Abstract
Brucella are highly infectious bacterial pathogens responsible for a severely debilitating zoonosis called brucellosis. Half of the human population worldwide is considered to live at risk of exposure, mostly in the poorest rural areas of the world. Prompt diagnosis of brucellosis is essential to prevent complications and to control epidemiology outbreaks, but identification of Brucella isolates may be hampered by the lack of rapid and cost-effective methods. Nowadays, many clinical microbiology laboratories use Matrix-Assisted Laser Desorption Ionization–Time Of Flight mass spectrometry (MALDI-TOF MS) for routine identification. However, lack of reference spectra in the currently commercialized databases does not allow the identification of Brucella isolates. In this work, we constructed a Brucella MALDI-TOF MS reference database using VITEK MS. We generated 590 spectra from 84 different strains (including rare or atypical isolates) to cover this bacterial genus. We then applied a novel biomathematical approach to discriminate different species. This allowed accurate identification of Brucella isolates at the genus level with no misidentifications, in particular as the closely related and less pathogenic Ochrobactrum genus. The main zoonotic species (B. melitensis, B. abortus and B. suis) could also be identified at the species level with an accuracy of 100%, 92.9% and 100%, respectively. This MALDI-TOF reference database will be the first Brucella database validated for diagnostic and accessible to all VITEK MS users in routine. This will improve the diagnosis and control of brucellosis by allowing a rapid identification of these pathogens.
Author summary
Brucella are bacteria that mainly infect animals. They can also be transmitted to humans and cause a serious disease called brucellosis. Half the world's population is considered exposed, especially in the poorest rural areas. Experts agree that prompt identification of Brucella isolates is essential to provide appropriate treatment to patients and to control epidemiological outbreaks. Mis-identification of these highly infectious pathogens may lead to delays in diagnosis, but also to increased risks of accidental exposure for laboratory workers. MALDI-TOF mass spectrometry is now the first line of bacterial identification in many routine diagnostic laboratories. However, not all clinical mass spectrometers can identify Brucella. In this work, we updated a database with Brucella spectra to improve the performance of MALDI-TOF mass spectrometers. These instruments will now be able to identify accurately Brucella isolates. This will greatly improve the diagnosis of brucellosis.
Introduction
Brucella are important pathogens in medical and veterinary context. These Gram-negative bacteria can be transmitted from their animal reservoir to humans, usually by ingestion of contaminated milk products or direct contact, causing brucellosis. This zoonosis causes a severely debilitating illness characterized by intermittent fever, chills, sweats, weakness, myalgia, osteoarticular or obstetrical complications and endocarditis.
This disease is largely unreported and the true incidence of human brucellosis is thus unknown [1]. According to the World Health Organization (WHO), half a million new cases are reported each year, most of them in the poorest rural areas of the world [2]. Indeed, while the disease has been successfully prevented in most industrialized countries, it remains a significant burden in the Mediterranean region, all over Asia, sub-Saharan Africa, and certain areas in Latin America. Approximately half of the human population worldwide is considered to live at risk of exposure [3]. Moreover, due to the low dose required to cause infection (10–100 colony-forming units) and the potential for aerosol dissemination, Brucella was considered a potential bioterrorism agent early in the 20th century [1] and its possession and use is still strictly regulated in many countries.
Currently, the Brucella genus consists of eleven recognized species plus several isolates that have not yet been officially designated. The major zoonotic species are B. melitensis, B. abortus and B. suis which are subdivided into biovars by a set of phenotypic characteristics including lipopolysaccharide (LPS) epitopes, phage sensitivity, dye sensitivity and a battery of biochemical tests. These three species are also the most common in domestic livestock. B. melitensis is responsible for the majority of human cases in the Mediterranean basin, the Arab peninsula, Latin America countries and Asia, while B. abortus is more prevalent in the United States, Northern Europe and Africa [4]. B. suis and B. canis infections are more sporadic in humans. Very rare human infections have also been reported with B. inopinata [5,6], B. ceti [7,8] and B. neotomae [9,10].
Clinical microbiology laboratories play a key role in the diagnosis and management of human brucellosis and should be able to provide a rapid and exact identification of Brucella spp. Currently, the most suitable tool for identification of bacteria is Matrix Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS). This method provides rapid, sensitive and cost-effective identification and is currently replacing phenotypic microbial identification. Its accuracy however largely depends on the coverage of the database of the commercially available MALDI-TOF MS systems. With regards to Brucella, identification was not possible because this genus was not represented in the databases of the two main MALDI-TOF MS system manufacturers (i.e. bioMérieux and Bruker) [11–13]. Only the Bruker Security Relevant (SR) database, or custom databases developed in some laboratories, can identify these highly pathogenic bacteria, but access to these databases is not possible in some countries due to export restriction regulations [13–15]. Moreover, only B. melitensis is included in the SR database.
Material and methods
Bacterial strains
The bacterial strains used for the construction of the database are listed in Tables 1 and S1. Each of these strains was cultivated on several different media (S1 Table). The bacterial isolates used for the external evaluation and their culture conditions are listed in Tables 2 and S2. All strains used in this study were previously characterized using an established workflow (phenotypic assays, Multiple-Locus Variable number tandem repeat Analysis -or MLVA-, whole-genome sequencing) [16].
Table 1. Brucella strains and isolates retained to generate the MALDI-TOF MS database.
Strains highlighted in grey are reference or type strains.
| Name | Species (biovar) | Origin |
|---|---|---|
| 16M | B. melitensis (1) | Veterinary isolate (goat) |
| 95-5009-1 | B. melitensis (1) | Veterinary isolate (sheep) |
| 95-2426-961 | B. melitensis (1) | Veterinary isolate (sheep) |
| 13-2582-4590 | B. melitensis (1) | Clinical isolate |
| 05–0737 | B. melitensis (1) | Clinical isolate (blood) |
| 08-2437-5214 | B. melitensis (1) | Veterinary isolate (goat) |
| 63/9 | B. melitensis (2) | Clinical isolate (blood) |
| 11-939-2005 | B. melitensis (2) | Clinical isolate (blood) |
| 04–1553 | B. melitensis (2) | Clinical isolate (blood) |
| Ether | B. melitensis (3) | Clinical isolate |
| 11-441-982 | B. melitensis (3) | Clinical isolate (blood) |
| 10–394 | B. melitensis (3) | Clinical isolate (blood) |
| 07-1184-2852 | B. melitensis (3) | Clinical isolate (blood) |
| 02–1213 | B. melitensis (3) | Veterinary isolate (cow) |
| 05–0682 | B. melitensis (3) | Clinical isolate (blood) |
| 11-2159-4003 | B. melitensis (3) | Clinical isolate (blood) |
| B115 | B. melitensis | Veterinary isolate (goat) |
| BT020216 | B. melitensis | Clinical isolate |
| BT071315-0001 | B. melitensis | Clinical isolate |
| BT072914 | B. melitensis | Clinical isolate |
| BT1202150001 | B. melitensis | Clinical isolate |
| 544 | B. abortus (1) | Veterinary isolate (cow) |
| 01–673 | B. abortus (1) | Veterinary isolate (cow) |
| 03-2770-3 | B. abortus (1) | Veterinary isolate (cow) |
| 05-147-200 | B. abortus (1) | Veterinary isolate (cow) |
| 93–12101 | B. abortus (1) | Clinical isolate (blood) |
| 2000031295 | B. abortus (1) | Clinical isolate |
| 86/8/59 | B. abortus (2) | Veterinary isolate (cow) |
| 03-2770-11 | B. abortus (2) | Veterinary isolate (cow) |
| 92–601 | B. abortus (2) | Veterinary isolate (cow) |
| Tulya | B. abortus (3) | Clinical isolate |
| 12–1745 | B. abortus (3) | Veterinary isolate (cow) |
| 03–2055 | B. abortus (3) | Veterinary isolate (sheep) |
| 99–4566 | B. abortus (3) | Veterinary isolate (cow) |
| 03–4278 | B. abortus (3) | Clinical isolate (blood) |
| 92-7369-2 | B. abortus (3) | Veterinary isolate (cow) |
| 292 | B. abortus (4) | Veterinary isolate (cow) |
| 99–9473 | B. abortus (4) | Veterinary isolate (cow) |
| B3196 | B. abortus (5) | Veterinary isolate (cow) |
| 870 | B. abortus (6) | Veterinary isolate (cow) |
| C68 | B. abortus (9) | Veterinary isolate (cow) |
| 1330 | B. suis | Veterinary isolate (pig) |
| 13-896-1815 | B. suis (1) | Clinical isolate |
| 12-2826-5972 | B. suis (1) | Clinical isolate |
| 04-1361Djakovo-1 | B. suis (1) | Veterinary isolate (boar) |
| 11-2920-5143 | B. suis (1) | Clinical isolate (blood) |
| 05–4266 | B. suis (1) | Clinical isolate (blood) |
| Thompsen | B. suis (2) | Veterinary isolate (hare) |
| 12-4327-8815 | B. suis (2) | Veterinary isolate (hare) |
| 12-2885-6046 | B. suis (2) | Clinical isolate |
| 11-3301-6219 | B. suis (2) | Veterinary isolate (pig) |
| 11-2942-5156 | B. suis (2) | Veterinary isolate (boar) |
| 11-028-111 | B. suis (2) | Veterinary isolate (pig) |
| 09-372-779 | B. suis (2) | Veterinary isolate (sheep) |
| 05–3495 | B. suis (2) | Clinical isolate (hip prosthesis) |
| 00–4898 | B. suis (2) | Veterinary isolate (cow) |
| 686 | B. suis (3) | Veterinary isolate (reindeer) |
| 40 | B. suis (4) | Veterinary isolate (reindeer) |
| 513 | B. suis (5) | Veterinary isolate (rodent) |
| 03-2770-12 | B. canis | Veterinary isolate (dog) |
| 04-2330-1 | B. canis | Veterinary isolate (dog) |
| 09-369-776(2) | B. canis | Veterinary isolate (dog) |
| 11-1961-3694(1) | B. canis | Veterinary isolate (dog) |
| 08-1276-2270 | B. canis | Clinical isolate (blood) |
| 63/290 | B. ovis | Veterinary isolate (sheep) |
| 12-1497-b | B. ovis | Veterinary isolate (sheep) |
| 11-868-1991 | B. ovis | Veterinary isolate (sheep) |
| 12-3480-79 | B. ovis | Veterinary isolate (sheep) |
| B1/94 | B. ceti | Veterinary isolate (porpoise) |
| 34/94 | B. ceti | Veterinary isolate (porpoise) |
| 97/0776 | B. ceti | Veterinary isolate (dolphin) |
| B202R | B. ceti | Veterinary isolate (whale) |
| 47/94 | B. ceti | Veterinary isolate (dolphin) |
| B14/94 | B. ceti | Veterinary isolate (dolphin) |
| 98/230 | B. ceti | Veterinary isolate (dolphin) [17] |
| 5/95 | B. ceti | Veterinary isolate (dolphin) |
| B2/94 | B. pinnipedialis | Veterinary isolate (seal) |
| 39/94 | B. pinnipedialis | Veterinary isolate (seal) |
| 55/94 | B. pinnipedialis | Veterinary isolate (otter) |
| 61/94 | B. pinnipedialis | Veterinary isolate (seal) |
| BO1 | B. inopinata | Clinical isolate (breast implant) [5] |
| BO2 | B. inopinata-like | Clinical isolate (lung) [6] |
| F8/08-60 | B. papionis | Veterinary isolate (baboon) [18] |
| F8/08-61 | B. papionis | Veterinary isolate (baboon) [18] |
Table 2. Bacterial strains and isolates used for external validation of the database.
| Name | Species (biovar) | Description |
|---|---|---|
| 2308 | B. abortus (1) | Veterinary isolate (cow) |
| S19 | B. abortus (1) | Vaccine strain (spontaneous attenuation) |
| RB51 | B. abortus (1) | Vaccine strain (rough mutant) |
| 97-4775-11 | B. abortus (1) | Veterinary isolate (cow) |
| 75–17 | B. abortus (3) | Veterinary isolate (cow) |
| 83–227 | B. abortus (3) | Veterinary isolate (cow) |
| 83–233 | B. abortus (3) | Veterinary isolate (cow) |
| 79–153 | B. abortus (3) | Veterinary isolate (cow) |
| 82–41 | B. abortus (3) | Veterinary isolate (cow) |
| 99-9971-135 | B. abortus (7) | Veterinary isolate (cow) [21] |
| 03-4923-239-D | B. abortus (7) | Veterinary isolate (cow) [21] |
| 77–9 | B. abortus (9) | Veterinary isolate (cow) |
| 80–133 | B. abortus (9) | Veterinary isolate (dog) |
| Mex 51 | B. canis | Veterinary isolate (dog) |
| 36/94 | B. ceti | Veterinary isolate (porpoise) [22] |
| F5/99 | B. ceti | Veterinary isolate (dolphin) [23] |
| UK3/05 | B. ceti | Veterinary isolate (dolphin) [24] |
| 75–3 | B. melitensis (1) | Clinical isolate |
| 78–158 | B. melitensis (1) | Veterinary isolate (sheep) |
| 88–44 | B. melitensis (1) | Clinical isolate |
| 1109 | B. melitensis (1) | Clinical isolate |
| Rev1 | B. melitensis (1) | Vaccine strain |
| 16M+GFP | B. melitensis (1) | Fluorescent 16M strain |
| 72–59 | B. melitensis (3) | Veterinary isolate (goat) |
| 77–47 | B. melitensis (3) | Clinical isolate |
| 78–13 | B. melitensis (3) | Veterinary isolate (goat) |
| 81–44 | B. melitensis (3) | |
| 81–140 | B. melitensis (3) | Clinical isolate |
| 82–73 | B. melitensis (3) | Clinical isolate |
| 82–87 | B. melitensis (3) | Clinical isolate |
| 90–129 | B. melitensis (3) | Veterinary isolate (cow) |
| 91–244 | B. melitensis (3) | Veterinary isolate (sheep) |
| 79–185 | B. melitensis (3) | Clinical isolate |
| CCM4915 | B. microti | Type strain (BCCN 07–01) [25] |
| 5K33 | B. neotomae | Type strain (ATCC 23459) [26] |
| 76250 | B. ovis | |
| 91268 | B. ovis | |
| 91212 | B. ovis | |
| 56/94 | B. pinnipedialis | Veterinary isolate (seal) [22] |
| 96/408 | B. pinnipedialis | Veterinary isolate (seal) [23] |
| UK9/99 | B. pinnipedialis | Veterinary isolate (seal) [24] |
| 04-1361Sisak-4 | B. suis (1) | Veterinary isolate (boar) |
| 15/95 | Brucella sp. | Veterinary isolate (seal) |
| 49/94 | Brucella sp. | Veterinary isolate (dolphin) |
| NF2637 | Brucella sp. | Veterinary isolate (rodents) [27] |
| NF2653 | Brucella sp. | Veterinary isolate (rodents) [27] |
| 02/611 | Brucella sp. | Clinical isolate (osteomyelitis) [7]. |
| B13-0095 | Brucella sp. | Veterinary isolate (frog) [28] |
| ATCC48188 | Ochrobactrum anthropi | Reference strain |
| LMG3301 | Ochrobactrum intermedium | Type strain |
MALDI-TOF MS samples
Samples used to build the spectra database were prepared according to a previously established inactivation protocol [19] consisting in resuspending two full loops of bacteria (i.e. multiple colonies) in 200 μL of solvent mix, vortexing (10 sec), centrifuging (10,000 g, 2 min) at room temperature, removing 190 μL and resuspending in the 10 μL of solvent left in the tube. For the external evaluation study, this protocol was simplified by suspending only one loop of bacteria in 100 μL of solvent mix, vortexing (10 sec) and incubating at room temperature (20–25°C, 3 minutes). Bacteria were efficiently inactivated by this method and the biomass concentration of the samples allowed identification by MALDI-TOF MS, demonstrating that the centrifugation step in the original protocol was not required.
MALDI-TOF MS analysis
One μL of each sample was applied to a single well of a disposable, barcode-labeled target slide (VITEK MS-DS, bioMérieux), overlaid with 1 μL of a saturated solution of alpha-cyano-4-hydroxycinnamic acid matrix in 50% acetonitrile and 2.5% trifluoroacetic acid (VITEK MSCHCA, bioMérieux) then air dried. For the database construction, several independent measurements were recorded for each strain (see S1 Table for the different culture conditions).
For instrument calibration, an Escherichia coli reference strain (ATCC 8739) was directly transferred to designated spots on the target slide using the procedure recommended by the manufacturer.
Mass spectra were acquired using a VITEK MS Plus (bioMérieux, Marcy l’Etoile) and the Launchpad v2.8 software program (Kratos, Shimadzu group Compagny, Manchester, UK). Dendrograms showing taxonomic relationships between strains were constructed using the SARAMIS software (bioMérieux, Marcy l’Etoile, France).
Construction and optimization of the database
The database was built as previously described [20]. Briefly, peak lists were binned by assigning each peak within the mass range of 3.000–17.000 Da to one of 1,300 bins. A predictive model was then established for each species using the Advanced Spectra Classifier (ASC) algorithm developed by bioMérieux (La Balme les Grottes, France). The outcome of this procedure provided an assignment of a dimensionless weight for each bin and for each species. As a result, a specific pattern of weights for the 1,300 bins was obtained and combined for all species in a weighted bin matrix.
For optimization, the spectral data were partitioned into 5 complementary subsets. One round of cross-validation involved a learning phase on 4 subsets (“training set”) and a validation of the predictive model on the remaining subset (“testing set”). Five rounds of cross-validation were performed by permutation, and the results from the five rounds combined.
To assess the accuracy of the database and calculate its performance in cross-validation, individual spectra were re-used as template for identification. The ASC algorithm compares the acquired spectrum to the specific pattern of each organism/organism group in the database and calculates a percent probability, or confidence value (%ID), which represents the similarity in terms of presence/absence of specific peaks between spectra. A perfect match provides a %ID of 99.9%. %ID >60 to 99.8% are considered as good. Scores <60% are considered to have no valid identification. The VITEK MS system renders the following types of identification results: “Single Choice”, when the spectrum acquired presents a high level of similarity (%ID >60 to 99.9%) with only one specific pattern in the database; “Low discrimination”, when the spectrum acquired presents a high level of similarity with 2 to 4 specific patterns in the database; or “No Identification”, when the spectrum acquired either does not match with any pattern in the database, or presents a high level of similarity to more than 4 specific patterns. During cross-validation, identification was considered as correct when the result was consistent with the reference identification. Low discrimination results were considered as correct if the expected identification was included in the matches. A misidentification was defined as discordant organism identification between the cross-validation result and the reference identification.
Evaluation of performances by external validation
External spectra were generated from bacteria cultivated with different growth conditions (media, incubation time, etc) to mimic possible inter-laboratory variations. To reflect clinical laboratory practice, inactivated samples were spotted in duplicate, and analyzed with the updated database. If only one of the two spectra allowed a correct identification, the isolate was considered correctly identified. The cut-off for identification confidence was as described above.
Results
To update the MALDI-TOF MS VITEK database, we used 84 Brucella strains, either reference strains or well characterized clinical/veterinary isolates (Tables 1 and S1), to generate independent spectra covering the Brucella genus. After initial selection based on quality criteria such as peak resolution, signal to noise ratio, number of peaks, absolute signal intensity, and intra-specific similarity, 590 spectra were retained and submitted for biomathematical analyses using an iterative system (bioMérieux patented ASC algorithm).
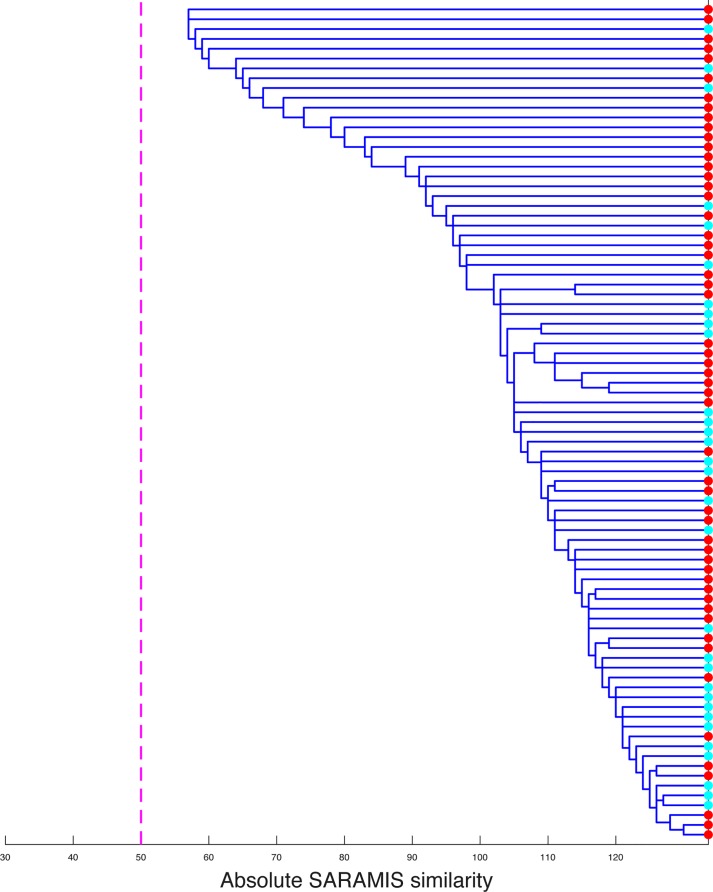
Using an optimization process, we next evaluated the possibility to discriminate between different Brucella species and biovars. Discrimination between the different species was obtained, with the exception of B. ceti and B. pinnipedialis. These two species could not be clearly separated, as illustrated by the intertwining of their spectra on a dendrogram (Fig 1). Distinguishing the different biovars of B. melitensis and B. abortus was not possible. Discrimination between several of B. suis biovars was obtained (S1 Fig), but biovars 1 and 4 gave cross-identifications.
Fig 1. Proximity of B. ceti and B. pinnipedialis MALDI-TOF MS spectra.
Cluster analysis, using correlation-based dissimilarity, was performed to assess the discriminating power of MALDI-TOF MS between the spectra corresponding to the species B. ceti (in red) or B. pinnipedialis (in blue). The threshold of 50 common peaks, which is considered as a minimum for considering that spectra are different, is shown as a dotted line.
Classes representing the different Brucella species were thus created by grouping together the different biovars of B. melitensis, of B. abortus and of B. suis, and the two species B. ceti and B. pinnipedialis. The eight species represented in the MALDI-TOF database are thus: B. melitensis (biovar 1, 2 or 3), B. abortus (biovar 1, 2, 3, 4, 5, 6 or 9), B. suis (biovar 1, 2, 3, 4 or 5), B. canis, B. ovis, B. ceti/B. pinnipedialis, B. inopinata and B. papionis.
After optimization, cross validation was performed to evaluate the performance of the updated database, which contains 37,902 spectra covering 1,095 bacterial species including Brucella. This mathematical method is used to assess how accurately the database can perform. Correct identification at the genus level was obtained in 97.29% of cases (Table 3). Importantly, the remaining 2.71% of results corresponded to “no ID”, but never to an incorrect identification. At the species level, the performance varied between the different classes. For the three main zoonotic species (B. melitensis, B. abortus and B. suis), correct identification was obtained with 96.06%, 100% or 89.34% of spectra, respectively.
Table 3. Results of identification in cross validation studies.
Identifications (ID) results, either at genus or species/class level, were classified as correct (either “Single Choice” or “Low Discrimination”), discordant or no-identification (No ID). This table gives the % of each type of identification results for the indicated species/classes using the updated database.
| CLASS | Species | # of strains | # of spectra | CORRECT ID | DISCORDANT ID | No ID | OVERALL % by genus |
OVERALL % by class |
||
|---|---|---|---|---|---|---|---|---|---|---|
| % Single choice |
% Low discrimination |
% Correct genus |
% Incorrect genus | |||||||
| B. melitensis | B. melitensis | 5 | 22 | 90.91 | 4.55 | 0 | 0 | 4.55 | 97.29 | 96.06 |
| B. melitensis biovar 1 | 8 | 35 | 85.71 | 8.57 | 2.86 | 0 | 2.86 | |||
| B. melitensis biovar 2 | 3 | 24 | 95.83 | 4.17 | 0 | 0 | 0 | |||
| B. melitensis biovar 3 | 7 | 46 | 84.78 | 10.87 | 2.17 | 0 | 2.17 | |||
| B. abortus | B. abortus biovar 1 | 7 | 32 | 93.75 | 6.25 | 0 | 0 | 0 | 100 | |
| B. abortus biovar 2 | 3 | 25 | 100 | 0 | 0 | 0 | 0 | |||
| B. abortus biovar 3 | 7 | 36 | 100 | 0 | 0 | 0 | 0 | |||
| B. abortus biovar 4 | 3 | 10 | 100 | 0 | 0 | 0 | 0 | |||
| B. abortus biovar 5 | 1 | 7 | 100 | 0 | 0 | 0 | 0 | |||
| B. abortus biovar 6 | 2 | 22 | 100 | 0 | 0 | 0 | 0 | |||
| B. abortus biovar 9 | 2 | 24 | 100 | 0 | 0 | 0 | 0 | |||
| B. suis | B. suis biovar 1 | 7 | 47 | 89.36 | 2.13 | 0 | 0 | 8.51 | 89.34 | |
| B. suis biovar 2 | 9 | 47 | 82.98 | 0 | 0 | 0 | 17.02 | |||
| B. suis biovar 3 | 1 | 8 | 100 | 0 | 0 | 0 | 0 | |||
| B. suis biovar 4 | 1 | 12 | 91.67 | 0 | 0 | 0 | 8.33 | |||
| B. suis biovar 5 | 1 | 8 | 100 | 0 | 0 | 0 | 0 | |||
| B. canis | B. canis | 5 | 19 | 84.21 | 15.79 | 0 | 0 | 0 | 100 | |
| B. ovis | B. ovis | 4 | 30 | 93.33 | 6.67 | 0 | 0 | 0 | 100 | |
| B. ceti/B. pinnipedialis | B. ceti | 8 | 54 | 98.15 | 1.85 | 0 | 0 | 0 | 100 | |
| B. pinnipedialis | 4 | 31 | 100 | 0 | 0 | 0 | 0 | |||
| B. inopinata | B. inopinata | 2 | 23 | 95.65 | 4.35 | 0 | 0 | 0 | 100 | |
| B. papionis | B. papionis | 2 | 28 | 100 | 0 | 0 | 0 | 0 | 100 | |
Finally, as an external validation, the database was challenged with the MALDI-TOF spectra from 48 independent Brucella isolates, and 2 strains of Ochrobactrum, which are “near neighbors” of the Brucella genus (Tables 2 and S2).
The implemented database allowed correct identification at the genus level in 88.4% of cases, all the other results being “No-identification” but never misidentification as another genus (Tables 4 and 5). At the species level, the performances varied. For B. melitensis, B. abortus, and B. suis, correct identification was obtained for 100%, 92.3% or 100% of strains, respectively. It should be noted however that only one extra B. suis isolate was available to be tested in the external validation.
Table 4. Identification results in external validation.
Identification (ID) results, either at genus or species/class level, were classified as correct, discordant or no-identification (No ID). This table gives the % of strains for which each type of identification result was obtained using the updated database. The number (n) of strains tested for each species/class is also indicated in parenthesis. N/A = not applicable.
| IDENTIFICATION AT THE GENUS LEVEL |
IDENTIFICATION AT THE SPECIES LEVEL |
||||||
|---|---|---|---|---|---|---|---|
| % Correct ID |
% Discordant ID |
% No ID |
% Correct ID |
% Discordant ID |
% No ID |
||
| Brucella species present in the database | B. melitensis (n = 16) | 100% (16) | 0% (0) | 0% (0) | 100% (16) | 0% (0) | 0% (0) |
| B. abortus (n = 13) | 92.3% (12) | 0% (0) | 7.7% (1) | 92.3% (12) | 0% (0) | 7.7% (1) | |
| B. suis (n = 1) | 100% (1) | 0% (0) | 0% (0) | 100% (1) | 0% (0) | 0% (0) | |
| B. canis (n = 1) | 100% (1) | 0% (0) | 0% (0) | 0% (0) | 100% (1) | 0% (0) | |
| B. ovis (n = 3) | 66.7% (2) | 0% (0) | 33.3% (1) | 66.7% (2) | 0% (0) | 33.3% (1) | |
| B. ceti/B. pinnipedialis (n = 9) | 66.7% (6) | 0% (0) | 33.3% (3) | 66.7% (6) | 0% (0) | 33.3% (3) | |
| Total (n = 43) | 88,4% (38) | 0% (0) | 11.6% (5) | 86% (37) | 2.3% (1) | 11.6% (5) | |
| Brucella species not present in the database | B. microti (n = 1) | 0% (0) | 0% (0) | 100% (1) | N/A | N/A | N/A |
| B. neotomae (n = 1) | 0% (0) | 0% (0) | 100% (1) | N/A | N/A | N/A | |
| Brucella strains with genus level characterization | Brucella spp (n = 3) | 66.7% (2) | 0% (0) | 33.3% (1) | N/A | N/A | N/A |
| Ochrobactrum strains | O. anthropi (n = 1) | 100% (1) | 0% (0) | 0% (0) | 100% (1) | 0% (0) | 0% (0) |
| O. intermedium (n = 1) | 100% (1) | 0% (0) | 0% (0) | 100% (1) | 0% (0) | 0% (0) | |
Table 5. Detailed results of external validation.
For each strain, the identification results for the two deposits are given. %ID are confidence values given by the VITEK MS system for each identification result. COS-B = Columbia Blood Agar (bioMérieux, 43 041); BBA = Brucella blood agar (bioMérieux); COS-O = Columbia agar with 5% sheep blood (Oxoïd).
| SAMPLE INFORMATION | IDENTIFICATION RESULTS | |||||
|---|---|---|---|---|---|---|
| Strain name | Species (biovar) | Culture condition | # of peaks | Identification type | Identification | % ID |
| 75–3 | B. melitensis (1) | COS-B (48h) | 120 | Single choice | Brucella melitensis | 98.15 |
| 99 | Single choice | Brucella melitensis | 99.99 | |||
| 78–158 | B. melitensis (1) | COS-B (48h) | 125 | Single choice | Brucella melitensis | 83.06 |
| 118 | Single choice | Brucella melitensis | 99.95 | |||
| 88–44 | B. melitensis (1) | COS-B (48h) | 165 | Single choice | Brucella melitensis | 99.99 |
| 141 | Single choice | Brucella melitensis | 99.99 | |||
| 1109 | B. melitensis (1) | COS-B (48h) | 121 | Single choice | Brucella melitensis | 99.99 |
| 127 | Single choice | Brucella melitensis | 99.99 | |||
| Rev1 | B. melitensis (1) | COS-B (48h) | 134 | Single choice | Brucella melitensis | 99.8 |
| 136 | Single choice | Brucella melitensis | 99.99 | |||
| 16M+GFP | B. melitensis (1) | COS-B (48h) | 129 | Single choice | Brucella melitensis | 99.99 |
| 158 | Single choice | Brucella melitensis | 98.44 | |||
| 72–59 | B. melitensis (3) | COS-B (48h) | 150 | Single choice | Brucella melitensis | 98.1 |
| 129 | Single choice | Brucella melitensis | 99.76 | |||
| 77–47 | B. melitensis (3) | COS-B (48h) | 127 | Single choice | Brucella melitensis | 99.99 |
| 124 | Single choice | Brucella melitensis | 99.99 | |||
| 78–13 | B. melitensis (3) | COS-B (48h) | 111 | Single choice | Brucella melitensis | 63.65 |
| 102 | Single choice | Brucella melitensis | 99.64 | |||
| 81–44 | B. melitensis (3) | COS-B (48h) | 122 | No identification | ||
| 101 | Single choice | Brucella melitensis | 99.51 | |||
| 81–140 | B. melitensis (3) | COS-B (48h) | 148 | Single choice | Brucella melitensis | 99.99 |
| 147 | Single choice | Brucella melitensis | 99.96 | |||
| 82–73 | B. melitensis (3) | COS-B (48h) | 99 | Single choice | Brucella melitensis | 87.07 |
| 101 | Single choice | Brucella melitensis | 99.97 | |||
| 82–87 | B. melitensis (3) | COS-B (48h) | 121 | Single choice | Brucella melitensis | 99.79 |
| 123 | Single choice | Brucella melitensis | 89.74 | |||
| 90–129 | B. melitensis (3) | COS-B (48h) | 110 | Single choice | Brucella melitensis | 99.99 |
| 125 | Single choice | Brucella melitensis | 99.99 | |||
| 91–244 | B. melitensis (3) | COS-B (48h) | 91 | No identification | ||
| 101 | Single choice | Brucella melitensis | 99.99 | |||
| 79–185 | B. melitensis (3) | COS-B (48h) | 163 | Single choice | Brucella melitensis | 89.68 |
| 162 | No identification | |||||
| 2308 | B. abortus (1) | COS-B (48h) | 165 | Single choice | Brucella abortus | 99.99 |
| 154 | Single choice | Brucella abortus | 99.99 | |||
| S19 | B. abortus (1) | BBA (72h) | 66 | Low Discrimination | Brucella abortus | 50.14 |
| 80 | Single choice | Brucella abortus | 99.99 | |||
| RB51 | B. abortus (1) | BBA (96h) | 173 | No identification | ||
| 104 | No identification | |||||
| 97-4775-11 | B. abortus (1) | COS-B (48h) | 134 | Single choice | Brucella abortus | 99.97 |
| 136 | Single choice | Brucella abortus | 81.79 | |||
| 75–17 | B. abortus (3) | COS-B (48h) | 127 | Single choice | Brucella abortus | 99.99 |
| 125 | Single choice | Brucella abortus | 99.99 | |||
| 83–227 | B. abortus (3) | COS-B (48h) | 137 | Single choice | Brucella abortus | 98.34 |
| 101 | Single choice | Brucella abortus | 99.87 | |||
| 83–233 | B. abortus (3) | COS-B (48h) | 136 | Single choice | Brucella abortus | 99.99 |
| 117 | Single choice | Brucella abortus | 99.99 | |||
| 79–153 | B. abortus (3) | COS-B (48h) | 130 | Single choice | Brucella abortus | 99.99 |
| 136 | Single choice | Brucella abortus | 99.99 | |||
| 82–41 | B. abortus (3) | COS-B (48h) | 130 | Single choice | Brucella abortus | 99.99 |
| 138 | Single choice | Brucella abortus | 99.99 | |||
| 99-9971-135 | B. abortus (7) | COS-B (72h) | 81 | Single choice | Brucella abortus | 99.99 |
| 105 | Single choice | Brucella abortus | 99.99 | |||
| 03-4923-239-D | B. abortus (7) | COS-B (48h) | 148 | Single choice | Brucella abortus | 99.99 |
| 149 | Single choice | Brucella abortus | 99.99 | |||
| 77–9 | B. abortus (9) | COS-B (48h) | 94 | Single choice | Brucella abortus | 99.99 |
| 118 | Single choice | Brucella abortus | 99.99 | |||
| 80–133 | B. abortus (9) | COS-B (48h) | 98 | Single choice | Brucella abortus | 99.99 |
| 115 | Single choice | Brucella abortus | 87.95 | |||
| 04-1361Sisak-4 | B. suis (1) | COS-O (72h) | 87 | Single choice | Brucella suis | 99.99 |
| 78 | Single choice | Brucella suis | 99.99 | |||
| Mex 51 | B. canis | COS-B (48h) | 140 | Single choice | Brucella suis | 99.98 |
| 158 | No identification | |||||
| 76250 | B. ovis | COS-B + 5% CO2 (96h) | 125 | No identification | ||
| 107 | No identification | |||||
| 91268 | B. ovis | COS-B + 5% CO2 (96h) | 116 | No identification | ||
| 104 | Single choice | Brucella ovis | 99.99 | |||
| 91212 | B. ovis | COS-B + 5% CO2 (96h) | 130 | Single choice | Brucella ovis | 99.99 |
| 101 | Single choice | Brucella ovis | 99.96 | |||
| F5/99 | B. ceti | COS-B + 5% CO2 (96h) | 106 | No identification | ||
| 91 | No identification | |||||
| UK3/05 | B. ceti | COS-B + 5% CO2 (96h) | 127 | No identification | ||
| 121 | Single choice | Brucella ceti/pinnipedialis | 99.98 | |||
| UK9/99 | B. pinnipedialis | COS-B + 5% CO2 (96h) | 118 | Single choice | Brucella ceti/pinnipedialis | 83.28 |
| 126 | Single choice | Brucella ceti/pinnipedialis | 99.99 | |||
| 36/94 | B. ceti | COS-B + 5% CO2 (96h) | 112 | No identification | ||
| 119 | No identification | |||||
| 49/94 | Brucella sp. (B. ceti ?) | COS-B + 5% CO2 (96h) | 119 | Single choice | Brucella ceti/pinnipedialis | 99.99 |
| 128 | Single choice | Brucella ceti/pinnipedialis | 99.96 | |||
| 02/611 | Brucella sp. (B. ceti-like) | COS-B + 5% CO2 (96h) | 148 | Single choice | Brucella ceti/pinnipedialis | 99.99 |
| 133 | Single choice | Brucella ceti/pinnipedialis | 100 | |||
| 15/95 | Brucella sp. (B. pinnipedialis ?) | COS-B + 5% CO2 (96h) | 112 | No identification | ||
| 114 | No identification | |||||
| 56/94 | B. pinnipedialis | COS-B + 5% CO2 (96h) | 116 | No identification | ||
| 126 | Single choice | Brucella ceti/pinnipedialis | 99.77 | |||
| 96/408 | B. pinnipedialis | COS-B + 5% CO2 (96h) | 131 | Single choice | Brucella ceti/pinnipedialis | 99.99 |
| 97 | No identification | |||||
| CCM4915 | B. microti | COS-B (24h) | 162 | No identification | ||
| 181 | No identification | |||||
| 5K33 | B. neotomae | COS-B (48h) | 145 | No identification | ||
| 127 | No identification | |||||
| NF2637 | Brucella sp. | COS-B (24h) | 114 | Single choice | Brucella inopinata | 99.99 |
| 144 | Single choice | Brucella inopinata | 77.86 | |||
| NF2653 | Brucella sp. | COS-B (24h) | 141 | No identification | ||
| 157 | No identification | |||||
| B13-0095 | Brucella sp. | COS-B (24h) | 136 | Single choice | Brucella inopinata | 100 |
| 129 | Single choice | Brucella inopinata | 99.99 | |||
| ATCC48188 | Ochrobactrum anthropi | COS-B (48h) | 127 | Single choice | Ochrobactrum anthropi | 100 |
| 113 | No identification | |||||
| LMG3301 | Ochrobactrum intermedium | COS-B (48h) | 122 | Single choice | Ochrobactrum intermedium | 100 |
| 118 | Single choice | Ochrobactrum intermedium | 100 | |||
Interestingly, the rare clinical isolate 02/611, described as B. ceti-like after molecular characterization [7], was indeed identified within the B.ceti/B. pinnipedialis class. Also, both the Bullfrog (B13-0095) and the Australian rodent (NF2637) isolates were identified as B. inopinata, in agreement with previous work showing that these belong to the atypical Brucella clade of this genus [27,28]. The two isolates belonging to “B. abortus biovar 7”, a rare biovar of this species, were identified as B. abortus. Finally, the recombinant 16M strain overexpressing the green fluorescent protein (GFP) was correctly identified as B. melitensis using this database. Moreover, using different culture conditions for 16M did not affect its identification by MALDI-TOF MS (S3 Table).
Discussion
A major asset of this MALDI-TOF MS database is its ability to identify Brucella isolates at the species level, which is essential for following epidemiological outbreaks. Obtaining such a resolution was very challenging for this genus, as highlighted in previous studies [29], because of the high similarity between species at the genetic level [30]. Discrimination between species was made possible using a patented approach to differentiate closely related species using internal calibration and a two-step algorithm. This was not sufficient to distinguish the two species of Brucella from marine mammals (B. ceti and B. pinnipedialis). This is in agreement with a recent Multi-Locus Sequence Analysis (MLSA) showing that the taxonomy is inconsistent with the phylogeny of these two species, and that taxonomic rearrangement should be envisaged [31]. This MALDI-TOF MS database is however able to discriminate eight different Brucella species, which include the most common in human or animal disease.
The updated database allowed correct identification of Brucella isolates at the genus level in 88.4% of cases. It is important to mention that none of them was identified as Ochrobactrum spp., a misidentification that is common with other standard identification methods [32–34] and recently reported using the VITEK MS database currently available [35]. Analysis at the species level gave only one discordant result, corresponding to cross identification between two Brucella species. Such result would have no consequence for human medicine, as identification at the genus level is sufficient to prescribe the appropriate treatment. As for all MALDI-TOF databases, the limitation of this system is its inability to identify non-clinically validated species or species not included in the database. However, the large coverage of the Brucella genus (in particular the most common species) in this database makes this risk is very minor.
Diminution of the performance at the genus and/or species level was due to “no ID” results for some rare and/or atypical Brucella spp. (B. neotomae strain 5K33, B. microti strain CCM4915, and the rodent isolate NF2653), several strains from marine mammals, and the vaccine strain B. abortus RB51. These results were not due to the quality of MALDl-TOF spectra, which was good (based on the number of spectral peaks, Table 5). In the spectra for RB51, we found that several masses characteristics of the B. abortus class were less frequently present, in particular the masses of 5,920.63, 6,040.32 and 7,467.89 Da were present in only 14.3% of spectra (vs. in 75–95% of the spectra of other B. abortus isolates, with a tolerance of 800 ppm). The only discordant result in our assay was obtained with B. canis Mex51, which was identified as B. suis. This was due to the presence in its spectra of additional masses that are common with the B. suis class in addition to the major peaks characteristics of the B. canis class. This finding is consistent with an exhaustive MLSA showing that B. canis strains are very close to B. suis biovars 3 and 4 [31].
Importantly, the MALDI-TOF database allowed the correct identification as Brucella of several recently discovered “atypical” isolates [5,6,28,36]. These strains represent a serious problem for diagnosis laboratories, as they are not identified as Brucella using classical phenotypic tests. It is possible that similar strains have been isolated in the past but misidentified. Very little is known concerning the ability of these new species to cause disease in humans or livestock. The possibility to identify these isolates as Brucella will thus be important for both human and animal health.
Overexpression of an exogenous protein (GFP) did not affect the identification of B. melitensis 16M. This is important since recombinant Brucella strains are common tools in research laboratories and could potentially infect lab workers. Moreover, the use of such Brucella strains as vaccines was proposed, since the presence anti-GFP antibodies would allow distinguishing vaccinated animals from naturally infected ones [37].
In conclusion, this updated MALDI-TOF MS database is a new diagnostic tool that allows the identification of Brucella. It combines precision of identification (broad coverage of the Brucella genus together with species-level identification) and widespread availability. After integration in the VITEK MS (v3.2), this will be the first Brucella database validated for diagnostic with CE accreditation and accessible to all users in routine. This will allow accurate diagnosis and timely treatment in brucellosis. These highly infectious pathogens also causing one of the most frequent laboratory-acquired infection [38], their rapid identification by MALDI-TOF MS will decrease the risk of accidental infection of laboratory workers. A paradox of global health however is that the countries where brucellosis is endemic may not have access to MALDI-TOF MS. This could be circumvented by the use of the in-tube inactivation method described earlier [19], which will allow the shipment of erstwhile infectious samples to mass spectrometry platforms.
Supporting information
Multidimensional Scaling (MDS) analysis of MALDI-TOF spectra obtained with B. suis isolates. The similarity between spectra is represented as distances, which depend on the presence/absence of peaks and their intensity in compared spectra. Results are presented on the three first dimensions. The color code used for each biovar is indicated in the figure.
(TIF)
Strains highlighted in grey are reference or type strains. The different culture conditions used for each strain (time of incubation in hours, media, ± 5% CO2) are indicated. BAS = Brucella blood agar with 5% sheep blood, Hemin and Vitamin K1 (Becton Dickinson PA-255509.05A), BBA = Brucella blood agar (bioMérieux, 411 968), CHOC-H = Chocolate agar (Hardy Diagnostic, E14), COS-B = Columbia Blood Agar (bioMérieux, 43 041), COS-D = Columbia agar with 5% sheep blood (Becton Dickinson, 90006 166), COS-O = Columbia agar with 5% sheep blood (Oxoïd, PB5039A).
(DOCX)
(DOCX)
The culture conditions (media, time of incubation in hours, ± 5% CO2) are indicated. BAS = Brucella agar with 5% sheep blood, hemin & vitamin K1 (Becton Dickinson); BBA = Brucella blood agar (bioMérieux); CHOC-O = Chocolate agar plate with vitox (Oxoïd); COS-B = Columbia Blood Agar (bioMérieux, 43 041); COS-O = Columbia agar with 5% sheep blood (Oxoïd); MHB = Mueller Hinton agar with 5% sheep blood (Biorad); MHF = Mueller Hinton agar with 5% horse blood and β-NAD (Biorad); TSA-S = Trypticase soy agar with 5% sheep blood (Becton Dickinson).
(DOCX)
Acknowledgments
We are grateful to the Emerging Infectious Diseases Laboratory (APHL-CDC, North Carolina State Laboratory of Public Health) for sharing strain collections and for data acquisitions and to Maryne Jay (ANSES, France) for administrative support with ANSM and shipment procedures.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Institut National de la Sante et de la Recherche Medicale (INSERM), the Université de Montpellier and Agence Santé Publique France. It was also partially sponsored by bioMérieux (Marcy l’Etoile, France). The funders had no influence on data collection, interpretation of this study or decision to publish.
References
- 1.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos E V. The new global map of human brucellosis. Lancet Infect Dis. 2006;6: 91–99. 10.1016/S1473-3099(06)70382-6 [DOI] [PubMed] [Google Scholar]
- 2.Cleaveland S, Sharp J, Abela-Ridder B, Allan KJ, Buza J, Crump JA, et al. One Health contributions towards more effective and equitable approaches to health in low- and middle-income countries. Philos Trans R Soc B Biol Sci. 2017;372: 20160168 10.1098/rstb.2016.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossetti CA, Arenas-Gamboa AM, Maurizio E. Caprine brucellosis: A historically neglected disease with significant impact on public health. PLoS Negl Trop Dis. 2017;11: 1–17. 10.1371/journal.pntd.0005692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garofolo G, Di Giannatale E, Platone I, Zilli K, Sacchini L, Abass A, et al. Origins and global context of Brucella abortus in Italy. BMC Microbiol. 2017;17: 28 10.1186/s12866-017-0939-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De BK, Stauffer L, Koylass MS, Sharp SE, Gee JE, Helsel LO, et al. Novel Brucella Strain (BO1) Associated with a Prosthetic Breast Implant Infection. J Clin Microbiol. 2008;46: 43–49. 10.1128/JCM.01494-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiller R V, Gee JE, Lonsway DR, Gribble S, Bell SC, Jennison A V, et al. Identification of an unusual Brucella strain (BO2) from a lung biopsy in a 52 year-old patient with chronic destructive pneumonia. BMC Microbiol. 2010;10: 23 10.1186/1471-2180-10-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald WL, Jamaludin R, Mackereth G, Hansen M, Humphrey S, Short P, et al. Characterization of a Brucella sp. strain as a marine-mammal type despite isolation from a patient with spinal osteomyelitis in New Zealand. J Clin Microbiol. 2006;44: 4363–4370. 10.1128/JCM.00680-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohn AH, Probert WS, Glaser CA, Gupta N, Bollen AW, Wong JD, et al. Human neurobrucellosis with intracerebral granuloma caused by a marine mammal Brucella spp. Emerg Infect Dis. 2003;9: 485–488. 10.3201/eid0904.020576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suárez-Esquivel M, Ruiz-Villalobos N, Jiménez-Rojas C, Barquero-Calvo E, Chacón-Díaz C, Víquez-Ruiz E, et al. Brucella neotomae infection in humans, Costa Rica. Emerg Infect Dis. 2017;23: 997–1000. 10.3201/eid2306.162018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villalobos-Vindas JM, Amuy E, Barquero-Calvo E, Rojas N, Chacón-Díaz C, Chaves-Olarte E, et al. Brucellosis caused by the wood rat pathogen Brucella neotomae: Two case reports. J Med Case Rep. Journal of Medical Case Reports; 2017;11: 1–4. 10.1186/s13256-016-1164-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudrik JT, Soehnlen MK, Perry MJ, Sullivan M, Reiter-Kintz W, Lee PA, et al. Safety and Accuracy of Matrix-Assisted Laser Desorption Ionization—Time of Flight Mass Spectrometry (MALDI-TOF MS) to Identify Highly Pathogenic Organisms. J Clin Microbiol. 2017;55: JCM.01023-17. 10.1128/JCM.01023-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tracz DM, Tyler AD, Cunningham I, Antonation KS, Corbett CR. Custom database development and biomarker discovery methods for MALDI-TOF mass spectrometry-based identification of high-consequence bacterial pathogens. J Microbiol Methods. Elsevier B.V.; 2017;134: 54–57. 10.1016/j.mimet.2017.01.009 [DOI] [PubMed] [Google Scholar]
- 13.Keller PM, Bruderer V, Müller F. Restricted Identification of Clinical Pathogens Categorized as Biothreats by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. Munson E, editor. J Clin Microbiol. 2016;54: 816–816. 10.1128/JCM.03250-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham SA, Patela R. Importance of using Bruker’s security-relevant library for biotyper identification of Burkholderia pseudomallei, Brucella species, and Francisella tularensis. J Clin Microbiol. 2013;51: 1639–1640. 10.1128/JCM.00267-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker SL, Zange S, Brockmeyer M, Grün U, Halfmann A. Rapid MALDI-TOF-based identification of Brucella melitensis from positive blood culture vials may prevent laboratory-acquired infections. J Hosp Infect. The Healthcare Infection Society; 2018; 10.1016/j.jhin.2018.04.008 [DOI] [PubMed] [Google Scholar]
- 16.Alton GG, Jones LM, Angus RD, Verger JM. Techniques for the brucellosis laboratory. Institut National de la Recherche Agronomique (INRA, Paris: ); 1988. [Google Scholar]
- 17.Miller WG, Adams LG, Ficht TA, Cheville NF, Payeur JP, Harley DR, et al. Brucella-induced abortions and infection in bottlenose dolphins (Tursiops truncatus). J Zoo Wildl Med. 1999;30: 100–10. [PubMed] [Google Scholar]
- 18.Schlabritz-Loutsevitch NE, Whatmore AM, Quance CR, Koylass MS, Cummins LB, Dick EJ Jr, et al. A novel Brucella isolate in association with two cases of stillbirth in non-human primates—first report. J Med Primatol. 2009;38: 70–73. 10.1111/j.1600-0684.2008.00314.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mesureur J, Ranaldi S, Monnin V, Girard V, Arend S, Welker M, et al. A simple and safe protocol for preparing Brucella samples for Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry analysis. J Clin Microbiol. 2016;54: 449–52. 10.1128/JCM.02730-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard V, Mailler S, Welker M, Arsac M, Cellière B, Cotte-Pattat PJ, et al. Identification of Mycobacterium spp. and Nocardia spp. from solid and liquid cultures by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Diagn Microbiol Infect Dis. 2016;86: 277–283. 10.1016/j.diagmicrobio.2016.07.027 [DOI] [PubMed] [Google Scholar]
- 21.Garin-Bastuji B, Mick V, Le Carrou G, Allix S, Perrett LL, Dawson CE, et al. Examination of taxonomic uncertainties surrounding Brucella abortus bv. 7 by phenotypic and molecular approaches. Appl Environ Microbiol. 2014;80: 1570–9. 10.1128/AEM.03755-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whatmore AM, Dawson C, Muchowski J, Perrett LL, Stubberfield E, Koylass M, et al. Characterisation of North American Brucella isolates from marine mammals. Roop RM, editor. PLoS One. 2017;12: 1–17. 10.1371/journal.pone.0184758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewalt DR, Payeur JB, Martin BM, Cummins DR, Miller WG. Characteristics of a Brucella Species from a Bottlenose Dolphin (Tursiops Truncatus). J Vet Diagnostic Investig. 1994;6: 448–452. 10.1177/104063879400600408 [DOI] [PubMed] [Google Scholar]
- 24.Groussaud P, Shankster SJ, Koylass MS, Whatmore AM. Molecular typing divides marine mammal strains of Brucella into at least three groups with distinct host preferences. J Med Microbiol. 2007;56: 1512–1518. 10.1099/jmm.0.47330-0 [DOI] [PubMed] [Google Scholar]
- 25.Scholz HC, Hubalek Z, Sedláček I, Vergnaud G, Tomaso H, Al Dahouk S, et al. Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int J Syst Evol Microbiol. 2008;58: 375–382. 10.1099/ijs.0.65356-0 [DOI] [PubMed] [Google Scholar]
- 26.Stoenner HG, Lackman DB. A new species of Brucella isolated from the desert wood rat, Neotoma lepida Thomas. Am J Vet Res. 1957;18: 947–51. [PubMed] [Google Scholar]
- 27.Tiller R V, Gee JE, Frace MA, Taylor TK, Setubal JC, Hoffmaster AR, et al. Characterization of novel Brucella strains originating from wild native rodent species in North Queensland, Australia. Appl Environ Microbiol. 2010;76: 5837–5845. 10.1128/AEM.00620-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soler-Lloréns PF, Quance CR, Lawhon SD, Stuber TP, Edwards JF, Ficht TA, et al. A Brucella spp. Isolate from a Pac-Man Frog (Ceratophrys ornata) Reveals Characteristics Departing from Classical Brucellae. Front Cell Infect Microbiol. 2016;6: 116 10.3389/fcimb.2016.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira L, Castaño SV, Sánchez-Juanes F, González-Cabrero S, Menegotto F, Orduña-Domingo A, et al. Identification of Brucella by MALDI-TOF mass spectrometry. Fast and reliable identification from agar plates and blood cultures. PLoS One. 2010;5 10.1371/journal.pone.0014235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wattam AR, Foster JT, Mane SP, Beckstrom-sternberg SM, Beckstrom-sternberg JM. Comparative Phylogenomics and Evolution of the Brucellae Reveal a Path to Virulence. 2014;196: 920–930. 10.1128/JB.01091-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whatmore AM, Koylass MS, Muchowski J, Edwards-Smallbone J, Gopaul KK, Perrett LL. Extended Multilocus Sequence Analysis to Describe the Global Population Structure of the Genus Brucella: Phylogeography and Relationship to Biovars. Front Microbiol. 2016;7: 1–14. 10.3389/fmicb.2016.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elsaghir AAF, James EA. Misidentification of Brucella melitensis as Ochrobactrum anthropi by API 20NE. Journal of Medical Microbiology. 2003. pp. 441–442. 10.1099/jmm.0.05153-0 [DOI] [PubMed] [Google Scholar]
- 33.Horvat RT, El Atrouni W, Hammoud K, Hawkinson D, Cowden S. Ribosomal RNA sequence analysis of Brucella infection misidentified as Ochrobactrum anthropi infection. J Clin Microbiol. 2011;49: 1165–8. 10.1128/JCM.01131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vila A, Pagella H, Vera Bello G, Vicente A. Brucella suis bacteremia misidentified as Ochrobactrum anthropi by the VITEK 2 system. J Infect Dev Ctries. 2016;10: 10–14. 10.3855/jidc.7532 [DOI] [PubMed] [Google Scholar]
- 35.Poonawala H, Marrs Conner T, Peaper DR. The Brief Case: Misidentification of Brucella melitensis as Ochrobactrum anthropi by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS). Burnham C-AD, editor. J Clin Microbiol. 2018;56: e00914–17. 10.1128/JCM.00914-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whatmore AM, Davison N, Cloeckaert A, Al Dahouk S, Zygmunt MS, Brew SD, et al. Brucella papionis sp. nov., isolated from baboons (Papio spp.). Int J Syst Evol Microbiol. 2014;64: 4120–8. 10.1099/ijs.0.065482-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chacón-Díaz C, Muñoz-Rodríguez M, Barquero-Calvo E, Guzmán-Verri C, Chaves-Olarte E, Grilló MJ, et al. The use of green fluorescent protein as a marker for Brucella vaccines. Vaccine. 2011;29: 577–582. 10.1016/j.vaccine.2010.09.109 [DOI] [PubMed] [Google Scholar]
- 38.Traxler RM, Lehman MW, Bosserman EA, Guerra MA, Smith TL. A literature review of laboratory-acquired brucellosis. J Clin Microbiol. 2013;51: 3055–3062. 10.1128/JCM.00135-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multidimensional Scaling (MDS) analysis of MALDI-TOF spectra obtained with B. suis isolates. The similarity between spectra is represented as distances, which depend on the presence/absence of peaks and their intensity in compared spectra. Results are presented on the three first dimensions. The color code used for each biovar is indicated in the figure.
(TIF)
Strains highlighted in grey are reference or type strains. The different culture conditions used for each strain (time of incubation in hours, media, ± 5% CO2) are indicated. BAS = Brucella blood agar with 5% sheep blood, Hemin and Vitamin K1 (Becton Dickinson PA-255509.05A), BBA = Brucella blood agar (bioMérieux, 411 968), CHOC-H = Chocolate agar (Hardy Diagnostic, E14), COS-B = Columbia Blood Agar (bioMérieux, 43 041), COS-D = Columbia agar with 5% sheep blood (Becton Dickinson, 90006 166), COS-O = Columbia agar with 5% sheep blood (Oxoïd, PB5039A).
(DOCX)
(DOCX)
The culture conditions (media, time of incubation in hours, ± 5% CO2) are indicated. BAS = Brucella agar with 5% sheep blood, hemin & vitamin K1 (Becton Dickinson); BBA = Brucella blood agar (bioMérieux); CHOC-O = Chocolate agar plate with vitox (Oxoïd); COS-B = Columbia Blood Agar (bioMérieux, 43 041); COS-O = Columbia agar with 5% sheep blood (Oxoïd); MHB = Mueller Hinton agar with 5% sheep blood (Biorad); MHF = Mueller Hinton agar with 5% horse blood and β-NAD (Biorad); TSA-S = Trypticase soy agar with 5% sheep blood (Becton Dickinson).
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.