mean ± SD. One-way ANOVA was used to compare the differences between groups. Chi-square test was performed to evaluate the correlation between the expression of MTHFD1 and the clinicopathological features of patients diagnosed with lung cancer. The value of P<0.05 was considered a significant difference.
Results
High expression of MTHFD1 was associated with the progress of lung cancer
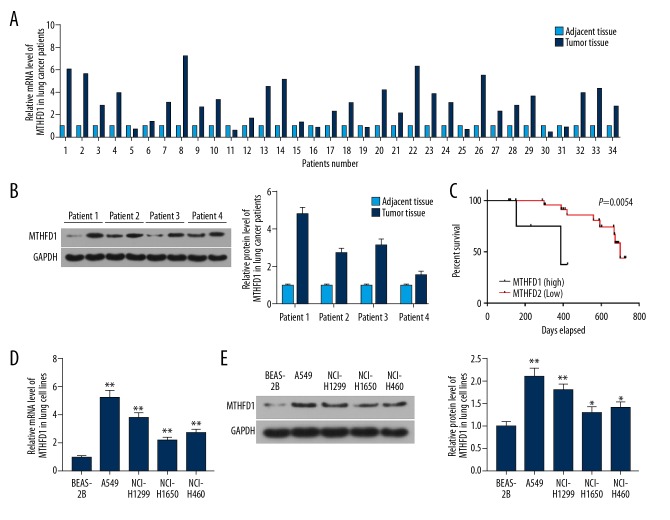
To study the influence of MTHFD1 in lung tumor, qRT-PCR and western bolt assays were performed. As the qRT-PCR and western blot results showed, the mRNA and proteins levels of MTHFD1 in tumor tissues were significantly higher compared to adjacent normal tissues (Figure 1A, 1B). At the same time, the mRNA and proteins levels of MTHFD1 in human normal lung epithelial cell line (BEAS-2B) were lower compared to NSCLC cell lines (A549, NCI-H1299, NCI-H1650, and NCI-H460) (Figure 1D, 1E, P<0.05). In addition, our data revealed that the survival percent of lung tumor patients with MTHFD1 high expression was lower than that of MTHFD1 low expression. Meanwhile, the survival time of patients with high MTHFD1 expression was shorter than those with low MTHFD1 expression (Figure 1C, P=0.0054). In addition, the chi-square test results observed that the expression level of MTHFD1 was linked to tumor size, TNM stage, histologic grade, and metastasis, but it was not linked to gender and age (Table 2).
Figure 1.
High expression of MTHFD1 associates with the progression of lung cancer. (A) The mRNA expression of MTHFD1 in lung tumor and adjacent normal tissues was analyzed by qRT-PCR assay. (B) The protein expression of MTHFD1 in lung tumor and adjacent normal tissues was assessed by western blot. (C) The correlation between MTHFD1 expression and the survival percent of the patients was quantified using GraphPad prism 7. (D) The mRNA levels of MTHFD1 in BEAS-2B, A549, NCI-H1299, NCI-H1650, and NCI-H460 cells were tested by qRT-PCR assay. (E) The protein level of MTHFD1 in cells was evaluated by western blot. GAPDH was considered as an internal control. The quality one was performed to measure and calculate the gray value. * P<0.05; ** P<0.01, versus BEAS-2B.
Table 2.
Correlation between the expression of MTHFD1 and the clinicopathological features of lung carcinoma patients.
| Features | Number of patients | Low MTHFD1 expression | High MTHFD1 expression | P value |
|---|---|---|---|---|
| Gender | 0.784 | |||
| Male | 26 | 10 (76.9%) | 16 (72.7%) | |
| Female | 9 | 3 (23.1%) | 6 (27.3%) | |
| Age(years) | 0.229 | |||
| <60 | 16 | 6 (35.3%) | 10 (66.7%) | |
| ≥60 | 19 | 11 (64.7%) | 5 (33.3%) | |
| Tumor size (cm) | 0.002* | |||
| <5 | 15 | 10 (76.9%) | 5 (22.7%) | |
| ≥5 | 20 | 3 (23.1%) | 17 (77.3%) | |
| TNM stage | 0.011* | |||
| I/II | 10 | 7 (53.8%) | 3 (13.6%) | |
| III/IV | 25 | 6 (46.2%) | 19 (86.4%) | |
| Histologic grade | 0.048* | |||
| Well | 3 | 2 (13.3%) | 1 (5%) | |
| Moderate | 12 | 8 (53.3%) | 4 (20%) | |
| Poor | 20 | 5 (33.3%) | 15 (75%) | |
| Metastasis | 0.024* | |||
| No | 23 | 15 (83.3%) | 8 (47.1%) | |
| Yes | 12 | 3 (16.7%) | 9 (52.9%) |
P<0.05, Chi-square test.
si-MTHFD1 represses cell viability and promotes apoptosis in NCI-H1299 cells
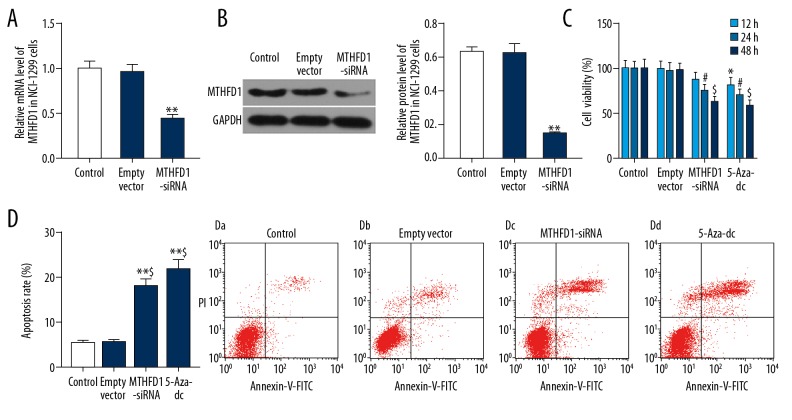
To interfere with the MTHFD1 gene in NCI-H1299 cells, the transfection efficiency of MTHFD1 was validated by qRT-PCR and western blot. The qRT-PCR data found that when cells were transfected with MTHFD1-siRNA vector, the mRNA level of MTHFD1 was markedly downregulated, compared to empty vector (Figure 2A, P<0.05). Meanwhile, the expression trend of MTHFD1 protein was consistent with the expression trend of MTHFD1 mRNA in cells that was transfected with MTHFD1-siRNA vector (Figure 2B, P<0.05).
Figure 2.
si-MTHFD1 represses cell viability and promotes apoptosis in NCI-H1299 cells. (A) NCI-H1299 cells were respectively treated with 0.1%PBS (control), human unspecific scrambled siRNA vector (empty vector), and MTHFD1-target siRNA (MTHFD1-siRNA) vector. The mRNA level of MTHFD1 was tested by qRT-PCR analysis. (B) The protein expression of MTHFD1 was examined by western blot. * P<0.05; ** P<0.01, versus empty vector. (C) NCI-H1299 cells were respectively exposed to 0.1%PBS (control), human unspecific scrambled siRNA vector (empty vector), MTHFD1-target siRNA (MTHFD1-siRNA) vector, and 5-Aza-dc (positive control). CCK-8 was carried out to assess cell viability for 12, 24, and 48 hours. * P<0.05, versus 12-hour empty vector. # P<0.05, versus 24-hour empty vector. @ P<0.05, $ P<0.01, versus 48-hour empty vector. (D) Annexin V-FITC/PI apoptosis detection kit was used to determine the cell apoptosis. * P<0.05, ** P<0.01, versus control. @ P<0.05, $ P<0.01, versus empty vector.
The viability of NCI-H1299 cells was analyzed by CCK-8. The results revealed that 5-Aza-dc obviously decreased cell viability in time-dependent manner. In the same way, si-MTHFD1 also conspicuously reduced the viability of NCI-H1299 cells in time-dependent manner (Figure 2C, P<0.05). Additionally, the cell apoptosis was assessed by flow cytometry. As flow cytometry data revealed, si-MTHFD1 and 5-Aza-dc apparently increased the apoptosis rate of NCI-H1299 cells (Figure 2D, P<0.05).
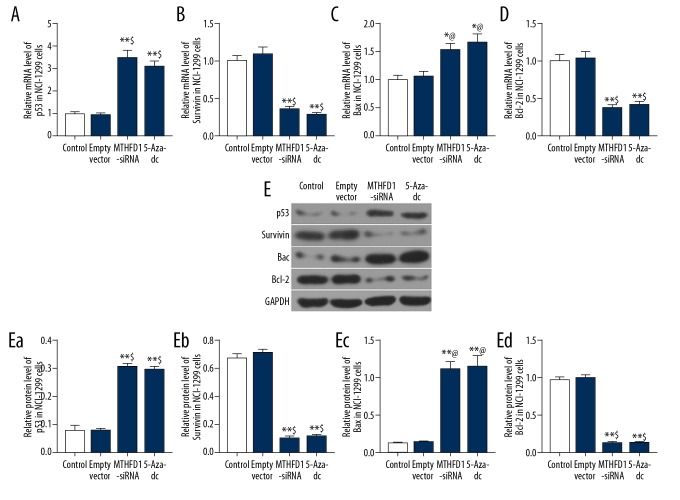
si-MTHFD1 regulated the expression of apoptosis-associated factors
In order to examine the apoptosis mechanism of MTHFD1 in NCI-H1299 cells, the levels of p53, survivin, Bax, and Bcl-2 were evaluated using qRT-PCR and western blot analysis. Our qRT-PCR results observed that si-MTHFD1 and 5-Aza-dc dramatically enhanced the mRNA levels of p53 and Bax, whereas attenuating survivin and Bcl-2 mRNA levels (Figure 3A–3D, P<0.05). Moreover, in comparison with empty vector, the protein levels of p53 and Bax in MTHFD1-siRNA and 5-Aza-dc were remarkably upregulated, and survivin and Bcl-2 levels were downregulated (Figure 3E, P<0.05).
Figure 3.
si-MTHFD1 regulates the expression of apoptosis-associated factors. (A–D) qRT-PCR was carried to examine the mRNA levels of p53 (A), survivin (B), Bax (C), and Bcl-2 (D). (E) Western blot was used to assess the protein levels of p53 (Ea), survivin (Eb), Bax (Ec), and Bcl-2 (Ed). * P<0.05, ** P<0.01, versus control. @ P<0.05, $ P<0.01, versus empty vector.
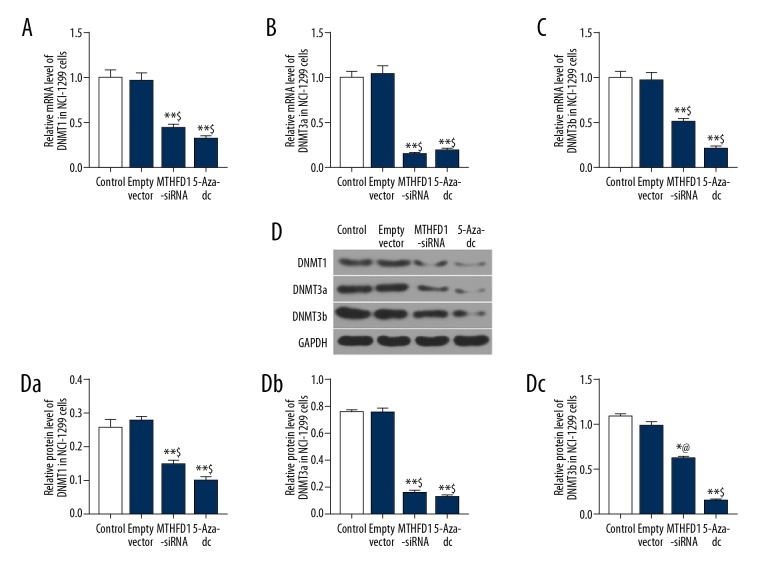
si-MTHFD1 downregulated DNA methylation level in NCI-H1299 cells
To explore the molecular mechanism of MTHFD1 in NCI-H1299 cells, the mRNA and protein levels of DNMT1, DNMT3a and DNMT3b were detected by qRT-PCR and western blot analysis. The qRT-PCR data showed that si-MTHFD1 and 5-Aza-dc significantly suppressed the expression of DNMT1, DNMT3a and DNMT3b (Figure 4A–C, P<0.05). Meanwhile, the expression tendency of DNMT1, DNMT3a and DNMT3b proteins in si-MTHFD1 and 5-Aza-dc was similar to the mRNA expression tendency of DNMT1, DNMT3a and DNMT3b (Figure 4D, P<0.05).
Figure 4.
si-MTHFD1 downregulates DNA methylation level in NCI-H1299 cells. (A–C) qRT-PCR was performed to examine the mRNA expression of DNMT1 (A), DNMT3a (B), and DNMT3b (C). (D) Western blot was used to evaluate the protein expression of DNMT1 (Da), DNMT3a (Db), and DNMT3b (Dc). (* P<0.05, ** P<0.01, versus control. @ P<0.05, $ P<0.01, versus empty vector.
Discussion
According to a large number of reports, it has been found that the MTHFD1 polymorphisms are related to the risk of various cancers [25–27]. Specifically, MTHFD1 G401A is linked to decrease risk of colon tumor, and MTHFD1 G1958A has protective effect on the lymphoblastic leukemia [27] and head and neck squamous tumors [26]. In addition, Jin et al. have found that MTHFD1 rs1950902 G>A is obviously related to with poor prognosis of NSCLC [28]. Hence, we suspected that MTHFD1 might be highly expressed in the lung cancer and be linked to the unfavorable prognosis of lung cancer. Our results showed that the expression levels of MTHFD1 in tumor tissues and cells were significantly higher than that of adjacent normal tissues and cells. In addition, MTHFD1 expression was associated with tumor size, TNM stage, histologic grade, and metastasis, but not related to gender and age. The survival time of patients with high MTHFD1 expression was shorter than that of lower expression. It was confirmed that high expression of MTHFD1 was relate to the progression of lung cancer.
It is well known that antitumor drugs and related genes generally inhibit the growth of cancer cells and promote apoptosis. Proliferation and apoptosis are 2 important processes of tumor development [29–32]. Yang et al. have confirmed that MTHFD1L silencing can repress viability of esophageal squamous cancer cells and expedite apoptosis [33]. At present, there is no report on the effect of MTHFD1 on the proliferation and apoptosis of NSCLC cells. We supposed that silence of MTHFD1 might decrease proliferation and increase apoptosis of NSCLC cells. Not surprisingly, our data revealed that si-MTHFD1 predominantly suppressed the viability of NCI-H1299 cells and promoted apoptosis. Hence, it was suggested that MTHFD1 silencing could repress proliferation and induce apoptosis of NSCLC.
Subsequently, the apoptosis mechanism of si-MTHFD1 in NCI-H1299 cells was investigated by detecting the apoptosis-related factors, including p53, survivin, Bax, and Bcl-2. Samarakoon et al. showed that triterpenoid saponin significantly facilitated the apoptosis of NSCLC cells via enhancing p53 and Bax expression and decreasing survivin levels [34]. Another study revealed that miR-145 upregulation promoted the apoptosis of NSCLC cells through augmenting the Bax/Bcl-2 ratio [35]. Homoplastically, our results showed that si-MTHFD1 notably enhanced the levels of p53 and Bax, whereas it reduced survivin and Bcl-2 levels in NCI-H1299 cells. This phenomenon explained si-MTHFD1 induced apoptosis of NSCLC cells by upregulating p53 and Bax expression, and downregulating survivin and Bcl-2 expression.
A recent study elucidated that the MTHFD1 1958GG genotype markedly decreased the risks of liver and colon tumors through associating with DNA hypomethylation [23]. Therefore, we considered that the role of si-MTHFD1 in proliferation and apoptosis of NSCLC cells was by mediating DNA methylation. DNMTs play an important role in the initial methylation and maintenance of methylation. DNMT1 is responsible for accurately replicating the DNA methylation form, playing the role of maintaining methylation [36]. DNMT3a and DNMT3b are mainly responsible for the methylation of non-methylated CpG sites [37,38]. Herein, our results found that si-MTHFD1 drastically inhibited the expression of DNMT1, DNMT3a, and DNMT3b in NCI-H1299 cells. These results suggest that si-MTHFD1 repressed DNA methylation in NSCLC.
Conclusions
Our study results proved that the silence of MTHFD1 could decrease proliferation and enhance apoptosis of NSCLC via suppressing DNA methylation. This study provides a theoretical basis for further exploring the influence of lung cancer in vitro. Furthermore, MTHFD1 may be a target for the treatment of lung cancer.
Footnotes
Conflict of interest
None.
Source of support: This work was supported by Zhejiang Chinese Medical Technology Project [Grant Number 2017ZA041]
References
- 1.Jain D, Roy-Chowdhuri S. Molecular pathology of lung cancer cytology specimens: A concise review. Arch Pathol Lab Med. 2018 doi: 10.5858/arpa.2017-0444-RA. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Huang W, Wang S, Zhang H, et al. Prognostic significance of combined fibrinogen concentration and neutrophil-to-lymphocyte ratio in patients with resectable non-small cell lung cancer. Cancer Biol Med. 2018;15(1):88–96. doi: 10.20892/j.issn.2095-3941.2017.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan BM. Lung cancer health disparities. Carcinogenesis. 2018;39(6):741–51. doi: 10.1093/carcin/bgy047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stone CJL, Vaid HM, Selvam R, et al. Multidisciplinary clinics in lung cancer care: A systematic review. Clin Lung Cancer. 2018 doi: 10.1016/j.cllc.2018.02.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Xue X, Wang L, et al. Suppressing autophagy enhances disulfiram/copper-induced apoptosis in non-small cell lung cancer. Eur J Pharmacol. 2018;827:1–12. doi: 10.1016/j.ejphar.2018.02.039. [DOI] [PubMed] [Google Scholar]
- 6.Chiba H, Kakuta Y, Kinouchi Y, et al. Allele-specific DNA methylation of disease susceptibility genes in Japanese patients with inflammatory bowel disease. PLoS One. 2018;13(3):e0194036. doi: 10.1371/journal.pone.0194036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Y, Wu R, Gaspar JM, et al. DNA methylome and transcriptome alterations and cancer prevention by curcumin in colitis-accelerated colon cancer in mice. Carcinogenesis. 2018;39(5):669–80. doi: 10.1093/carcin/bgy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richmond RC, Sharp GC, Herbert G, et al. The long-term impact of folic acid in pregnancy on offspring DNA methylation: Follow-up of the Aberdeen Folic Acid Supplementation Trial (AFAST) Int J Epidemiol. 2018 doi: 10.1093/ije/dyy032. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mutize T, Mkandla Z, Nkambule BB. Global and gene-specific DNA methylation in adult type 2 diabetic individuals: A protocol for a systematic review. Syst Rev. 2018;7(1):46. doi: 10.1186/s13643-018-0708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeshurun S, Hannan AJ. Transgenerational epigenetic influences of paternal environmental exposures on brain function and predisposition to psychiatric disorders. Mol Psychiatry. 2018 doi: 10.1038/s41380-018-0039-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Gan Y, Ma W, Wang X, et al. Coordinated transcription of ANRIL and P16 genes is silenced by P16 DNA methylation. Chin J Cancer Res. 2018;30(1):93–103. doi: 10.21147/j.issn.1000-9604.2018.01.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Zhang R, Chen Z, et al. Differential DNA methylation profiles of human B lymphocytes and Epstein-Barr virus-immortalized B lymphocytes. Chin J Cancer Res. 2018;30(1):104–11. doi: 10.21147/j.issn.1000-9604.2018.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordahl KM, Randolph TW, Song X, et al. Genome-wide DNA methylation in pre-diagnostic blood and bladder cancer risk in the Women’s Health Initiative. Cancer Epidemiol Biomarkers Prev. 2018;27(6):689–95. doi: 10.1158/1055-9965.EPI-17-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun W, Bunn P, Jin C, et al. The association between copy number aberration, DNA methylation and gene expression in tumor samples. Nucleic Acids Res. 2018;46(6):3009–18. doi: 10.1093/nar/gky131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Schnitzler KL, Ma Y, et al. The clinical influence of autophagy-associated proteins on human lung cancer. Dis Markers. 2018;2018 doi: 10.1155/2018/8314963. 8314963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Huang Y, Mu X, et al. DNA methylation is a common molecular alteration in colorectal cancer cells and culture method has no influence on DNA methylation. Exp Ther Med. 2018;15(4):3173–80. doi: 10.3892/etm.2018.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Xu J, Hu Z, et al. Quantitative DNA methylation analysis of paired box gene 1 and LIM homeobox transcription factor 1 alpha genes in cervical cancer. Oncol Lett. 2018;15(4):4477–84. doi: 10.3892/ol.2018.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacques PF, Bostom AG, Williams RR, et al. Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation. 1996;93(1):7–9. doi: 10.1161/01.cir.93.1.7. [DOI] [PubMed] [Google Scholar]
- 19.Galbiatti AL, da Silva LM, Ruiz-Cintra MT, et al. Association between 11 genetic polymorphisms in folate-metabolising genes and head and neck cancer risk. Eur J Cancer. 2012;48(10):1525–31. doi: 10.1016/j.ejca.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 20.Lautner-Csorba O, Gezsi A, Erdelyi DJ, et al. Roles of genetic polymorphisms in the folate pathway in childhood acute lymphoblastic leukemia evaluated by Bayesian relevance and effect size analysis. PLoS One. 2013;8(8):e69843. doi: 10.1371/journal.pone.0069843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parle-McDermott A, Kirke PN, Mills JL, et al. Confirmation of the R653Q polymorphism of the trifunctional C1-synthase enzyme as a maternal risk for neural tube defects in the Irish population. Eur J Hum Genet. 2006;14(6):768–72. doi: 10.1038/sj.ejhg.5201603. [DOI] [PubMed] [Google Scholar]
- 22.Zee RY, Rose L, Chasman DI, Ridker PM. Genetic variation of fifteen folate metabolic pathway associated gene loci and the risk of incident head and neck carcinoma: The Women’s Genome Health Study. Clin Chim Acta. 2013;418:33–36. doi: 10.1016/j.cca.2012.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moruzzi S, Guarini P, Udali S, et al. One-carbon genetic variants and the role of MTHFD1 1958G>A in liver and colon cancer risk according to global DNA methylation. PLoS One. 2017;12(10):e0185792. doi: 10.1371/journal.pone.0185792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Jin G, Wang H, et al. Association of polymorphisms in one-carbon metabolizing genes and lung cancer risk: A case-control study in Chinese population. Lung Cancer. 2008;61(1):21–29. doi: 10.1016/j.lungcan.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 25.da Silva LM, Galbiatti AL, Ruiz MT, et al. MTHFD1 G1958A, BHMT G742A, TC2 C776G and TC2 A67G polymorphisms and head and neck squamous cell carcinoma risk. Mol Biol Rep. 2012;39(2):887–93. doi: 10.1007/s11033-011-0813-3. [DOI] [PubMed] [Google Scholar]
- 26.Silva LM, Silva JN, Galbiatti AL, et al. Head and neck carconogenesis: Impact of MTHFD1 G1958A polymorphism. Rev Assoc Med Bras. 2011;57(2):194–99. doi: 10.1590/s0104-42302011000200018. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Ma H, Li L, et al. Association of methylenetetrahydrofolate dehydrogenase 1 polymorphisms with cancer: A meta-analysis. PLoS One. 2013;8(7):e69366. doi: 10.1371/journal.pone.0069366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin G, Huang J, Hu Z, et al. Genetic variants in one-carbon metabolism-related genes contribute to NSCLC prognosis in a Chinese population. Cancer. 2010;116(24):5700–9. doi: 10.1002/cncr.25301. [DOI] [PubMed] [Google Scholar]
- 29.Cui F, Hu J, Fan Y, et al. Knockdown of spindle pole body component 25 homolog inhibits cell proliferation and cycle progression in prostate cancer. Oncol Lett. 2018;15(4):5712–20. doi: 10.3892/ol.2018.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsis M, Alexiou GA, Vartholomatos E, et al. N-(p-coumaroyl) serotonin induces cell cycle arrest and apoptosis in breast cancer cells. J BUON. 2018;23(1):129–33. [PubMed] [Google Scholar]
- 31.Wang Y, Du C, Zhang N, et al. TGF-beta1 mediates the effects of aspirin on colonic tumor cell proliferation and apoptosis. Oncol Lett. 2018;15(4):5903–9. doi: 10.3892/ol.2018.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z, Zhu Q, Yin Y, et al. Traditional Chinese Medicine CFF-1 induced cell growth inhibition, autophagy, and apoptosis via inhibiting EGFR-related pathways in prostate cancer. Cancer Med. 2018;7(4):1546–59. doi: 10.1002/cam4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang YS, Yuan Y, Hu WP, et al. The role of mitochondrial folate enzyme MTHFD1L in esophageal squamous cell carcinoma. Scand J Gastroenterol. 2018;53(5):533–40. doi: 10.1080/00365521.2017.1407440. [DOI] [PubMed] [Google Scholar]
- 34.Samarakoon SR, Ediriweera MK, Nwokwu CDU, et al. A study on cytotoxic and apoptotic potential of a triterpenoid saponin (3-O-alpha-L-arabinosyl oleanolic acid) isolated from Schumacheria castaneifolia vahl in human non-small-cell lung cancer (NCI-H292) cells. BioMed Res Int. 2017;2017 doi: 10.1155/2017/9854083. 9854083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Y, Ye C, Tian Q, et al. miR-145 suppresses the proliferation, invasion and migration of NSCLC cells by regulating the BAX/BCL-2 ratio and the caspase-3 cascade. Oncol Lett. 2018;15(4):4337–43. doi: 10.3892/ol.2018.7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye D, Zhang L, Fan W, et al. Genipin normalizes depression-like behavior induced by prenatal stress through inhibiting DNMT1. Epigenetics. 2018;13(3):310–17. doi: 10.1080/15592294.2018.1450033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang Y, Deng S, Zhang J, et al. Melatonin-mediated development of ovine cumulus cells, perhaps by regulation of DNA methylation. Molecules. 2018;23(2) doi: 10.3390/molecules23020494. pii: E494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hervouet E, Peixoto P, Delage-Mourroux R, et al. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenetics. 2018;10:17. doi: 10.1186/s13148-018-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]