Abstract
DNA methylation patterns change with age and can be used to derive an estimate of “epigenetic age,” an indicator of biological age. Several studies have shown associations of posttraumatic stress disorder (PTSD) with worse somatic health and early mortality, raising the possibility of accelerated biological aging. This study examined associations between estimated epigenetic age and various variables in 160 male combat-exposed war veterans with (n = 79) and without PTSD (n = 81). DNA methylation was assessed in leukocyte genomic DNA using the Illumina 450K DNA methylation arrays. Epigenetic age was estimated using Horvath's epigenetic clock algorithm and Δage (epigenetic age-chronological age) was calculated. In veterans with PTSD (Δage = 3.2), Δage was on average lower compared to those without PTSD (Δage = 5.0; p = 0.02; Cohen's d = 0.42). This between-group difference was not explained by race/ethnicity, lifestyle factors or childhood trauma. Antidepressant use, however, explained part of the association. In the PTSD positive group, telomerase activity was negatively related to Δage (β = −0.35; p = 0.007). In conclusion, veterans with PTSD had significantly lower epigenetic age profiles than those without PTSD. Further, current antidepressant use and higher telomerase activity were related to relatively less epigenetic aging in veterans with PTSD, speculative of a mechanistic pathway that might attenuate biological aging-related processes in the context of PTSD.
Keywords: Aging, Antidepressants, Epigenetics, Posttraumatic stress disorder
Introduction
Aging can be seen as a time-dependent functional decline that affects living organisms [1]. Whereas chronological aging is indexed by calendar time, biological aging is indexed by multiple cellular and molecular processes such as genomic instability, telomere attrition and mitochondrial dysfunction [1]. A recent indicator of biological aging is based on DNA methylation patterns [2]. Essentially, DNA methylation is characterized by the addition of a methyl group to cytosine at cytosine-phosphate-guanine (CpG) sites, thereby regulating gene expression. Recent studies identified several genes and genomic regions that become either hypomethylated or hypermethylated with age [2, 3], which, together, can be used to develop a so-called epigenetic clock. In 2013, Horvath [3] developed such an epigenetic age predictor using 8,000 samples from multiple tissues, identifying 353 CpG sites that predict chronological age with high accuracy [3]. Since then, several studies have applied this epigenetic clock to a variety of conditions, showing associations between accelerated epigenetic aging and insomnia [4], Down syndrome [5], obesity [6], Parkinson's disease [7], and all-cause mortality [8, 9].
Indicators of biological age, rather than chronological age, are of increased interest in the study of stress-related psychiatric disorders, because of their associations with increased risks for somatic illnesses commonly seen with advanced age [10, 11] and an almost twofold higher risk of mortality [12]. In recent years, telomere length, one of the most robust indicators of cellular aging, has been extensively studied and was found to be shorter across a variety of psychiatric disorders [13, 14], including posttraumatic stress disorder (PTSD) [15]. Associations with the epigenetic clock, which may also be linked to the telomere/telomerase system [16], remain relatively unexplored in psychiatric settings. So far, few studies have examined the epigenetic age in PTSD. Boks et al. [17] found that in 96 male veterans, combat trauma exposure was associated with accelerated epigenetic aging based on Horvath's algorithm, but in contrast, the development of PTSD symptoms during follow-up was associated with less epigenetic aging. In a study comprising 281 male and female veterans, Wolf et al. [18] found no associations between Horvath's epigenetic age estimate and lifetime PTSD, but alternatively they found that accelerated epigenetic age was associated with lifetime PTSD based on an another epigenetic estimate using 71 CpG sites by Hannum et al. [19]. In a follow-up study, Wolf et al. [20] found an association between accelerated epigenetic age (as estimated by Hannum et al. [19]) and PTSD hyperarousal symptoms in 339 trauma-exposed veterans but not with total PTSD severity. Last of all, Zannas et al. [21] found that lifetime stressors, but not current PTSD symptomatology, were associated with accelerated epigenetic aging in 392 highly traumatized African American individuals.
Overall, the literature examining associations of epigenetic age with PTSD symptomatology or diagnosis is scarce and mixed [17, 18, 20, 21]. The current study examined epigenetic age in white blood cells based on Horvath's algorithm in 79 male combat-exposed war veterans with PTSD and 81 male combat-exposed war veterans without PTSD. To our knowledge, this is the first study to compare trauma-exposed veterans with a clinically verified current PTSD diagnosis with a control sample of veterans matched for age and sex without PTSD. Furthermore, data of a subsample (n = 44) were available for follow-up longitudinal testing, which allowed us to examine the stability of epigenetic age estimates over time.
Patients and Methods
Ethical Statement
The Institutional Review Boards of Icahn School of Medicine at Mount Sinai (ISMMS; New York, NY, USA), the James J. Peters Veterans Administration Medical Center (JJPVAMC; Bronx, New York, NY, USA), New York University Medical Center (NYU; New York, NY, USA), the US Army Medical Research and Materiel Command, and the University of California, San Francisco, Medical Center (UCSF; San Francisco, CA, USA) approved this study. Study participants gave their written informed consent to participate. Participants were compensated for their participation. The study was conducted in accordance with the provisions of the Helsinki Declaration.
Recruitment Procedures and Study Sample
One hundred sixty-six veterans from Operation Iraqi Freedom and Operation Enduring Freedom were recruited by NYU and ISMMS/JJPVAMC. Participants were recruited from the Mental Health Services of the Manhattan, Bronx and Brooklyn Veterans Affairs Medical Centers, other regional VA medical centers, Veterans Service Organizations, National Guard, reservist agencies and organizations and from the general community. Recruitment methods included flyers, in-person presentations, media advertisements, Internet postings (e.g., Craigslist), and referral from clinicians. Criteria for inclusion were (a) having served in war zones; (b) current age between 20 and 60; (c) males; and (d) proficient in the English language. Exclusion criteria included: (a) history of alcohol dependence within the past 8 months; (b) history of drug abuse or dependence (except nicotine dependence) within the past year; (c) lifetime history of any psychiatric disorder with psychotic features, bipolar disorder, or obsessive-compulsive disorder; (d) those who were currently being exposed to recurrent trauma or had been exposed to a traumatic event within the past 3 months; (e) prominent suicidal or homicidal ideation; (f) neurologic disorder or systemic illness affecting central nervous system function; (g) history of anemia or recent blood donation in the past 2 months; (i) veterans who were not stable for at least 2 months on psychiatric medication, anticonvulsants, antihypertensive medication, or sympathomimetic medication; (j) veterans who were classified with a moderate or severe traumatic brain injury (TBI) on the Ohio State University TBI Identification Method-Short Form [22].
All study participants had exposure to a combat-related PTSD Criterion A trauma and participants in the PTSD+ group were also required to have a current (past month) Clinician Administered PTSD Scale for DSM-IV (CAPS) score [23] ≥40 and to meet full DSM-IV criteria for PTSD; in the non-PTSD group, all veterans were required to have a current CAPS score ≤20 and they could never have met criteria for PTSD in the past. The CAPS cut-off scores of ≤20 and ≥40 were selected to create stringent and distinct groups of PTSD negative and positives participants. The average duration from any self-reported PTSD symptoms was 72 months for PTSD positive and 29 months for PTSD negative veterans (p < 0.001). The 166 combat-exposed male veterans were recruited for a larger overall study examining biomarkers of PTSD. Of these, 6 were removed because there were no DNA methylation data available (n = 4), hepatitis was diagnosed (n = 1) and a psychiatric diagnosis in the control group was made (n = 1), respectively, leaving 79 participants with PTSD and 81 without PTSD. Forty-four of the veterans with PTSD and none of the combat-exposed controls fulfilled the criteria for major depressive disorder (MDD). Additionally, longitudinal follow-up data were available for a subsample of 44 participants (28 PTSD+ veterans and 16 controls). The average follow-up duration was 3.3 years (SD = 1.1; range 1.1–5.1).
Clinical Assessments
To establish psychiatric diagnoses, a Structured Clinical Interview for DSM-IV disorders (SCID) was conducted by doctoral-level psychologists, was audio recorded and calibrated weekly with a senior clinician [24]. Current (past month) and lifetime PTSD symptom severity was assessed with the CAPS. Depression symptom severity was assessed with the self-rated Beck Depression Inventory-II (BDI-II) [25]. The Perceived Stress Scale was used to measure the perception of psychological stress in the past month [26]. Exposure to early life trauma was evaluated using the Early Trauma Inventory-Self Report Short Form [27]. Antidepressant use was assessed by self-report.
Data from the Deployment Risk and Resilience Inventory-2 (DRRI-2) were available for 90 participants (35 with and 55 without PTSD) [28]. The DRRI-2 is a suite of 17 individual scales that assesses key deployment-related risk and resilience factors. For the purpose of assessing the degree of combat exposure in this sample, we used the Combat Experiences subscale (section D). Further, the number of tours of deployment was assessed, including deployment to regions other than Iraq and Afghanistan.
Blood Draw and DNA Methylation Assessment
Blood was drawn in the morning after a night of fasting. Genomic DNA was isolated from blood collected in PAXgene Blood DNA Tubes using the PAXgene Blood DNA Kit according to the manufacturer's protocol (Qiagen, Germantown, MD, USA). The DNA yield was assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher, Wilmington, DE, USA). The A260/280 and A260/230 ratios were measured by spectrophotometric assay, and quality controlled samples were selected if both ratios were larger than 1.75. The methylated DNA fraction was selected from the whole genome via immunocapture performed on a 200 μL fragmented pooled DNA sample.
The whole blood genomic DNA (500ng) from each sample was bisulfite-converted using the EZ96 DNA methylation kit (Zymo Research, Orange, CA, USA) following the manufacturer's standard protocol with instructions specific for Illumina arrays. The bisulfite-treated DNA was then analyzed using HumanMethylation450 BeadChip (Illumina Inc., USA) according to the manufacturer's protocol. To minimize the potential batch effects, the samples were fully randomized in each experimental step, and all samples were processed with the same technician and machines. The samples were de-identified, the labs that performed the assays were blind to all other measurements, and case and control samples were randomly distributed over 4 plates.
Epigenetic Age
Epigenetic age was calculated from DNA methylation data using the online calculator by Horvath [3]. In short, normalization of the relevant CpG methylations levels was conducted using a mixed effect model and based on the DNA methylation levels. We used a customized R code, according to the calculator's pipeline and used the “BMIQ method using a Gold standard” [3] method for normalization. Horvath predicted ages of tissue based on regression coefficients obtained from training sets encompassing over 13,000 subjects, described in the original article [3]. This epigenetic profile correlated strongly with chronological age assessed across tissues and cell types, and across multiple studies and populations.
Cell Counts
Cell counts were estimated by Horvath's epigenetic clock software [29]. We also used complete blood count to confirm the estimations. The correlations between cell type percentages using Complete Blood Count and the estimations are presented in online Supplementary Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000491431). Leukocyte cell type counts had no significant associations with any of the CAPS scores, PTSD status or combat exposure. To adjust for cell count in analyses with epigenetic age, Horvath's estimates were used (see Statistical Analyses).
Telomere Length and Telomerase Activity
The assay of telomere length from leukocytes was adapted from the published original method [30] as reported elsewhere [31]. The same reference DNA was used for all PCR runs. Telomerase activity in leukocytes was assessed with an optimized assay on the basis of the commercially available kit TRAPeze (Chemicon, Temecula, CA, USA) reported in more detail elsewhere [32]. In short, 2 concentrations (5,000 and 10,000 cells) were used for Telomeric Repeat Amplification Protocol (TRAP) reactions to ensure that the assay for each sample is in the linear range. In all, 293T cells are used as positive controls. In addition to the 293T cancer cell extract as the standard for telomerase activity quantification, we also include 2 positive control extracts from the human cancer cell line Hela. The average intra assay variability of PBMC samples (n = 6, assayed in triplicates) is 8% and the inter-assay variability of PBMC samples (n = 24, assayed on 2 different days) is 6.7%. More details of both methods can be found in the study by Lin et al. [33].
Statistical Analyses
Baseline characteristics were compared between PTSD+ and PTSD– veterans using chi-square statistics and analyses of variance. ΔAge was calculated for all participants by subtracting actual chronological age from estimated epigenetic age (epigenetic age minus chronological age) to reflect accelerated (positive Δage values) or decelerated (negative Δage values) epigenetic aging. PTSD+ and PTSD– groups were matched for chronological age on a group level and therefore age was not added as a covariate in subsequent analyses. Analyses of covariance were used to determine differences in Δage between veterans with and without PTSD. All analyses with Δage were controlled for abundance measures of cell types estimated by Horvath's epigenetic clock software: naïve CD8 and CD4 T-cells, CD8+CD28-CD45RA T-cells, plasma blast cells, natural killer cells, monocytes, and granulocytes (model 1); additionally, analyses were controlled for race/ethnicity, alcohol use, smoking status, and BMI (model 2). In secondary analyses, we tested the extent to which between-group differences were explained by antidepressant use, childhood trauma, comorbid MDD and number of deployment tours. Regression analyses further tested associations between ∆age and symptom severity scales, antidepressant use, childhood trauma and number of deployment tours in the total sample. To gain insight into relationships of epigenetic age with other measures of biological aging, we tested associations between Δage and telomere length and telomerase activity.
As a validation check for using Δage as our main outcome variable, we performed additional analyses in which we regressed DNA methylation age on chronological age and saved the unstandardized residuals. Positive DNA methylation age residuals reflect accelerated epigenetic age, while negative residuals reflect decelerated epigenetic age. All analyses were performed with both ∆age and unstandardized residuals as outcome variables, both analyses yielded highly similar results (results not shown).
Results
Sample Characteristics
The mean chronological age of the total study sample was 32.8 years (SD = 7.9, range 20–59). Characteristics for veterans with and without PTSD are presented in Table 1. The groups did not differ significantly in age, education, race/ethnicity, and alcohol use. Veterans with PTSD had a higher average BMI, reported more childhood trauma and were more often current smokers. Further, the groups differed significantly on all clinical variables, with the PTSD positive group having more severe symptoms, comorbid MDD diagnoses and antidepressant users, as expected. The groups did not differ in the number of deployment tours (range 1–4).
Table 1.
Sample characteristics (mean ± SD or % [n]) by PTSD status (n = 160)
| PTSD negative (n = 81) | PTSD positive (n = 79) | p value* | |
|---|---|---|---|
| Sociodemographic characteristics | |||
| Age | 32.6 (8.0) | 33.0 (7.8) | 0.790 |
| Years of education | 13.4 (5.1) | 13.0 (3.9) | 0.649 |
| Race/ethnicity, % (n) | 0.170 | ||
| Hispanic | 32.1 (26) | 45.6 (36) | |
| Non-hispanic black | 23.5 (19) | 26.6 (21) | |
| Non-hispanic white | 33.3 (27) | 24.1 (19) | |
| Asian | 6.2 (5) | 1.3 (1) | |
| Other | 4.9 (4) | 2.5 (2) | |
| Lifestyle characteristics | |||
| Body mass index | 28.5 (4.8) | 30.1 (5.1) | 0.042 |
| Hazardous alcohol use (AUDIT) | 2.9 (2.4) | 3.2 (3.8) | 0.612 |
| Smoking (% yes, n) | 11.1 (9) | 34.2 (27) | <0.001 |
| Clinical characteristics | |||
| Clinician administered PTSD scale (CAPS), current | 3.7 (5.2) | 68.8 (16.9) | <0.001 |
| Clinician administered PTSD scale (CAPS), lifetime | 9.3 (8.5) | 91.1 (15.7) | <0.001 |
| Beck depression inventory (BDI-II) | 5.6 (6.2) | 25.1 (10.9) | <0.001 |
| Perceived stress scale | 1.8 (0.6) | 3.1 (0.7) | <0.001 |
| Comorbid major depressive disorder (% yes, n) | 0 (0) | 55.7 (44) | <0.001 |
| Current antidepressant use (% yes, n) | 4.9 (4) | 27.8 (22) | <0.001 |
| Childhood trauma inventory score | 2.2 (1.9) | 2.8 (2.2) | 0.079 |
| Deployment characteristics | |||
| Combat experiences (DRRI-D score; n = 90) | 31.5 (12.4) | 53.2 (18.4) | <0.001 |
| Number of military tours (range 1–4) | 1.8 (0.9) | 1.8 (0.8) | 0.882 |
p values are based in one-way ANOVA for continuous variables and chi's square test for categorical variables.
Epigenetic age was strongly correlated with chronological age across the total sample (r = 0.812; p < 0.001); see online Supplementary Figure 1. ΔAge ranged from −10.8 to 19.5, and the average Δage across the entire sample was 4.1 (SD = 4.9). ΔAge was negatively related to chronological age (r = −0.261; p = 0.001), but was not associated with race/ethnicity (p = 0.192), alcohol use (p = 0.564), smoking status (p = 0.846) or BMI (p = 0.453). Those participants who currently used antidepressant medication (n = 26) had lower Δage than those without antidepressants (p = 0.017) across the total sample.
Between-Group Comparisons of ΔAge
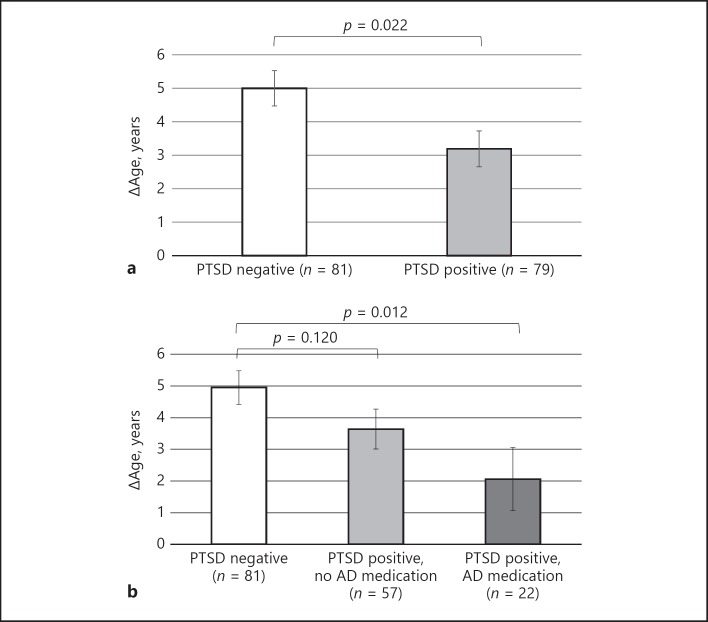
The PTSD positive group had an average Δage of 3.2 (SD = 4.5), while the PTSD negative group had a Δage of 5.0 (SD = 4.6). This showed that those with a PTSD diagnosis had an average lower Δage than that of the group without PTSD (model 1: F[1,150] = 6.85; p = 0.022; Cohen's d = 0.42, Fig. 1a), indicating significantly less acceleration of epigenetic age in those with PTSD. Adjusting for race/ethnicity (including Hispanic status), smoking, alcohol, and BMI did not impact the group difference (model 2: p = 0.019).
Fig. 1.
Mean Δage (epigenetic age – chronological age) by (a) PTSD status and (b) PTSD and current antidepressant medication use. Note. Estimated means (and standard error) of Δage from analysis of covariance, adjusted for abundance measures of cell types (model 1); AD medication = antidepressant medication.
In sensitivity analyses, we tested whether clinical and deployment characteristics were explanatory factors in the between-group comparison. The between-group difference remained significant when adjusting for childhood trauma (p = 0.032) or number of tours (p = 0.023). When adjusting for current antidepressant use, the between-group association between PTSD and ∆age became non-significant (p = 0.105). Interestingly, dividing the PTSD positive group into 2 groups based on antidepressant medication use showed that compared to the PTSD negative controls (Δage = 5.0), those with PTSD that used antidepressant medication use (∆age = 2.1; p = 0.012), but not those without medication use (Δage = 3.6; p = 0.120) had significantly lower Δage (model 1: overall effect: F[2,150] = 3.59; p = 0.030, Fig. 1b; model 2: p = 0.015). Within the PTSD group, there was no significant difference in ∆age between those with and without antidepressant medication (p = 0.187).
As a post hoc analysis, we tested whether the association between PTSD status and ∆age was mediated by antidepressant medication, using the Sobel method [34], which estimates the direct and indirect effects of the independent variable on the dependent variable through the mediator variable. Results showed that current antidepressant use was – albeit reducing the direct effect of PTSD on ∆age from c = −1.8 (se = 0.8) to c’ = −1.3 (se = 0.8) – not a significant mediator (p = 0.11).
Associations of ΔAge and Clinical Characteristics
Current and lifetime severity of PTSD symptoms, measured with the CAPS, was negatively associated with Δage across all participants (Table 2). Similar negative associations were found across all participants for the BDI-II and the Perceived Stress Scale. Associations were, however, not significant within the 2 groups separately, indicating that severity measures did not further differentiate within the PTSD group and results might thus reflect the group difference. Those with a MDD diagnosis, further, had significantly lower ∆age than those without MDD (p = 0.013). ΔAge was not associated with childhood trauma (Early Trauma Inventory; β = −0.05; p= 0.512) across all participants and within the 2 groups separately. We found no associations between ∆age and number of deployment tours (β = −0.05; p = 0.517) or severity of combat experiences (β = −0.08; p = 0.448). Adjusting for chronological age did not change any of the results.
Table 2.
Univariate linear regression analyses of Δage with various characteristics in the total sample (n = 160)
| B | SE | β | p value | |
|---|---|---|---|---|
| Clinical characteristics | ||||
| Clinician administered PTSD scale (CAPS), current | –0.03 | 0.01 | –0.21 | 0.007 |
| Clinician administered PTSD scale (CAPS), lifetime | –0.02 | 0.01 | –0.19 | 0.014 |
| Beck's depression inventory (BDI-II) | –0.08 | 0.03 | –0.21 | 0.008 |
| Perceived stress scale | –1.34 | 0.43 | –0.24 | 0.002 |
| Antidepressant use (no = reference) | –2.48 | 1.03 | –0.19 | 0.017 |
| Early Trauma Inventory score | –0.05 | 0.08 | –0.05 | 0.512 |
| Deployment characteristics | ||||
| Number of military tours | –0.30 | 0.46 | –0.05 | 0.517 |
| Combat experiences (DRRI-D; n = 90) | –0.02 | 0.02 | –0.08 | 0.448 |
| Other cellular aging characteristics | ||||
| Telomere length1 | 0.35 | 0.40 | 0.07 | 0.377 |
| Telomerase activity1 | –1.01 | 0.42 | –0.24 | 0.012 |
Univariate regression analyses adjusted abundance measures of cell types (model 1).
Additionally adjusted for chronological age.
Associations of ΔAge and Telomere Length and Telomerase Activity
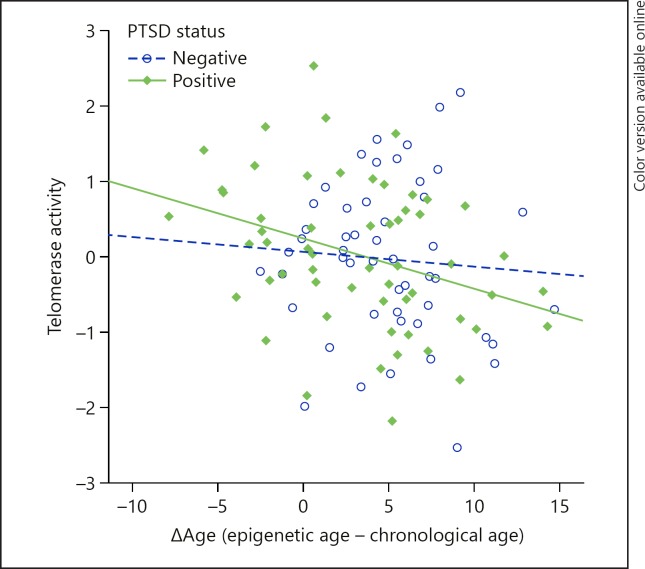
ΔAge was not related to telomere length in the whole sample (model 1: p = 0.377, Table 2) or in the PTSD positive (p = 0.592) or negative (p = 0.925) samples separately. However, we did find a significant negative association between Δage and telomerase activity in in the full sample (model 1: β = −0.238; p = 0.012, Table 2). This association was driven by the group with PTSD (β = −0.352; p = 0.007), while no association was found in the controls (β = −0.147; p = 0.371), showing that in the PTSD group, higher telomerase activity was associated with relatively less epigenetic age acceleration (Fig. 2). The interaction effect of PTSD-by-telomerase activity was trending to be significant (p = 0.83; n = 107). In the current study sample, neither telomere length (p = 0.263) nor telomerase activity (p = 0.761) was significantly associated with PTSD status, in analyses corrected for chronological age. More detailed associations with PTSD status and telomere length or telomerase activity in a larger study sample are being explored in a more comprehensive analysis and are beyond the scope of the current paper (Kang et al., in preparation).
Fig. 2.
Scatterplot of telomerase activity by Δage for PTSD groups separately. Telomerase activity is only significantly associated with Δage in the PTSD positive group (β = −0.35; p = 0.007) and not in the PTSD negative group (β = −0.15; p = 0.371).
Stability of ΔAge Over Time
Follow-up data were available for a subsample of 44 individuals, including 28 veterans with PTSD and 16 without PTSD. Epigenetic age at baseline correlated highly with epigenetic age at an average of 3-year follow-up in this subsample (r = 0.91; p < 0.001), demonstrating a high within-person stability. Correlations between Δage at baseline and follow-up were similarly high (r = 0.68; p < 0.001). Results of a between-group comparison of Δage at follow-up were in the same direction as at baseline, albeit non-significant (PTSD positive Δage = 3.3, PTSD negative Δage = 4.0).
Discussion
This study examined epigenetic age acceleration based on leukocyte DNA methylation levels in male combat-exposed war veterans. Contradicting our expectations, those with PTSD showed less accelerated epigenetic aging than those without PTSD. In other words, those with PTSD showed a relatively “younger” epigenetic profile than PTSD negative controls, according to Horvath's epigenetic age estimator [3]. The significant between-group difference in epigenetic aging was not explained by race/ethnicity, BMI, smoking or alcohol use, childhood trauma, or number of tours of duty. Differences were also not accounted for by proportional differences in leukocyte subtype composition. Interestingly, the between-group difference became non-significant when adjusting for current antidepressant use, suggesting that part of the association might be explained by mechanisms secondary to antidepressant use. Further, analyses suggested that veterans with the most severe psychiatric profiles, such as those with high PTSD symptom severity and MDD comorbidity, had the lowest ∆age. Lastly, in those with PTSD, but not in those without PTSD, telomerase activity was negatively related to Δage, indicating that the higher the telomerase activity, the younger the epigenetic age.
The finding of relatively less accelerated DNA methylation profiles in combat-exposed veterans with PTSD compared to veterans without PTSD was unexpected. Meta-analyses show that PTSD is associated with a higher risk of all-cause mortality [12, 35] as well as several aging-related diseases [36, 37]. Further, recent work showed that epigenetic age is independently associated with mortality [9], which would logically lead to the hypothesis of relatively more accelerated epigenetic aging in those with PTSD compared to those without PTSD. However, most previous studies on PTSD and somatic health or mortality compared PTSD individuals to non-traumatized healthy controls [12, 36, 37], making it difficult to separate the effects of trauma exposure from those of PTSD status per se. Our data suggest that individuals with combat trauma-exposed PTSD showed less epigenetic acceleration than those without PTSD. While this finding was unexpected, another study also found indications of decelerated epigenetic aging in PTSD: Boks et al. [17] found that the development of more PTSD symptoms over 13 months was associated with decelerated DNA methylation aging, as estimated with Horvath's metric. In line with our results, they found that trauma exposure per se (i.e., combat-exposure) was associated with increased epigenetic aging, but the reverse was true for PTSD development in 96 war veterans who were evaluated pre-deployment and again 6 months later. On the other hand, Wolf et al. [20] found no associations in 281 male and female veterans between epigenetic age according to Horvath's algorithm and “latent lifetime PTSD severity,” a construct they developed with factor analysis from pre-military, post-military, and current PTSD symptoms. However, they did not report whether associations were present in the current PTSD group compared to those without PTSD, which makes it difficult to compare with the current study. It should be noted that Wolf et al. [20] did find an association between PTSD and another estimate of epigenetic age by Hannum et al. [19] In a follow-up study, Wolf et al. [20] found an association between accelerated epigenetic age according to Hannum's estimation and PTSD hyperarousal symptoms in 339 trauma-exposed veterans, but not with PTSD severity overall. In that study, Horvath's estimation was not examined. It is interesting to note that analyses with Horvath's and Hannum's estimates have shown mixed results in the literature up until now, indicating that both clocks might capture different (epigenetic) traits. Both clocks have only 6 CpG sites in common and are associated with different genetic loci affecting age acceleration [38]. Although beyond the scope of the present study, future studies should estimate whether the clocks reflect meaningful differences in the context of PTSD (see e.g., [39, 40]).
Currently, we can only speculate on the reasons why we find relatively less accelerated epigenetic age in PTSD compared to the non-PTSD group. One possible explanation is the generation of a compensatory mechanism in response to stress and subsequent PTSD development, an explanation also proposed by Boks et al. [17] Interestingly, we showed that high telomerase activity was associated with lower Δage only in the PTSD group, which is in line with earlier studies that found elevated telomerase in response to acute psychological stress in human caregivers [41, 42], in unmedicated individuals with MDD [32], and chronically stressed rats [43]. Telomerase has several important functions in addition to its role in telomere maintenance, including promotion of cell growth and cell longevity [44, 45], and might thus induce mechanisms that reverse or decelerate cellular aging. For example, elevated telomerase has been found to prevent age-related decrease in DNA 5-methylcytosine transferase I (DNMT1) expression that could contribute in the maintenance of a relatively “younger” epigenetic profile [45, 46]. Further, a recent GWAS revealed a critical role for the human TERT gene in epigenetic aging rates and showed TERT-specific and independent effects on both longer telomere length and epigenetic age acceleration [16]. Like other studies, we found no associations between epigenetic aging and telomere length [47]. A recent study, however, did find an inverse association between telomere length and advanced epigenetic aging, possibly as a result a high memory CD8+ T cells/naive CD8+ T cells ratio [48].
Another possible pathway is that epigenetic changes may interact with altered glucocorticoid responses in PTSD [49], as a recent study found that 85 of the 353 epigenetic clock CpG sites from Horvath's clock were located within glucocorticoid response genes [21]. Activation of the glucocorticoid receptor as a consequence of cortisol secretion might drive changes in DNA methylation and other epigenetic modifications [50]. This may be especially relevant here, since combat-veterans with PTSD showed lower methylation of the promoter of the glucocorticoid receptor gene [51]. Further, we found a suggestion that our results were partly explained by antidepressant medication use. Currently used antidepressants drugs may exert direct epigenetic effects which might impact epigenetic aging [52, 53]. It should be noted, however, that it remains to be explored if our finding regarding antidepressant medication is an effect of antidepressants per se, or rather of severity, since those who used antidepressants also had higher MDD comorbidity levels and CAPS and BDI scores (p values < 0.001).
The current study found no evidence of an association between epigenetic age and early life adversities. This is in line with 2 recent studies that found no associations between epigenetic age and childhood maltreatment in 96 male veterans [17] and 392 individuals from an African American population cohort [21]. Interestingly, the latter study found that in those with high childhood abuse, the associations between personal life stressors and accelerated epigenetic age were attenuated, which the authors proposed to be the consequence of some “resilience mechanisms” triggered by childhood trauma [21]. Kananen et al. [54] found that epigenetic age showed great stability over 4-year and 25-year follow-ups, with hardly any changes in individuals’ rank orders. This is consistent with the stability in epigenetic age that we found in a subset of participants tested at 2 time-points. It remains to be explored whether epigenetic age can be used as a pre-deployment vulnerability marker by future longitudinal studies, including the time before the occurrence of traumas and the onset of PTSD.
Study strengths are the relatively large sample size with extensive clinical phenotyping, including information on clinical severity, combat trauma exposure, childhood trauma, MDD, antidepressant use and a wide variety of lifestyle factors. Further, methylation patterns were all assayed simultaneously in the same lab and randomized over batches, thus minimizing potential batch-related noise. Some limitations of this study should, however, also be noted. First, both groups in this study consisted of male war veterans, selected as relatively young and healthy individuals without traumatic brain injury, thus the generalizability of our findings in samples with and without PTSD in both men and women remains to be explored. It should thus be noted that results might be different for older groups with more comorbidity. Also, those without PTSD in our sample had also experienced combat traumas but did not develop PTSD, which may indicate that they are highly resilient group, as opposed to a traditional healthy control group. Second, our suggestion of overall accelerated epigenetic aging in the current sample should be considered within the parameters of this specific population. We found a suggestion that both veteran groups on average showed older DNA methylation age compared to their chronological age. However, we withhold from interpreting this finding, since we cannot rule out other explanations such as a mean error in the estimation of epigenetic age causing the mean ∆age to be off by a certain constant. Third, the estimate of cellular aging used in this study is designed to provide an accurate predictor of chronological age, rather than to explore mechanistic pathways that lead to alterations in cellular aging. DNA methylation age predicts all-cause mortality in later life [9], suggesting an association with a process related to aging, but it is unknown if the methylation changes at the 353 CpGs used in the clock model play a direct causal role in the aging process. Fourth, we are, to our knowledge, the first to report a negative association between telomerase activity and ∆age in the PTSD, this finding should therefore be considered preliminary, and future research in independent samples should find if this association can be replicated. Fifth, DNA methylation data were collected from mixed blood cell types, while DNA methylation patterns are known to differ between cell types [3]. However, we controlled for abundance measures of cell types as provided by Horvath's online calculator. Lastly, we cannot rule out pre-deployment differences. Future studies could add substantial knowledge by exploring these associations longitudinally and, in the context of veterans, including pre-deployment data.
This study found that combat trauma-exposed veterans with PTSD, showed lower ∆age values, indicating relatively less accelerated epigenetic aging, based on Horvath's “epigenetic clock,” compared to a well-matched control group of combat trauma-exposed veterans without PTSD. This association was partly explained by antidepressant use. Higher telomerase activity was related to relatively lower ∆age only in PTSD patients, suggesting a mechanistic pathway that might attenuate cellular aging-related processes in response to stress. Future research remains to explore the long-term health consequences of these changes in epigenetic age.
Disclosure Statement
All authors declare that there are no conflicts of interest to disclose.
Funding Source
This study was supported by the following grants: U.S. Department of Defense, W81XWH-11-2-0223 (PI: Charles Marmar) and U.S. Department of Defense, W81XWH-10-1-0021 (PI: Owen Wolkowitz). USAMRMC Military Operational Medicine Research Program (MOMRP)/Defense Health Agency (DHA)/Congressional Special Interests (CSI) and MOMRP 190040. JE Verhoeven was supported by the Academy Van der Gaag Grant by the Royal Netherlands Academy of Arts and Sciences.
Disclaimer
The views, opinions, and/or findings contained in this report are those of the authors and should not be construed as an official Department of the Army position, policy, or decision, unless so designated by other official documentation.
Supplementary Material
Supplementary data
References
- 1.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell JT, Tsai PC, Yang TP, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;33:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;33:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll JE, Irwin MR, Levine M, et al. Epigenetic aging and immune senescence in women with insomnia symptoms: findings from the women's health initiative study. Biol Psychiatry. 2016;0:97–111. doi: 10.1016/j.biopsych.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horvath S, Garagnani P, Bacalini MG, et al. Accelerated epigenetic aging in down syndrome. Aging Cell. 2015;14:491–495. doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horvath S, Erhart W, Brosch M, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci. 2014:201412759. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson's disease patients. Aging (Albany, NY) 2015;7:1130–1142. doi: 10.18632/aging.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marioni RE, Shah S, McRae AF, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany, NY) 2016;8:1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viron MJ, Stern TA. The impact of serious mental illness on health and healthcare. Psychosomatics. 2010;51:458–465. doi: 10.1176/appi.psy.51.6.458. [DOI] [PubMed] [Google Scholar]
- 11.Penninx BWJH, Milaneschi Y, Lamers F, Vogelzangs N. Understanding the somatic consequences of depression: biological mechanisms and the role of depression symptom profile. BMC Med. 2013;11:1–14. doi: 10.1186/1741-7015-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker ER, McGee RE, Druss BG. Mortality in mental disorders and global disease burden implications: a systematic review and meta-analysis. JAMA Psychiatry. 2015;72:334–341. doi: 10.1001/jamapsychiatry.2014.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darrow SM, Verhoeven JE, Révész D, et al. The association between psychiatric disorders and telomere length: a meta-analysis involving 14,827 persons. Psychosom Med. 2016;78:776–787. doi: 10.1097/PSY.0000000000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindqvist D, Epel ES, Mellon SH, et al. Psychiatric disorders and leukocyte telomere length: underlying mechanisms linking mental illness with cellular aging. Neurosci Biobehav Rev. 2015;55:333–364. doi: 10.1016/j.neubiorev.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Wang J, Zhou J, Huang P, Li J. The association between post-traumatic stress disorder and shorter telomere length: a systematic review and meta-analysis. J Affect Disord. 2017;218:322–326. doi: 10.1016/j.jad.2017.03.048. [DOI] [PubMed] [Google Scholar]
- 16.Lu AT, Xue L, Salfati EL, et al. GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun. 2018;9:387. doi: 10.1038/s41467-017-02697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boks MP, Mierlo HC van, Rutten BPF, et al. Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology. 2015;51:506–512. doi: 10.1016/j.psyneuen.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Wolf EJ, Logue MW, Hayes JP, et al. Accelerated DNA methylation age: associations with PTSD and neural integrity. Psychoneuroendocrinology. 2016;63:155–162. doi: 10.1016/j.psyneuen.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf EJ, Logue MW, Stoop TB, et al. Accelerated DNA Methylation Age. Psychosom Med. 2017;80:1. doi: 10.1097/PSY.0000000000000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zannas AS, Arloth J, Carrillo-Roa T, et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 2015;16:266. doi: 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corrigan JD, Bogner J. Initial reliability and validity of the Ohio state university TBI identification method. J Head Trauma Rehabil. 2007;22:318–329. doi: 10.1097/01.HTR.0000300227.67748.77. [DOI] [PubMed] [Google Scholar]
- 23.Blake DD, Weathers FW, Nagy LM, et al. The development of a clinician-administered PTSD scale. J Trauma Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- 24.First MB, Spitzer RL, Miriam G, Williams JB. New York: Biometrics Research, New York State Psychiatric Institute; 2002. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition. [Google Scholar]
- 25.Beck AT, Steer RA, Ball R, Ranieri WF. Comparison of beck depression inventories-IA and-II in psychiatric outpatients. J Pers Assess. 1996;67:588–597. doi: 10.1207/s15327752jpa6703_13. [DOI] [PubMed] [Google Scholar]
- 26.Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. J Health Soc Behav. 1983;24:385–396. [PubMed] [Google Scholar]
- 27.Bremner JD, Bolus R, Mayer EA. Psychometric properties of the early trauma inventory-self report. J Nerv Ment Dis. 2007;195:211–218. doi: 10.1097/01.nmd.0000243824.84651.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogt D, Smith BN, King LA, King DW, Knight J, Vasterling JJ. Deployment risk and resilience inventory-2 (DRRI-2): an updated tool for assessing psychosocial risk and resilience factors among service members and veterans. J Trauma Stress. 2013;26:710–717. doi: 10.1002/jts.21868. [DOI] [PubMed] [Google Scholar]
- 29.Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212:1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;33:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolkowitz OM, Mellon SH, Epel ES, et al. Leukocyte telomere length in major depression: correlations with chronicity, inflammation and oxidative stress – preliminary findings. PLoS One. 2011;6:e17837. doi: 10.1371/journal.pone.0017837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolkowitz OM, Mellon SH, Epel ES, et al. Resting leukocyte telomerase activity is elevated in major depression and predicts treatment response. Mol Psychiatry. 2012;17:164–172. doi: 10.1038/mp.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin J, Epel E, Cheon J, et al. Analyses and comparisons of telomerase activity and telomere length in human T and B cells: Insights for epidemiology of telomere maintenance. J Immunol Methods. 2010;352:71–80. doi: 10.1016/j.jim.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobel ME. Asymptotic confidence intervals for indirect effects in structural equation Models. 1982;13:290–312. [Google Scholar]
- 35.Boscarino JA. Posttraumatic stress disorder and mortality among U.S. Army veterans 30 years after military service. Ann Epidemiol. 2006;16:248–256. doi: 10.1016/j.annepidem.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Rosenbaum S, Stubbs B, Ward PB, Steel Z, Lederman O, Vancampfort D. The prevalence and risk of metabolic syndrome and its components among people with posttraumatic stress disorder: a systematic review and meta-analysis. Metabolism. 2015;64:926–933. doi: 10.1016/j.metabol.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Edmondson D, Kronish IM, Shaffer JA, Falzon L, Burg MM. Posttraumatic stress disorder and risk for coronary heart disease: a meta-analytic review. Am Heart J. 2013;166:806–814. doi: 10.1016/j.ahj.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raina A, Zhao X, Grove ML, et al. Cerebral white matter hyperintensities on MRI and acceleration of epigenetic aging: the atherosclerosis risk in communities study. Clin Epigenetics. 2017;9:21. doi: 10.1186/s13148-016-0302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armstrong NJ, Mather KA, Thalamuthu A, et al. Aging, exceptional longevity and comparisons of the Hannum and Horvath epigenetic clocks. Epigenomics. 2017;9:689–700. doi: 10.2217/epi-2016-0179. [DOI] [PubMed] [Google Scholar]
- 40.Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. E Bio Medicine. 2017;21:29–36. doi: 10.1016/j.ebiom.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epel ES, Lin J, Dhabhar FS, et al. Dynamics of telomerase activity in response to acute psychological stress. Brain Behav Immun. 2010;24:531–539. doi: 10.1016/j.bbi.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Damjanovic AK, Yang Y, Glaser R, et al. Accelerated telomere erosion is associated with a declining immune function of caregivers of Alzheimer's disease patients. J Immunol. 2007;179:4249–4254. doi: 10.4049/jimmunol.179.6.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beery AK, Lin J, Biddle JS, Francis DD, Blackburn EH, Epel ES. Chronic stress elevates telomerase activity in rats. Biol Lett. 2012;8:1063–1066. doi: 10.1098/rsbl.2012.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith LL, Coller HA, Roberts JM. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 45.Bersani FS, Lindqvist D, Mellon SH, et al. Telomerase activation as a possible mechanism of action of psychopharmacological interventions. Drug Discov Today. 2015;20:1305–1309. doi: 10.1016/j.drudis.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Young JI, Sedivy JM, Smith JR. Telomerase expression in normal human fibroblasts stabilizes DNA 5-methylcytosine transferase I. J Biol Chem. 2003;278:19904–19908. doi: 10.1074/jbc.M301685200. [DOI] [PubMed] [Google Scholar]
- 47.Marioni RE, Harris SE, Shah S, et al. The epigenetic clock and telomere length are independently associated with chronological age and mortality. Int J Epidemiol. 2016;45:424–432. doi: 10.1093/ije/dyw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen BH, Carty CL, Kimura M, et al. Leukocyte telomere length, T cell composition and DNA methylation age. Aging (Albany NY) 2017;9:1983–1995. doi: 10.18632/aging.101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yehuda R, Seckl J. Minireview: Stress-related psychiatric disorders with low cortisol levels: a metabolic hypothesis. Endocrinology. 2011;152:4496–4503. doi: 10.1210/en.2011-1218. [DOI] [PubMed] [Google Scholar]
- 50.Gassen NC, Chrousos GP, Binder EB, Zannas AS. Life stress, glucocorticoid signaling, and the aging epigenome: implications for aging-related diseases. Neurosci Biobehav Rev. 2017;74((pt B)):356–365. doi: 10.1016/j.neubiorev.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Yehuda R, Flory JD, Bierer LM, et al. Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans with posttraumatic stress disorder. Biol Psychiatry. 2015;77:356–364. doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Hyman SE. Even chromatin gets the blues. Nat Neurosci. 2006;9:465–466. doi: 10.1038/nn0406-465. [DOI] [PubMed] [Google Scholar]
- 53.Menke A, Binder EB. Epigenetic alterations in depression and antidepressant treatment. Dialogues Clin Neurosci. 2014;16:395–404. doi: 10.31887/DCNS.2014.16.3/amenke. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kananen L, Marttila S, Nevalainen T, et al. The trajectory of the blood DNA methylome ageing rate is largely set before adulthood: evidence from two longitudinal studies. Age (Dordr) 2016;38:65. doi: 10.1007/s11357-016-9927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data