Abstract
Rationale:
Autosomal-dominant mutations in ryanodine receptor type-2 (RYR2) are responsible for ~60% of all catecholaminergic polymorphic ventricular tachycardia (CPVT). Dysfunctional RyR2 subunits trigger inappropriate calcium leak from the tetrameric channel resulting in potentially lethal ventricular tachycardia. In vivo CRISPR/Cas9-mediated gene editing is a promising strategy that could be used to eliminate the disease-causing Ryr2 allele and hence rescue CPVT.
Objective:
To determine if somatic in vivo genome editing using the CRISPR/Cas9 system delivered by adeno-associated viral (AAV) vectors could correct CPVT arrhythmias in mice heterozygous for RyR2 mutation R176Q (R176Q/+).
Methods and Results:
Guide RNAs (gRNA) were designed to specifically disrupt the R176Q allele in the R176Q/+ mice using the Staphylococcus aureus Cas9 (SaCas9) genome editing system. AAV vectors based on serotype 9 (AAV9) were used to deliver Cas9 and gRNA to neonatal mice by a single subcutaneous injection at postnatal day 10. Strikingly, none of the R176Q/+ mice treated with AAV-CRISPR developed arrhythmias, compared with 71% of R176Q/+ mice receiving control AAV9. Total Ryr2 mRNA and protein levels were significantly reduced in R176Q/+ mice, but not in wildtype littermates. Targeted deep sequencing confirmed successful and highly specific editing of the disease-causing R176Q allele. No detectable off-target mutagenesis was observed in the wildtype Ryr2 allele or the predicted putative off-target site, confirming high specificity for SaCas9 in vivo. In addition, confocal imaging revealed that gene editing normalized the enhanced Ca2+ spark frequency observed in untreated R176Q/+ mice without affecting systolic Ca2+ transients.
Conclusions:
AAV9-based delivery of the SaCas9 system can efficiently disrupt a disease-causing allele in cardiomyocytes in vivo. This work highlights the potential of somatic genome editing approaches for the treatment of lethal autosomal-dominant inherited cardiac disorders such as CPVT.
Keywords: Catecholaminergic polymorphic ventricular tachycardia, CRISPR/Cas9, gene editing, arrhythmia, electrophysiology, gene therapy, ryanodine receptor, AAV, Arrhythmias
INTRODUCTION
Catecholaminergic polymorphic ventricular tachycardia (CPVT) is a life-threatening inherited cardiac arrhythmia syndrome that can lead to stress-induced arrhythmias and sudden death beginning in childhood. If untreated, the mortality rate can reach up to 50% during the first 8 years following diagnosis.1 Despite the poor prognosis, currently available treatments are only partially effective and are often accompanied by side effects. For example, β-blockers which are commonly used for the treatment of CPVT can only control 30-50% of the symptoms.2 Significant adverse side effects such as bradycardia and reduced exercise tolerance can lead to poor adherence in particular in children.3 Implantable defibrillators can be a life-saving treatment for ventricular arrhythmias but repeated shocks or electrical storm leading to death may still occur.4 Therefore, there is a significant unmet need for permanent curative therapies for this devastating disease.
The most common form of CPVT is caused by autosomal-dominant mutations in the gene encoding ryanodine receptor 2 (RYR2), which is responsible for 60% of all CPVT cases.5 The RyR2 channel is activated by calcium (Ca2+) influx through plasmalemmal voltage-gated Ca channels. The consequent release of Ca2+ from the sarcoplasmic reticulum (SR) via RyR2 initiates myocyte contraction. Mutations in RYR2 that cause CPVT promote uncontrolled calcium leak from the SR, which may persist after depolarization, and can trigger lethal arrhythmias.6 In autosomal-dominant CPVT, the RyR2 homotetrameric channel is composed of wildtype and mutant subunits which can combine in varying stoichiometries (4:0, 3:1, 2:2, 1:3, 0:4). Since RyR2 channels containing at least one mutant monomer can promote calcium leak, we reasoned that removing even a fraction of these mutant subunits could increase the proportion of fully functional wildtype channels in the R176Q/+ mice.
The CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 system uses an RNA-guided nuclease (Cas9) to make targeted double-stranded breaks (DSB) in DNA.7 These DSB are most often repaired by the non-homologous end-joining (NHEJ) pathway in quiescent cell types, resulting in insertion and deletion mutations (indels). Since most indels are frameshifting, CRISPR/Cas9 editing can very efficiently knock out alleles through the introduction of premature stop codons, splicing abnormalities, and/or nonsense mediated decay (NMD) of edited transcripts.8 Since CPVT is often caused by gain-of-function missense mutations in RYR2, we reasoned that this autosomal-dominant disease may serve as an excellent candidate for allele-specific disruption with CRISPR/Cas9. Reduced RyR2 levels were found to be well tolerated in RyR2 heterozygous mutant mice with CPVT, suggesting that a reduction in (mutant) RyR2 levels per se would not be expected to be harmful.9 We designed guide RNAs (gRNA) to specifically target the disease-causing R176Q allele for disruption. The heterozygous R176Q/+ mice were previously shown to develop bidirectional or polymorphic VT in response to adrenergic stress.10 Adeno-associated viral vectors (AAV) were used to deliver the CRISPR/Cas9 machinery to cardiomyocytes in vivo to determine whether allele-specific disruption of Ryr2 expression could correct CPVT in live animals. Here we show that AAV-CRISPR is both efficient and specific in the heart, and that disruption of the R176Q mutant allele can prevent arrhythmogenesis in a CPVT mouse model.
METHODS
The data that support the findings of this study are available from the corresponding author upon reasonable request
Animal studies.
All studies were performed according to protocols approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine conforming to the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (NIH Publication No. 85-23, revised 1996). We used R176Q/+ knock-in mice on a C57Bl6/J background that are maintained as an in-house breeding colony, and genotyped as described.10,11 Similar numbers of male and female mice were used, and randomization to treatment groups took place at postnatal day 10. Inclusion criteria were successfully genotyped heterozygous R176Q/+ mice and wild-type littermates, 10 days of age; exclusion criteria were runt mice with a body weight <2SD below the litter average. All analyses were performed in a blinded manner.
AAV9 production and delivery.
The AAV-CRISPR vector, 1255_pAAV-U6-SA-BbsI-MluI-gRNA-CB-SACas9-HA-OLLAS-spA was generated by the Lagor lab (Addgene plasmid 109320) based on the SaCas9 transgene from px602, Addgene plasmid 61593 (gift of Dr. Feng Zhang).12 Cloning details and AAV9 production procedures are provided in the Online Supplement. For systemic delivery of virus, mice on postnatal day 10 (P10) were subcutaneously injected with AAV9. Doses ranged from 5e11-1e12 genome copies/mouse, delivered in a 50 μl volume of Ringer’s lactate solution. Mice were randomly allocated to receive the different AAV9 treatments.
Programmed intracardiac stimulation.
In vivo electrophysiology studies were performed at 5-6 weeks post-injection in similar numbers of male and female R176Q/+ mice and wildtype littermates,13 see Supplemental Methods for details. The experimenter was blinded to the genotype and type of AAV9 treatment.
Quantitative real-time PCR.
Total RNA was isolated from ventricular tissues by TRIzol™ (Life technologies, 15596) and Direct-zol™ (ZYMO Research, R2072), and was reverse transcribed by iScript™ (Bio-Rad, 1708841). The iTaq Universal SYBR Green Supermix was used for the qPCR analysis of diluted cDNA in 1:10. Primers targeting upstream (exon 3-6) and downstream (exon 11-13) of the gRNA target site in Ryr2 are listed in Online Table I. The ΔΔCT method was used to calculate relative quantities normalized to Gapdh as the housekeeping gene.
Western blot analysis.
Heart tissue was snap-frozen in liquid nitrogen. Western blotting was performed as described.14
Next generation deep sequencing to quantify CRISPR/Cas9 at on-/off-target sites.
To identify potential off-target sites in the mouse genome (mm9 and mm10), we applied COSMID (https://crispr.bme.gatech.edu/) to find any off-target sites with three mismatches without insertion or deletion and 2 base mismatches with an insertion or deletion.15 Genomic DNA was extracted from cardiac tissue by DNeasy kit (Qiagen, 69504) and was amplified by locus specific primers (Online Table I) targeting on-/off-target sites using AccuPrime™ (Thermo Fisher, 12346086). All amplicons were then purified using the PCR Clean-up system (Promega, A9282). P5 and P7 adapter sequences and indexes for each sample identification were added by PCR, all barcoded amplicons in an equal molar ratio were pooled together and processed on the Illumina MiSeq platform by 2 × 250 paired-end sequencing. Next gen sequencing data are available on the NCBI/ US National Library of Medicine website using the link: https://www.ncbi.nlm.nih.gov/sra/SRP155568.
Cellular Ca2+ imaging.
Isolated ventricular myocytes were loaded with 2 μM Fluo-4-AM (Invitrogen, Carlsbad, CA) in normal Tyrode buffer solution (pH 7.4 ± 0.05) containing 1.8 mM Ca2+ and prepared for Ca2+ sparks imaging, as described.16 Briefly, Ca2+ sparks were recorded using line-scan mode with 1024 pixels per line at 500 Hz on a LSM880 confocal microscope (Carl Zeiss, Thornwood, NY). 1-Hz steady-state Ca2+ transient pacing of myocytes were induced followed by a 20-s pause during which sparks were recorded. Ca2+ spark frequency (CaSF) was assessed with ImageJ software using SparkMaster plugin. Ca2+ transient amplitude (CaT) was quantified by normalizing peak fluorescence to basal fluorescence using clampfit. Additionally, SR Ca2+ load was measured as peak fluorescence to basal fluorescence after pacing followed by acute caffeine application (10 mM) to release SR Ca2+.
Statistical analysis.
Results are expressed as mean ± SEM. For clustered data in which continuous variables were observed, generalized estimating equation was used via SPSS version 24 (IBM) or Prism using an unpaired Student t test or ANOVA, after performing the D’Agostino-Pearson normality test for normal distribution of the data. Categorical variables were evaluated with Fisher’s exact test. For multiple group comparison, one-way ANOVA followed by Tukey’s post-test was used. P<0.05 was considered statistically significant.
RESULTS
Design of an AAV-CRISPR vector for disruption of a pathogenic Ryr2 allele.
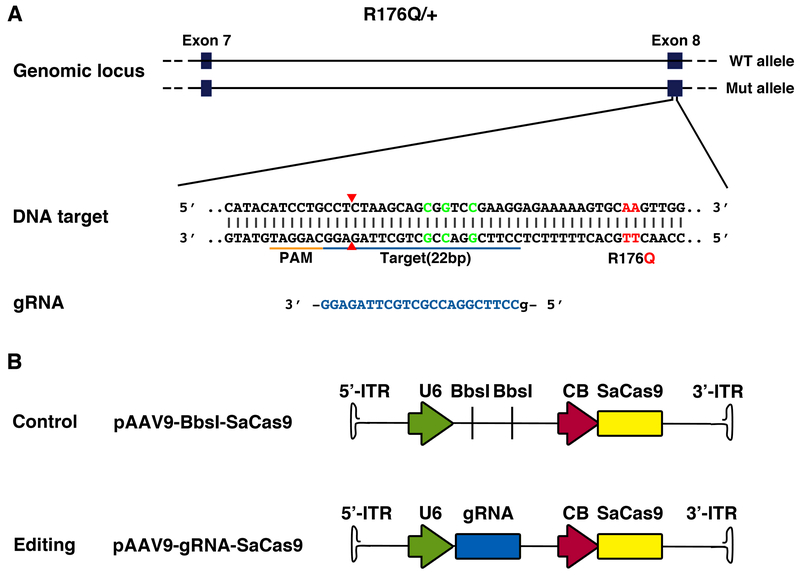
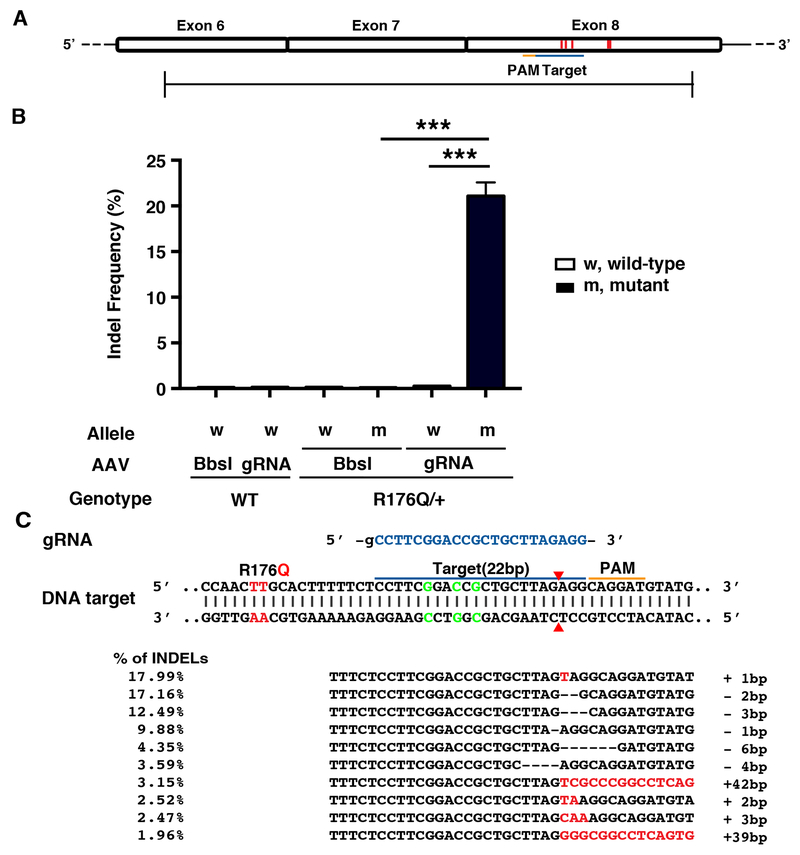
To determine whether the mutant R176Q allele in heterozygous R176Q/+ mice with CPVT could be disrupted using gene editing, we used the CRISPR/Cas9 system from Staphylococcus aureus, which is small enough to be packaged into AAV vectors together with the gRNA. A gRNA for SaCas9 was designed to target the mutant allele exclusively by binding to a silent restriction site present only in the knock-in allele, about 15 bp from the mutation site (Figure 1A). The gRNA was cloned into an AAV9 vector driven by the U6 promoter with a downstream SaCas9 sequence driven by the ubiquitous chicken beta actin (CB) promoter (Figure 1B). A similar construct containing an empty gRNA cloning sites (BbsI) was used as the non-targeting control.
Figure 1. Design of gRNA-SaCas9 to target the mutant Ryr2 allele encoding the R176Q mutation using AAV9.
A, Schematic of gRNA design to target R176Q mutant Ryr2 allele in genomic DNA of R176Q/+ mouse. The mutant codon resulting in the R176Q mutation (red) and the silent Rsr restriction site (green) are marked in the sequence. The guide sequence (22-nt, blue) pairs with the DNA target marked as blue bar upstream of a 5’-NNGRRT adjacent motif (PAM; orange). Cas9 will mediate a double-stranded break ~3 bp upstream of the PAM (red triangle). B, Schematic representation of the SaCas9 AAV9 constructs. Original backbone with BbsI cloning sites is used as control.
AAV9-mediated gene editing makes R176Q/+ mice resistant to arrhythmia induction.
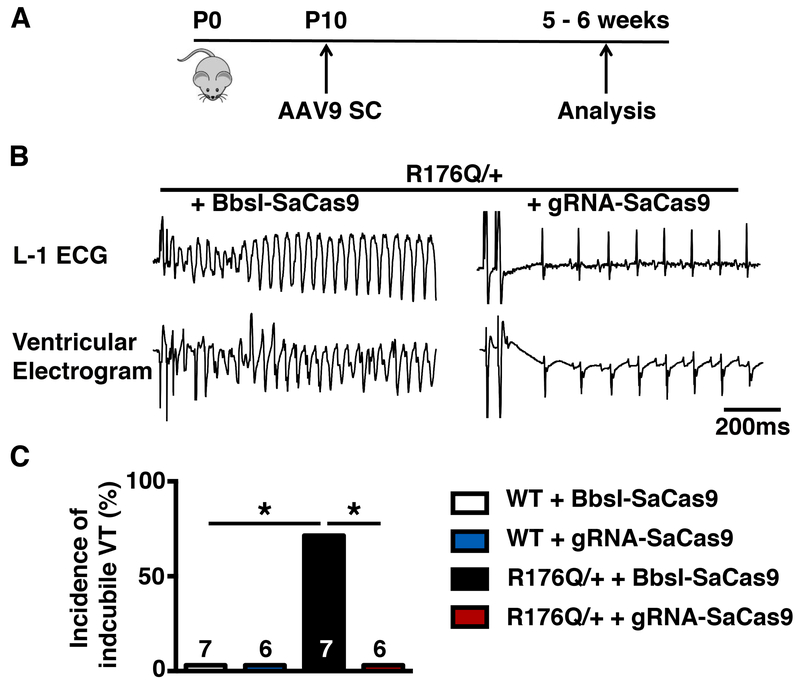
A single-dose systemic in vivo delivery of CRISPR/Cas9 was achieved by injecting 0.5 - 1e12 genome copies of AAV9 subcutaneously into R176Q/+ mice at postnatal day 10. To test for functional correction of CPVT, we tested for susceptibility of these mice to VT following programmed electrical stimulation (PES) at 5-6 weeks after AAV9 administration (Figure 2A). To mimic the adrenergic stimulation, mice were injected with isoproterenol (0.5 mg/kg, i.p.) and caffeine (120 mg/kg, i.p.). None of the WT mice treated with BbsI-SaCas9 (n=7) or gRNA-SaCas9 (n=6) exhibited sustained VT following PES. As expected, we found that 71% of the R176Q/+ mice treated with the BbsI-SaCas9 negative control virus (n=7) developed sustained VT including both polymorphic and bidirectional VT following PES (P<0.05 vs. WT). Remarkably, none of the R176Q/+ mice treated with gRNA-SaCas9 developed pacing-induced VT (0%, n=6) (P< 0.05; Figure 2B). These findings indicate that allele-specific disruption of the R176Q/+ allele using AAV-mediated gene editing offers significant protection from VT. To determine whether gene editing altered cardiac contractility, echocardiography was performed on R176Q/+ mice treated with gRNA-SaCas9 or BbsI-SaCas9. There was no significant difference in ejection fraction or end-diastolic left ventricular diameter comparing both groups (Online Figure I).
Figure 2. AAV9-mediated gene editing makes R176Q/+ mice resistant to arrhythmia induction.
A, Experimental timeline of AAV9 injections and subsequent phenotypical analysis B, Representative examples of simultaneous recording of surface ECG lead I (L-1) and intracardiac ventricular electrogram showing sustained polymorphic ventricular tachycardia in a R176Q/+ mouse treated with placebo (AAV9-Bbsl-SaCas9; left) and normal sinus rhythm in an AAV9-gRNA-SaCas9 treated R176Q/+ mouse (right). C, Bar graph summarizing the incidence of inducible sustained VT in R176Q/+ mice and WT littermates treated with AAV9-Bbsl-SaCas9 or AAV9-gRNA-SaCas9, respectively. Abbreviations: P, postnatal day; SC, subcutaneous. *p<0.05.
Evidence for removal of the mutant RyR2 subunit in mice treated with AAV-CRISPR.
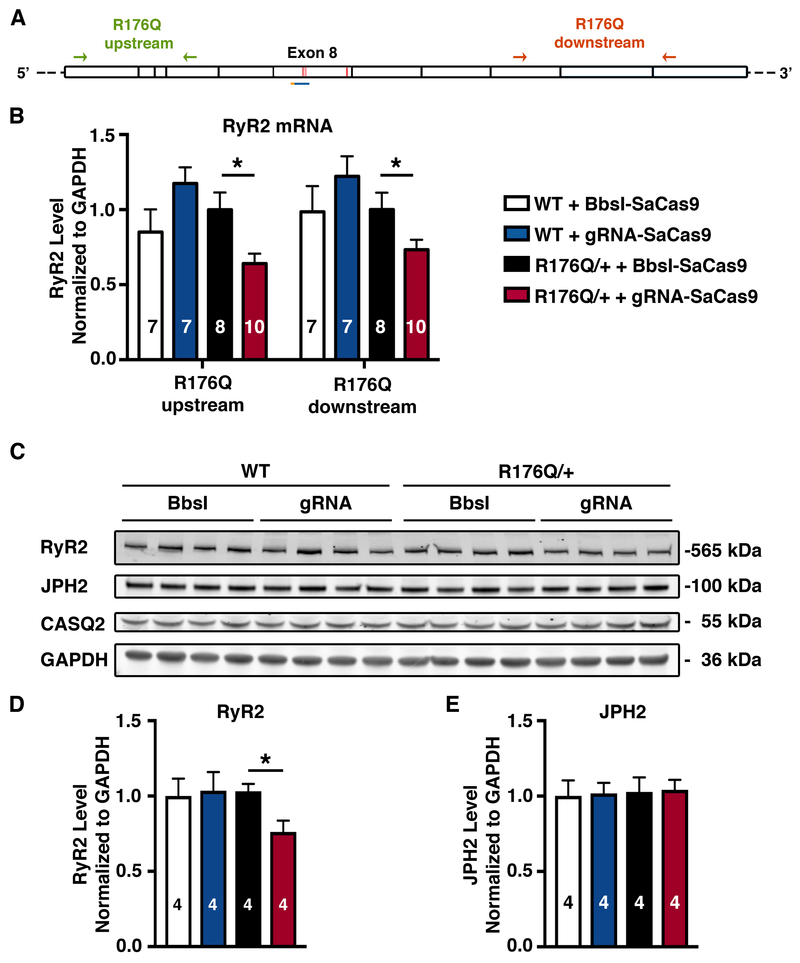
To determine the mechanism underlying the therapeutic effects of CRISPR/Cas9 gene editing in our mouse model, we designed qPCR primers targeting sequences upstream and downstream of Ryr2 exon 8, which contains the R176Q mutation (Figure 3A). Quantitative PCR was performed to measure the level of total Ryr2 mRNA in ventricular samples. R176Q/+ heterozygous mice treated with the gRNA-SaCas9 vector showed a roughly 30% decrease of total Ryr2 mRNA, compared to R176Q/+ mice receiving the non-targeting control virus (P<0.05; Figure 3B). Similar results were obtained with qPCR primers targeting sequences upstream and downstream of exon 8 (Figure 3B). In contrast, there was no change in Ryr2 mRNA levels in WT mice treated with the gRNA-SaCas9 vector compared with the control, suggesting a high degree of specificity of gene editing for the mutant allele.
Figure 3. Transcriptional and translational modifications of RyR2 in R176Q/+ mice mediated by CRISPR/Cas9 genome editing.
A, Diagram showing where PCR primers bind to sequences in exons 3-6 upstream of mutation R176Q in exon 8 (green) and primers targeting exons 11-13 downstream of R176Q (orange). B, Levels of RyR2 mRNA in cardiac tissue from R176Q/+ mice or WT littermates, treated with BbsI-SaCas9 or gRNA-SaCas9. C, Western blot analysis of RyR2 and JPH2 protein levels in heart tissue from R176Q/+ mice or WT littermates. D, Corresponding quantifications of RyR2, and E, JPH2 proteins levels normalized to GAPDH. *p<0.05.
Next, we examined possible splicing of exon 8 using reverse transcription PCR (RT-PCR). Interestingly, we detected exon 8-deleted mRNA in cardiac tissue from R176Q/+ mice that received the gRNA-SaCas9 CRISPR vector (Online Figure II-A). The 113 base-pair deletion associated with splicing-mediated removal of exon 8 causes a frameshift mutation predicted to cause a downstream stop codon in exon 9 after the translation of 29 aberrant amino acids. Amplicon sequencing of the lower molecular weight band confirmed splicing-mediated removal of exon 8 since the sequences of exon 7 and exon 9 were found to be contiguous (Online Figure II-B).
We used a RyR2 antibody with an epitope downstream of the editing site (amino acid 2367) to only detect the full-length RyR2 protein by western blotting (Figure 3C). In agreement with the mRNA data, the level of total RyR2 protein in R176Q/+ mice receiving the gRNA-SaCas9 vector was decreased by ~25% compared to mice injected with control virus (P<0.05; Figure 3D). Likewise, total RyR2 protein levels were not reduced in WT mice treated with the gRNA-SaCas9 vector, further supporting the specificity of our CRISPR editing strategy. Finally, we determined whether gene editing affected the levels of junctophilin-2 (JPH2), a key structural protein located near RyR2 within Ca2+ release units in cardiomyocytes. Western blotting revealed that JPH2 protein levels were not affected (Figure 3E), indicating that RyR2 knockdown was a specific result of CRISPR editing rather than any disturbance of junctional membrane complexes.
High allele-specificity of the CRISPR/Cas9 system targeting RyR2 exon 8.
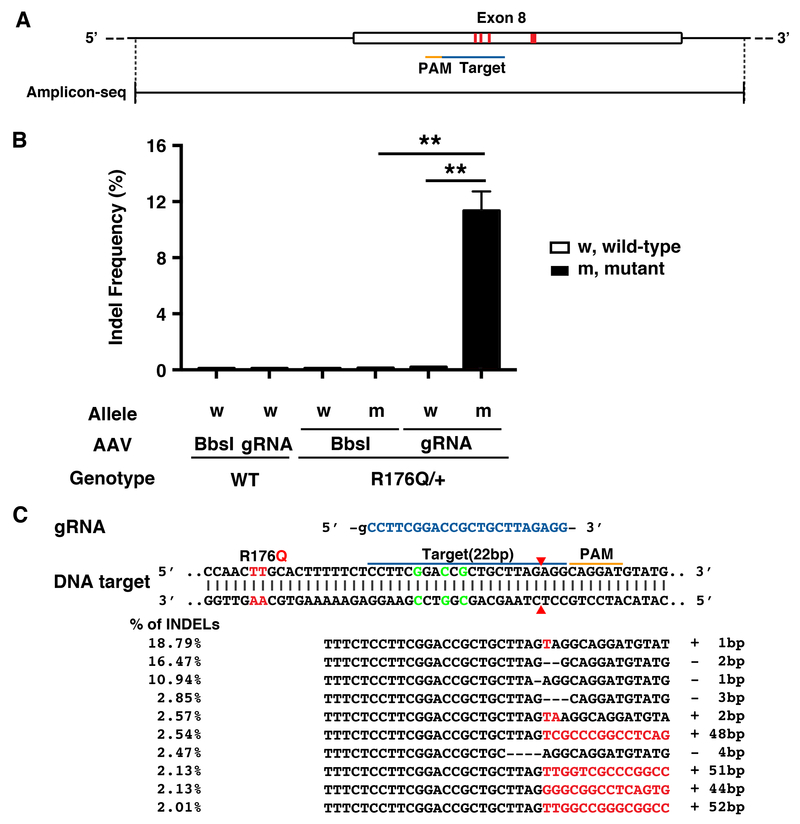
To the efficiency and specificity of our CRISPR editing approach, next generation sequencing (NGS) was performed using heart tissue obtained 5-6 weeks after AAV9 injection. Due to the high specificity of SaCas9 which has a complex PAM (NNGRRT), only one potential off-target site was predicted by COSMID in the mouse genome.15 Both the on-target site (Figure 4A) and off-target site were amplified and barcoded by PCR. There was no evidence of editing in wild-type mice receiving the gRNA-SaCas9 vector, since the background indel frequency was very low (0.10±0.01%) similar to WT mice receiving Bbsl-SaCas9 control vector (0.10±0.00%; P=NS; Figure 4B). Similarly, there was no non-specific editing of the WT allele in heterozygous R176Q/+ mice receiving Bbsl-SaCas9 (0.11±0.01%) or gRNA-SaCas9 vector (0.19±0.02%; P=NS), respectively. By contrast, there was a significantly higher indel frequency at the on-target site on the R176Q/+ allele in mice receiving gRNA-SaCas9 (11.3±1.39%) compared with Bbsl-Cas9 (0.12±0.01%; P<0.01). Importantly, the indel frequency at the off-target site was much lower and not significantly increased in WT or R176Q/+ mice receiving gRNA-SaCas9 (Online Figure III). Finally, we analyzed the top 10 indels taking an average of 4 heart samples from R176Q/+ mice treated with gRNA-SaCas9 (Figure 4C). The majority of indel mutations were 1-2 base pairs in size, centered around position −3 relative to the PAM. Such indels cause frameshifts that ultimately result in premature stop codons.
Figure 4. Deep sequencing validates the efficiency and specificity of gRNA-SaCas9 delivered by AAV9 to target the mutant RyR2 allele.
A, Sequencing amplicon covering exon 8. Thick red line within exon 8 marks the codon containing the R176Q mutation in the Ryr2 gene. Triplet of thin red lines mark the silent restriction site. The gRNA target is marked by the blue bar and the PAM sequence is marked by the orange bar. B, Frequency of indels in genomic DNA at the on-target RyR2 site in cardiac tissue from R176Q/+ mice and WT littermates (n=4 each). C, Common indels observed at the target site. Insertions are marked with red, and deletions are represented by dashed lines. Target sequence is highlighted in blue and PAM is marked with an orange line. ** p<0.01.
As shown in our recent report on S. pyogenes (SpCas9) gene editing,17 we also observed small insertion events containing sequences from the AAV inverted terminal repeats. Representative examples of AAV-based ITR insertions are shown in Online Figure IV. In addition to small ITR insertions, PCR using primers specific to the AAV-CRISPR vector revealed evidence of whole vector genome insertions at the cut site, which occurred in both forward and reverse orientations (Online Figures V and VI). Thus far, these results indicate that our AAV-CRISPR vector can disrupt Ryr2 with high efficiency and specificity, introducing a spectrum of different mutations, which in aggregate reduce the protein levels of the mutant RyR2 subunit.
Impact of CRISPR/Cas9 gene editing on mRNA transcripts.
To gain a better understanding of the impact of gene editing on the relative amounts of WT and R176Q mutant mRNA transcripts, we performed deep sequencing of cDNA that was reverse transcribed from the same tissue samples. One of the primers was designed to bind within exon 8 to avoid capturing the exon 8-deleted transcript (Figure 5A). The relative proportions of mRNA transcribed from the WT or mutant allele of R176Q/+ mice were significantly altered in gRNA-SaCas9 treated R176Q/+ mice compared to those receiving BbsI-SaCas9 control vector (Online Figure VII). There were significantly fewer cDNA reads containing the mutation (19.5%±1.1%) compared to WT reads (80.5%±1.1%; P<0.001), suggesting that the CRISPR-edited transcripts may exert differential effects on mRNA splicing and stability.
Figure 5. Validation of the efficiency and specificity of gene editing of the RyR2 mutation allele in cDNA using deep sequencing.
A, Sequencing amplicon covering exon 6 through 8. Red line marks the mutation in R176Q allele. Target sequence is marked with the blue bar and PAM sequence is marked using the orange bar. B, Frequency of indels in cDNA at the on-target RyR2 site in WT and R176Q/+ mice (n=4). C, Most common indels in cDNA observed at the target site. Insertions are marked with red and deletions are represented by dash line. Target sequence is highlighted in blue and PAM is marked as above. *** p<0.001.
The background indel frequency in the cDNA samples was also very low in WT mice receiving BbsI-SaCas9 (0.12±0.01%) or gRNA-SaCas9 (0.14±0.01%; P=NS), and R176Q/+ mice receiving BbsI-SaCas9 (0.13±0.01% of WT alleles; 0.11±0.01% of mutant alleles; P=NS; Figure 5B). In contrast, there was a high indel frequency in the mutant allele of gRNA-SaCas9 treated R176Q/+ mice (21.1±1.51%), which is significantly higher than in the WT allele (0.23±0.01%; P<0.001). These results provide clear evidence for allele specificity of gene editing in the R176Q/+ mice. Moreover, the higher indel frequency in mRNA (21%) compared with the genomic DNA (11%) consistent with the assumption that about 50% of the nuclei in cardiac tissue samples are derived from cardiomyocytes, which are the most abundant cell type in the heart expressing RyR2. Finally, we analyzed the top 10 indels in the amplified mRNA taking an average of 4 heart samples from R176Q/+ mice treated with gRNA-SaCas9. We found that the most common of these were generally consistent with the edits observed at the genomic level (Figure 5C).
AAV9-mediated normalized intracellular Ca2+ handling in R176Q/+ mice.
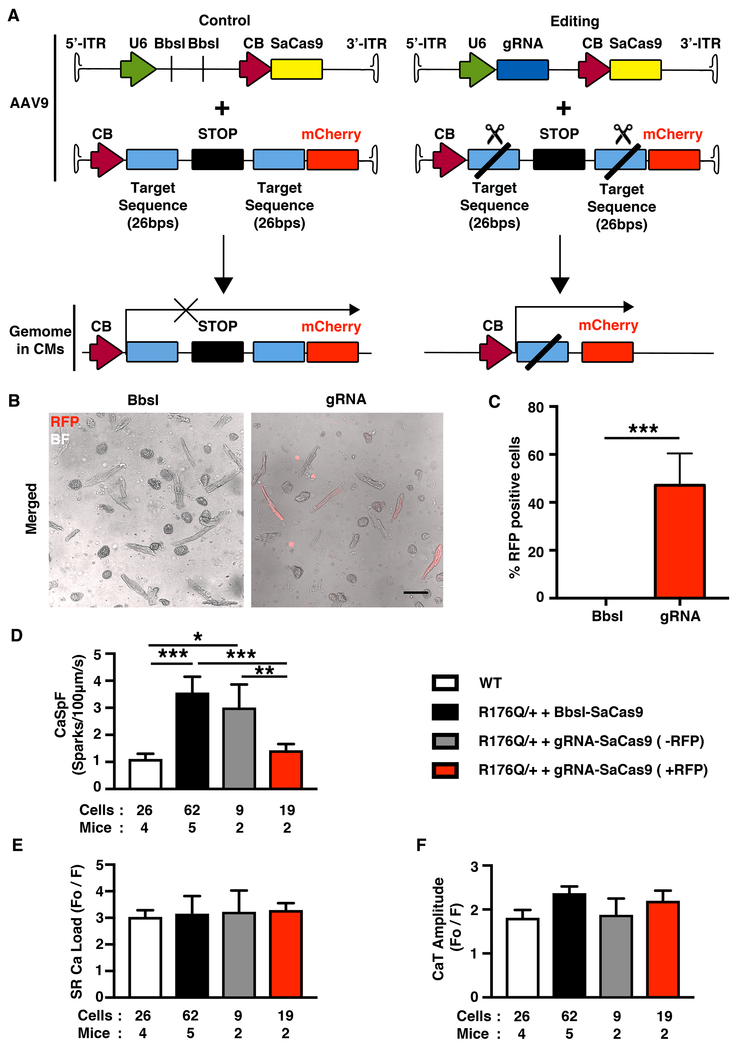
To determine the effects of AAV9-mediated gene editing on intracellular Ca2+ handling, gRNA-SaCas9 (or Bbsl-SaCas9 as control) and mCherry “reporter” vectors were co-injected in R176Q/+ mice. The reporter vector contains a STOP cassette flanked by two copies of the gRNA target sequence, which can be excised in the presence of gRNA and SaCas9. Hence, if both AAV9 successfully transduce cardiomyocytes in R176Q/+ mice, fluorescent mCherry expression can identify cells with gene editing activity (Figure 6A). Confocal microscopy of ventricular myocytes isolated from R176Q/+ mice 5-6 weeks after treatment with gRNA-SaCas9 and reporter AAV9 revealed that 47% of the cells were positive for mCherry (Figure 6B and C). In contrast, none of the myocytes were mCherry-positive in R176Q/+ mice that received the control Bbsl-SaCas9 vector together with reporter (Figure 6B and C).
Figure 6. AAV9-mediated normalized intracellular Ca2+ handling in R176Q/+ mice.
A, Schematic picture showing gRNA-SaCas9 and “reporter” AAV9 co-injected in R176Q/+ mice. The “reporter” virus with cloned targeted sequence was cut by gRNA-SaCas9 but not the control BbsI-SaCas9. The mCherry protein is only expressed in cardiomycytes with SaCas9 expression. B, Representative confocal images of ventricle myocytes from R176Q/+ mice co-injected with Bbsl-SaCas9 (control) + “reporter” and gRNA-SaCas9 (editing) + “reporter”, merged with bright field (BF) and red fluorescence (RFP) signal overlay. Scale bar, 100 μm. C, Bar graph showing percentages of mCherry-positive cardiomyocytes. D-F, Summary data for Ca2+ spark frequency, SR Ca2+ load, and SR Ca2+ transient amplitude. *p<0.05, **p<0.01, ***p<0.001.
Next, myocytes were loaded with Ca2+ fluorophore Fluo-4-AM and RyR2 activity was assessed by measuring the Ca2+ spark frequency (CaSpF) after a 1-Hz conditioning train. Consistent with prior studies,18 the CaSpF was significantly higher in myocytes from R176Q/+ mice (treated with Bbsl-SaCas9; 2.3±0.5 a.u.) compared with those from WT mice (1.1±0.2 a.u.; P<0.001; Figure 6D). The CaSpF was significantly lower in mCherry-positive myocytes from R176Q/+ mice + gRNA-SaCas9 (1.4±0.2 a.u.) in which gene editing occurred compared with mCherry-negative myocytes from the same animals (3.0±0.8 a.u.; P<0.01; Figure 6D). These differences in CaSpF were not a consequence of alterations in SR Ca2+ loading (Figure 6E). Finally, we measured the amplitude of SR Ca2+ transients during 1-Hz field stimulation and did not observe significant differences among the groups. These data demonstrate that gene editing as confirmed by the mCherry signal led to normalization of SR Ca2+ release in myocytes from R176Q/+ mice.
DISCUSSION
In this study, we applied somatic genome editing with CRISPR/Cas9 to correct a potentially lethal monogenic disorder of the heart. We demonstrated that a single AAV9 vector encoding both SaCas9 and gRNA caused efficient editing of cardiomyocytes in vivo. By specifically targeting the disease-causing allele in a model of autosomal-dominant CPVT, our approach reduced levels of the mutant RyR2 subunit and restored normal cardiac function. In addition, we performed confocal imaging to test if the AAV9-mediated gRNA-SaCas9 gene editing therapy normalized intracellular Ca2+ handling in R176Q/+ mice. The increased frequency of Ca2+ sparks seen in untreated R176Q/+ mice was reduced to levels seen in WT mice with gRNA-SaCas9 treatment. In addition, the systolic Ca2+ transients were not affected, demonstrating that the in vivo gene editing approach is effective and safe, and that a mild reduction in overall RyR2 protein levels does not affect cardiac contractility. Thus, our editing strategy was efficient and specific, and prevented catecholamine-induced VT in a relevant mouse model of this human disease.
Recent papers have shown that AAV9 vectors can deliver CRISPR/Cas9 to the heart to somatically knock out genes.19,20 These studies have relied on the commonly used S. pyogenes Cas9 (SpCas9) system, which presents delivery challenges with AAV due to its size. In these cases, the gRNA must either be delivered via a separate vector or be introduced into SpCas9 transgenic mice. Since the S. aureus Cas9 (SaCas9) is about 1 kb smaller than SpCas9, it can fit with together with the gRNA into a single AAV vector.12 AAV-SaCas9 has been used successfully for gene editing in liver12,21,22 skeletal muscle,23,24 and very recently in heart for excision of exon 23 from the mdx model of Duchenne muscular dystrophy.25 In related work, Xie et al.26 generated transgenic and knock-in mice to model the H530R mutation in the PRKAG gene, which causes non-sarcomeric familial hypertrophic cardiomyopathy associated with Wolff-Parkinson-White in humans. These authors used a dual vector AAV9-based system to deliver SpCas9 along with a gRNA to disrupt the mutant allele. Indel rates were in the range of ~2.6-6.5% in mice injected as neonates. In their study, disruption was very specific for the disease-causing PRKAG allele, and fully restored heart morphology and function. In our study, we were able to achieve much higher indel rates for the R176Q mutant allele (11.3% of genomic, 21.1% of mRNA) in R176Q/+ mice that received the gRNA-SaCas9 vector. To our knowledge, our study is the first example of such efficient somatic gene editing in cardiac myocytes, which effectively cured a lethal autosomal-dominant disease of the heart.
We examined allele-specific editing in detail at the DNA, mRNA, and protein levels. Our gRNA was very specific to the R176Q mutant Ryr2 allele, with no evidence for mutagenesis of the wildtype Ryr2. Allele-specificity is critical in the case of CPVT arising from RYR2 mutations, as the calcium release channel is mandatory for proper excitation-contraction coupling in the heart. It is currently unknown whether a 20-25% reduction in total RyR2 in humans would create new pathology, and this would need to be addressed through human genetics of rare variants and studies in relevant cell systems. The indels at the cut site were primarily frame-shifting insertions or deletions that are expected to promote nonsense-mediated decay, consistent with the reduced mRNA levels.
In this study we found fragments of the ITRs as well as whole AAV-CRISPR vector genomes inserted at the cut site in Ryr2. Whole vector genome insertions were an unexpected consequence which occurred only in the R176Q/+ mutant mice that received the AAV vector expressing the gRNA. It is noteworthy that these were not found in WT mice receiving the AAV-CRISPR vector, further emphasizing the specificity of the gRNA for the mutant allele. While it is not technically possible to quantify these insertions relative to the shorter indels, they may represent a substantial fraction of the mutant alleles. This may help explain why apparently modest rates of editing detected by NGS could cause such a striking correction of CPVT. All of these large insertions would prevent expression of the mutant RyR2 protein subunit. This does however present an additional safety concern of “on-target” insertional mutagenesis that merits further investigation.
In addition to a reduction in the total mRNA level for Ryr2, we also observed skipping of exon 8. Consistent with a previous report,27 this accounted for a relative small fraction of all the gRNA-modified sequences. In addition to allele-specificity, we found no evidence of off-target mutagenesis at predicted sites. This is logical given the very restrictive PAM of SaCas9 (NNGRRT), which minimizes potential off-target sites elsewhere in the genome, but also greatly limits target site selection. The development of refined genome editing tools such as KKH SaCas9 will be invaluable in expanding the targeting capability of this system.28 A limitation of our study is that the gRNA does not bind to the exact mutation site, but an artificial adjacent sequence in the R176Q model. Nonetheless, we provide compelling evidence that AAV-SaCas9 vectors can efficiently edit Ryr2 in an allele-specific manner, rescuing CPVT pathology. Moreover, we provide a platform for targeting other previous reported mutations also located in exon 8 including P164S, R169Q, V186M, E189D.29
Recently, Bongianino et al.9 performed allele-specific silencing of mutant Ryr2 through AAV9 expressed shRNA in another mouse model of CPVT. In this case, the R4496C mutant RyR2 subunit was selectively silenced at the mRNA level, resulting in a net 15% reduction in total RyR2 protein levels. This AAV9-shRNA vector corrected mitochondrial and junctional SR/T-tubule abnormalities, and reduced the incidence of adrenergically-stimulated VT in the mice.9 This study and ours support the concept of removing disease-causing Ryr2 alleles as a viable therapeutic approach to treat CPVT. However, RNA interference and antisense approaches still have significant specificity and delivery challenges for targeting human cardiomyocytes. In the case of CPVT, repeated and chronic dosing would be needed for life. In contrast, a single-dosed AAV-CRISPR genome editing approach is particularly attractive, and would be potentially curative if demonstrated to be safe and specific enough for clinical trials.
There is currently no consensus on what large animal preclinical models are best suited to address issues of CRISPR/Cas9 efficiency, specificity, efficacy and safety. In addition, off-target mutagenesis is likely to be dependent on the patient-specific gRNA, and will vary based on the potential sites present in the rest of their unique genomes. Recent studies have shown very promising results using AAV9 based vectors in a clinical study involving patients suffering from spinal muscular atrophy type 1.30 A single dose of intravenous AAV9 coding for the missing survival motor neuron 1 results in longer survival and superior motor function compared to a historic cohort of patients without such treatment. AAV9 has also been used extensively for gene delivery to heart and skeletal muscle in mice, include recent genome editing studies for Duchenne muscular dystrophy.23,24 Since RyR2 is also expressed in other cell types such as smooth muscle cells and neurons, AAV9-based approaches might be developed to correct RyR2 mutations in those cell types. This may even be important for the treatment of CPVT, since we have demonstrated that mutant RyR2 can contribute to neuronal deficits that contribute to arrhythmia development in CPVT mice.31 The editing of neuronal RyR2 mutants would be therapeutic rather than harmful in this context. However, it has been demonstrated that only high doses of AAV9 can cross the blood-brain barrier and may transduce neurons.32 Therefore, it is possible but unlikely (given the AAV9 dose used in this study) that editing of neurons contributed to the prevention of arrhythmias. Nevertheless, in future studies neuronal gene editing might be exploited for the treatment of cardiac arrhythmias in addition to direct cardiomyocyte targeting. In summary, our findings demonstrate that AAV9 mediated gene editing may be a promising treatment modality of inherited monogenic diseases such as CPVT.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Catecholaminergic polymorphic ventricular tachycardia (CPVT) is an inherited disease of cardiac rhythm.
More than 60% of CPVT cases are caused by autosomal-dominant missense mutations in RYR2.
The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 system can be used to edit the genome of living cells.
What New Information Does This Article Contribute?
A single injection of adeno-associated virus (AAV) was used to deliver CRISPR/Cas9 genome editing machinery to a mouse model of CPVT.
In vivo genome editing resulted in specific disruption of the disease-causing Ryr2 allele, while sparing the normal copy of the gene.
Gene editing reduced total RyR2 mRNA and protein levels and restored normal calcium handling.
In vivo gene editing protected the R176Q/+ mice from pacing-induced cardiac arrhythmias.
The findings demonstrate that AAV-CRISPR vectors can be used to treat a lethal inherited arrhythmia syndrome, through targeted removal of a pathogenic allele in vivo. Gene editing was specific for Ryr2, and reduced the amount of mutant but not the normal RyR2 allele. These studies highlight the promise of somatic genome editing for the treatment of CPVT and other lethal autosomal-dominant diseases of the heart.
ACKNOWLEDGEMENTS
We thank Dr. Susan Hamilton for providing the R176Q/+ mouse model.
SOURCES OF FUNDING
This work is supported by NIH grants R01-HL132840 (W.R.L.), R01-HL089598, R01-HL091947, R01-HL117641 (X.H.T.W.), R56-HL131649, and R01-HL136389 (N.L.), an Individual Investigator Research Grant Award from the RYR1 foundation (W.R.L.), an American Heart Association grant 14SDG20080008 (N.L.), and a grant from the Saving Tiny Hearts Foundation (X.H.T.W.).
Nonstandard Abbreviations and Acronyms:
- AAV9
adeno-associated virus serotype 9
- Cas9
CRISPR-associated nuclease 9
- CPVT
catecholaminergic polymorphic ventricular tachycardia
- CRISPR
clustered regularly interspaced short palindromic repeats
- gRNA
guide RNA
- indel
insertions or deletions
- PAM
protospacer adjacent motif
- RyR2
ryanodine receptor type 2
- SR
sarcoplasmic reticulum
- VT
ventricular tachycardia
- WT
wildtype
Footnotes
In June 2018, the average time from submission to first decision for all original research papers submitted to Circulation Research was 13.26 days.
DISCLOSURES
XHTW is a founding partner of Elex Biotech, a start-up company that developed drug molecules that target ryanodine receptors for the treatment of cardiac arrhythmia disorders. Other authors have no conflicts.
REFERENCES
- 1.Hayashi M, Denjoy I, Extramiana F, Maltret A, Buisson NR, Lupoglazoff JM, Klug D, Hayashi M, Takatsuki S, Villain E, Kamblock J, Messali A, Guicheney P, Lunardi J and Leenhardt A. Incidence and risk factors of arrhythmic events in catecholaminergic polymorphic ventricular tachycardia. Circulation. 2009;119:2426–34. [DOI] [PubMed] [Google Scholar]
- 2.Imberti JF, Underwood K, Mazzanti A and Priori SG. Clinical Challenges in Catecholaminergic Polymorphic Ventricular Tachycardia. Heart Lung Circ 2016;25:777–83. [DOI] [PubMed] [Google Scholar]
- 3.van der Werf C and Lieve KV. Beta-blockers in the treatment of catecholaminergic polymorphic ventricular tachycardia. Heart Rhythm 2016;13:441–2. [DOI] [PubMed] [Google Scholar]
- 4.Sumitomo N. Current topics in catecholaminergic polymorphic ventricular tachycardia. J Arrhythm. 2016;32:344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landstrom AP, Dobrev D and Wehrens XHT. Calcium Signaling and Cardiac Arrhythmias. Circ Res 2017;120:1969–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wehrens XH, Lehnart SE and Marks AR. Intracellular calcium release and cardiac disease. Annu Rev Physiol 2005;67:69–98. [DOI] [PubMed] [Google Scholar]
- 7.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA and Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popp MW and Maquat LE. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell. 2016;165:1319–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bongianino R, Denegri M, Mazzanti A, Lodola F, Vollero A, Boncompagni S, Fasciano S, Rizzo G, Mangione D, Barbaro S, Di Fonso A, Napolitano C, Auricchio A, Protasi F and Priori SG. Allele-Specific Silencing of Mutant mRNA Rescues Ultrastructural and Arrhythmic Phenotype in Mice Carriers of the R4496C Mutation in the Ryanodine Receptor Gene (RYR2). Circ Res 2017;121:525–536. [DOI] [PubMed] [Google Scholar]
- 10.Kannankeril PJ, Mitchell BM, Goonasekera SA, Chelu MG, Zhang W, Sood S, Kearney DL, Danila CI, De Biasi M, Wehrens XH, Pautler RG, Roden DM, Taffet GE, Dirksen RT, Anderson ME and Hamilton SL. Mice with the R176Q cardiac ryanodine receptor mutation exhibit catecholamine-induced ventricular tachycardia and cardiomyopathy. Proc Natl Acad Sci U S A. 2006;103:12179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathur N, Sood S, Wang S, van Oort RJ, Sarma S, Li N, Skapura DG, Bayle JH, Valderrabano M and Wehrens XH. Sudden infant death syndrome in mice with an inherited mutation in RyR2. Circ Arrhythm Electrophysiol 2009;2:677–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA and Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li N and Wehrens XH. Programmed electrical stimulation in mice. J Vis Exp 2010:1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li N, Chiang DY, Wang S, Wang Q, Sun L, Voigt N, Respress JL, Ather S, Skapura DG, Jordan VK, Horrigan FT, Schmitz W, Muller FU, Valderrabano M, Nattel S, Dobrev D and Wehrens XHT. Ryanodine receptor-mediated calcium leak drives progressive development of an atrial fibrillation substrate in a transgenic mouse model. Circulation. 2014;129:1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cradick TJ, Qiu P, Lee CM, Fine EJ and Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids. 2014;3:e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N, Wang T, Wang W, Cutler MJ, Wang Q, Voigt N, Rosenbaum DS, Dobrev D and Wehrens XH. Inhibition of CaMKII phosphorylation of RyR2 prevents induction of atrial fibrillation in FKBP12.6 knockout mice. Circ Res 2012;110:465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarrett KE, Lee CM, Yeh YH, Hsu RH, Gupta R, Zhang M, Rodriguez PJ, Lee CS, Gillard BK, Bissig KD, Pownall HJ, Martin JF, Bao G and Lagor WR. Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci Rep 2017;7:44624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li N, Wang Q, Sibrian-Vazquez M, Klipp RC, Reynolds JO, Word TA, Scott L Jr., Salama G, Strongin RM, Abramson JJ and Wehrens XHT. Treatment of catecholaminergic polymorphic ventricular tachycardia in mice using novel RyR2-modifying drugs. Int J Cardiol 2017;227:668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll KJ, Makarewich CA, McAnally J, Anderson DM, Zentilin L, Liu N, Giacca M, Bassel-Duby R and Olson EN. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc Natl Acad Sci U S A. 2016;113:338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, VanDusen NJ, Zhang L, Gu W, Sethi I, Guatimosim S, Ma Q, Jardin BD, Ai Y, Zhang D, Chen B, Guo A, Yuan GC, Song LS and Pu WT. Analysis of Cardiac Myocyte Maturation Using CASAAV, a Platform for Rapid Dissection of Cardiac Myocyte Gene Function In Vivo. Circ Res 2017;120:1874–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML and Wilson JM. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol 2016;34:334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Yasui K, Opthof T, Ishiki R, Lee JK, Kamiya K, Yokota M and Kodama I. Developmental changes of Ca(2+) handling in mouse ventricular cells from early embryo to adulthood. Life Sci 2002;71:1279–92. [DOI] [PubMed] [Google Scholar]
- 23.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM and Wagers AJ. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D and Gersbach CA. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Refaey M, Xu L, Gao Y, Canan BD, Adesanya TMA, Warner SC, Akagi K, Symer DE, Mohler PJ, Ma J, Janssen PML and Han R. In Vivo Genome Editing Restores Dystrophin Expression and Cardiac Function in Dystrophic Mice. Circ Res 2017;121:923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie C, Zhang YP, Song L, Luo J, Qi W, Hu J, Lu D, Yang Z, Zhang J, Xiao J, Zhou B, Du JL, Jing N, Liu Y, Wang Y, Li BL, Song BL and Yan Y. Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res 2016;26:1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharpe JJ and Cooper TA. Unexpected consequences: exon skipping caused by CRISPR-generated mutations. Genome Biol 2017;18:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z and Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 2015;33:1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medeiros-Domingo A, Bhuiyan ZA, Tester DJ, Hofman N, Bikker H, van Tintelen JP, Mannens MM, Wilde AA and Ackerman MJ. The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome: a comprehensive open reading frame mutational analysis. J Am Coll Cardiol 2009;54:2065–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, Lowes L, Alfano L, Berry K, Church K, Kissel JT, Nagendran S, L’Italien J, Sproule DM, Wells C, Cardenas JA, Heitzer MD, Kaspar A, Corcoran S, Braun L, Likhite S, Miranda C, Meyer K, Foust KD, Burghes AHM and Kaspar BK. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N Engl J Med 2017;377:1713–1722. [DOI] [PubMed] [Google Scholar]
- 31.Aiba I, Wehrens XH and Noebels JL. Leaky RyR2 channels unleash a brainstem spreading depolarization mechanism of sudden cardiac death. Proc Natl Acad Sci U S A. 2016;113:E4895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duque S, Joussemet B, Riviere C, Marais T, Dubreil L, Douar AM, Fyfe J, Moullier P, Colle MA and Barkats M. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther 2009;17:1187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.