Abstract
Convolutional neural networks (CNN) has become the state-of-the-art method for medical segmentation. However, repeated pooling and striding operations reduce the feature resolution, causing the loss of the detailed information. Additionally, tumors of different patients are of different sizes. Thus small tumors may be ignored while big tumors may exceed the receptive fields of convolutions. The purpose of this study is to further improve the segmentation accuracy using a novel CNN (named CAC-SPP) with cascaded atrous convolution (CAC) and spatial pyramid pooling module (SPP). This work was the first attempt to apply SPP for segmentation in radiotherapy. We improved the network based on ResNet-101 yielding accuracy gains from greatly increased depth. We added CAC to extract high-resolution feature map while maintaining the large receptive fields. We also adopted a parallel SPP with different atrous rates to capture the multi-scale features. The performance was compared with the widely adopted U-Net and ResNet-101 with independent segmentation of rectal tumors for two image sets, separately: (1) 70 T2-weighted MR images and (2) 100 planning CT images. The results show that the proposed CAC-SPP outperformed the U-Net and ResNet-101 for both image sets. The Dice similarity coefficient (DSC) values of CAC-SPP were 0.78±0.08 and 0.85±0.03, respectively, which were higher than those of U-Net (0.70±0.11 and 0.82±0.04) and ResNet-101 (0.76±0.10 and 0.84±0.03). The segmentation speed of CAC-SPP was comparable with ResNet-101, but about 36% faster than U-Net. In conclusion, the proposed CAC-SPP which could extract high-resolution features with large receptive fields and capture multi-scale context yields improve the accuracy of segmentation performance for rectal tumors.
Keywords: radiotherapy, automated segmentation, tumor target, convolutional neural networks, spatial pyramid pooling
1. Introduction
Segmenting a tumor target from patients’ images is a critical step for radiotherapy. The increasing usage and importance of multimodal medical images are placing heavier workloads on physicians. Moreover, considerable inter- and intra-observer variability (Van et al 2002, Caravatta et al 2014) in target delineation will lead to uncertainty and affect the accuracy and effectiveness of treatment. There is, therefore, a growing need for automation to segment the target quickly and accurately. The automated segmentation of tumor target can relieve physicians from the labor-intensive aspects of their work while increasing the accuracy, consistency, reproducibility, and efficiency of the delineations, in the meantime allowing them to focus more on other aspects of their job. As a solution, ‘atlas-based’ (Bondiau et al 2005, Young et al 2011, Anders et al 2012) automated segmentation software is often used in clinical applications. However, it needs to build different atlases according to the shapes and sizes of different patients and the deformable registration process is very time-consuming (Reed et al 2009).
Recently, deep learning, especially with regard to convolutional neural networks (CNN), has rapidly become the state-of-the-art and the most successful methodology for image analysis in computer vision (Krizhevsky et al 2012, Simonyan et al 2014, Szegedy et al 2015, Long et al 2015, Ronneberger et al 2015, He et al 2016). The biggest advantage of CNN is that it could learn hierarchical feature from the images automatically instead of handcrafted feature extractions. It is trained end-to-end in a supervised fashion that greatly simplifies the application process. The outstanding performance in a variety of artificial intelligence (AI) applications prompted the use of CNN in medical applications, such as disease diagnosis (Li et al 2014, Shin et al 2016), survival prediction (Zhu et al 2016, Christ et al 2017), image optimization (Dong et al 2014, Zhang et al 2017), tumor classification (Rouhi et al 2015, Pan et al, and organ segmentation (Pereira et al 2016, Hu et al 2017). There is also considerable interest in applying CNN in radiotherapy for automated segmentation. Ibragimov et al (2017) used CNN for organs-at-risk (OARs) segmentation in head and neck CT images and obtained Dice similarity coefficient (DSC) values from 37.4% to 89.5%. Lustberg et al (2018) showed that CNN contouring had promising results in lung cancer compared with the atlas-based method. Lavdas et al (2017) also demonstrated CNN performed favorably in MR images. The author (Men et al 2017) previously reported a dilated CNN for segmentation of the target and OARs of rectal cancer and achieved acceptable results.
On the structure, a standard CNN for segmentation consists of convolutional layers interspersed with pooling layers, followed by fully convolutional layers to make the pixel-wise prediction (Long et al 2015). There are certain networks that have high performance and become widely known standards. VGG-16 (Simonyan et al 2014) which is composed of 16 weight layers, employs small, fixed size kernels in each layer to reduce the parameters and introduce more nonlinearities. The levels of feature can be enriched by the number of stacked layers (depth). The ResNet-101 (He et al is famous due to its increased depth (101 layers) and the residual blocks that guarantee it performs better than previous networks. The tumor segmentation needs to identify the category of each pixel in the image. However, there are two main challenges in the application of CNN to tumor segmentation. The first one is that the repeated operations of pooling and striding in CNN reduce the resolution of the feature map (up to 1/32 of the input image), causing the loss of detailed information. To deal with the reduced feature resolution, a common solution is using the encoder-decoder architecture like U-Net (Ronneberger et al 2015), in which the encoder gradually reduces the spatial dimension with pooling layers and the decoder gradually recovers the full spatial resolution with deconvolutions. But the deconvolutions increase the computation dramatically and reduce the efficiency of segmentation. The second challenge is that the tumors of different patients are at multiple scales. Thus small objects may be ignored while big objects may exceed the receptive fields of convolutions. One approach (Chen et al 2016) involves providing multiple rescaled images as input to the network and aggregating the multiple-feature for prediction, but it also increases the cost of computation due to the analysis of multiply scaled versions of the input image.
To handle these problems, we trained a CNN named CAC-SPP with cascaded atrous convolution (CAC) and spatial pyramid pooling (SPP) module (He et al 2014, Chen et al 2018) for more accurate tumor segmentation. There are three main advantages. First, we added these improvements to a very deep network which can take advantage of the accuracy gains from greatly increased depth. Second, we novelly adopted the CAC to extract high-resolution feature map. Compare with traditional down-sampling structure, CAC has the strong ability to increase the receptive filed of convolution kernels without reducing the resolution of feature map. Furthermore, we adopted a parallel four-level SPP module in our networks. Although SPP has been used in computer vision recently (Chen et al 2018), it has been only tested on some databases (PASCAL VOC 2012, PASCAL-Context, PASCAL-Person-Part, and Cityscapes) commonly used in computer vision. To the best of our knowledge, this work was the first attempt to apply SPP for auto-segmentation of the tumor and CTV in radiotherapy. The proposed SPP consisted of a parallel four-level atrous convolution module that could capture the features containing more useful information with different scales. Moreover, to deal with segmentation of smaller regions, we proposed applying smaller atrous rates to improve from the traditional methods. In a word, the CAC module kept the resolution while the SPP module captured multi-scale information. Based on this architecture, the CAC-SPP can extract denser and multi-scale features that are essential for accurate localization. We compared its performance with the U-Net and ResNet-101 on two segmentation tasks with different medical image modalities.
2. Methods
2.1. Patients data and pre-processing
The proposed network was evaluated on two tumor target segmentation tasks, separately, including tumor segmentation on T2-weighted MR data and CTV segmentation on planning CT data from patients with rectal cancer. Quantitative evaluation and comparisons with state-of-the-art (U-Net and ResNet-101) were reported for each of the tasks.
Seventy patients with rectal cancer from our institution underwent MR imaging on a variety of scanners (Siemens, Philips, and GE) with different magnetic field strengths (1.5 T or 3.0 T). The T2-weighted MR data were used to detect the visible tumor. The imaging protocols included turbo spin echo (TSE) and proprietary name for periodically rotated overlapping parallel lines with enhanced reconstruction (PROPELLER/BLADE). MR scan thickness was 3–7 mm with a median value of 5 mm. The image resolution ranges from 256×256 to 512×512 while the pixel size 0.33–1.72 mm with a median value of 0.75 mm. The physicians contoured the tumor manually on all the axial MR slices in which the tumor exists. A total of 3,598 two-dimensional (2D) MR images were used, of which 1,023 were labeled with tumors (GTV). The tumor covered an area of 65.63 to 4,835.51 mm2 in the 2D axial image with a median value of 642.36 mm2.
One hundred patients with rectal cancer receiving radiotherapy in our institution were used as the second dataset in this study. CT simulation was done with the patient in the supine or the prone position. A custom immobilization device was used to minimize setup variability. CT images were acquired on different systems (Siemens, Philips, and GE). The X-ray tube voltage was set to 120 kVp for all the patients and the X-ray tube current varied from 200 to 440 mA according to the patient’s size, with a median value of 325 mA. CT images were reconstructed with a 512×512 matrix and a slice thickness of 3 mm. The pixel size was 0.97–1.27 mm with a median value of 1.17 mm. Physicians contoured the CTV on the planning CT as part of clinical care. In total, there are 20,977 CT images, of which 5,057 were labeled with CTV. The CTV area delineated in the 2D CT image was from 651.33 to 15,607.70 mm2 with a median value of 5,446.47 mm2.
The image data were pre-processed in MATLAB R2017b (MathWorks, Inc., Natick, Massachusetts, United States). A custom-built script was used to extract and label all the voxels that belonged to the manual contours from the DICOM structure files. All MR and CT images were resampled to isotropic 0.75×0.75 mm2 and 1.00×1.00 mm2 pixel resolution, respectively. It was done using MATLAB’s ‘imresize’ function with bi-cubic interpolation and antialiasing. Due to the limitation of GPU memory, our hardware device can optimize only images of size 417×417. Thus each 2D slice was transformed to a dimension of 417×417 for both MR and CT. More specifically, the resampled image was centrally and symmetrically cut to 417×417 if it was larger than 417×417 or filled with zero around if it was smaller than 417×417. After the pre-processing, the finial input images had a uniform dimension of 417×417. We performed histogram matching on the MR images using MATLAB’s ‘imhistmatchn’ function in the Image Processing Toolbox to normalize the rectum-region intensity and contrast of different sequences. We used a contrast-limited adaptive histogram equalization (Reza et al 2004) algorithm to pre-process the CT images for image enhancement. For the patients with ‘supine’ position, the images were rotated 180 degrees clockwise to create the corresponding ‘virtual prone’ images. This is to make the images of patients in the dataset the same orientation. The final data used for CAC-SPP were the 2D MR or CT slices and the corresponding 2D tumor or CTV labels. The process and the additional image pre-processing were fully automated.
2.2. CAC-SPP for tumor segmentation
Similar to semantic image segmentation, tumor recognition and localization can also be solved by pixel-wise classification. The fully convolutional neural networks (Long et al 2015) has become the state-of-the-art method for medical image segmentation due to its high accuracy and efficiency. In this study, we introduced an end-to-end segmentation framework that could predict pixel-wise class labels in MR and CT images.
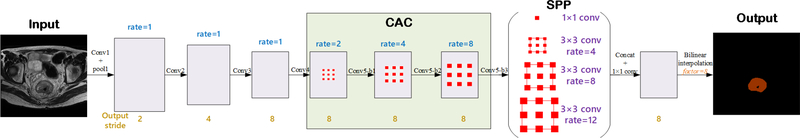
The network architecture of the proposed CAC-SPP is illustrated in Figure 1. The network depth is of central importance and the very deep models lead the performance in many visual recognition tasks. A very deep neural network could extract the multi-level features from low-level to high-level. The proposed CAC-SPP model was based on the popular ResNet-101 (He et al 2016) that has 101 weighted layers but is easy to train. We expected the segmentation accuracy could greatly benefit from the very deep model by enriching the levels of features. The max-pooling (down-sampling) was used to reduce the amount of parameters of the network, increase the receptive fields, and control overfitting. However, one drawback is that the repeated down-sampling reduced spatial resolution of the feature maps significantly. In order to avoid the loss of the detailed information, we replaced the down-sampling operator from the last few blocks of ResNet-101 with CAC of different rates (2, 4, and 8). The atrous convolutions in CAC could effectively enlarge the receptive fields of filters without reducing feature resolution or increasing the amount of computation. Another issue about tumor segmentation is that the tumors of different patients are of different sizes. In order to overcome this problem and efficiently extract multi-scale feature maps, we adopted a parallel four-level SPP
Figure 1.
The architecture of the proposed CAC-SPP
module on top of CAC. The SPP consists of one 1×1 convolution and three 3×3 convolutions with atrous rates = (1, 4, 8, and 12). It could capture the features containing more useful information with four different receptive fields. The original SPP module (Chen et al 2018) uses very large atrous rates, which are inappropriate to our problem. This is because the tumor in MR/CT image is much smaller with respect to the whole image compared to the case in common object segmentation in computer vision. It will fail to capture long range information due to image boundary effects when applying a 3×3 atrous convolution with a too large rate. So we adjusted the atrous rates to make sure all the filter weights are applied to most of the valid region on feature map. Compared with the traditional multi-scale context feature extraction methods, the SPP can obtain local and global context without changing the image resolution. The four-scale features extracted with different receptive fields were fused to create the final global feature. A fully convolution (FC) layer and a bilinear interpolation operation following the layers above generated the pixel-wise prediction. With these modifications, the proposed CAC-SPP can take advantage of accuracy gains from the deep layers, high-resolution features, and multi-scale information.
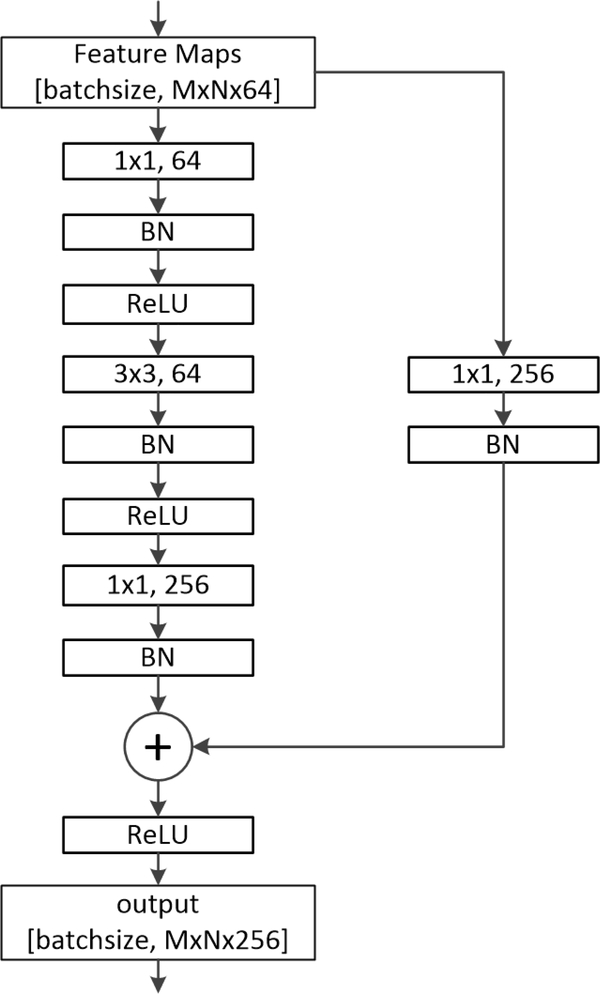
In detail, Table 1 shows the architecture of the CAC-SPP. The first convolution layer (Convl) used filters with size of 3×3, a stride of 2 and a padding of 3 to generate 64 feature maps. A max-pooling operation with window size of 3×3, a stride of 2, and a padding of 1 followed for down-sampling. The deeper bottleneck architecture (DBA) is the core of deep residual network. As illustrated in Figure 2, each DBA consisted of a block of 3 layers that are 1×1, 3×3 and another 1×1 convolutions. The two 1×1 layers were responsible for reducing and then increasing dimensions, leaving the 3×3 layer a bottleneck with smaller input/output dimensions. Conv2, Conv3, and Conv4 had 3, 4, and 23 such blocks, respectively. We removed the max-pooling operations at Conv3_1, Conv4_1, and Conv5_1 of the original ResNet-101. Conv5 (including Conv5-b1, Conv5-b2, and Conv5-b3) was the proposed CAC module that used similar blocks but adopted cascaded atrous convolutions with atrous rates of 2, 4, and 8, respectively. The corresponding receptive field sizes were 9×9, 17×17, and 33×33 pixels. This modification could effectively enlarge the receptive fields without increasing the convolution kernel size and parameters. In the following SPP module, it included four parallel atrous convolutions with different atrous rates of 1, 4, 8, and 12, and so had multiple receptive fields of size 3×3, 15×15, 31×31, and 47×47 pixels. They generated multi-scale feature maps through one 1×1 convolution and three 3×3 convolutions, with a stride size of 1 pixel, and a padding size of 0, 4, 8, and 12 pixels, respectively. Then, a Concat operation fused the multi-scale features. The final 1×1 convolution options in FC carried on the pixel-level classification. A bilinear interpolation with factor of 8 was used to restore the prediction map to the size of the original image.
Table 1.
The detailed architecture of CAC-SPP.
| Layers | Shape | ||||||
|---|---|---|---|---|---|---|---|
| kernel | padding | atrous | stride | output | number of parameters | ||
| Input data | - | - | - | - | 417×417×1 | ||
| conv1 | [7 × 7] | 3 | 1 | 2 | 209×209×64 | 3,200 | |
| max-pool 1 | [3 × 3] | 1 | 1 | 2 | 105×105×64 | ||
| conv2 | 105×105×64 105×105×64 105×105×256 |
173,184 | |||||
| conv3 | 53×53×128 53×53×128 53×53×512 |
986,112 | |||||
| conv4 | 53×53×256 53×53×256 53×53×1024 |
22,645,248 | |||||
| CAC | conv5-b1 | 53×53×512 53×53×512 53×53×2048 |
11,805,696 | ||||
| conv5-b2 | 53×53×512 53×53×512 53×53×2048 |
13,378,560 | |||||
| conv5-b3 | 53×53×512 53×53×512 53×53×2048 |
13,378,560 | |||||
| SPP | spp-conv1 | [1 × 1] | 0 | 1 | 1 | 53×53×1024 | 2,098,176 |
| spp-conv2 | [3 × 3] | 4 | 4 | 1 | 53×53×1024 | 18,875,392 | |
| spp-conv3 | [3 × 3] | 8 | 8 | 1 | 53×53×1024 | 18,875,392 | |
| spp-conv4 | [3 × 3] | 12 | 12 | 1 | 53×53×1024 | 18,875,392 | |
| concat | spp-conv1 + spp-conv2 + spp-conv3 + spp-conv4 | ||||||
| fc | [1 × 1] | 0 | 1 | 1 | 53×53×2 | 4,098 | |
| interpolation | factor = 8 | 417×417×2 | |||||
| Output data | 417×417×1 | ||||||
Figure 2.
An example of the deeper bottleneck architecture (DBA).
Like other CNN, there was a batch-normalized (BN) operation following each convolutional layer. The rectified linear unit (ReLU) was used as the nonlinear activation function to introduce non-linearity in CNN by replacing all negative pixel values in the feature map by zero, i.e. output = max (0, input).
2.3. Experiments
The performance of the network was evaluated on two independent segmentation tasks of rectal cancer: (1) segmentation of tumor on 70 T2-weighted MR data, and (2) segmentation of CTV on 100 planning CT data. The model training and testing of the two tasks were undertaken separately. We performed 5-fold cross-validation for evaluation, where the dataset was randomly divided into 5 equal-sized subsets. Firstly, we trained the model on the first 4 subsets (80% of the data) and tested on the 5th subset (20% of the data). Subsequently, we chose another subset as the test set and trained a second model on the remaining 4 subsets. We repeated this step until we trained 5 models for each task.
2.3.1. Implementation details
We implemented the training, testing, analysis and visualization pipeline of our model using Caffe (Jia et al 2014), which is a publicly available deep learning platform. The training set was used to “tune” the parameters of the networks. In detail, the original 2D MR or CT slices were the inputs and the corresponding maps about the manual segmentation labels were the outputs. The model parameters for each network were initialized using the weights from the corresponding model trained on ImageNet (Deng et al 2009). Data augmentation is a very useful method to extend the training set that is also widely used in the training of CNN. In this work, we adopted an on-line augmentation approach. The augmented images were not created on disk. We wrote the ‘data augmentation operation’ in the training file while randomly scaling the input images (from 0.5 to 1.5), randomly cropping, and randomly left-right flipping. The scaling factors were 0.5, 0.75, 1, 1.25, and 1.5. The randomly cropping size was 321^321 due to the limitation of the GUP memory. In detail, during the training phase,both MR/CT image and its corresponding label were randomly scaled by a factor listed above. Then the cropping extracted random sub-patch (321×321) from the scaled image. An additional flipping randomly flips the input image horizontally. This data augmentation scheme makes the network avoid overfitting. While during the testing phase, no data augmentation was applied and the original image with size of 417×417 was used as the input for prediction. The optimization algorithm of training used backpropagation with the stochastic gradient descent (SGD) implementation of Caffe. We use the “poly” learning rate policy where current learning rate equals to the iter base one multiplying . In this study, we set the base learning rate to 0.001 and power to 0.9. The batch size was set to 1 due to the limitation of physical memory on GPU card. Every model was trained using 40 epochs. This value was determined by exploratory experiments that evaluated test scores every epoch. The momentum and weight decay were set to 0.9 and 0.0005, respectively.
The training and testing phases were fully automated with no manual interaction. All computations were undertaken on a personal computer with two Intel® Xeon E5 processors (2.1 GHz) and an NVidia Quadro M5000 GPU.
2.3.2. Performance evaluation
Once the model was trained with the training set, the independent test set was used to evaluate the performance of the networks. The input was the 2D MR or CT slice and the final output was 2D pixel-wise classification label. Standard reference segmentation was defined as the manual segmentation. The spatial consistency between the automated segmentation and the manual reference segmentation was quantified using two metrics: the Dice similarity coefficient (DSC) (Crum et al 2006) and the Hausdorff distance (HD) (Huttenlocher et al 1993). Both of them measure the degree of mismatch between the automated segmentation (A) and the manual segmentation (B). The DSC is calculated as DSC = 2TP/(2TP+FP+FN) using the definition of true positive (TP), false positive (FP), and false negative (FN). It ranges from 0, indicating no spatial overlap between the two segmentations, to 1, indicating complete overlap. The HD is the greatest of all the distances from a point in A to the closest point in B. Smaller value usually represents better segmentation accuracy.
The accuracy of CAC-SPP was compared with U-Net and ResNet-101 that are used widely for segmentation tasks in medical images. We also evaluate the time cost for auto-segmentation with these different models. There were about 28.2M, 75.5M, and 121.1 weighted parameters in U-Net, ResNet-101, and CAC-SPP, respectively. We have listed the detailed architecture of U-Net in the appendix for reference.
3. Results
3.1. Accuracy
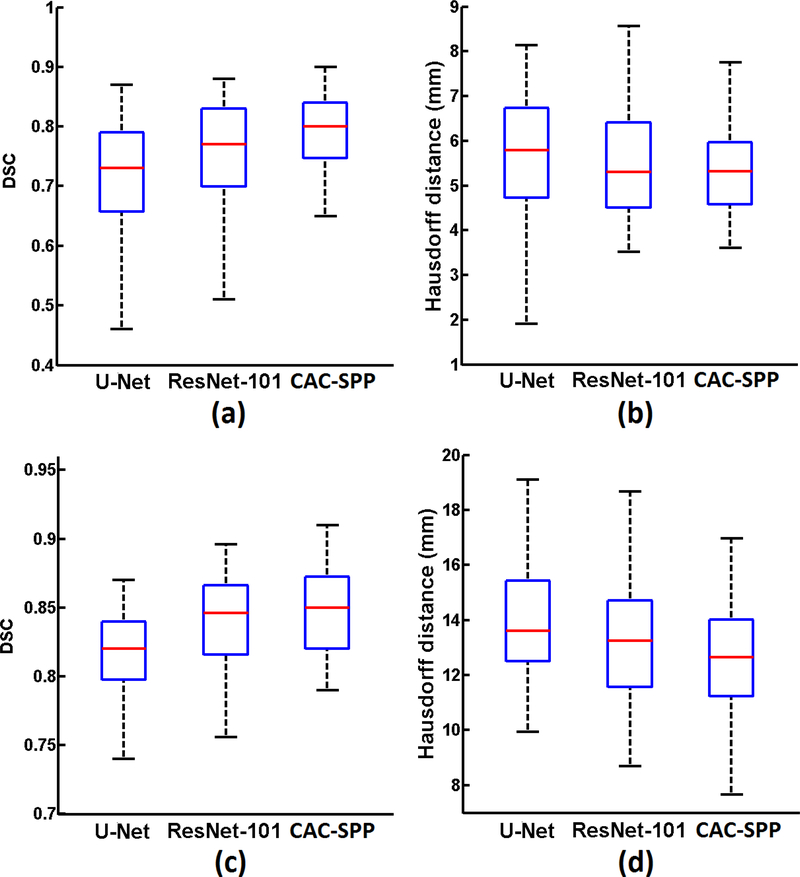
The results of the segmentation of (1) tumor on T2-weighted MR dataset and (2) CTV on planning CT dataset are summarized in Figure 3 (a-d), respectively. The proposed CAC-SPP method outperformed the U-Net and ResNet-101 on both datasets. For the MR dataset, CAC-SPP had DSC value of 0.78 ±0.08, which was 0.08 and 0.02 higher than U-Net (0.70 ±0.11) and ResNet-101 (0.76 ±0.10), respectively. The HD value of CAC-SPP (5.4 + 1.0 mm) was better than the other two models (U-Net: 5.8 + 1.3 mm, ResNet-101: 5.6 + 1.2 mm). For the CT dataset, CAC-SPP also had the best evaluation metrics, with DSC value of 0.85 + 0.03 (U-Net: 0.82 + 0.04, ResNet-101: 0.84 + 0.03) and HD value of 13.2 + 2.3 mm (U-Net: 14.3 + 2.5 mm, ResNet-101: 13.6 +2.4mm).
Figure 3.
Boxplots showing the measured metrics: (a) DSC, (b) Hausdorff distance on T2-weighted MR dataset; (c) DSC, (d) Hausdorff distance on planning CT dataset.
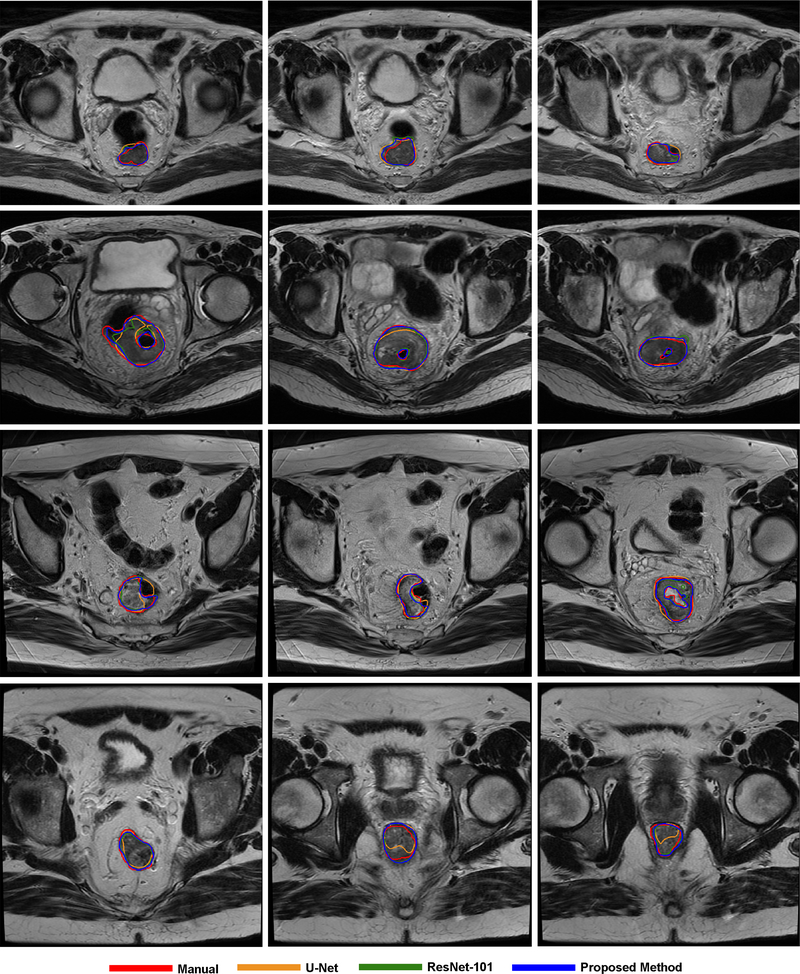
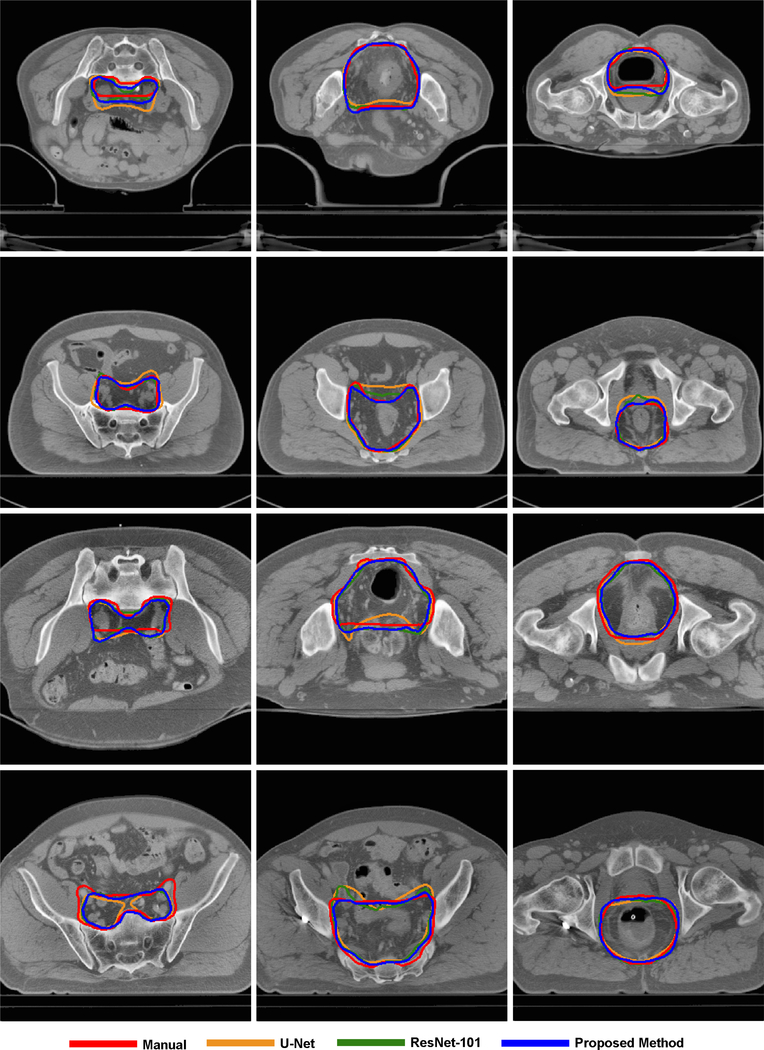
Figure 4 and Figure 5 show some visual comparisons of the automated segmentation and manual segmentation. These images come from different patients with various tumor sizes. The auto-segmented contours with the proposed CAC-SPP were in better agreement with the manual reference contours than the two models. The automated segmentation was close to the manually segmented contours in shape, volume and location and only slight corrections would have been necessary to validate the automated segmentation.
Figure 4.
Examples of tumor segmentation on T2-weighted MR, each row belongs to the same patient.
Figure 5.
Examples of CTV segmentation on planning CT, each row belongs to the same patient.
3.2. Time cost
We tested the time cost for segmentation using a personal computer with two Intel® Xeon E5 processors (2.1 GHz) and an NVidia Quadro M5000 GPU. The mean time for automated segmentation on MR dataset was about 10 s (CAC-SPP), 15 s (U-Net) and 9 s (ResNet-101) per patient, respectively, while the time on CT dataset was about 35 s (CAC-SPP), 55 s (U-Net) and 34 s (ResNet-101) per patient, respectively.
4. Discussion
The segmentation of structures of interest in medical images allows further analysis and guides the clinical intervention. In this study, we trained a model named CAC-SPP for fast and more accurate segmentation. There are three advantages of the proposed network: first, it takes advantage of the deeper residual network to learn multi-level features; second, it adopts CAC, rather than down-sampling to increase the receptive fields while not reduce spatial resolution of the feature map or increase the amount of parameters of the network; third, the SPP module could extract multi-scale feature. These advantages make it meet the requirements of automated segmentation methods: a) robust to deal with multi-modality image, b) accurate enough so that further analysis can be carried out based on the results, and c) fast enough to relieve clinicians’ workflow.
The robustness of this segmentation method is demonstrated by applying it to two different image modalities. Both experiments demonstrate that there are advantages to be gained by incorporating the CAC and SPP in the CNN architecture. The first, visible tumor segmentation from T2-MR data, demonstrated the robustness of the algorithm by applying it to multiple-intensity data. The second, invisible CTV segmentation from CT data, demonstrated the model can learn the non-explicit physician’s knowledge hidden in the data. We used DSC and Hausdorff distance as two metrics to evaluate the accuracy quantitatively. The values of two metrics depend on the data and the segmentation task in question. Compared with the segmentation of CTV on CT, the segmentation of tumor (GTV) on MR gave worse spatial overlap results (DSC) for all methods; however it had better (lower) Hausdorff distance for all methods. This difference comes from that the tumor on MR was smaller than CTV on CT; hence small deviations in Hausdorff distance can result in larger changes in the spatial overlap measures.
Even though current CNN-based methods demonstrate a promising performance, the higher accuracy is worth studying. The comparisons reveal that the proposed CAC-SPP algorithm results in more accurate segmentation than the U-Net and ResNet-101 algorithms. Compared to U-Net, CAC-SPP deploys much deeper layers to enrich the levels of features. Moreover, CAC-SPP can extract multi-scale contextual features with the spatial pyramid pooling module. Compared to ResNet-101, CAC-SPP employs cascaded atrous convolutions and a parallel four-level pyramid pooling module to extract high-resolution and multi-scale features without reducing receptive fields. With these advantages, CAC-SPP can capture global features containing abundant contextual information, which improves performance.
The important advantages of deep learning-based segmentation over other methods are its universality and efficiency. We don’t need to divide the patients into different groups according to their volume, shape or position because the CNN can “learn” such knowledge by itself. Once the model has been trained, the segmentation process is very fast. Compared with the manual contouring, all the CNN techniques greatly improve efficiency. The proposed CAC-SPP has a comparable speed with the ResNet-101, but reduces the segmentation time by 36% compared with the U-Net. That is because the introduction of the CAC and SPP does not increase the computation while the deconvolution operation in U-Net is very time-consuming.
We adopted image pre-processing of histogram matching and contrast-limited adaptive histogram equalization to MR and CT images, respectively. Comparative experiments and quantitative evaluation demonstrated a considerable improvement with the pre-processing operations. The average DSC values increased (MR: 0.78±0.08 vs. 0.70±0.13, CT: 0.85±0.03 vs. 0.83±0.05) and the Hausdorff distance values also became better (MR: 5.4±1.0 mm vs. 5.8±1.4 mm, CT: 13.2±2.3 mm vs. 14.0±2.6 mm). After histogram matching, the standard deviation decreased, which indicates that the variation between images of different patients decreases. The improvement in CT image was smaller than that in MR image, which can be explained as follows: The CT scans used the same X-ray tube voltage (120 kVp), and HU values remain consistent between patients. However, the MR scan protocols differ, resulting in a large variation in intensity. In fact, the difference in MR images between patients remains significant even with the same scanning parameters. For the above reasons, histogram matching operation can greatly reduce the variation and improve the results for MR images.
Delineation of the target and OARs in radiotherapy is still associated with considerable inter- and intra-observer variations. These variations will result in inconsistent dose calculation and evaluation, therefore, mislead the treatment planning. Another potential application of the proposed model is to check and control the quality of the contours which is usually delineated manually and dependent on the physicians’ knowledge and experience. Since our CAC-SPP model was found able to accurately and efficiently predict the segmentation, it could be used in radiotherapy workflow to assist physicians in their daily work of contouring and/or quality assurance, although careful human review and editing will be required for some details.
Limited by experimental conditions, we don’t have very large datasets. However, what we proposed was a 2D network, and there were a lot of slices per patient that can be used to train the model. Moreover, we applied data augmentation techniques such as random scaling, random cropping, and random left-right flipping which could enlarge the training dataset greatly and thus avoid overfitting. We were able to obtain promising results with our model although the datasets were limited. With increased quantity and improved accuracy of the datasets, the model can be more robust and the segmentation accuracy is likely to further improve.
While the proposed model focused on 2D images, the same principle can be extended to 3D CNN with 3D convolution and pooling. The 3D model could extract features from the spatial dimensions, thereby capturing not only transverse but also sagittal and coronal information encoded in volumetric data. But the introduction of 3D convolutions dramatically increases the number of parameters in the network, the computations, as well as the need for more powerful GPU. Moreover, 3D CNN also needs more training data; however, the medical image datasets are often small. The performance of a 3D model with larger datasets merits further investigation.
It is important to mention that this work was not pioneering in the use of SPP for segmentation. Chen et al (2018) has used the SPP in computer vision recently but they only tested on some common databases in computer vision. Nevertheless, our work may be of interest for the medicine community because it is the first work that applied SPP for auto-segmentation of the tumor and CTV in radiotherapy. Our work has novelty because this is an important area of research in Medical Physics that needs to be fully explored with the current success with deep learning techniques. We adjusted the atrous rates according to the characteristics of our dataset and improved the segmentation accuracy through this SPP module more suitable to the medical problem in this study. It is conceivable that, in the near future, the application of SPP in radiotherapy will improve the accuracy of segmentation, and advance the automation of radiotherapy so that radiation dose could be focused more on tumor while sparing adjacent normal tissue.
5. Conclusions
Accurate segmentation is an essential step of radiotherapy. Its automation and precision improvement are worthy of investigation. In this study, we are the first applying cascaded atrous convolution and spatial pyramid pooling to segmentation of tumor and CTV in radiotherapy. These modifications can extract high-resolution features with large receptive fields and capture multi-scale contextual information. Our experimental results show that the accuracy is improved with high speed.
Acknowledgments
This project was supported by U24CA180803 (IROC) from the National Cancer Institute (NCI).
Footnotes
Conflicts of interest
The authors report no conflicts of interest with this study.
References
- Anders LC, Stieler F, Siebenlist K, Schäfer J, Lohr F and Wenz F. 2012. Performance of an atlas-based autosegmentation software for delineation of target volumes for radiotherapy of breast and anorectal cancer. Radiotherapy & Oncology 102 (1):68–73. [DOI] [PubMed] [Google Scholar]
- Bondiau PY, Malandain G, Chanalet S, Marcy PY, Habrand JL, Fauchon F, Paquis P, Courdi A, Commowick O and Rutten I. 2005. Atlas-based automatic segmentation of MR images: validation study on the brainstem in radiotherapy context. International Journal of Radiation Oncology Biology Physics 61 (1):289–98. [DOI] [PubMed] [Google Scholar]
- Caravatta L, Macchia G, Mattiucci GC, Sainato A, Cernusco NL, Mantello G, Tommaso MD, Trignani M, Paoli AD and Boz G. 2014. Inter-observer variability of clinical target volume delineation in radiotherapy treatment of pancreatic cancer: a multi-institutional contouring experience. Radiation Oncology 9 (1):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LC, Yang Y, Wang J, Xu W and Yuille AL. 2016. Attention to Scale: Scale-Aware Semantic Image Segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition 3640–9. [Google Scholar]
- Chen LC, Papandreou G, Kokkinos I, Murphy K, and Yuille AL. 2018. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE transactions on pattern analysis and machine intelligence 40(4): 834–48. [DOI] [PubMed] [Google Scholar]
- Christ PF, Ettlinger F, Kaissis G, Schlecht S, Ahmaddy F, Grün F, Valentinitsch A, Ahmadi S A, Braren R and Menze B. 2017. SurvivalNet: Predicting patient survival from diffusion weighted magnetic resonance images using cascaded fully convolutional and 3D convolutional neural networks. In Biomedical Imaging (ISBI 2017), 2017 IEEE; 14th International Symposium 839–43. [Google Scholar]
- Crum WR, Camara O and Hill DLG. 2006. Generalized Overlap Measures for Evaluation and Validation in Medical Image Analysis. IEEE Transactions on Medical Imaging 25 (11):1451–61. [DOI] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li LJ, Li K and Li FF. 2009. ImageNet: A large-scale hierarchical image database. Computer Vision and Pattern Recognition, 2009. CVPR 2009. IEEE; Conference on 248–55. [Google Scholar]
- Dong C, Loy CC, He K and Tang X. 2014. Learning a Deep Convolutional Network for Image Super-Resolution European Conference on Computer Vision 2014. Springer, Cham: 8692:184–99. [Google Scholar]
- He K, Zhang X, Ren S, and Sun J. 2014. Spatial pyramid pooling in deep convolutional networks for visual recognition. In European Conference on Computer Vision 346–61 [DOI] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S and Sun J. 2016. Deep Residual Learning for Image Recognition. Proceedings of the IEEE conference on computer vision and pattern recognition 770–8. [Google Scholar]
- Hu P, Wu F, Peng J, Bao Y, Chen F and Kong D. 2017. Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets. International Journal of Computer Assisted Radiology and Surgery 12 (3):399–411 [DOI] [PubMed] [Google Scholar]
- Huttenlocher DP, Klanderman GA and Rucklidge WJ. 1993. Comparing images using the Hausdorff distance. IEEE Transactions on Pattern Analysis and Machine Intelligence 15 (9):850–63. [Google Scholar]
- Ibragimov B and Xing L. 2017. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Medical Physics 44 (2):547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S and Darrell T. 2014. Caffe: Convolutional Architecture for Fast Feature Embedding. Proceedings of the 22nd ACM international conference on Multimedia 675–8. [Google Scholar]
- Krizhevsky A, Sutskever I and Hinton GE. 2012. ImageNet classification with deep convolutional neural networks. In Advances in neural information processing systems 1097–105. [Google Scholar]
- Lavdas I, Glocker B, Kamnitsas K, Rueckert D, Mair H, Taylor SA, Aboagye EO and Rockall AG. 2017. Fully automatic, multi-organ segmentation in normal whole body magnetic resonance imaging (MRI), using classification forests (CFs), convolutional neural networks (CNNs) and a multi-atlas (MA) approach. Medical Physics 44 (10):5210–20. [DOI] [PubMed] [Google Scholar]
- Li R, Zhang W, Suk HI, Wang L, Li J, Shen D and Ji S. 2014. Deep Learning Based Imaging Data Completion for Improved Brain Disease Diagnosis. In International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, Cham 305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Shelhamer E and Darrell T. 2015. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition 3431–40. [DOI] [PubMed] [Google Scholar]
- Lustberg T, van Soest J, Gooding M, Peressutti D, Aljabar P, van der Stoep J, van Elmpt W and Dekker A. 2018. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiotherapy and Oncology, 126(2), 312–7. [DOI] [PubMed] [Google Scholar]
- Men K, Dai J and Li Y. 2017. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Medical Physics 44(12):6377–89. [DOI] [PubMed] [Google Scholar]
- Pan Y, Huang W, Lin Z, Zhu W, Zhou J, Wong J and Ding Z. 2015. Brain tumor grading based on Neural Networks and Convolutional Neural Networks. In Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE 699–702. [DOI] [PubMed] [Google Scholar]
- Pereira S, Pinto A, Alves V, and Silva CA. 2016. Brain Tumor Segmentation using Convolutional Neural Networks in MRI Images. IEEE Transactions on Medical Imaging 35 (5):1240–51. [DOI] [PubMed] [Google Scholar]
- Reed VK, Woodward WA, Zhang L, Strom EA, Perkins GH, Tereffe W, Oh JL, Yu TK, Bedrosian I, Whitman GJ, Buchholz TA and Dong L. 2009. Automatic segmentation of whole breast using atlas approach and deformable image registration. International Journal of Radiation Oncology Biology Physics 73 (5):1493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza AM 2004. Realization of the Contrast Limited Adaptive Histogram Equalization (CLAHE) for Real-Time Image Enhancement. Journal of VLSI Signal Processing Systems for Signal Image and Video Technology 38 (1):35–44. [Google Scholar]
- Ronneberger O, Fischer P and Brox T. 2015. U-Net: Convolutional Networks for Biomedical Image Segmentation In International Conference on Medical image computing and computer-assisted intervention, Springer, Cham: 9351:234–41. [Google Scholar]
- Rouhi R, Jafari M, Kasaei S and Keshavarzian P. 2015. Benign and malignant breast tumors classification based on region growing and CNN segmentation. Expert Systems with Applications An International Journal 42 (3):990–1002. [Google Scholar]
- Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D and Summers RM. 2016. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics, and Transfer Learning. IEEE Transactions on Medical Imaging 35 (5):1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K and Zisserman A. 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv preprint arXiv:1409.1556.
- Szegedy C, Liu W, Jia Y, Sermanet P, Reed C, Anguelov D, Erhan D, Vanhoucke V and Rabinovich A. 2015. Going deeper with convolutions. In IEEE Conference on Computer Vision and Pattern Recognition. [Google Scholar]
- Van de Steene J, Linthout N, de Mey J, Vinh-Hung V, Claassens C, Noppen M, Bel A and Storme G. 2002. Definition of gross tumor volume in lung cancer: inter-observer variability. Radiotherapy and Oncology 62 (1):37–49. [DOI] [PubMed] [Google Scholar]
- Young AV, Wortham A, Wernick I, Evans A and Ennis RD. 2011. Atlas-based segmentation improves consistency and decreases the time required for contouring postoperative endometrial cancer nodal volumes. International Journal of Radiation Oncology Biology Physics 79 (3):943–7. [DOI] [PubMed] [Google Scholar]
- Zhang K, Zuo W, Chen Y, Meng D, and Zhang L. 2017. Beyond a Gaussian Denoiser: Residual Learning of Deep CNN for Image Denoising. IEEE Transactions on Image Processing 26 (7):3142–55. [DOI] [PubMed] [Google Scholar]
- Zhu X, Yao J and Huang J. 2016. Deep convolutional neural network for survival analysis with pathological images. In Bioinformatics and Biomedicine (BIBM), 2016 IEEE International Conference 544–7. [Google Scholar]