Abstract
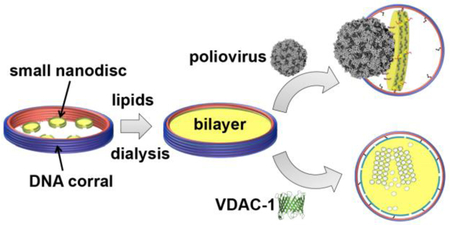
Here we present a modular method for manufacturing large-sized nanodiscs using DNA-origami barrels as scaffolding corrals. Large-sized nanodiscs can be produced by first decorating the inside of DNA barrels with small lipid-bilayer nanodiscs, which open up when adding extra lipid to form large nanodiscs of diameters ~45 or ~70 nm as prescribed by the enclosing barrel dimension. Densely packed membrane protein arrays are then reconstituted within these large nanodiscs for potential structure determination. Furthermore, we demonstrate the potential of these nanodiscs as model membranes to study poliovirus entry.
Graphical Abstract
Phospholipid-bilayer nanodiscs have attracted great interest as a membrane model that provides native-like environment for the study of embedded membrane proteins.1–6 A nanodisc can be stabilized by polymers7–9, peptides10–12 or proteins13. A popular version of nanodiscs is stabilized with Apolipoprotein A1-derived membrane-scaffold proteins (MSPs).13–14 Typically two copies of MSPs wrap around a lipid-bilayer patch, where the length of the MSP determines the diameter of the captured lipid nanodisc.
Up until recently, only up to ~16–17 nm in diameter or smaller MSP-based nanodiscs could be assembled.15–16 Therefore, the utility of this system for structural studies, especially for large membrane protein complexes/arrays or in vitro reconstitution for biological processes has been limited. More recently, we engineered covalently circularized nanodiscs (cNDs) where a phospholipid bilayer is surrounded by amphipathic helical polypeptides, derived from Apolipoprotein A1, that can be circularized by sortase A.3 With this approach, very homogeneous, covalently closed nanodiscs have been engineered with diameters of ~ 8, 11, 15, and 50 nm. These cNDs not only serve as excellent tools for the studies of small- or medium-size membrane proteins17–18 but could also act as a surrogate membrane to study the interactions of small virus particles with their membrane-bound receptors.3, 19 This latter application was demonstrated by poliovirus (30 nm diameter) engagement with CD155 decorated cND, which showed formation of a putative pore in the nanodisc and RNA ejection through the pore.3 Motivated by these results, we attempted to make even larger nanodiscs that could be used to study the early steps of cell entry for larger viruses. However, the attempts to make stable cNDs larger than 50 nm in diameter turned out to be difficult, as these large nanodiscs are susceptible to aggregation. Moreover, it is challenging to express and purify the large scaffold proteins required to make these nanodiscs using E. coli expression systems.
To overcome these issues, we applied external DNA-origami barrels as scaffolding corrals. Scaffolded DNA origami employs folding of a long scaffold strand with hundreds short staple strands to achieve self-assembly of custom shapes.20–22 Using DNA origami, each component strand can be uniquely addressed, providing a molecular billboard to arrange biomolecules with prescribed compositions and stoichiometries for biofunctional study.23–24 The mechanical stiffness of self-assembled DNA nanostructures also can confine the precise morphology and dimensions of nanomaterials, including hard inorganic and soft biomaterials, through casting growth.25 Each DNA barrel we used in this study recruits a number of ~11-nm diameter nanodiscs that are circumscribed by a pair of non-circularized, oligonucleotide-functionalized scaffold proteins (3C-NW11), and directs their reconstitution into a single large nanodisc with a diameter prescribed by the dimension of the enclosing barrel (Figure 1). We employed two different sized barrels: 90 and 60 outer diameter to reconstitute ~70 or ~45 nm nanodiscs, respectively (Figure 1, Figures S2, S4 and S5). The final yield for the properly formed DCND is between 40–70 % for both nanodiscs.
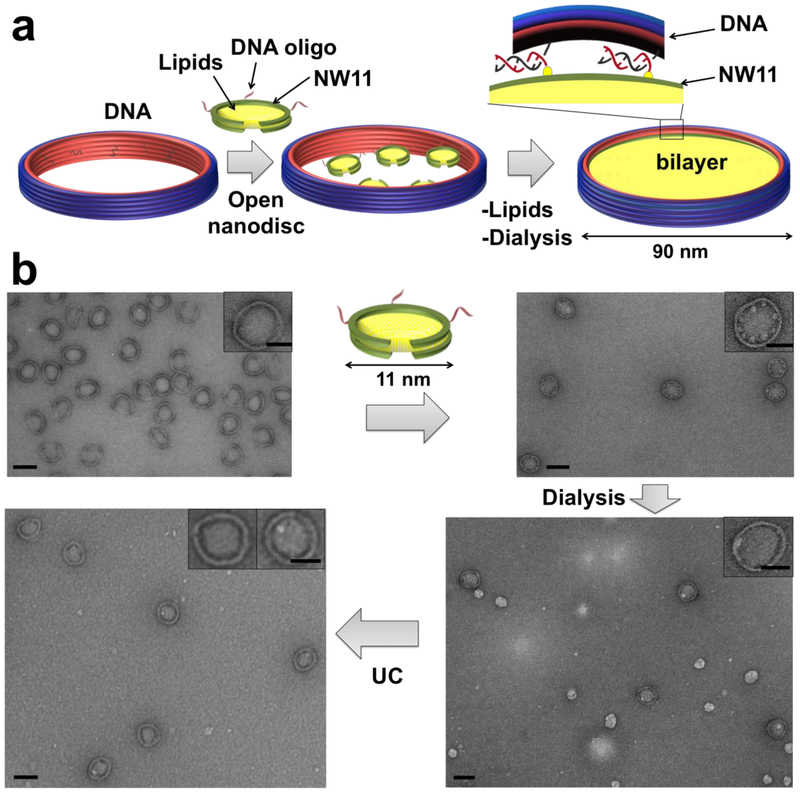
Figure 1. DNA-corralled nanodisc reconstitution.
(a) Addition of detergent and lipids (POPC/POPG plus 10% cholesterol) to small nanodisc-decorated barrels (90 nm outer diameter) followed by dialysis results in the reconstitution of DCND. Each DNA barrel is decorated with ssDNA overhang as handles to hybridize with ssDNA chemically conjugated small nanodisc. (b) Negative-stain TEM images of DNA-origami barrel, with 70 nm inner diameter, before (top left) and after (top right) assembly with small nanodiscs. TEM images after dialysis to remove detergent, forming integrated large sized nanodiscs (bottom right) and after ultracentrifugation (UC) to remove free lipid vesicles (bottom left). Scale bar, 100 nm (top insets with 50 nm scale bar). Each step is imaged at lower magnification in Figures S1-S5.
Our DNA-origami barrels were designed using the software caDNAno26 (Figures S6 and S7) and folded in a one-pot reaction using a 20-hour thermal annealing program. Correctly folded structures were isolated by glycerol-gradient centrifugation and verified by negative-stain transmission electron microscopy (TEM) (Figure 1b and Figure S3) and native agarose gel electrophoresis (Figures S8). The 11 nm nanodiscs were first covalently coupled to oligonucleotides through Sulfo-SMCC crosslinkers (Figure S9), and then assembled onto the DNA-origami barrels through hybridization to the single-stranded DNA handles (36, 24 handles for 90-nm, 60-nm barrel respectively) pre-immobilized onto the nanostructure (Figure 1a). The successful hybridization with 11-nm sized nanodiscs was confirmed by negative stain-EM. As shown in Figure 1b and Figure S1, the DNA-origami barrel inner face is lined with small white disk-shaped structures along the interior face of the barrels. Most of the internalized nanodiscs have sizes around 11-nm in diameter, while some nanodiscs have size bigger than 11-nm, which can result from the heterogeneity of uncircularized nanodiscs.
In order to direct the reconstitution of the 11-nm nanodiscs into one single large nanodisc, we added excess lipids solubilized in detergent followed by dialysis. After dialysis, we performed isopycnic ultracentrifugation through a sucrose gradient to separate the DNA-barrel-scaffolded lipid nanodiscs from free vesicles on the basis of their differences in density (Figure 1b).
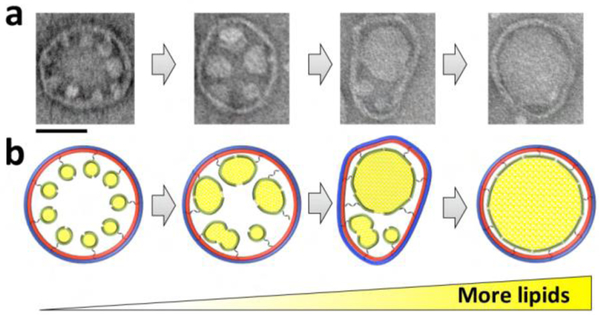
In Figure 2, we propose a model to explain the formation of the large bilayer. We hypothesize that bringing the 11-nm nanodiscs to within several nanometers, achieved here by DNA-origami templating, is an important first step for the successful assembly of the larger bilayer. Secondly, a destabilization must be developed at some point between the neighboring 11-nm nanodiscs to induce a highly localized rearrangement/fusion of the adjacent bilayers, resulting in one interconnected structure. This destabilization step is facilitated by the detergent that is used to solubilize the lipid mixture. The detergent destabilizes the interaction of MSP with its enclosed lipids27 thus allowing the merge of neighboring nanodiscs. Thirdly, fusion of smaller nanodiscs should initially create ellipsoid larger nanodiscs, until additional lipids can be recruited. Therefore, flexibility in the DNA-origami barrel (i.e. ability to distort into an ellipsoid as in Figure 2a,b, third panels) may help facilitate later fusion events. Lastly, additional lipids are needed to inflate ellipsoid larger nanodiscs into circular larger nanodiscs. All these factors together contribute to the success of the reconstitution of large bilayer nanodiscs.
Figure 2. A proposed model for the assembly of the large bilayer within DNA-origami barrels (90 nm outer diameter).
Bringing the 11-nm nanodiscs close to each other to within several nanometers is a critical first step. Adding excess lipids solubilized in detergent destabilizes and induces fusion of the neighboring nanodiscs. Next, the intermediate-sized nanodiscs fuse, perhaps aided by transient deformation of the barrel into an ellipsoid contour, to yield a single large bilayer nanodisc. Scale bar, 50 nm.
We were able to assemble DCNDs using different lipids including the neutral lipid DOPC and the negatively charged lipid mixture POPC/POPG with or without cholesterol (Figure 1b, Figure S2). We obtained the best yield when we used POPC/POPG with 10% cholesterol as the cholesterol molecule is believed to increase the membrane mechanical stability.28 Interestingly, the yield did not improve after adding more cholesterol molecules. With the barrel acting as a bumper case to prevent aggregation, the enclosed nanodiscs were stable for at least 6 weeks when stored at 4 ºC (Figures S10 and S14). Also, these nanodiscs show good tolerance to a broad range of pH and divalent ion concentration (Figures S11 and S12), representing a great improvement in large nanodisc stability.
Having established a modular method to reconstitute large stable nanodiscs using DNA barrel, we proceeded to examine whether this nanodisc platform could be used for membrane-protein incorporation for further structural and/or functional studies. Here we chose two model proteins that are known to establish functional assemblies with or without other proteins and co-factors: human Voltage-Dependent Anion Channel 1 (hVDAC-1) and the Rhodobacter sphaeroides photosynthetic reaction center protein (RC). hVDAC-1, a beta barrel protein, is known to oligomerize on the surface of the outer mitochondrial membrane in apoptosis induction.29 Early EM and AFM studies of mitochondria from Neurospora grassa30–31 or potato tubers32 showed dense packing of VDAC pores or even hexagonal spindle-like arrangements. It is therefore hypothesized that oligomer interactions of VDAC may play a role for its function. On the other hand, RC is a mostly alpha-helical protein that plays a key role in the photochemical conversion of light into chemical energy.33
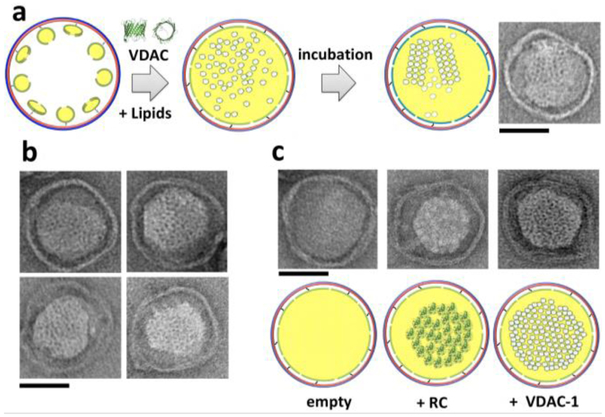
Purified hVDAC1 and RC in detergent were added to the 11-nm nanodisc-decorated DNA origami along with lipids before the dialysis step (Figure 3a). After dialysis and purification of hVDAC1- and RC-loaded DCND by density gradients, we performed EM and SDS-PAGE analysis (silver-stain) to confirm the incorporation. As shown in Figure 3b,c and Figure S13, EM images and SDS-PAGE analysis verify the incorporation of both RC and hVDAC1 respectively into nanodiscs. We also showed that multimeric assemblies are formed (Figure 3a) within many of the nanodiscs when we added excess amount of either protein before the dialysis step. Presenting membrane proteins at high density, both in freely diffusing as well as crystalline contexts, may prove useful to investigate both structural and functional rearrangements directly from the very same membranes. Often proteins are found functional in 2D crystals 34–36 allowing a number of studies to collect electron diffraction data and yielding high resolution structures.37 Our method could be further developed to a general platform for membrane-protein structure determination by subatomic averaging using cryo-electron tomography38, electron diffraction or by X-ray free electron laser (XFEL).
Figure 3. Reconstruction of hVDAC-1 and RC clusters within DCND.
(a) Addition of lipids solubilized in detergent and hVDAC-1 to small nanodisc-decorated DNA-origami barrels (90 nm outer diameter), followed by dialysis, leads to reconstitution of hVDAC1 clusters within DCND. Multimeric assemblies of VDAC can be formed within the nanodiscs (right). (b) Typical DCND containing multimeric assemblies of VDAC. (c) Comparison of empty, RC and VDAC containing DCND. Scale bar, 50 nm.
We next explored whether our nanodiscs could be used as a model membrane system to study early steps in viral entry. Poliovirus (∼30-nm diameter) is the prototype member of the enterovirus genus of the picornavirus family, which are positive-sense, single-stranded RNA viruses with ∼7,500-base genomes enclosed by an icosahedral capsid, missing an envelope.39 CD155 (also known as the poliovirus receptor, PVR) is the receptor to induce poliovirus infection, which catalyzes a conformational rearrangement and expansion of the virus particle40, eventually leading to RNA release across the membrane.
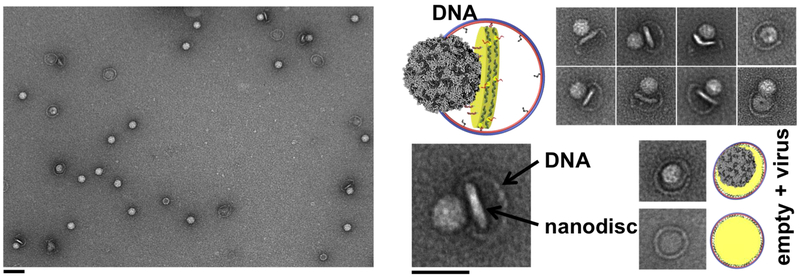
We prepared the 45-nm DCNDs (hosted by 60 nm outer-diameter DNA-origami barrels) containing lipids derivatized with a nitrilotriacetate (NTA) nickel-chelating head group. These DCND were functionalized with the His-tagged CD155 ectodomain. Next, the receptor-decorated nanodiscs were incubated with poliovirus for 5 minutes at 4 °C. The complex was then heated to 37°C for 15 minutes to initiate receptor-mediated viral uncoating. Negative-stain EM confirmed the binding of the virus to DCND (Figure 4, Figure S14); in some cases, we observed apparent pore formation in the nanodiscs after the virus initial binding. We also observed by negative-stain and cryo-EM that many of the nanodiscs were partially released from their DNA scaffolds after binding the virus (Figure 4 and Figure S14 and S15). One potential explanation is the creation of extra membrane tension after receptor binding, which causes dsDNA dissociation between 3C-NW11 and DNA barrel. These partially detached nanodiscs are likely stabilized due to the hydrophobic interactions between 3C-NW11 and the acyl chains of the lipid bilayer (i.e the 3C-NW11 get detached from the DNA barrel and stay with the bilayer).
Figure 4. Poliovirus interactions with DCND containing CD155 receptor.
(Left) Negative-stain TEM images of DCND (60 nm outer diameter) containing CD155 ectodomain interacting with poliovirus. Scale bar, 100 nm. (Right) TEM images showing individual viral particles tethered to DCND. Some images show the bending of the bilayer and the creation of a pore in nanodisc by the poliovirus (top right). Scale bar, 50 nm.
In conclusion, we have developed a modular method to produce stable nanodiscs with sizes up to ~70 nm diameter using DNA-origami barrel as scaffold. We believe our system can potentially create much larger nanodiscs, with more complicated geometry. Additionally, site-specific functionalization of DNA-origami provides a tool to precisely control membrane protein insertion, potentially with precisely controlled stoichiometry and transmembrane orientation. Therefore, our platform could provide an excellent tool for producing homogenous membrane protein complexes in native-like environment with designed composition, stoichiometry, and orientation. We have demonstrated the utility of this model system to reconstitute membrane protein 2D clusters for potential structure determination. We used two membrane proteins, hVDAC-1 and RC, which are known to form functional oligomeric arrays in native membranes. Furthermore, we have established this system to probe an outstanding virology problem of viral entry through host membranes.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to thank Dr. Su Lin from ASU for providing RC protein. We thank Dr. Shelley Wickham and John Min for the design and validation of the DNA-origami barrels. We thank Dr. Mike Strauss for helpful discussion.
Funding Sources
This work was supported by NIH grant F32GM113406 to M.L.N and grants: GM047467 and AI037581 to G. W. and Wyss Institute at Harvard Faculty Award, NSF Expeditions Award (CCF-1317291), and ARO MURI Award W911NF-12-1-0420 to W.M.S. Grant AI20566 to J.M.H
Footnotes
ASSOCIATED CONTENT
Supporting Information.
The supporting information is available free of charge via the Internet at http://pubs.acs.org. Experimental details and EM data for the 90 and 60 nm DCND. Also, SMCC coupling, DNA design illustration and staples sequences for 90-nm, 60-nm DNA origami barrel are included.
Notes
The Harvard University Office of Technology Development has filed a patent application on technologies described herein.
REFERENCES
- 1.Denisov IG; Grinkova YV; Lazarides AA; Sligar SG, Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc 2004, 126 (11), 3477–87. [DOI] [PubMed] [Google Scholar]
- 2.Raschle T; Lin C; Jungmann R; Shih WM; Wagner G, Controlled Co-reconstitution of Multiple Membrane Proteins in Lipid Bilayer Nanodiscs Using DNA as a Scaffold. ACS Chem Biol 2015, 10 (11), 2448–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasr ML; Baptista D; Strauss M; Sun ZJ; Grigoriu S; Huser S; Pluckthun A; Hagn F; Walz T; Hogle JM; Wagner G, Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods 2017, 14 (1), 49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouck JE; Krapf JE; Roy J; Huff HC; Das A, Recent advances in nanodisc technology for membrane protein studies (2012–2017). FEBS Lett 2017, 591 (14), 2057–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frey L; Lakomek NA; Riek R; Bibow S, Micelles, Bicelles, and Nanodiscs: Comparing the Impact of Membrane Mimetics on Membrane Protein Backbone Dynamics. Angew Chem Int Ed Engl 2017, 56 (1), 380–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang M; Huang R; Ackermann R; Im SC; Waskell L; Schwendeman A; Ramamoorthy A, Reconstitution of the Cytb5-CytP450 Complex in Nanodiscs for Structural Studies using NMR Spectroscopy. Angew Chem Int Ed Engl 2016, 55 (14), 4497–9. [DOI] [PubMed] [Google Scholar]
- 7.Ravula T; Ramadugu SK; Di Mauro G; Ramamoorthy A, Bioinspired, Size-Tunable Self-Assembly of Polymer-Lipid Bilayer Nanodiscs. Angew Chem Int Ed Engl 2017, 56 (38), 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 8.Knowles TJ; Finka R; Smith C; Lin YP; Dafforn T; Overduin M, Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. Journal of the American Chemical Society 2009, 131 (22), 7484–5. [DOI] [PubMed] [Google Scholar]
- 9.Tonge SR; Tighe BJ, Responsive hydrophobically associating polymers: a review of structure and properties. Adv Drug Deliv Rev 2001, 53 (1), 109–22. [DOI] [PubMed] [Google Scholar]
- 10.Midtgaard SR; Pedersen MC; Kirkensgaard JJ; Sorensen KK; Mortensen K; Jensen KJ; Arleth L, Self-assembling peptides form nanodiscs that stabilize membrane proteins. Soft Matter 2014, 10 (5), 738–52. [DOI] [PubMed] [Google Scholar]
- 11.Epand RM; Gawish A; Iqbal M; Gupta KB; Chen CH; Segrest JP; Anantharamaiah GM, Studies of synthetic peptide analogs of the amphipathic helix. Effect of charge distribution, hydrophobicity, and secondary structure on lipid association and lecithin:cholesterol acyltransferase activation. The Journal of biological chemistry 1987, 262 (19), 9389–96. [PubMed] [Google Scholar]
- 12.Zhao Y; Imura T; Leman LJ; Curtiss LK; Maryanoff BE; Ghadiri MR, Mimicry of high-density lipoprotein: functional peptide-lipid nanoparticles based on multivalent peptide constructs. J Am Chem Soc 2013, 135 (36), 13414–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayburt TH; Grinkova YV; Sligar SG, Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett 2002, 2 (8), 853–856. [Google Scholar]
- 14.Ritchie TK; Grinkova YV; Bayburt TH; Denisov IG; Zolnerciks JK; Atkins WM; Sligar SG, Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 2009, 464, 211–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grinkova YV; Denisov IG; Sligar SG, Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng Des Sel 2010, 23 (11), 843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagn F; Etzkorn M; Raschle T; Wagner G, Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc 2013, 135 (5), 1919–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao H; Das D; Courtney NA; Jiang Y; Briguglio JS; Lou X; Roston D; Cui Q; Chanda B; Chapman ER, Dynamics and number of trans-SNARE complexes determine nascent fusion pore properties. Nature 2018, 554 (7691), 260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGoldrick LL; Singh AK; Saotome K; Yelshanskaya MV; Twomey EC; Grassucci RA; Sobolevsky AI, Opening of the human epithelial calcium channel TRPV6. Nature 2018, 553 (7687), 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasr ML; Wagner G, Covalently circularized nanodiscs; challenges and applications. Current opinion in structural biology 2018, 51, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothemund PW, Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440 (7082), 297–302. [DOI] [PubMed] [Google Scholar]
- 21.Dietz H; Douglas SM; Shih WM, Folding DNA into twisted and curved nanoscale shapes. Science 2009, 325 (5941), 725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douglas SM; Dietz H; Liedl T; Hogberg B; Graf F; Shih WM, Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459 (7245), 414–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu J; Yang YR; Johnson-Buck A; Liu M; Liu Y; Walter NG; Woodbury NW; Yan H, Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat Nanotechnol 2014, 9 (7), 531–6. [DOI] [PubMed] [Google Scholar]
- 24.Derr ND; Goodman BS; Jungmann R; Leschziner AE; Shih WM; Reck-Peterson SL, Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 2012, 338 (6107), 662–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun W; Boulais E; Hakobyan Y; Wang WL; Guan A; Bathe M; Yin P, Casting inorganic structures with DNA molds. Science 2014, 346 (6210), 1258361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douglas SM; Marblestone AH; Teerapittayanon S; Vazquez A; Church GM; Shih WM, Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res 2009, 37 (15), 5001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih AY; Freddolino PL; Sligar SG; Schulten K, Disassembly of nanodiscs with cholate. Nano Lett 2007, 7 (6), 1692–6. [DOI] [PubMed] [Google Scholar]
- 28.Redondo-Morata L; Giannotti MI; Sanz F, Influence of cholesterol on the phase transition of lipid bilayers: a temperature-controlled force spectroscopy study. Langmuir 2012, 28 (35), 12851–60. [DOI] [PubMed] [Google Scholar]
- 29.Shoshan-Barmatz V; Mizrachi D, VDAC1: from structure to cancer therapy. Front Oncol 2012, 2, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mannella CA, Structure of the outer mitochondrial membrane: ordered arrays of porelike subunits in outer-membrane fractions from Neurospora crassa mitochondria. J Cell Biol 1982, 94 (3), 680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mannella CA, Structure of the mitochondrial outer membrane channel derived from electron microscopy of 2D crystals. J Bioenerg Biomembr 1989, 21 (4), 427–37. [DOI] [PubMed] [Google Scholar]
- 32.Hoogenboom BW; Suda K; Engel A; Fotiadis D, The supramolecular assemblies of voltage-dependent anion channels in the native membrane. J Mol Biol 2007, 370 (2), 246–55. [DOI] [PubMed] [Google Scholar]
- 33.Ermler U; Fritzsch G; Buchanan SK; Michel H, Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 A resolution: cofactors and protein-cofactor interactions. Structure 1994, 2 (10), 925–36. [DOI] [PubMed] [Google Scholar]
- 34.Miyazawa A; Fujiyoshi Y; Unwin N, Structure and gating mechanism of the acetylcholine receptor pore. Nature 2003, 423 (6943), 949–55. [DOI] [PubMed] [Google Scholar]
- 35.Zheng H; Taraska J; Merz AJ; Gonen T, The prototypical H+/galactose symporter GalP assembles into functional trimers. J Mol Biol 2010, 396 (3), 593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murata K; Mitsuoka K; Hirai T; Walz T; Agre P; Heymann JB; Engel A; Fujiyoshi Y, Structural determinants of water permeation through aquaporin-1. Nature 2000, 407 (6804), 599–605. [DOI] [PubMed] [Google Scholar]
- 37.Gonen T; Cheng Y; Sliz P; Hiroaki Y; Fujiyoshi Y; Harrison SC; Walz T, Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 2005, 438 (7068), 633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schur FK; Obr M; Hagen WJ; Wan W; Jakobi AJ; Kirkpatrick JM; Sachse C; Krausslich HG; Briggs JA, An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 2016, 353 (6298), 506–8. [DOI] [PubMed] [Google Scholar]
- 39.Tuthill TJ; Groppelli E; Hogle JM; Rowlands DJ, Picornaviruses. Curr Top Microbiol Immunol 2010, 343, 43–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendelsohn CL; Wimmer E; Racaniello VR, Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 1989, 56 (5), 855–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.