Abstract
Metformin is a first-line drug for the treatment of individuals with type 2 diabetes, yet its precise mechanism of action remains unclear. Metformin exerts its anti-hyperglycemic action primarily through lowering of hepatic glucose production (HGP). This suppression is thought to be mediated through inhibition of mitochondrial respiratory complex I, and thus elevation of 5′-adenosine monophosphate (AMP) levels and the activation of AMP-activated protein kinase (AMPK), though this proposition has been challenged given results in mice lacking hepatic AMPK. Here, we report that the AMP-inhibited enzyme fructose-1,6-bisphosphatase-1 (FBP1, EC 3.1.3.11), a rate-controlling enzyme in gluconeogenesis, functions as a major contributor to the therapeutic action of metformin. We identified a point mutation in FBP1 that renders it insensitive to AMP while sparing regulation by fructose-2,6-bisphosphate (F-2,6-P2) and knockin (KI) of this mutant into mice significantly reduces their response to metformin treatment. We observe this during a metformin tolerance test and in a metformin-euglycemic clamp that we have developed. The anti-hyperglycemic effect of metformin in high fat diet-fed diabetic FBP1 KI mice was also significantly blunted compared to wild-type controls. Collectively, we show a new mechanism of action of metformin, while providing further evidence that molecular targeting of FBP1 can have anti-hyperglycemic effects.
Diabetes is characterized by impaired glucose homeostasis partly due to abnormally elevated hepatic glucose production (HGP). The biguanide drug metformin (N,N-dimethylbiguanide) works principally through inhibition of HGP, although enhanced glucose disposal has also been reported in some studies1. It is widely accepted that metformin inhibits mitochondrial respiration through complex I2–4, reducing hepatocellular energy charge. A previous study examined if metformin-mediated AMP-activated protein kinase (AMPK) activation is responsible for its therapeutic effects and the reported data supporting a mechanism involving AMPK-dependent inhibition of HGP and lipogenesis5 albeit using an inhibitor of questionable selectivity6. Indeed a recent study has demonstrated that inhibitory phosphorylation of acetyl-CoA carboxylase (ACC) by AMPK plays an important role in metformin-induced improvements in insulin action by maintaining hepatic lipid homeostasis7. However, the significance of AMPK in metformin action on HGP has been challenged in experiments using mice lacking hepatic AMPK8. Recent studies report that metformin inhibits HGP through hepatic AMPK-independent mechanisms, either by attenuating the ability of glucagon to increase 3’,5’-cyclic adenosine monophosphate (cAMP) levels and promote HGP9 or direct inhibition of mitochondrial glycerol-3-phosphate dehydrogenase and subsequent increase in cytosolic free [NADH]:[NAD+] leading to impaired utilization of lactate for gluconeogenesis10. These findings suggest that the underlying mechanisms responsible for the HGP- and glucose-lowering effects of metformin in diabetes may not be explained by any single target or pathway. Interestingly, a widely-used pharmacological AMPK activator, 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR), an AMP mimetic, profoundly suppressed glucose output in hepatocytes lacking AMPK8,11, indicating AMP per se but not AMPK plays a vital role in suppressing HGP. Similarly, a tight correlation between the magnitude of increase in [AMP]:[ATP] and inhibition of glucose output in hepatocytes has been noted8.
Given that the anabolic process of gluconeogenesis is energetically costly, hepatocytes must balance this energy demand with production thereby maintaining energy homeostasis. Hepatocytes are equipped with a mechanism to control the rate of hepatic gluconeogenesis in response to energy status and fructose bisphosphatase 1 (FBP1) has long been recognized as a key component12. FBP1 catalyzes the irreversible hydrolysis of fructose-1,6-bisphosphate (F-1,6-P2) to fructose-6-phosphate (F6P) and inorganic phosphate (Pi) in the presence of divalent cations. FBP1 is a key rate-controlling enzyme in the gluconeogenic pathway and individuals with FBP1 deficiency present with hypoglycemia and metabolic acidosis due to impaired gluconeogenesis13. FBP1 activity is regulated synergistically by the allosteric inhibitors AMP and F-2,6-P2. AMP inhibits noncompetitively by binding to a unique allosteric site whereas F-2,6-P2 binds to the active site in competition with F-1,6-P2. While levels of F-2,6-P2 are largely under hormonal control, AMP concentration is a function of the energy status of the tissue and contributes to autoregulation of gluconeogenesis. We hypothesized that acute inhibition of gluconeogenesis by metformin is due to inhibition of FBP1, secondary to increases in the AMP concentration through reducing hepatocellular energy charge.
Results
Identification of an AMP insensitive FBP1 mutant
Demonstrating the importance of allosteric regulation of a rate-controlling enzyme in metabolic flux control in vivo is difficult partly due to the lack of an established experimental strategy. The only definitive approach is to generate a knockin (KI) animal model that specifically renders the target enzyme insensitive to the ligand of interest, while leaving all other modes of regulation intact14. Such a mutant cannot be designed from basic principles and can only be chosen on the basis of available or predicted structure combined with detailed in vitro enzyme kinetic analysis. Before designing such a point mutant of FBP1, we first established that metformin does not have a direct inhibitory effect on FBP1 up to 10 mM (Supplementary Fig. 1a). Neither does it appear to inhibit the reported target AMP deaminase 1 (AMPD1)15 (Supplementary Fig. 1a).
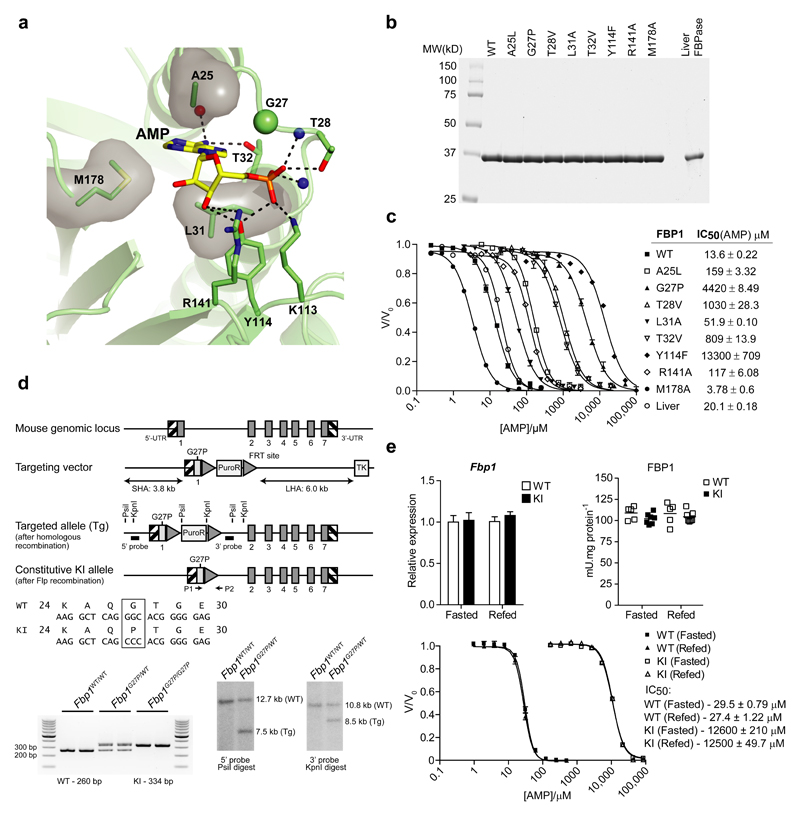
Next, to identify an AMP-resistant FBP1 mutant, we performed structure-guided mutagenesis based on the reported structure of the human FBP1-AMP complex16 (PDBID 1FTA) (Fig. 1a) and evolutionary conservation of key AMP-contacting residues (Supplementary Fig. 1b). Several point mutants designed to disrupt AMP binding to mouse FBP1 were prepared using an Fbp1-null E.coli strain. Native FBP1 from mouse liver was purified and used as a reference material to validate recombinant mouse 6HIS-FBP1. We obtained high-purity recombinant and native FBP1 as judged by Coomassie-stained SDS-PAGE gels (Fig. 1b). IC50 values for AMP were comparable between recombinant wild-type (WT) and native mouse liver FBP1 at ~14 and ~20 μM, respectively (Fig. 1c). Among the mutants tested, we found that G27P and Y114F showed markedly higher IC50 values for AMP (4420 and 13300 μM, respectively) and were essentially unaffected by up to 1 mM AMP, a concentration expected to greatly exceed in vivo limits (Fig. 1c). We subjected G27P and Y114F to further analysis using untagged preparations and ultimately rejected further exploration of Y114F due to an increased IC50 for F-2,6-P2 (data not shown), while we determined the detailed kinetic properties of G27P (Supplementary Table 1). Core properties including specific activity and the apparent affinity/Hill coefficient for ligands (F-1,6-P2, Mg2+ and F-2,6-P2) were essentially identical between WT and G27P mutant. In contrast, IC50 for G27P compared to WT protein was drastically higher for AMP and related compounds, including the active form of AICAR, 5’-AICAR monophosphate (ZMP), and two nucleotide-mimetic commercial FBPase inhibitors.
Figure 1. Generation of an AMP-insensitive FBP1 knockin mouse model.
(a) Human FBP1 structure (PDBID 1FTA) represented as ribbons and AMP and interacting residues (numbered from the initiator methionine) are shown as sticks. Dashed lines represent hydrogen bonding interactions, whereas residues making hydrophobic contracts are illustrated as sticks and transparent surfaces. Red and blue spheres represent backbone oxygen and nitrogen atoms respectively. (b) Coomassie-stained SDS-PAGE of mouse liver FBPase and recombinant mouse 6HIS-FBP1 preparations with single point mutations designed to disrupt AMP binding. (c) AMP inhibition curves of mouse 6HIS-FBP1 mutants. FBPase activity is expressed as a ratio of the maximum activity in the absence of AMP (V/Vo). IC50 values represent the mean ± SD of three independent measurements on two enzyme preparations. The line graph is representative of the results from a single preparation. (d) Schematic illustrating the targeting strategy used to generate C57BL/6NTac FBP1G27P knockin (KI) mice. Exons and FRT recombination sites are represented by dark grey boxes and triangles respectively. The KI allele containing the G27P mutation in exon 1 is shaded pale grey. Correct recombination was confirmed by Southern blotting of PsiI and KpnI digests of genomic DNA isolated from targeted embryonic stem cells with the corresponding 5’ and 3’ probes (black boxes). Genotyping of the constitutive KI allele was performed by PCR of genomic DNA using primers P1 and P2. (e) Liver biopsies from overnight fasted (16 h) or refed (4 h) FBP1WT/WT (WT) or FBP1G27P/G27P (KI) mice were assayed for Fbp1 mRNA expression by qPCR (left chart) or FBPase activity (right chart) by spectrophotometric assay. The line graph (below) represents AMP inhibition curves of FBPase activity in liver homogenates expressed as a ratio of the activity in the absence of AMP (V/Vo). Results represent mean ± SD, n = 5-7 per group.
Interestingly, inhibition by 5-inosine monophosphate (IMP) was unaffected. While not a physiologically relevant ligand for FBP1, this highlights that the substitution has not disrupted the function of the allosteric pocket, but merely reduced the affinity for AMP beyond the physiological range. Indeed, titration with the fluorescent AMP analogue, TNP-AMP demonstrated a significant reduction in binding affinity (Supplementary Table 1), which was further confirmed by the inability of G27P FBP1 to bind AMP immobilized on a solid support (Supplementary Fig. 1c). Finally, we assessed thermal stability as an indicator of the stability of the folded state of the mutant (Supplementary Fig. 1d). Melting temperature (Tm) was essentially unaffected by the G27P substitution (69.2 ± 0.2 °C vs. 68.6 ± 0.2 °C). Interestingly, saturating AMP concentration did not shift Tm for WT but substantially reduced resting fluorescence at ambient temperature revealing a clear biphasic transition from the more closed tense (T) state upon heating. As expected, this was not observed for G27P but the stabilizing effect of F-1,6-P2 was conserved.
Generation and characterization of an AMP insensitive FBP1 G27P KI mouse model
To establish if AMP-mediated inhibition of FBP1 activity contributes to the anti-hyperglycemic action of metformin in vivo, we generated an FBP1 KI mouse model in which the codon for glycine 27 of Fbp1 was modified to encode proline (Fig. 1d). FBP1 is predominantly expressed in liver and kidney and to a much lesser extent in testes and small intestine (Supplementary Fig. 2a). Although expression of FBP1 in islets has been reported17, it was undetectable in our hands using a highly specific antibody (Supplementary Fig. 2a). We confirmed that expression and activity of FBP1 were comparable between homozygous FBP1G27P/G27P KI and control WT mice in liver and kidney, although FBP1 expression in KI mice was modestly higher in small intestine and lower in testes compared to WT (Fig. 1e and Supplementary Fig. 2b,c). Assayed in crude liver extracts, FBP1 G27P exhibited > 400-fold higher IC50 for AMP compared to WT FBP1 (Fig. 1e), which is far beyond the physiological range of cellular AMP concentrations18.
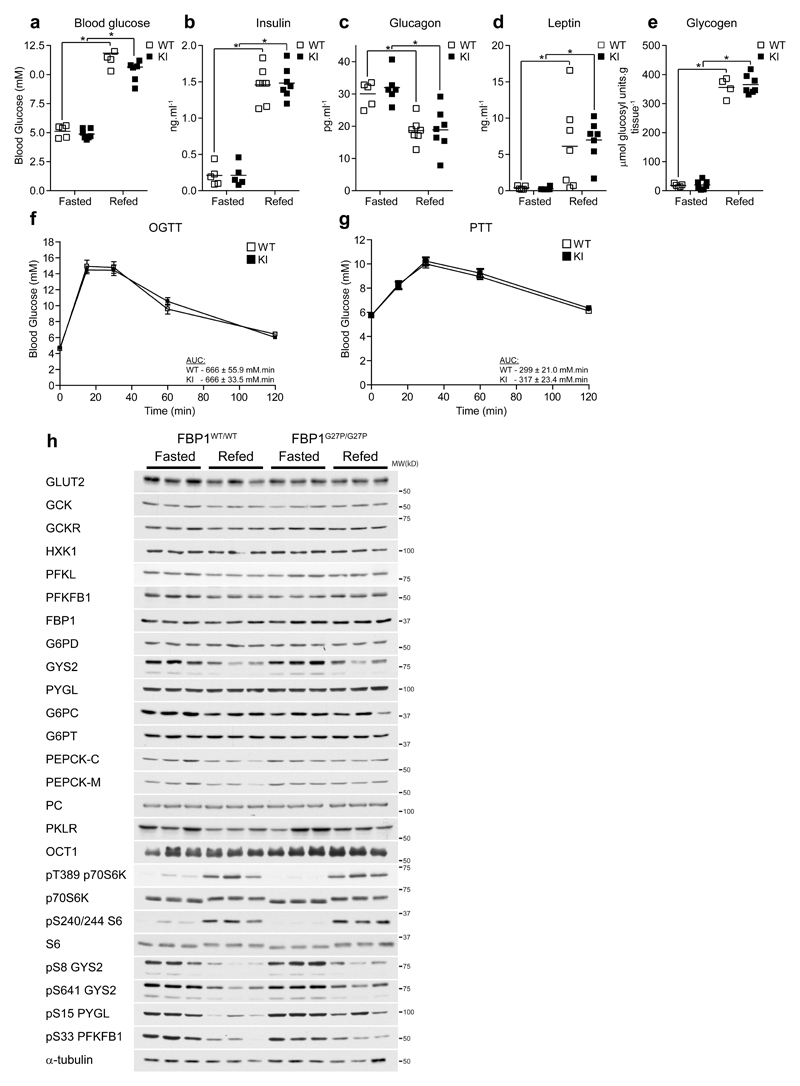
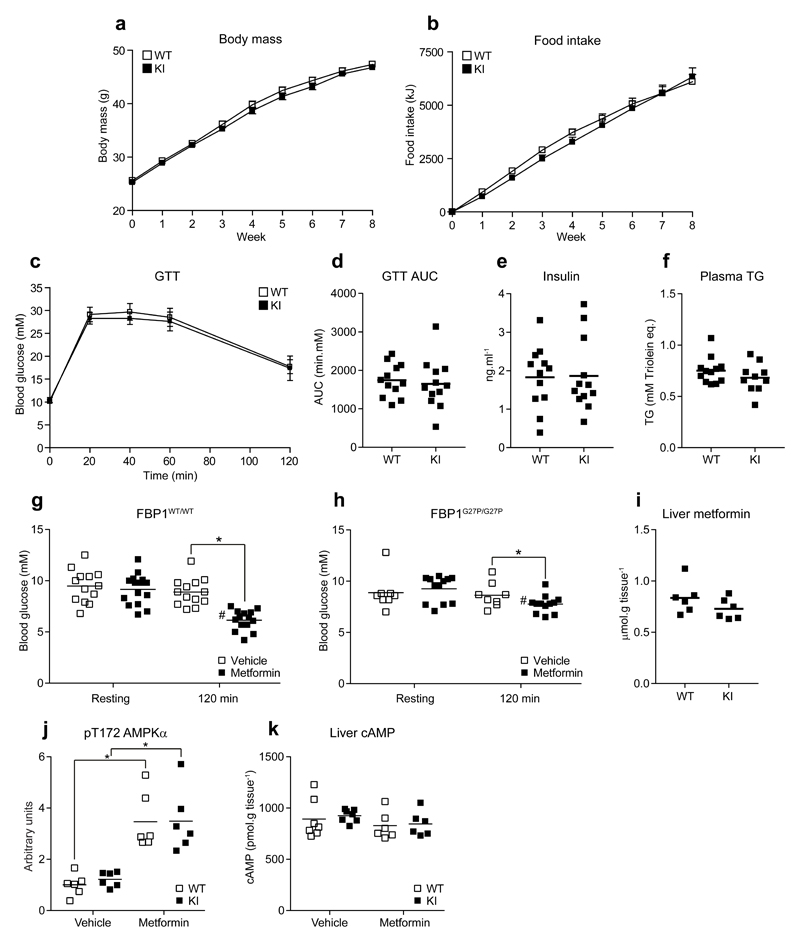
FBP1G27P/G27P mice were born at the expected Mendelian frequency and displayed similar body weight and growth curves (data not shown), food intake and respiratory exchange ratio, as well as locomotor activity compared to WT mice (Supplementary Fig. 3a-f). Compared to WT, FBP1 KI mice exhibited similar blood glucose, plasma insulin, glucagon and leptin levels, as well as hepatic glycogen content under fasted and refed (4 h ad libitum following overnight fast) conditions (Fig. 2a-e). KI mice also displayed normal blood glucose tolerance (Fig. 2f) and gluconeogenic capacity assessed by pyruvate tolerance test (Fig. 2g). Consistent with these observations, immunoblot analysis revealed comparable expression of major metabolic proteins involved in hepatic glucose metabolism (e.g. GLUT2, glucokinase (GCK) and its regulator protein GCKR, hexokinase 1 (HXK1), phosphofructokinase (PFKL), 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB1), FBP1 and glucose-6-phosphate dehydrogenase (G6PD)), glycogen metabolism (e.g. glycogen synthase (GYS2) and glycogen phosphorylase (PYGL)) and gluconeogenesis (e.g. catalytic and transporter subunit of glucose-6 phosphatase (G6PC/T), cytosolic/mitochondrial isoforms of phosphoenolpyruvate carboxykinase (PEPCK-C/M), pyruvate carboxylase (PC), pyruvate kinase (PKLR)) as well as the major metformin transporter organic cation transporter 1 (OCT1) between WT and KI under fasted and refed conditions (Fig. 2h and Supplementary Fig. 4). Moreover, the activity of the major gluconeogenic/glycolytic enzymes were similar between WT and KI under both fasted and refed conditions (Supplementary Fig. 5a-i). At transcript level, mRNA expression of genes involved in gluconeogenesis (Pck1, Pparg1c, Foxo1a) and lipogenesis (Fasn) were similar between WT and KI mice (Supplementary Fig. 5j-m). Conversely, G6pc mRNA level was significantly lower in KI mice under both fasted and refed conditions (Supplementary Fig. 5n), although this did not translate into a difference at protein level (Fig. 2h and Supplementary Fig. 4b) or activity (Supplementary Fig. 5b). Similarly, Gck mRNA was higher in refed KI mice (Supplementary Fig. 5o); however, again, protein levels and enzyme activity were unchanged (Supplementary Fig. 4d and Supplementary Fig. 5a). Phosphorylation states of hormone- and/or nutrient-regulated proteins showed the anticipated changes in response to refeeding such as increased phosphorylation of p70S6K and S6, downstream components of mTOR, and decreased phosphorylation of GYS2 and PYGL, as well as PFKFB1, the enzyme that synthesizes F-2,6-P2 and a major substrate of cAMP-dependent protein kinase (Fig. 2h and Supplementary Fig. 4). Hepatic lactate and pyruvate are the major three carbon precursors for gluconeogenesis and their levels were unchanged between WT and KI mice under both fasted and refed conditions (Supplementary Table 2). Likewise, hepatic glucose and G6P concentrations were similar between WT and KI. Furthermore, there was no difference in the levels of F6P and F-1,6-P2, the substrate and product of FBP1, respectively (Supplementary Table 2). The loss of regulation by AMP of FBP1 does not have an apparent impact on gluconeogenic flux under normal conditions. The mutant does remain sensitive to hormonal regulation by F-2,6-P2, whose levels were also unchanged between WT and KI. Hepatic adenine nucleotide levels and energy charge were similar in fasted and refed conditions and comparable between WT and KI mice (Supplementary Table 3). Taken together, these results demonstrated that FBP1 KI mice possess normal hepatic energy and metabolic homeostasis, as well as whole body glucose homeostasis.
Figure 2. FBP1 G27P knockin mice display normal glucose homeostasis.
(a-e) FBP1WT/WT (WT) or FBP1G27P/G27P (KI) mice were fasted overnight for 16 h (Fasted) or subsequently given free access to standard chow for 4 h (Refed). Blood and liver biopsies were taken and the following parameters determined: blood glucose (a), plasma insulin (b), plasma glucagon (c), plasma leptin (d) and liver glycogen (e). n = 5-7 (WT) and 5-7 (KI) per group. (f) Glucose (2 g.kg-1 p.o.) and (g) pyruvate (1 g.kg-1 i.p.) tolerance was assessed on mice fasted for 16 h. Results represent mean ± SE, n = 10 per group. (h) Expression of the major enzymes and regulatory components of the gluconeogenic, glycogenic and glycogenolytic pathways in liver samples from fasted or refed animals was determined by Western blotting. Representative results from three mice per group are shown. *P < 0.05 (Fasted vs. refed). Statistical significance was determined using unpaired, two-tailed Student’s t-test and an alpha level of 0.05.
FBP1 G27P KI mice are resistant to the hypoglycemic action of AMP mimetic compounds
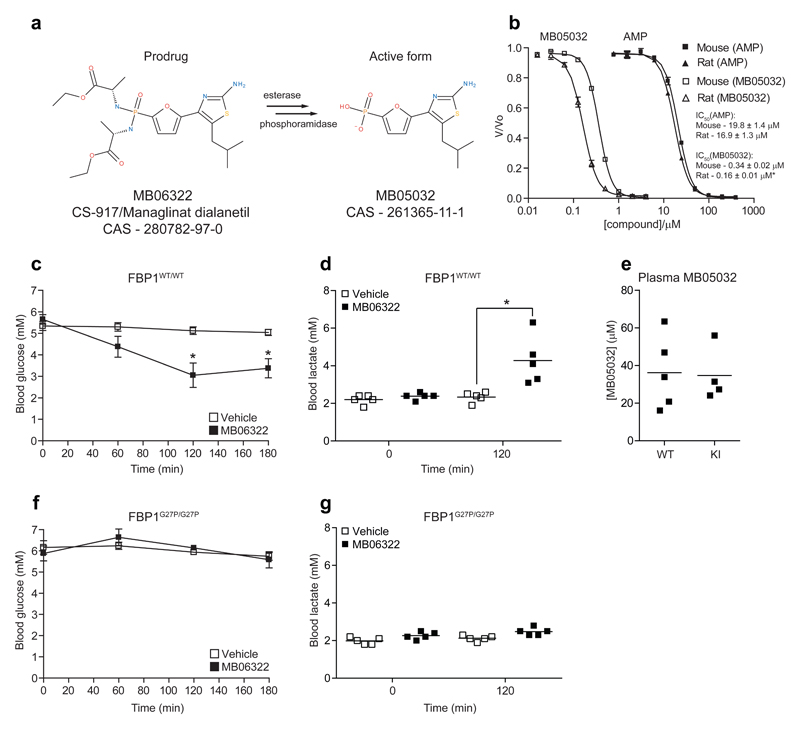
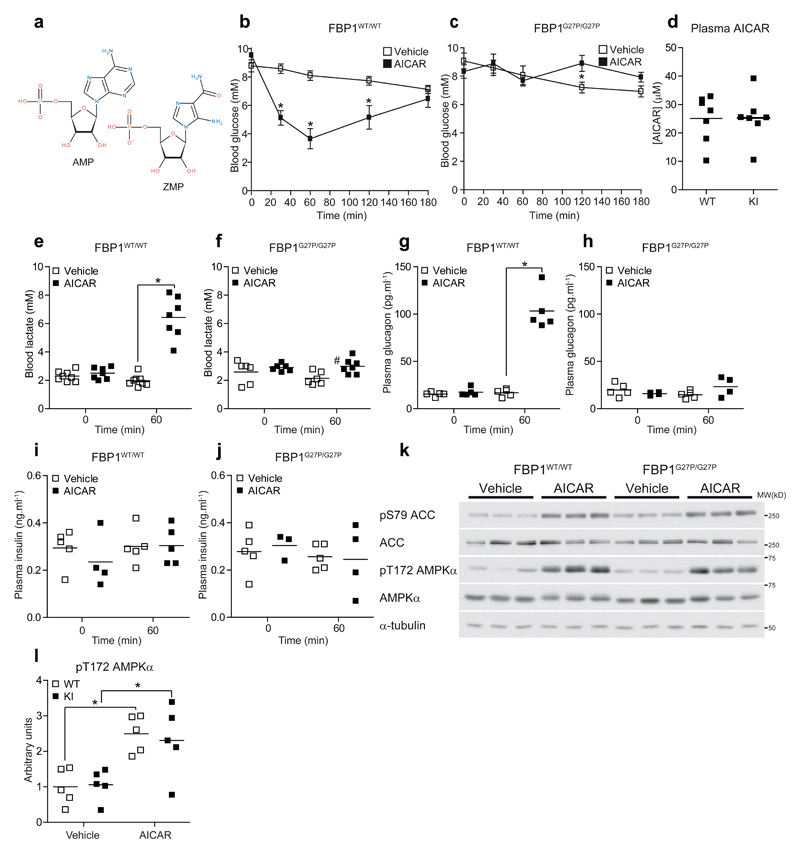
Prior to testing the effect of metformin, we wanted to confirm if FBP1G27P/G27P KI mice are resistant to AMP-mediated blood glucose-lowering in vivo. For this purpose, we initially used the AMP mimetic FBPase inhibitor MB06322 (ref. 19), the pro-drug of MB05032 (Fig. 3a and Supplementary Table 1). We observed that recombinant mouse FBP1 was ~2-fold less sensitive to MB05032 compared to rat FBP1 in vitro, while sensitivity to AMP was comparable between the two species (Fig. 3b). As all available pre-clinical data was only performed in rats, we took this species difference in drug response into account and 75 mg.kg-1 of MB06322 was administered (i.p.) to WT and KI mice. In WT animals MB06322 treatment resulted in a robust and sustained decrease (~40 % at 2-3 h post injection) in blood glucose levels (Fig. 3c), which was accompanied by an increase in blood lactate levels (Fig. 3d). In contrast, even though plasma concentration of the drug was comparable between genotypes (Fig. 3e), MB06322 had no significant effect on both blood glucose and lactate concentration in KI mice (Fig. 3f, g). We next sought to determine if the well-documented hypoglycemic effect of AICAR20, which is converted intracellularly to the AMP-mimetic ZMP (Fig. 4a), is mediated through ZMP-dependent inhibition of FBP1 in vivo. Administration of AICAR (250 mg.kg-1, i.p.) resulted in a profound (up to ~60 %) decrease in blood glucose levels in WT (Fig. 4b), but not in KI (Fig. 4c) mice, while plasma concentration of AICAR was similar between the two genotypes (Fig. 4d). Consistent with the results observed with MB06322, AICAR induced a marked increase in blood lactate levels in WT (Fig. 4e), but only modestly in KI (Fig. 4f) mice. Plasma glucagon levels were increased only in AICAR-treated WT (Fig. 4g), but not in KI (Fig. 4h) mice, most likely to counteract the rapid induction of hypoglycemia (Fig. 4b). There was no significant change in plasma insulin levels (Fig. 4i,j). As anticipated, AICAR robustly stimulated phosphorylation of liver AMPK and its bona fide substrate ACC in both WT and KI mice (Fig. 4k,l). This was accompanied by a profound increase in liver ZMP and ZTP concentrations in both genotypes (Supplementary Table 4). As previously reported in both intact animals11,20 and isolated hepatocytes12,21 AICAR administration in WT mice resulted in a substantial decrease in the total adenine nucleotide pool (Supplementary Table 4). The conversion of AICAR to ZMP by adenosine kinase consumes ATP and acts as a trap for phosphate, similar to the metabolic consequences of a large fructose bolus. This effect is further enhanced by significant substrate cycling between AICAR and ZMP due to dephosphorylation by 5’-nucleotidase22. This leads to depletion of Pi resulting in de-inhibition of AMP deaminase and the loss of adenine nucleotides. However, this mechanism cannot be fully responsible as AICAR had minimal effect on adenine nucleotides in KI mice despite similar accumulation of ZMP (Supplementary Table 4). Furthermore, AICAR was previously observed to have no significant effect on ATP levels in fed mice20. The common factor here is likely the absence of an acute glucose-lowering effect and inhibition of a high gluconeogenic flux which leads to the accumulation of additional phosphorylated species (e.g. F-1,6-P2 and the triose phosphates) as additional sinks for Pi (data not shown). Interestingly, AICAR treatment lead to a similar increase in hepatic NAD+ in both WT and KI mice (Supplementary Table 4), which is likely due to AMPK-dependent inhibition of fatty acid synthesis and subsequent increase in β-oxidation.
Figure 3. FBP1 G27P KI mice are resistant to the hypoglycemic action of an AMP-mimetic FBPase inhibitor.
(a) Diagram showing the structure of MB06322 and the active metabolite, MB05032. (b) Mouse and rat FBP1 preparations were assayed for inhibition by AMP (closed symbols) and MB05032 (open symbols). Results represent mean ± SD, n = 3. *P < 0.05 (Mouse vs. rat) (c-g) Vehicle (10:10:80 Solutol HS 15:PEG 400:water) or MB06322 (75 mg.kg-1 i.p.) was administered to fasted (16 h) WT (c) or KI (f) mice and blood glucose monitored at the indicated intervals for 3 h. (d, g) Lactate was measured just prior to drug administration and at t = 120 min using a lactate meter. (e) Plasma levels of MB05032 were assayed from blood samples drawn at the end of the protocol (t = 180 min). Results represent mean ± SE, n = 4-5 per treatment group. *P < 0.05 (Vehicle vs. MB06322). Statistical significance was determined using unpaired, two-tailed Student’s t-test and an alpha level of 0.05.
Figure 4. FBP1 G27P KI mice are resistant to the hypoglycemic action of AICAR.
(a) Diagram illustrating the structure of AMP and ZMP. (b-l) AICAR tolerance was determined by administering vehicle (0.9 % saline) or AICAR (250 mg.kg-1- i.p.) to fasted (16 h) WT (b, e, g, i) or KI (c, f, h, j) mice. Blood glucose was measured at the indicated timepoints for a period of 3 h. (d) Plasma AICAR, (e, f) blood lactate, (g, h) plasma glucagon and (i, j) plasma insulin were assayed from blood samples drawn at t = 60 min. Results represent mean ± SE, n = 4-8 per treatment group. *P < 0.05 (Vehicle vs. AICAR). #P < 0.05 (WT vs. KI). (k, l) Vehicle (0.9 % saline) or AICAR (250 mg.kg-1- i.p.) was administered to fasted (16 h) mice and after 60 min exposure, liver biopsies were taken and assayed for AMPK activation by Western blotting. The blot image depicts three representative mice from each treatment group and a quantitative analysis of pT172 AMPKα phosphorylation from the entire sample set is shown in (l). Results are expressed as pT172 AMPKα/AMPKα ratio normalized to the WT-vehicle group. n = 4-5 per treatment group. *P < 0.05 (Vehicle vs. AICAR). Statistical significance was determined using unpaired, two-tailed Student’s t-test and an alpha level of 0.05.
In skeletal muscle, AICAR failed to increase AMPKα T172 phosphorylation and activity in both genotypes (Supplementary Fig. 6a-c), although it enhanced phosphorylation of ACC and TBC1 domain family member 1 (TBC1D1) most likely via ZMP-dependent allosteric activation of AMPK (Supplementary Fig. 6d, e). Notably, AICAR was unable to stimulate phosphorylation of RAPTOR which is a marker of more robust AMPK activation23 (Supplementary Fig. 6f). Indeed ZMP concentration and [ZMP]:[ATP] ratio in skeletal muscle were > 50-fold and > 150-fold less, respectively, compared to that detected in the liver following AICAR treatment, a consequence of the significant first-pass metabolism of AICAR and low plasma concentration (Supplementary Table 4 and Supplementary Fig. 6g). Thus, it is presumed that the magnitude of AMPK activation following AICAR treatment in skeletal muscle was below the threshold to cause blood glucose-lowering in KI mice through promoting glucose uptake in this tissue. Collectively, using two AMP mimetic drugs we have confirmed that the FBP1 KI model is suitable to investigate the effect of an AMP-elevating agent (i.e. metformin) on blood glucose in vivo.
FBP1 G27P KI mice are metformin intolerant
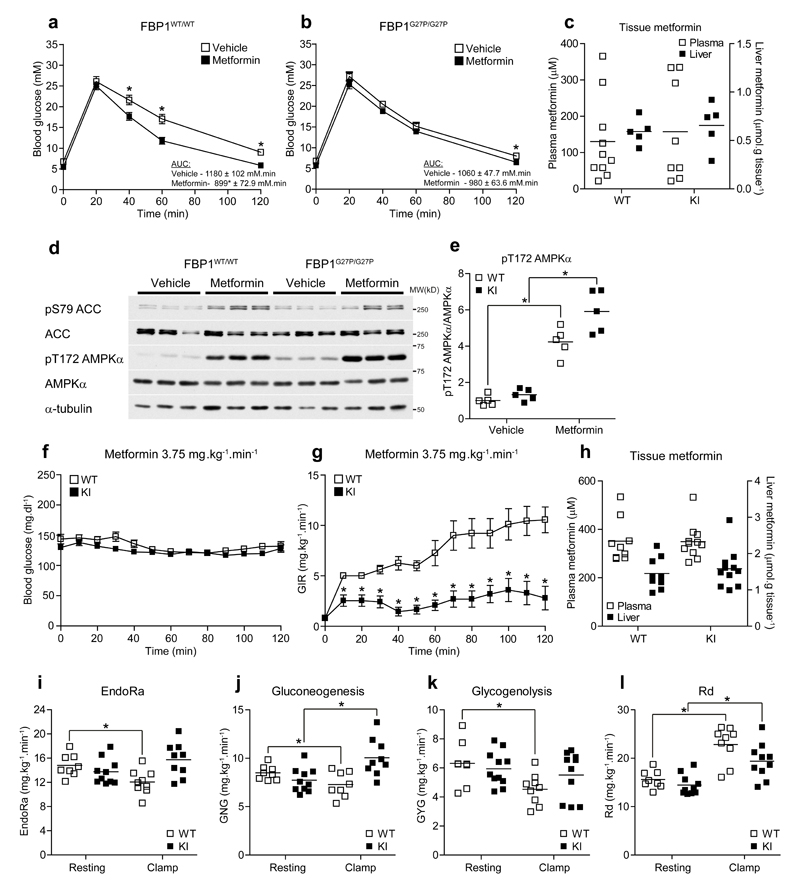
To determine if FBP1 KI mice exhibited altered responses to an acute dose of metformin, we performed a metformin tolerance test. Since metformin affects intestinal glucose absorption24, we intraperitoneally injected glucose (2 g.kg-1) following an oral administration of 250 mg.kg-1 metformin (Supplementary Fig. 7a), a commonly used dose in rodents to elicit an acute glucose-lowering effect7–9,25. Prior administration of metformin promoted significantly faster disappearance of blood glucose compared to vehicle-treated control in FBP1 WT mice (Fig. 5a). In contrast, metformin’s glucose-lowering effect was significantly lower in KI mice (Fig. 5b) even though plasma and liver metformin levels (~125-150 μM and ~0.7 μmol.g-1 respectively) were comparable between genotypes (Fig. 5c). Metformin caused a comparable increase in hepatic [AMP]:[ATP] (~2-fold) and decreased energy charge (~10 %) in both genotypes (Supplementary Table 5). This change in energy status was associated with a robust increase in phosphorylation of liver AMPK and ACC (Fig. 5d,e).
Figure 5. FBP1 G27P KI mice exhibit resistance to the acute glucose-lowering effect of metformin.
(a, b) Vehicle (water) or metformin (250 mg.kg-1 p.o.) was administered to fasted (16 h) mice and after 45 min, glucose tolerance (2 g.kg-1 i.p.) was assessed by monitoring blood glucose over a period of 2 h. Results represent mean ± SE, n = 18 (WT-vehicle), 17 (WT-metformin), 15 (KI-vehicle) and 15 (KI-metformin). *P < 0.05 (vehicle vs. metformin). (c-e) Mice were fasted for 16 h and dosed with vehicle (water) or metformin (250 mg.kg-1 p.o.). After 1 h exposure, blood and liver biopsies were taken and assayed for metformin (c). (d) Western blotting of ACC and AMPKα phosphorylation in livers from vehicle and metformin-treated mice. Representative results from three mice per group are shown. (e) Quantitative analysis of pT172 AMPKα. Results are expressed as pT172 AMPKα/AMPKα ratio normalized to the WT-vehicle group. n = 5. (f) Arterial blood glucose and glucose infusion rate (GIR) (g) during metformin-euglycemic clamps in FBP1WT/WT (WT) or FBP1G27P/G27P (KI) mice. Animals were fasted for 5 h and infused i.v. with metformin (3.75 mg.kg-1.min-1) and a variable infusion of 50 % glucose to maintain euglycemia at 120 mg.dl-1 over a period of 120 min. *P < 0.05 (WT vs. KI). (h) Plasma and liver metformin concentrations at the end of the clamp period. (i-l) Rates of endogenous glucose production (EndoRa) (i), gluconeogenesis (GNG) (j), glycogenolysis (GYG) (k) and glucose disappearance (Rd) (l) during the resting period (5 h fasted) and steady state of the metformin clamp (average from 100-120 min). Results represent mean ± SE, n = 8 (WT-resting), 8-9 (WT-clamp), 10 (KI-resting) and 9-11 (KI-clamp). *P < 0.05. Statistical significance was determined using unpaired, two-tailed Student’s t-test and an alpha level of 0.05.
To further investigate whether FBP1 KI mice were resistant to the glucose-lowering effect of metformin in vivo, we developed and optimized a “metformin-euglycemic clamp” protocol in the conscious, unrestrained mouse (Supplementary Fig. 7b). A similar technique has been used previously to assess the effect of metformin on HGP in mice26. Two different doses of metformin (1.875 mg.kg-1.min-1 and 3.75 mg.kg-1.min-1) were tested. Metformin was infused intravenously at a constant rate, while the glucose infusion rate (GIR) was adjusted to maintain euglycemia (Fig. 5f). We observed that the lower dose (1.875 mg.kg-1.min-1) failed to increase GIR significantly from baseline in WT (and also KI) mice under euglycemic condition (Supplementary Fig. 7c,d), even though metformin had reached ~170 μM and ~0.8 μmol.g-1 in plasma and liver, respectively (Supplementary Fig. 7e). These concentrations were comparable to those observed following an acute oral administration of 250 mg.kg-1 (Fig. 5c). This is likely due to differences in the route of administration. The first-pass of metformin from the gastrointestinal tract, via the portal vein and liver, into systemic circulation is crucial for the glucose-lowering effect. Firstly, administration of metformin via the portal-hepatic pathway produces more profound glucose-lowering than direct systemic infusion27. Secondly, it has been proposed that there is also a direct effect of metformin on the gut itself. For example, it has been recently suggested that metformin reduces HGP through a gut-brain-liver neuronal network via activation of AMPK in the duodenum resulting in release of GLP-128. Another study has shown that metformin reshapes the gut microbiota through interacting with different bacteria, possibly via metal homeostasis29.
Infusion of a higher dose of metformin (3.75 mg.kg-1.min-1) resulted in a substantial increase in GIR leading to steady state by the end of the 120 min clamp in WT mice (Fig. 5g). In contrast, KI mice displayed only a modest increase in GIR (Fig. 5g), even though plasma and liver/muscle metformin, as well as plasma insulin concentrations were similar between genotypes (Fig. 5h and Supplementary Fig. 7f,g). Combining the metformin-euglycemic clamp with administration of 2H stable isotope tracers enabled the quantification of endogenous glucose production (EndoRa) including the relative contribution of glycogenolysis and gluconeogenesis. EndoRa was significantly suppressed (~20 %) in WT during the clamp (Fig. 5i). This was due to reduction of both gluconeogenesis and glycogenolysis (Fig. 5j,k). In contrast, metformin-induced suppression of EndoRa, gluconeogenesis and glycogenolysis, was ablated in FBP1 KI mice (Fig. 5i-k). Overall, the clamp study revealed that KI mice were largely insensitive to the glucose-lowering effect of metformin due to ablation of metformin-induced suppression of HGP. However, it should be noted that due to the systemic route of delivery (as mentioned above), much higher and supra-pharmacological doses were needed to elicit a robust glucose-lowering effect resulting in artificially higher glucose disposal rate (~40-50 % increase in both WT and KI) than is seen at therapeutic doses (Fig. 5l). It has been shown that metformin can stimulate glucose uptake in isolated rat skeletal muscle at supra-pharmacological doses (but not at clinical doses due to the absence of OCT130) by activating AMPK5, which could sensitize insulin action and further promote glucose uptake in muscle31. In support of this premise, metformin concentration in skeletal muscle was increased 2-fold when infused at the higher rate (3.75 mg.kg-1.min-1) compared to the lower rate (1.875 mg.kg-1.min-1) (Supplementary Fig. 7g). The molecular basis underlying reduced glycogenolysis in WT mice during the clamp is unknown, as we observed no significant difference in the levels of phosphorylation/activity of glycogen synthase (GS) and glycogen phosphorylase (GPa), as well as hepatic glycogen content between genotypes at the end of the clamp (Supplementary Fig. 7h-m). We monitored tissue distribution and pharmacokinetics of metformin via systemic route and performed positron emission tomography (PET) analysis following an acute intravenous infusion of [11C]-metformin. We found that the kinetics and total metformin uptake in liver was comparable between WT and KI mice (Supplementary Fig. 8) consistent with the snapshot measurements of hepatic metformin concentration as shown in Fig. 5c and Supplementary Fig. 7e. In addition, PET analysis highlighted a rapid and marked accumulation of metformin in the bladder, corroborating the need for a much higher dose to achieve a glucose-lowering effect via systemic route than gastrointestinal route.
FBP1 G27P KI mice are resistant to the acute hypoglycemic action of metformin in an obesity-induced model of diabetes
We next assessed if FBP1 KI mice were resistant to the glucose-lowering effect of metformin under hyperglycemic/diabetic condition. WT and KI mice were fed with high-fat diet (HFD) for 10 weeks and both genotypes had similar profiles of weight gain and food intake over the period of dietary intervention (Fig. 6a,b). HFD-fed WT and KI mice showed hallmark features of type 2 diabetes, including glucose intolerance, hyperglycemia, hyperinsulinemia as well as hypertriglyceridemia (Fig. 6c-f). At the end of HFD intervention, we orally treated WT and KI mice with metformin (250 mg.kg-1) or vehicle (water) and monitored blood glucose levels two hours post treatment. We found that metformin, but not vehicle, produced a significant reduction of blood glucose levels in WT (~30-40 %), but the effect was significantly blunted in KI mice (P = 0.047, Fig. 6g,h). We verified that hepatic metformin levels (Fig. 6i) and the magnitude of changes in [AMP]:[ATP], energy charge (Supplementary Table 6), as well as associated increases in AMPK phosphorylation (Fig. 6j) were comparable between genotypes. Inhibition of gluconeogenesis by metformin in WT mice could also be supported by a modest fall in hepatic glucose and G6P levels, which was blunted in KI animals. Similarly, inhibition of the step catalyzed by FBP1 was suggested by a decrease in F6P (P = 0.053) and a concomitant increase in F-1,6-P2, in livers from metformin-treated mice (P = 0.091) that was lower in KI animals (Supplementary Table 7). However, given that there was a modest (~10 %), but significant effect for metformin to decrease blood glucose in KI mice (Fig. 6h), there must be additional mechanisms, independent of AMP-mediated FBP1 inhibition, to lower blood glucose. This is unsurprising given that metformin has multiple proposed primary and secondary targets1, including mitochondrial complex I2–4 and glycerol-3-phosphate dehydrogenase10 in the liver, as well as duodenal AMPK28. While it has been proposed that one of the mechanisms of action of metformin involves a reduction in hepatic cAMP9 (a key mediator of glucagon signaling), there was no significant effect (Fig. 6k) and an actual increase in downstream phosphorylation of PKA substrates (i.e., pS33 PFKFB1 and pS133 cAMP response element–binding protein (CREB)) (Supplementary Fig. 9a,b) under the conditions of our model system, (it has been reported that the reduction in cAMP is only apparent at substantially higher dose [i.e. 400 mg.kg-1]7), although we cannot rule out the possibility that we have missed the time point where cAMP-PKA signaling was suppressed following metformin treatment.
Figure 6. FBP1 G27P KI mice are resistant to the glucose lowering effects of metformin in an obesity-induced model of diabetes.
Body mass (a) and cumulative food intake (b) for FBP1WT/WT (WT) and FBP1G27P/G27P (KI) fed a 60 % Kcal high fat diet ad libitum over a period of eight weeks. (c-f) Glucose tolerance (1.5 g.kg-1 i.p.) (Glycemia in c and corresponding AUC in d), plasma insulin (e) and triglyceride (TG) (f) were assessed after eight weeks of dietary intervention. Results represent mean ± SE (a-c), n = 10-12 per group. (g, h) After 10 weeks of dietary intervention mice were fasted for 16 h, administered vehicle (water) or metformin (250 mg.kg-1 p.o.) and blood glucose measured after 2 h. n =13 (WT-vehicle), 15 (WT-metformin), 8 (KI-vehicle) and 12 (KI-metformin). *P < 0.05 (Vehicle vs. metformin). #P < 0.05 (Resting vs. 120 min) (i-k) After 12 weeks of dietary intervention mice were fasted for 16 h, administered vehicle (water) or metformin (250 mg.kg-1 p.o.) and liver biopsies were taken after 2 h of drug treatment. Liver metformin (i), pT172 AMPKα phosphorylation (expressed as pT172 AMPKα/AMPKα ratio normalized to the WT-vehicle group) (j), and cAMP (k) are shown. n = 6-7 (WT-vehicle), 6 (WT-metformin), 6-7 (KI-vehicle) and 6 (KI-metformin). *P < 0.05 (Vehicle vs. metformin). #P < 0.05 (WT vs KI). Statistical significance was determined using unpaired, two-tailed Student’s t-test and an alpha level of 0.05.
Discussion
Metformin has been in use for more than 50 years as an antihyperglycemic agent for the treatment of diabetes. Despite the clinical success of metformin, there is no clear consensus as to its mode of action and multiple, seemingly contradictory mechanisms, have been proposed. However, a common narrative emerges when a clear distinction is drawn between acute vs. chronic effects of metformin that can be mediated by either direct or indirect effects on HGP by metabolic or genic means. We have focused specifically on the acute effect of metformin on HGP, where it is clear that AMPK is dispensable. Hepatic AMPK-null mice do not exhibit a defect in either steady state glycemia, glucose/pyruvate tolerance or the acute glucose-lowering effect of metformin8. Furthermore, treatment of cultured hepatocytes8 or i.v. infusion of the specific AMPK activator A769662 has no effect on glucose production10. While hepatic LKB1-null mice present with severe hyperglycemia and hyperlipidemia due to upregulation of CREB/peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC1α) transcriptional targets (Pck1 and G6pc)32, recent work suggests that this is due to impaired activity of the AMPK-related kinase, salt-inducible kinase (SIK)33, and not AMPK. Much is often made of the short-term changes in Pck1 and G6pc mRNA in response to pharmacological activation of AMPK, but when protein levels are assessed they are invariably unaltered. Indeed, a poor correlation has been noted between the expression of gluconeogenic genes and HGP34. In short, although biguanides can clearly activate AMPK, it is neither sufficient nor necessary for acute inhibition of HGP.
Obesity-induced diabetes is a standard model in rodent studies and it is generally accepted that the associated hyperglycemia is a consequence of hepatic insulin resistance in fatty livers. It is well established that AMPK is a critical regulator of lipogenesis and chronic treatment of diabetic mice with metformin significantly improves glucose tolerance by reducing hepatic steatosis and improving insulin resistance. Of note, mice expressing non-phosphorylatable mutants of ACC1 and ACC2 (ACC1/2 knockin), were resistant to the lipid and glucose-lowering effect of chronic metformin treatment. Significantly, the acute hypoglycemic effect of a single dose of metformin was unaffected7. The same mechanism likely underpins the glucose-lowering effect of chronic A769662 treatment in obese ob/ob mice where dramatic reductions in hepatic and plasma lipid were observed35. Consequently, AMPK plays a role in the chronic, indirect inhibition of HGP by alleviating hepatic insulin resistance.
Plasma metformin concentration in humans is markedly variable, due in part to the complex pharmacokinetics of the drug and profound inter-subject variations in absorption and elimination36. It is difficult to quote a meaningful elimination half-life (t½) because the time course of plasma concentrations of metformin follows a multiphasic pattern, but values in the range from 1.7 to 4.5 hours have been reported36. Plasma metformin concentration in patients treated with a normally prescribed dose of metformin (1 g, twice a day) has been reported within 0.4-32 μM range (plasma levels obtained 14 hours after last drug administration) in 159 type 2 well-regulated diabetic subjects under controlled conditions37. In contrast, plasma metformin levels between 0 and 113 mg/L (868 μM) have been measured in random blood samples from diabetic subjects38. Both studies have provided useful information, as the former37 gives an estimate of nadir during multiple dosing, whereas the latter38 provides a valuable estimate of Cmax of metformin during clinical use. However, overall it has been to date a challenge to establish the therapeutic range of metformin concentrations in plasma.
The current debate on metformin has turned towards the issue of the validity of the concentrations and doses used in rodent models. It has long been recognized that the effective hypoglycemic dose exhibits a marked species dependence and researchers rightly use doses that produce robust, reproducible effects. In this regard, it is difficult to compare studies that have not measured plasma and/or liver accumulation as the drug formulation, route of administration and degree of fasting will impact the pharmacokinetics. Species differences in OCT1 expression have also been identified; while both rodent and human liver express OCT1, its expression in human intestines appears much lower than in mice39. As metformin is administered orally, differences in intestinal OCT1 expression may affect portal vein levels of metformin and thereby hepatic uptake without detectable effects on metformin levels in peripheral veins. In addition, hepatic exposure to the drug depends not only on OCT1 but also multidrug and toxin extrusion (MATE) isoforms acting as influx and efflux transporters, respectively. Differential hepatic expression of MATE1 between humans and rodents has been reported40, which may also affect metformin kinetics. It has been observed that a glucose-lowering effect occurs in response to a single dose of 50 mg.kg-1 in rats10. However, it should be noted that metformin (50 mg.kg-1) was given intravenously, which led to plasma metformin concentration of ~74 μM 30 min after administration, while 100 mg.kg-1 and 250 mg.kg-1 doses increased plasma metformin concentration of 345 μM and 1300 μM, respectively10. Further, the study exploring the effect of metformin in ACC1/2 KI mice used a chronic dose of 50 mg.kg-1 (i.p.) for 12 weeks, however, a dose of 200 mg.kg-1 was needed (50 mg.kg-1 had no effect) to observe a glucose-lowering effect in single-dose experiments7. Taken together, the dose range 200-350 mg.kg-1 has consistently been used7–9,25 and indeed the dose used in our study is similar to the maximum daily human dose (2 g) when allometric scaling is applied (dose for 25 g mouse = 2000×(0.025/65)0.75 = 5.5 mg = 220 mg.kg-1).
With cultured hepatocytes it is typical to see claims of enhanced justification on the basis of using lower doses but for greatly increased periods of time (often 16-24 h). Given that the uptake of metformin in isolated hepatocytes is relatively slow2 (in contrast to the rapid uptake in human and rodent livers in vivo41,42) and potentially antagonized by competing OCT substrates in complex media43, intracellular accumulation is both time and concentration dependent such that higher drug concentrations will reach effective intracellular levels within more relevant time periods. The apparent discrepancy in reported efficacy of metformin on isolated hepatocytes is likely due to the wide range of culture conditions used with respect to media (complex vs. balanced salt solutions), carbon sources and hormones. Indeed, under commonly used conditions for glucose production assays where the sole carbon sources are typically lactate and pyruvate, metformin produces precipitous decreases in intracellular ATP concentration (~85 % reduction at 1 mM) that closely follow apparent changes in glucose production or gene expression8. Such extremes in adenylate energy charge are not observed in any physiological setting in vivo. Consequently, the lack of standardization in hepatocyte protocols and the ease with which extreme energy deprivation can be induced with metformin renders the many reported results difficult to consolidate.
The final area of controversy concerns the role of changes in adenine nucleotides in metformin action. Several early studies reported activation of AMPK in the apparent absence of increases in AMP concentration44, which has been used to argue that metformin does not alter cellular energetics. However, experiments using AMP-resistant AMPKγ2 mutants have clarified that AMPK activation by metformin and essentially all xenobiotic compounds is mediated by increases in AMP concentration45. Technical errors in many contemporary studies have resulted in misleading values reported for hepatic adenine nucleotides ([ATP]:[AMP] <<10, often approaching 1). Hepatic AMP concentration is a very sensitive indicator of stress and increases by ~10-fold within just 30 seconds of hypoxia18. Consequently, tissues must be freeze-clamped in-situ to enable accurate measurements and reveal subtle changes in energy status. Even then, AMPK can easily be activated by increases in free AMP concentrations that are below the limit of quantification. Consequently, AMPK is a sensitive reporter of subtle changes in [AMP]:[ATP] and AMPKα T172 phosphorylation is a more reliable indicator of changes in the AMP concentration than technically demanding direct measurements. Hence, it can be argued that a consensus emerges that metformin induces changes in hepatic energy status which is sufficient to modify the activity of sensors such as AMPK and FBP1. Nonetheless, we observed here a modest, but significant reduction in energy charge in response to metformin treatment in the liver, which may have partly contributed to suppression of energy-demanding gluconeogenic flux8.
Understanding the mechanism by which metformin reduces HGP and normalizes blood glucose levels in hyperglycemic type 2 diabetics is of considerable importance. Our results show that metformin induces a mild energy stress in liver, leading to an increase in AMP concentration that allosterically inhibits FBP1 to lower HGP. This is potentially a powerful mechanism as the subsequent increase in F-1,6-P2 will activate PK and increase glycolytic flux46,47. Our study further supports the advancement of FBP1 as a key target for the treatment of type 2 diabetes, either directly using targeted inhibition48 or indirectly, as a consequence of inducing energy stress. The later mechanism may contribute significantly to the apparent glucose lowering effect of many biologically active secondary metabolites and to novel antidiabetic drugs exploiting the emerging concept of mild mitochondrial uncoupling49.
Online Methods
Materials
E.coli strain DF657 was sourced from the CGSC (Coli Genetic Stock Centre, Yale University) and BL21-CodonPlus(DE3)-RIL were from Agilent. Casamino acids were from BD Biosciences. Talon® cobalt IMAC resin was from Clontech. P11 and P81 phosphocellulose and BA-85 nitrocellulose were from Whatman. Leupeptin, pepstatin A and isopropyl-β-D-thiogalactopyranoside (IPTG) were from Serva. GST-HRV3C protease was from the Division of Signal Transduction Therapy (DSTT, Dundee). Fructose-2,6-bisphosphate (F-2,6-P2) (30 % purity) was from Toronto Research Chemicals. Higher purity F-2,6-P2 was kindly provided by Mark Rider (Université catholique de Louvain, Belgium). FBPase-1 inhibitor was from Santa Cruz. MB06322 was synthesised by SpiroChem (Zurich, Switzerland) as previously described1. MB05032 was from MedChem Express. 1-methoxy-5-methylphenazinium methyl sulfate was from Applichem. Sypro Orange and sterile 20 % (w/v) glucose were from Life Technologies. 2’,3’-O-trinitrophenyl-adenosine-5’-monophosphate (TNP-AMP), 2’/3’-O-(2-aminoethyl-carbamoyl)-adenosine-5’-monophosphate (2’/3’-EDA-AMP)-agarose and Z (AICAR) nucleotide standards were from Jena Bioscience. KAPA2G Fast HotStart genotyping mix was from Kapa Biosystems. Metformin-HCl, 5,5-diphenylhydantoin, Solutol (Kolliphor) HS 15, PEG (Kollisolv) 400 and all HPLC-grade solvents and additives were from Sigma. Microcystin-LR was from Enzo Life Sciences. NADH and AICAR (5-amino-1-(β-D-ribofuranosyl)-1H-imidazole-4-carboxamide) were from Apollo Scientific Ltd. Acetyl-CoA, trilithium salt was from Roche Life Science. Immobilon-P PVDF membrane was from Merck Millipore. Sephadex G-25, Blue Sepharose 6 FF, Superdex 10/300 GL and enhanced chemiluminescent reagent were from GE Healthcare. [γ-32P]-ATP, [U-14C]-uridine diphosphate glucose and [U-14C]-glucose-1-phosphate were from Perkin Elmer. 2H2O and [6,6-2H2]-glucose were from Cambridge Isotope Laboratories (Tewksbury, MA). AMARA substrate peptide (NH2-AMARAASAAALARRR-COOH) was synthesized by GL Biochem (Shanghai). Chicken muscle was sourced from a local supermarket and rabbit muscle was purchased from Harlan UK Ltd. Unless otherwise stated, all other reagents were from Sigma.
Antibodies
PEPCK-M (#6924), pS641 GYS (#3891), GYS (#3886), pT389 p70S6K (#9234), p70S6K (#2708), pS240/244 S6 (#2215), S6 (#2217), pS79/S212 ACC1/2 (#3661), ACC1/2 (#3676), pT172 AMPKα1/2 (#2535), AMPKα1/2 (#2532), AMPKα1/2 mAb (#2793), pT792 RAPTOR (#2083), RAPTOR (#2280), Fas (#3180), HXK1 (#2024), G6PD (#12263), TBC1D1 (#4629), pS133 CREB (#9198) and streptavidin-HRP (#3999) were from Cell Signaling Technology. G6PT (sc-135479) and FBP1 (sc-32435) were from Santa Cruz Biotechnology. PFKL (ab181064), PKLR (ab171744) and PEPCK-C (ab28455) were from Abcam. GAPDH (G8795) and α-tubulin (T6074) were from Sigma. SLC22A1/OCT1 (#ACT-011) antibody was from Alomone Labs. pS237 TBC1D1 was from Merck Millipore (07-2268). PYGL (15851-1-AP) was from Proteintech. pS15 PYGL (S961A) was from DSTT. AMPKα1 and AMPKα2 antibodies used for immunoprecipitation were raised in sheep against C-355TSPPDSFLDDHHLTR369 and C-352MDDSAMHIPPGLKPH366 (human sequences) respectively. GLUT2 antibody was provided by Bernard Thorens (University of Lausanne, Switzerland). GCK/HXK4 antibody was provided by Mark Magnuson (Vanderbilt University, TN). GCKR antibody was from Masakazu Shiota (Vanderbilt University, TN). G6PC antibody was provided by Giles Mithieux (University of Lyon, France). pS33 PFKFB1 antibody was provided by Jianxin Xie (Cell Signaling Technology). PFKFB1 antibody was provided by Simone Baltrusch (University of Rostock, Germany). pS8 GYS2 antibody was provided by Joan Guinovart (University of Barcelona, Spain). HRP-coupled and Alexa Fluor 680/Alexa Fluor 790-labelled secondary antibodies were from Jackson Immunoresearch.
Primers
Oligonucleotides were synthesized by Life Technologies.
Genotyping primers for C57BL/6NG27P mice: for-TGACAGTTAAGATTCTGCTCTGC, rev-TTAGGGATGATACTGAATTAGAAGC
| Target | Forward | Reverse |
|---|---|---|
| Fasn | AGCGGCCATTTCCATTGCCC | CCATGCCCAGAGGGTGGTTG |
| Fbp1 | GTGTCAACTGCTTCATGCTG | GAGATACTCATTGATGGCAGGG |
| Foxo1a | CTACGAGTGGATGGTGAAGAGC | CCAGTTCCTTCATTCTGCACTCG |
| G6pc | ACTGTGGGCATCAATCTCCTC | CGGGACAGACAGACGTTCAGC |
| Gck | GCATCTCTGACTTCCTGGACAAG | CTTGGTCCAGTTGAGCAGGATG |
| Pck1 | CCATCACCTCCTGGAAGAACA | ACCCTCAATGGGTACTCCTTCTG |
| Pparg1c | ATACCGCAAAGAGCACGAGAAG | CTCAAGAGCAGCGAAAGCGTCACAG |
| 18S | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
Cloning and mutagenesis
Mouse Fbp1 (NCBI reference AJ132693.1) was amplified from IMAGE EST 5054854 using KOD Hot Start DNA Polymerase (Merck Millipore) and cloned into the BamHI NotI sites to produce pET28a 6HIS-FBP1 and pET15 6HIS-HRV3C-FBP1. Mutations were created following the QuikChange method (Agilent) but using KOD Hot Start DNA Polymerase. The phosphatase domain of mouse 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (NCBI reference NM_008824.3) covering amino acids 251-440 was amplified from mouse liver RNA (Agilent #736009-41) using GoTaq 1-step RT-qPCR kit (Promega). The resulting PCR product was ligated into pGEX-6P-1 vector (GE Healthcare) as a Bamh1-Not1 fragment. Spinach chloroplast fructose-1,6-bisphosphatase 58-415 was cloned from a synthetic fragment (GeneArt Strings, based on Uniprot P22418) and ligated into a modified pET-15b plasmid as a Bamh1-Not1 fragment. The sequence of all constructs was verified by in-house sequencing using the BigDye® Terminator 3.1 kit on a 3500xL Genetic analyzer (ABI-Invitrogen).
Preparation of rFBP1
6HIS-FBP1 was expressed in the Fbp-null E.coli strain, DF657(DE3) as described by Giroux2. DF657 was sourced from the CGSC (Yale University, CT) and the DE3 lysogen prepared using the λDE3 lysogenization kit (Novagen #69734). Cells were transformed with pET28a FBP1 and cultured in minimal media (M9 salts, 2 mM MgSO4, 0.4 % (w/v) glycerol, 5 μg/ml thiamine-HCl, 0.5 % (w/v) casamino acids and 50 μg/ml kanamycin) overnight at 37°C. Minimal media (0.5 – 1 L containing 25 μg/ml kanamycin) was inoculated 1:40 with the starter culture and induced with 0.4 mM IPTG at OD600 ~ 0.4 for 16 h at 37°C. Cells were lysed in 5 ml/g 50 mM phosphate pH 7, 150 mM NaCl, 0.5 mM TCEP, 0.2 mM PMSF and 5 μg/ml leupeptin by sonication (1 min, 40 % amplitude) and clarified at 20,000 g for 20 min at 4°C. 6HIS-FBP1 was batch bound to Co+-charged IMAC resin (Talon®) for 30 min at 4°C, washed with 10 vol. lysis buffer, 10 vol. 5 mM imidazole and eluted with 5 vol. 150 mM imidazole. Preparations were exchanged into 50 mM imidazole pH 7.4, 0.3 M KCl, 0.2 mM EDTA and 0.5 mM TCEP over Sephadex G-25, concentrated using 10 kD MWCO centrifugal devices (Sartorius Vivaspin) and stored at –20°C in 50 % (v/v) glycerol. The presence of imidazole in concentrated solutions of 6HIS-FBP1 was essential to prevent aggregation. Untagged FBP1 was prepared by on-column cleavage of 6HIS-LEVLFQ*GPGS-FBP1 (constructed in pET15 and prepared as described above) with 50 μg HRV-3C protease per mg 6HIS-FBP1 in 50 mM TES pH 7.4, 150 mM KCl, 0.5 mM TCEP. Preparations were polished over a Superdex 200 10/300GL column equilibrated with 50 mM TES pH 7.4, 0.3 M KCl, 1 mM DTT and stored at –20°C in 50 % (v/v) glycerol. Preparations were stable for at least 6 months. As a reference, FBPase was purified from mouse liver essentially as described by Tashima3. Briefly, 20 g liver from C57BL/6N mice was homogenized in four volumes of 20 mM phosphate pH 7, 150 mM KCl, 1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 2 μg/ml leupeptin and clarified at 10,000 g for 20 min at 4°C. The supernatant was subjected to heat denaturation at 60°C for 1 min and centrifuged at 20,000 g for 20 min at 4°C. The supernatant was subjected to a 55-75 % ammonium sulfate cut and the resulting pellet dissolved in 1 mM EDTA and dialyzed overnight against 10 mM sodium malonate pH 6.2, 1 mM EDTA (Buffer A). FBPase was batch bound to 30 g P11 phosphocellulose, washed with 300 ml Buffer A under suction and transferred to an XK26/20 column. The resin was washed with buffer A containing 50 mM NaCl until A280 < 0.01 and FBPase was eluted with 2 mM fructose-1,6-bisphosphate (F-1,6-P2) and 20 μM 5’-adenosine monophosphate (AMP). Positive fractions were dialyzed against Buffer A, the pH adjusted to 5.8 with malonic acid and applied to a 1.6 × 10 cm column of Blue Sepharose FF (C16/20). The column was washed with 10 mM sodium malonate pH 5.8, 1 mM EDTA and eluted with 1 mM F-1,6-P2 and 1 mM AMP. The preparation was polished over a Superdex 200 10/300GL column equilibrated with 20 mM phosphate pH 7, 150 mM KCl, 1 mM EDTA and stored in 50 % (v/v) glycerol at –20°C.
FBPase assay
Fructose-1,6-bisphosphatase (EC 3.1.3.11) activity was determined by monitoring the formation of fructose-6-phosphate (F6P) using a coupled spectrophotometric assay. The specific activity of rFBP1 was determined in 1 ml reactions containing 50 mM TES pH 7.4, 0.2 mM NADP+, 0.1 M KCl, 0.05 mM EDTA, 2 mM (NH4)2SO4, 2 mM MgCl2, 0.05 % (w/v) BSA, 2 mM 2-mercaptoethanol, 0.8 U/ml phosphoglucose isomerase and 0.5 U/ml glucose-6-phosphate dehydrogenase. Reactions were started by the addition of 35 μM F-1,6-P2 and A340 recorded in a Cary 100 spectrophotometer at 30°C. Reaction rates were calculated from the linear phase assuming ε(NADPH) = 6.22 mM-1.cm-1. 1 U is defined as 1 μmol F6P formed per min at 30°C. Where appropriate, 1 U/ml AMP deaminase (purified from chicken muscle using P11 phosphocellulose essentially as described by Smiley4) was included to remove contaminating AMP from NADP+ as described by Han5. The activity ratio at pH 7.2/9.4 was determined under similar conditions in reactions buffered with 50 mM bis-tris propane at the appropriate pH. Other kinetic properties were determined in a 96 well format in non-binding black microplates (Greiner #655900), where the quantity of NADP+ was reduced to 0.15 mM and reactions were monitored by the increase in fluorescence (λex = 345 nm, λem = 465 nm) calibrated by the addition of 5 nmol F6P. Km(F-1,6-P2) was determined at 2 mM Mg2+ and fitted to equation 1:
| (Eq.1) |
Where: V = initial velocity, Vm = maximum velocity, [S] = F-1,6-P2 conc., Km = Michaelis constant for S, Ks = apparent substrate inhibition constant and b = factor determining maximum activity at high [S].
Km(Mg2+) was determined at 35 μM F-1,6-P2 and fitted to equation 2:
| (Eq.2) |
Where: V = initial velocity, Vm = maximum velocity, [S] = Mg2+ conc., Km = Michaelis constant for S and h = hill coefficient.
IC50 for inhibitory compounds was determined at 2 mM Mg2+, 35 μM F-1,6-P2 and fitted to equation 3:
| (Eq.3) |
Where: V = initial velocity, V0 = maximum velocity in the absence of inhibitor, I = conc. inhibitor, I0.5 = conc. of inhibitor that gives 50 % inhibition and h = hill coefficient.
Stock solutions of F-1,6-P2 were standardized by enzymatic assay in reactions containing 50 mM imidazole pH 7, 0.15 mM NADH, 0.02 U/ml aldolase, 1.2 U/ml triosephosphate isomerase and 0.16 U/ml glycerol-3-phosphate dehydrogenase. Stock solutions of AMP, 5’-inosine monophosphate (IMP) and 5’-AICAR monophosphate (ZMP) were prepared in 20 mM TES pH 7.4, neutralized with NaOH and standardized by UV absorbance in 0.1 M phosphate pH 7 at A259 (εAMP = 15.4 mM-1.cm-1), A249 (εIMP = 12 mM-1.cm-1) and A265 (εZMP = 12.5 mM-1.cm-1) respectively. F-2,6-P2 is difficult to obtain commercially and the crude product available from Toronto Research Chemicals was solubilized in 10 mM NaOH and standardized in reactions containing 50 mM HEPES pH 7.1, 5 mM MgCl2, 0.1 mM EDTA, 0.15 mM NADP+ by sequential addition of 0.1 U/ml glucose-6-phosphate dehydrogenase, 0.2 U/ml phosphoglucose isomerase and 0.25 U/ml GST-FBPase-2 P251-N440 while recording the increase in A340.
Thermal Stability Assays
TSA was performed on untagged FBP1 preparations using a Roche LightCycler 480 II. FBP1 was diluted to 0.2 mg/ml in 20 mM HEPES pH 7.4, 100 mM KCl, 1 mM MgCl2 and the indicated ligands and 5× Sypro Orange added sequentially. Solutions (20 μl) were dispensed in quadruplicate into 384-well white plates, sealed with optical tape and centrifuged at 200 g for 1 min. Fluorescence (λex – 465 nm, λem – 580 nm, Melt factor = 1, Quant factor = 10) was monitored while the block temperature was ramped from 20°C to 95°C at ~1°C/min (24 acquisitions/°C in continuous mode). The melting temperature (Tm) was determined from the maximum of the first derivative of the raw data using Roche Protein Melting software.
TNP-AMP fluorescence spectroscopy
Titration of 2’,3’-O-trinitrophenyl-adenosine-5’-monophosphate (TNP-AMP) was determined as described by Nelson6 using 1 μM FBP1 in 50 mM tris-acetate pH 7.4, 5 mM F6P, 5 mM Pi and 2 mM MgCl2 at 25°C. Raw fluorescence (λex = 410 nm, λem = 535 nm) was corrected for dilution and inner filter effects using equation 4:
| (Eq.4) |
Where Fcorr is the corrected fluorescence, V is the volume at a specific titration point and V0 is the initial volume.
Data were analyzed by non-linear regression to equation 5:
| (Eq.5) |
Where ΔF is the change in fluorescence caused by the addition of ligand (L), Fo is the initial fluorescence in the absence of ligand, Kd is the dissociation constant of L and h is the hill coefficient.
Immobilized ligand affinity binding
FBP1 (2 μg) in 20 mM TES pH 7.4, 100 mM KCl, 1 mM MgCl2, 0.1 mM F-1,6-P2 and 0.01 % (w/v) BRIJ-35 was mixed with 5 μl 2’/3’-O-(2-aminoethyl-carbamoyl)-adenosine-5’-monophosphate (2’/3’-EDA-AMP)-agarose or unconjugated agarose for 30 min at 4°C. Excess free ligand (0.5 mM AMP) was included as a negative control. Resin was pelleted at full-speed for 5 s, washed 3× 0.5 ml binding buffer and eluted with 20 μl Laemmli sample buffer. Samples were denatured at 95°C for 2 min, fractionated by SDS-PAGE and stained with colloidal Coomassie G-250.
Animals
Animal studies were approved by the local ethics committee and all protocols were approved by the Service Vétérinaire Cantonal (Lausanne, Switzerland) under license VD2841. C57BL/6NTac FBP1G27P mice were generated by Taconic Biosciences GmbH as described in Fig. 1d. Animals were kept in a standard temperature and humidity controlled environment on a 12/12 h light/dark cycle and had free access to water and standard chow or 60 kcal% fat diet (Research Diets Inc. D12492) as described. [11C]-metformin positron emission tomography (PET) was performed in accordance with the Danish Animal Experimentation Act and the European convention for the protection of vertebrate animals used for experimental and other purposes and was approved by the Animal Experiments Inspectorate, Denmark. Metformin-euglycemic clamps were completed with the approval of the Vanderbilt Animal Care and Use Committee. Animals were housed on a 12/12 h light/dark cycle in a temperature (23°C) and humidity-stable environment. Mice were maintained on a standard chow diet (5L0D LabDiet, St. Louis, MO). Male mice between the ages of 14-22 weeks were used for all procedures. Basic phenotyping was performed by PhenoPro (Illkirch, France) in a licensed animal facility (agreement #A67-218-40). All experiments were approved by the local ethical committee (Com’Eth, accreditations #2014-011), and were supervised by B.P.D. or M.F.C. who are qualified in compliance with the European Community guidelines for laboratory animal care and use (2010/63/UE). For glucose tolerance test, pyruvate tolerance test, metformin tolerance test, AICAR injection and MB06322 injection, the appropriate sample size was estimated to be 8-10 based on a power calculation assuming α = 0.05, power = 0.8 and variance from previous studies. However, based on Pilot experiments, responses to some of the compounds were much larger than anticipated and less animals were required as indicated in figure legends. Animals were arbitrarily but not randomly assigned to experimental groups and investigators were unblinded.
Tissue homogenization
Liver biopsies were powdered in a liquid nitrogen cooled mortar and pestle and homogenized in 10 volumes of extraction buffer using a rotor-stator homogenizer (Polytron, Kinematica AG). For Western blotting and assay of G6PC, PK and FBP1, tissues were homogenized in 50 mM tris-HCl pH 7.5, 0.27 M sucrose, 1 mM EDTA, 1 % (w/v) Triton X-100, 20 mM glycerol-2-phosphate, 50 mM NaF, 5 mM Na4P2O7, 0.5 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, 1 μM microcystin-LR, 1 mM DTT and clarified at 3500 g for 5 min at 4°C. Glycerol-2-phosphate was omitted for extracts used for assay of GS and GP. DTT was omitted for extracts prepared for the assay of CS and PC. For GCK, PFK and PEPCK-C assays, tissues were gently homogenized in 50 mM HEPES-KOH pH 7.4, 100 mM KF, 15 mM EGTA, 5 % (w/v) glycerol, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml aprotinin and 5 mM DTT and centrifuged at 100,000 g for 30 min to prepare a cytosolic fraction free of mitochondrial and microsomal fragments. Skeletal muscle biopsies were powdered in a liquid nitrogen cooled Bessman pulverizer and homogenized in 10 volumes of Triton X-100 extraction buffer supplemented with 50 mM KCl to prevent gelling.
Western blotting
Tissue extracts were denatured in Laemmli buffer at 95°C for 2 min, fractionated by tris-glycine SDS-PAGE and transferred to PVDF membrane at 100 V for 1 h in Towbin buffer (25 mM tris, 192 mM glycine, 10 % (v/v) methanol). Membranes were blocked with 5 % (w/v) skimmed milk in TBS-T (20 mM tris-HCl pH 7.5, 137 mM NaCl, 0.1 % (v/v) Tween-20) for 1 h at room temperature and incubated in primary antibodies prepared in TBS-T containing 5 % (w/v) BSA overnight at 4°C. Membranes were developed using HRP-conjugated secondary antibodies and ECL reagent. For integral membrane proteins (GLUT2, G6PC and G6PT) samples were not boiled, but heated at 37°C for 30 min. Pyruvate carboxylase was detected using HRP-conjugated streptavidin. OCT1 (SLC22A1) was detected by immunoprecipitation from 200 μg detergent extracts with 2 μg anti-OCT1 (Alomone ACT-011) and 5 μl protein G-Sepharose for 2 h at 4°C. Immune complexes were washed 3× 1 ml lysis buffer and eluted with 20 μl Urea-SDS sample buffer (62.5 mM tris-HCl pH 6.8, 2 % (w/v) SDS, 0.5 mM EDTA, 6 M urea, 0.01 % (w/v) bromophenol blue and 10 % (w/v) glycerol) for 1 h at RT. Specificity of detection was confirmed using liver extracts from OCT1 KO mice (provided by Niels Jessen, Aarhus University Hospital, Denmark). Quantitative blotting was performed using detection with either infrared fluorescent secondary antibodies (AF680 and AF790) on nitrocellulose membranes using an Odyssey CLx infrared imaging system (Li-COR) or developed films following ECL detection were scanned and quantitated by densitometry using ImageJ.
Metabolic phenotyping
Mice were weighed and food consumption monitored on the same day. Mice were starved for 16 h, starting at 17:00 on the previous day so that all procedures commenced at 09:00 the following morning. Blood glucose was monitored using a glucometer (AlphaTRAK 2, Abbott Logistics B.V.) on venous blood drawn from the tail. Blood lactate was assayed using a meter (Lactate Pro 2, Arkray Inc.). The genotype of experimental animals was confirmed by PCR on 1 μl blood lysed in 20 μl 20 mM NaOH, 60 % (w/v) PEG-2007 using KAPA2G polymerase. Glucose tolerance was determined by administration of 2 g.kg-1 D-glucose p.o. or i.p. from a 20 % (w/v) glucose solution after 16 h fast. Pyruvate tolerance was determined by administration of 1 g.kg-1 pyruvate (free acid) i.p. from a 12.7 % (w/v) sodium pyruvate solution (pH 6) after a 16 h fast. The pyruvate stock solution was standardized by assay in 50 mM phosphate pH 7, 0.15 mM NADH and 0.1 U/ml lactate dehydrogenase. AICAR tolerance was determined by administration of 250 mg.kg-1 i.p. from a 12.5 mg.ml-1 solution in 0.9 % (w/v) saline after a 16 h fast. The acute glucose lowering effect of MB06322 was assessed by administration of 75 mg.kg-1 i.p. from a 7.5 mg.ml-1 solution in a vehicle composed of 10:10:80 Solutol HS 15:PEG 400:water (Compound was dissolved in PEG 400 by gentle heating/sonication, combined with warm liquid Solutol HS 15 and dispersed in water). Energy expenditure, food intake and spontaneous activity (beam-break) was determined by indirect calorimetry (Labmaster, TSE Systems GmbH, Germany). Following a 3 h acclimatization period, mice were monitored for a 21 h period from 14:00 on day 1 to 23:00 on day 2 (12/12 h light/dark cycle at 21±2°C).
Plasma metabolites
Blood was drawn by tail bleeding into lithium heparin coated capillaries (Sarstedt Microvette CB-200) and plasma prepared by centrifugation at 3,000 g for 5 min at 4°C. Plasma was stored at -80°C prior to analysis. Insulin and glucagon were determined by sandwich ELISA using kits from Mercodia (#10-1249-01 and #10-1281-01). Leptin was measured using an ELISA from Merck Millipore (#EZML-82K). Triglyceride was determined using an enzymatic assay from Sigma (TR0100).
Metformin tolerance test
Animals were starved for 16 h and 250 mg.kg-1 metformin-HCl administered p.o. by gavage. After 45 min, resting blood glucose (t = 0) was recorded and 2 g.kg-1 glucose was administered i.p. Blood glucose was monitored at t = 20, 40, 60 and 120 min. At the end of the procedure ~30 μl of blood was drawn into heparinized capillaries for determining plasma metformin concentration.
Metformin assay
Metformin was assayed in plasma and tissues by ion-pair reverse phase chromatography on a Dionex Ultimate-3000 RS HPLC essentially as described by Zarghi8. Plasma was deproteinized with three volumes 80 % (v/v) acetonitrile containing 5 μg/ml 5,5-diphenylhydantoin as internal standard. Protein was pelleted at 10,000 g for 5 min and the supernatant used directly. Standards were prepared by spiking metformin (1 mg/ml standard prepared in methanol and stored at 4°C) into drug-free heparinized plasma (Innovative Research Inc. #IMS-C57BL6-N) so that the final concentration after solvent extraction ranged from 0.3 to 5 μg/ml. Tissue samples were powdered in a liquid nitrogen cooled mortar and pestle and homogenized with 10 volumes 10 mM NaOH using a bead mill (Qiagen TissueLyser II, 2×2 min at 30 Hz) and 100 μl was extracted with 1 ml 1:1 acetonitrile:methanol containing 5 μg/ml 5,5-diphenylhydantoin. Samples were centrifuged at 16,000 g for 10 min and 0.3 ml supernatant was evaporated in a Speedvac at 45°C. The residue was dissolved in 100 - 200 μl 40 % (v/v) acetonitrile and particulate material removed by centrifugation before analysis. Standards were prepared by spiking metformin into alkaline tissue extracts from drug-free animals so that the final concentration ranged from 0.3 to 5 μg/ml. Samples and standards (20 μl) were injected onto a Syncronis 150×4.6 mm, 5 μm C18 column (Thermo 97105-154630) with a 10×4.0 mm guard column (Thermo 97105-014001) equilibrated with mobile phase containing 10 mM NaH2PO4, 10 mM SDS pH 5.1 and 40 % (v/v) acetonitrile at 1.3 ml/min at 26°C. Metformin and 5,5-diphenylhydantoin were resolved isocratically with retention times of 4.0 and 4.4 min and monitored by UV absorbance (235 nm). Chromatograms were acquired and integrated using Chromeleon v7.1. Results for tissue metformin are uncorrected for blood contamination. For correction of apparent drug tissue concentrations for residual blood volume, tissues were homogenized in 10 volumes ice-cold 40 mM potassium phosphate pH 8.1 and blood content determined by assaying the pseudoperoxidase activity of haemoglobin9 in reactions containing 85 mM NaCl, 5.8 M acetic acid, 0.83 mM EDTA, 33 mM chlorpromazine and 0.6 % H2O2 assuming an average haematocrit of 46.6 %. Reactions were monitored at A525 and a standard curve prepared using a reference sample of whole mouse blood collected in 4 mM EDTA. Metformin was extracted from homogenates in phosphate buffer and assayed as described above.
AICAR assay
Blood was drawn into EDTA capillaries and immediately supplemented with 25 μM dipyridamole and 25 nM A13457 to prevent further uptake and metabolism of AICAR by erythrocytes10. Plasma was deproteinized with three volumes 0.4 N PCA and pelleted at 16,000 g for 5 min at 4°C. Supernatant was neutralized with 0.16 volumes 2 N KHCO3 and solid KClO4 removed at 16,000 g for 5 min. Standards were prepared by spiking AICAR into drug-free plasma in the range 0 – 100 μM. Samples/standards (20 μl) were injected onto a Syncronis 150×4.6 mm, 5 μm C18 column with a 10×4.0 mm guard column equilibrated with mobile phase containing 95:5 10 mM phosphate pH 8.2:acetonitrile at 1 ml.min-1 at 26°C. AICAR was resolved isocratically with a retention time of 3.2 min and monitored at A260. Chromatograms were acquired and integrated using Chromeleon v7.1
MB05032 assay
Plasma was deproteinized with two volumes methanol and pelleted at 16,000 g for 10 min at 4°C. Standards were prepared by spiking MB06322 and MB05032 (1 mM solutions in methanol) into the methanol precipitant so that the effective plasma concentration ranged from 0 to 80 μM. Samples and standards (20 μl) were injected onto a Syncronis 150×4.6 mm, 5 μm C18 column with a 10×4.0 mm guard column equilibrated with mobile phase (A – 20 mM phosphate pH 6.2, 10 % acetonitrile) at 1.5 ml.min-1 at 40°C. The column was resolved with an acetonitrile gradient (B – 80 % acetonitrile): 0 min – 0 % B, 1 min – 0 % B, 13 min – 100 % B, 15 min – 0 % B and re-equilibrated with A for 8 min. Peaks were detected at A300. Water used for mobile phase preparation was filtered through Empore SBD-XC cartridge filters to remove impurities and improve baseline stability. Chromatograms were acquired and integrated using Chromeleon v7.1.
Metformin-euglycemic clamp
Prior to the onset of the study an estimation of the required sample size was determined according to:
where the level of significance was α = 0.05 and desired power was 1-β = 0.8. The quantities Zα/2 and Z1−β are critical values from the normal distribution being 1.96 and 0.8416, respectively. The sampled standard deviation, s, has a value of 2 mg.kg-1.min-1. This quantity was used as initial studies in our laboratory using stable isotopes to quantify in vivo EndoRa in the fasted mouse provided a standard deviation of 2 mg.kg-1.min-1. δ (2 mg.kg-1.min-1) represents the difference we aimed to identify for EndoRa. As such, the resulting sample size rounded off at n = 8. Experimenters were blinded to the genotype until the conclusion of the study. Values were excluded from means reported based on the following pre-determined exclusion criteria:
Following completion of metformin clamps for all mice designated for study the glucose infusion rates (GIR) were assessed. Any mouse that displayed GIR outliers during the steady state sampling period (100-120 minutes) was removed from any further analysis. Outliers were designated as those with values ± 1.5 standard deviations from the group mean of a specific time point.
Estimated glucose fluxes (EndoRa, Rd, gluconeogenesis and glycogenolysis) were excluded from reported means if the value was ± 2 standard deviations from the group mean.
Mice were chronically catheterized approximately seven days prior to study, as described previously11. Briefly, catheters were implanted in the carotid artery and jugular vein for sampling and infusing, respectively. Animals were housed individually post-surgery and monitored for distress. Prior to study, mice were within 10 % of pre-surgery weight. On the day of study, mice were placed in bedded containers without food or water at 07:00 (t = -300 min), five hours prior to initiation of the clamp. An arterial blood sample (80 μl) was drawn for evaluating the unlabeled, natural isotopic abundance of glucose after three hours of fasting (t = -120 min). Subsequently, a bolus of 2H2O (99.9 %) was delivered over 25 minutes to enrich total body water to 4.5 %. A [6,6-2H2]glucose prime (80 mg.kg−1) was dissolved in the bolus. Following the prime, [6,6-2H2]glucose was continuously infused (0.8 mg.kg−1.min−1) for the remainder of the fasting period. An arterial blood sample (110 μl) was taken to determine basal glucose kinetics, arterial glucose and insulin (t = -5 min). Metformin was delivered as a continuous infusion (1.875 or 3.75 mg.kg−1.min−1) followed by a variable infusion of 50 % dextrose (8 % [6,6-2H2]glucose) to clamp blood glucose levels at 120 mg.dl−1. All infused solutions were prepared in a 4.5 % 2H2O-saline solution. Blood glucose was monitored (AccuCheck; Roche Diagnostics, Indianapolis, IN) every 10 minutes and donor erythrocytes were infused to maintain hematocrit levels during the study. Three arterial blood samples (~100 μl each) were obtained during the clamp steady-state period, 90 min after metformin infusion was initiated for determination of glucose fluxes as well as arterial glucose, insulin and/or metformin levels. Plasma was stored at −20°C until analysis. Mice were rapidly euthanized through cervical dislocation immediately after the final steady-state sample. Tissues were rapidly dissected (within 30 s), freeze-clamped in liquid nitrogen and stored at -80°C until further analysis.
Preparation of glucose derivatives for GC-MS analysis
Plasma samples were separated into three aliquots. Each aliquot was derivatized separately to obtain di-O-isopropylidene propionate, aldonitrile pentapropionate, and methyloxime pentapropionate derivatives of glucose as previously described12,13.
GC-MS analysis
GC-MS analysis employed an Agilent 7890A gas chromatography system with an HP-5ms capillary column (Agilent J&W Scientific) interfaced with an Agilent 5975C mass spectrometer and was executed as previously described12 with minor modifications. Injection volumes were 1 μl with purge flow times between 20 and 120 s. A custom MATLAB function was used to integrate each derivative peak in order to obtain mass isotopomer distributions (MIDs) for the following ion ranges: aldonitrile, m/z 173–178, 259–264, 284–289, and 370–375; methyloxime, m/z 145–149; di-O-isopropylidene, m/z 301–308. MIDs of each fragment were averages of two injections per sample. Root mean square error was determined to provide uncertainty and was calculated by comparing the MIDs of unlabeled glucose samples to the theoretical MIDs obtained from the known abundances of naturally occurring isotopes.
Glucose positional deuterium enrichment analysis
The positional deuterium enrichment at each carbon of glucose was determined by least-squares regression as previously described13 using the six glucose fragments, all glucose isotopomers up to M+2, and INCA software14 (available at http://mfa.vueinnovations.com/mfa). Goodness of fit was assessed by a chi-square test and confidence intervals of 95 % were determined as previously described12,15. Fits were accepted according to a chi-square test (P = 0.05) with nine degrees of freedom.
Glucose Kinetics
The infusion rate of [6,6-2H2]glucose and model-derived, plasma [6,6-2H2]glucose enrichment were used to determine glucose turnover (Rt). Assuming steady state conditions, glucose disappearance (Rd; mg.kg−1.min−1) is equivalent to Rt. Endogenous glucose production (EndoRa; mg.kg−1.min−1) was calculated by subtracting the glucose infusion rate (GIR) from total Rt. The model-derived positional deuterium enrichment at carbon 5 (D5) and carbon 2 (D2) of plasma glucose allowed the fractional contribution of gluconeogenesis and glycogenolysis to be determined as previously outlined16,17. Briefly, fractional contribution of gluconeogenesis (GNG) was obtained by the ratio between D5 and D2 (GNG=D5/D2). Fractional contribution of glycogenolysis (GYG) to EndoRa was determined from the equation, GYG = 1-GNG. Multiplying by EndoRa allowed for absolute rates of glycogenolysis and gluconeogenesis to be calculated. Glucose flux rates for the three clamp steady-state samples were averaged to obtain representative values for each mouse.
[11C]-Metformin MicroPET
[11C]-Metformin was synthesized by methylation of 1-methylbiguanide with [11C] methyl triflate18 and prepared as a solution (0.1-0.5 μg/ml) in 100 mM (NH4)2HPO4, pH 5. Animals were anesthetized in a chamber filled with 5 % isoflurane in a mixture of O2 (0.4 L.min-1) and air (1.5 L.min-1). After induction of anesthesia, the head of the animal was placed in an acrylic glass holder and anesthesia maintained with isoflurane (1.8–2.0 %) in O2 (0.4 L.min-1) and air (1.5 L.min-1). A single bolus of [11C]-metformin (4.6 ± 0.3 MBq/mouse) was injected via a catheter inserted into the tail vein, followed by 60 min dynamic PET- and 15 min MR-imaging in a Mediso nanoScan PET/MR (Mediso Ltd, Hungary). Respiratory frequency was monitored and body temperature maintained at 36-37°C. Animals were euthanized at the end of the procedure by cervical dislocation. Data obtained from the dynamic PET was reconstructed with a 3D OSEM algorithm (Tera-Tomo 3D, full detector model and normal regularization; Mediso Ltd, Hungary) with four iterations and six subsets, voxel size 0.4×0.4×0.4 mm3. Corrections were made for randoms, dead-time and decay using a delayed coincidence window. Attenuation and scatter was not corrected. The 60 min dynamic PET-scans were reconstructed as 30 frames increasing in duration from 5 s to 10 min. Multiple regions of interest (ROIs) were placed on coronal slices in the organ of interest using PMOD version 3.6 (PMOD Technologies Ltd, Zurich, Switzerland) creating a volume of interest (VOI). An image-derived input function was generated by averaging images from the first 20 s and placing a circle with a diameter of 15 Pixels on the six slices with the highest activity in the heart (68 μl), representing primarily the blood-pool in left ventricle. Hepatic VOIs were drawn in the anterior part of the liver on PET-images averaged from 0-60 min in which it can be easily identified. Positioning of all VOIs was controlled in each time frame. MR-images were used for defining size and demarcation of liver and left ventricle. Time-activity curves were generated from the VOIs. Results are expressed as tissue-to-blood ratio by dividing the tissue concentration of [11C]-metformin by the blood concentration at each time point for each animal. Area under the curve (AUC) of the tissue-to-blood ratio is a reflection of the tissue extraction ratio and represents the distributional relationship between uptake and elimination from the tissue of interest.
RT-PCR
RNA was extracted from powdered livers using Trizol reagent and silica columns (ThermoFisher #12183-555) using the standard protocol with the exception that Trizol homogenates were centrifuged at 12,000 g for 10 min at 4°C prior to phase separation and the RNA fraction was loaded onto silica columns in 25 % (v/v) ethanol to prevent co-precipitation of RNA with glycogen. RNA integrity was monitored by electrophoresis. Samples were denatured in formamide/formaldehyde loading buffer (Sigma R1386) containing 0.1 % (v/v) SYBR® Safe and separated on 1 % (w/v) agarose gels in TAE. cDNA was synthesized using random hexamers and oligo(dT) primers using iScript cDNA synthesis kit (Bio-Rad #1725038). RT-PCR was performed using SYBR green detection on a Roche LightCycler 480 II. Reactions contained 50 ng template, 0.5 μM primers and 1×LightCycler 1536 DNA Green master reagent (Roche #05573092001) and amplification was performed using a hot-start, touchdown protocol: 95°C, 7 min followed by 35 cycles of 95°C for 10s, 63°C > 58°C (0.5°C/cycle) for 10s and 72°C for 10s. Specificity was determined by melting curves and agarose gel electrophoresis of reaction products. Relative quantification of target genes and propagation of error was determined using 18S as a reference gene and the ΔΔCt method of Livak19. Statistical significance of relative expression ratios was tested using REST© 2009 (http://www.gene-quantification.com/rest-2009.html).
Tissue Metabolites
To avoid even transient hypoxia which results in severe disturbances in tissue metabolites20, mice were anaesthetized using isoflurane, the abdomen was exposed and the left lobe of the liver was freeze-clamped in-situ using liquid nitrogen cooled Wollenberger tongs. Prior to analysis, powdered livers were stored in cryovials in vapour phase liquid nitrogen. Glycogen was assayed by a modification of the method of Keppler and Decker21. Briefly, samples were digested with 10 volumes 1 M KOH at 80°C for 20 min, adjusted to pH 4.8 with 0.5 volumes 4 N acetic acid and incubated with 5 U/ml amyloglucosidase for 2 h at 40°C. Samples were clarified at 16,000 g for 10 min and free glucose assayed in reactions containing 50 mM tris-HCl pH 8.1, 1 mM MgCl2, 0.1 % (w/v) BSA, 0.5 mM ATP, 0.5 mM NADP+, 0.5 mM iodonitrotetrazolium chloride (INT), 10 μM 1-methoxy-5-methylphenazinium methyl sulfate, 0.5 U/ml hexokinase and 0.1 U/ml glucose-6-phosphate dehydrogenase by following the increase in A495. Glucose in perchloric acid extracts was assayed as described above but omitting coupling to INT, which forms an insoluble perchlorate precipitate. Adenine and Z nucleotides were assayed by ion-pair, reverse phase chromatography on an Ultimate 3000-RS HPLC essentially as described by Ryll22. Powdered liver was homogenized in 6 volumes ice-cold 0.6 N perchloric acid, 0.5 mM EGTA and centrifuged at 16,000 g for 3 min at 4°C. Protein pellets were dissolved in 10 volumes 0.5 M NaOH and [protein] determined using Bradford reagent. Perchloric acid was extracted from the supernatant by shaking with two volumes 1:3 trioctylamine:chloroform (~0.6 N trioctylamine) and centrifugation at 2000 g for 1 min at 4°C to induce phase separation. The upper aqueous phase was recovered and neutralized by the addition of 5 mM phosphate pH 7. Samples were analyzed immediately by injection (20 μl) on a Supelcosil LC-18 T 150×4.6 mm, 3 μm C18 column equilibrated with mobile phase (A - 100 mM potassium phosphate pH 5.5, 8 mM tetrabutylammonium hydrogen sulfate) at 26°C. The column was resolved with a methanol gradient (B = A:methanol 70:30, pH 6): 0 min – 0 % B, 2 min – 0 % B, 16 min – 40 % B, 17 min – 100 % B, 23 min – 100 % B, and re-equilibrated with A for 8 min. Peaks were detected at A254. Skeletal muscle was powdered in a liquid nitrogen-cooled Bessman pulverizer and homogenized with 10 volumes 0.6 N PCA, 0.5 mM EGTA using a rotor-stator homogenizer. Samples were clarified at 16,000 g for 5 min at 4°C, the supernatant neutralized with two volumes 1:3 trioctylamine:chloroform as described above and 10 nM P1,P5-di(adenosine 5') pentaphosphate (A5pA) added to inhibit any residual myokinase activity. Samples were analyzed immediately by injection (10 μl) on an Accucore 100×3.0 mm, 2.6 μm C18 column (Thermo 17126-103030) with a 10×3.0 mm guard column (Thermo 17126-013005) equilibrated with mobile phase (A - 100 mM potassium phosphate pH 5.5, 5 mM tetrabutylammonium hydrogen sulfate) at 0.6 ml.min-1 at 26°C. The column was resolved with an acetonitrile gradient (B = A:acetonitrile 75:25, pH 6): 0 min – 0 % B, 1.5 min – 0 % B, 5 min – 10 % B, 9 min – 50 % B, 10 min – 100 % B, 13 min – 100 % B and re-equilibrated with A for 6 min. Peaks were detected at A254. Water used for mobile phase preparation was filtered through Empore SBD-XC cartridge filters to remove impurities and improve baseline stability. Chromatograms were acquired and integrated using Chromeleon v7.1 and calibrated using standards prepared in water and standardized at A259 (ε = 15.4 mM-1 in 0.1 M phosphate pH 7). Remaining metabolites were assayed in 0.6 N perchloric acid, 1 mM EDTA extracts neutralized with 0.25 volumes 2 M KOH, 0.4 M KCl, 0.4 M imidazole. To prevent excessive loss of pyruvate it was necessary to add 500 U/ml catalase immediately upon neutralization to remove H2O2, which forms spontaneously in neutralized perchloric acid extracts of blood rich tissues and decarboxylates pyruvate to acetate. Lactate was assayed using a modification of the method of Noll23 in reactions containing 0.1 M 2-amino-2-methyl-1-propanol pH 9.2, 20 mM glutamate, 1.5 mM NAD+, 10 μM 1-methoxy-5-methylphenazinium methyl sulfate, 0.5 mM WST-1, 20 U/ml lactate dehydrogenase and 5 U/ml glutamate-pyruvate transaminase. A440 was recorded and [lactate] determined by interpolation of a standard curve prepared using lithium lactate. Pyruvate was assayed as described by Passonneau and Lowry24 in reactions containing 50 mM phosphate pH 7, 20 μM NADH and 0.04 U/ml lactate dehydrogenase. The decrease in fluorescence (λex = 345 nm, λem = 465 nm) was recorded and calibrated using known quantities of sodium pyruvate. G6P, F6P and F-1,6-P2 were assayed using a modification of the method of Racker25 in reactions containing 50 mM tris-HCl pH 8.5, 5 mM MgCl2, 0.1 mM EDTA, 100 μM NADP+, 0.2 U/ml diaphorase and 20 μM resazurin. Glucose-6-phosphate dehydrogenase (0.1 U/ml), phosphoglucose isomerase (0.2 U/ml) and recombinant spinach chloroplast fructose-1,6-bisphosphatase (1 U/ml, prepared in E.coli) were added sequentially and the increase in resorufin fluorescence (λex = 540 nm, λem = 590 nm) recorded and calibrated by addition of known quantities of NADH. Fructose-2,6-bisphosphate was assayed using PPi-dependent fructose-6-phosphate 1-phosphotransferase from potato exactly as described by Van Schaftingen26. 3’,5’-cAMP was assayed in trichloroacetic acid (TCA) extracts using a commercial enzyme immunoassay from Sigma (CA-201). Briefly, powdered tissue was homogenized in 10 volumes ice-cold 5 % (w/v) TCA and centrifuged at 16,000 g for 10 min at 4°C. The supernatant was extracted 4× 3 volumes water-saturated diethyl ether, frozen and lyophilized in a Speedvac. Samples were reconstituted in the supplied assay buffer and analyzed according to the manufacturer’s instructions.
Enzyme assays
GCK (EC 2.7.1.2) activity was assayed in cytosolic fractions (which substantially improves the assay by removing G6PC) prepared using the recommendations of Davidson27 in reactions containing 50 mM HEPES pH 7.4, 0.1 M KCl, 0.1 mM EDTA, 7.5 mM ATP, 0.5/100 mM glucose, 0.5 mM NAD+, 2.5 mM DTT, 1 % (w/v) BSA, 2 U/ml glucose-6-phosphate dehydrogenase and 10 μM rotenone at 30°C. Activity at 0.5 mM glucose was subtracted from that at 100 mM glucose to correct for hexokinase-1 activity. Blanks were performed in the absence of glucose and/or ATP. 6-phosphofructo-1-kinase (EC 2.7.1.11) activity was determined by the method of Castano28. Extracts (40 μg) were incubated in 50 mM HEPES pH 7.1, 0.1 M KCl, 6.5 mM MgCl2, 1.5 mM ATP, 0.25 mM fructose-6-phosphate, 0.75 mM glucose-6-phosphate, 0.1 mM AMP, 5 mM Pi, 1 mM NH4Cl, 0.2 mM NADH, 0.05 % (w/v) BSA, 2 mM 2-mercaptoethanol, 10 μM rotenone, 1 U/ml aldolase, 10 U/ml triosephosphate isomerase, 2 U/ml glycerol-3-phosphate dehydrogenase and 1 U/ml phosphoglucose isomerase at 30°C. Coupling enzymes were buffer exchanged over Sephadex G-25 equilibrated with 10 mM tris-HCl pH 7.1 to remove sulfate. Total PK (EC 2.7.1.40) activity and ratio at 1.3/6.6 mM phosphoenolpyruvate at 66 mM KCl was determined as described by Blair29. Lysates (10 μg) were incubated in reactions containing 100 mM tris-HCl pH 7.5, 66 mM KCl, 10 mM MgSO4, 2.5 mM ADP, 0.2 mM NADH, 10 μM rotenone, 1 μM microcystin-LR, 0.05 % (w/v) BSA, 3.2 U/ml lactate dehydrogenase and 1.3 or 6.6 mM phosphoenolpyruvate. PEPCK-C (EC 4.1.1.32) was assayed using the method of Petrescu30. Cytosolic extracts (100 μg) were incubated in reactions containing 50 mM tris-HCl pH 7.4, 1 mM MnCl2, 0.1 mM EGTA, 0.05 % (w/v) BSA, 0.5 mM PEP, 0.2 mM NADH, 10 μM rotenone, 0.2 mM 2-deoxy-GDP, 2 U/ml malate dehydrogenase and either 20 mM NaCl or 20 mM NaHCO3 (saturated with CO2) at 30°C. FBP1 (EC 3.1.3.11) activity was assayed as described above. AMPD1 (EC 3.5.4.6) was assayed in reactions containing 50 mM MOPS pH 7.2, 100 mM KCl, 1 mM DTT, 0.05 % (w/v) BSA, 1 mM ATP, 0.2 mM AMP, 7.5 mM 2-oxoglutarate, 0.15 mM NADH and 5 U/ml glutamate dehydrogenase. All assays were performed in a final volume of 200 μl and monitored by changes in A340. Initial rates were determined from the linear phase and activity calculated assuming εNAD(P)H = 6.22 mM-1.cm-1. G6PC (EC 3.1.3.9) activity was determined by monitoring the release of Pi using the compleximetric method of Saheki31. Extracts (20 μg) were incubated in 20 mM MOPS pH 7.2, 100 mM NaCl, 2 mM 2-mercaptoethanol containing 10 mM glucose-6-phosphate or glycerol-2-phosphate (to correct for background due to non-specific phosphatases) for 20 min at 30°C. Reactions were quenched by the addition of 1 % (w/v) SDS and an aliquot (25 μl) was removed for determination of released phosphate by sequential addition of 180 μl 15 mM zinc acetate, 100 mM ammonium molybdate and 45 μl 10 % (w/v) ascorbic acid (adjusted to pH 5 with 10 N NaOH). Reactions were left to develop for 15 min at 30°C and A850 recorded. Pi was calculated by interpolation of a standard curve prepared using desiccated KH2PO4. CS (EC 2.3.3.1) was assayed using the method of Srere32. Lysates (10 μg) were incubated in reactions containing 50 mM tris-HCl pH 8.1, 0.1 mM EDTA, 0.1 mM 5’5-dithiobis(2-nitrobenzoic acid) (DTNB), 0.3 mM acetyl-CoA and 0.5 mM oxaloacetate at 30°C. Blanks were performed in the absence of oxaloacetate. A412 was monitored and activity calculated from the linear phase assuming εTNB2- = 14.15 mM-1.cm-1. PC (EC 6.4.1.1) was assayed by coupling the formation of oxaloacetate to the reduction of DTNB using CS. Lysates (20 μg) were incubated in reactions containing 50 mM tris-HCl, 50 mM NaHCO3, 5 mM MgCl2, 2.5 mM ATP, 0.1 mM acetyl-CoA, 0.2 mM DTNB, 5 mM pyruvate and 5 U/ml citrate synthase at 30°C. Blanks were performed in the absence of pyruvate and A412 was monitored as described for CS. GS (EC 2.4.1.11) was assayed by the method of Thomas33. Lysates (50 μg) were incubated in reactions containing 25 mM tris-HCl pH 7.8, 50 mM NaF, 1 mM EDTA, 0.9 % (w/v) glycogen, 1 mM DTT, 20 μM 1-deoxynojirimycin, 4.4 mM [U-14C] UDP-glucose (0.1-0.2 mCi.mmol-1) in the absence and presence of 10 mM G6P for 20 min at 30°C. Reactions were stopped by spotting on squares of 3MM filter paper and immersion in ice-cold 66 % (v/v) ethanol. Filters were washed 3×20 min with 66 % (v/v) ethanol, rinsed with acetone and [14C] incorporation into glycogen determined by scintillation counting in Emulsifier Safe (Perkin Elmer). GPa (EC 2.4.1.1) was assayed in the reverse direction using the method of Gilboe34 following the recommendations of Stalmans35. Lysates (50 μg) were incubated in reactions containing 50 mM MES pH 6.5, 50 mM [U-14C] glucose-1-phosphate (0.02 mCi.mmol-1), 150 mM NaF, 5 mM EDTA, 1 % (w/v) glycogen, 20 μM 1-deoxynojirimycin, 15 mM 2-mercaptoethanol and 0.5 mM caffeine at 30°C for 20 min. Reactions were spotted on filters and processed as described for GS with the exception that room temperature 66 % (v/v) ethanol was used for quenching to prevent high blanks caused by the co-precipitation of glucose-1-phosphate. AMPK phosphotransferase activity (EC 2.7.11.1) was assayed using immunoprecipitates. Briefly, lysates (50 μg) were incubated with 2 μg anti-AMPKα1 or anti-AMPKα2 and 5 μl protein G Sepharose for 2 h at 4°C. Immune complexes were pelleted at 500 g for 1 min and washed 3× 1 ml lysis buffer and 2× 1 ml 50 mM tris-HCl pH 8, 0.1 mM EGTA. Phosphotransferase activity was determined in reactions containing 50 mM HEPES pH 7.5, 10 mM MgCl2, 0.1 mM EGTA, 0.1 mM [γ-32P] ATP (250 CPM.pmol-1), 0.1 mM AMARA (NH2-AMARAASAAALARRR-COOH). Reactions were quenched by spotting onto P81 filters and immersion in 75 mM phosphoric. Filters were washed 3× 10 min with 75 mM phosphoric acid, rinsed with acetone and [32P] incorporation determined by Cherenkov counting. With the exception of AMPK, 1 U = 1 μmol product formed per min at 30°C. For AMPK, 1 U = 1 nmol phosphate incorporated per min at 30°C.
Data Analysis
Fitting to models was performed by least squares non-linear regression using Levenberg-Marquardt minimization using Graphpad Prism v5.0. Area under the curve was calculated using the trapezoidal rule with subtraction of the area below baseline (t = 0). Statistical significance was determined using unpaired, two-tailed Student’s t-test and an alpha level of 0.05. All data were normally distributed based on D'Agostino-Pearson omnibus tests and sample variance was similar between groups being compared.
Supplementary Material
Acknowledgments
We thank M. Deak for molecular biology assistance and S. Jakobsen and J. Frøkiær for support in method development of the [11C]-metformin uptake study. We also thank E. Heikkilä for performing islet isolation, S. Ducommun for performing pTBC1D1 blot, and S. Cotting for constructing Wollenberger tongs. This study was supported by Vanderbilt Mouse Metabolic Phenotyping Center Grant DK059637 (D.H.W.) and R37 DK050277 (D.H.W.), a Foundation Grant (FND 143277) from the Canadian Institutes of Health Research (F.S.), the Danish Council for Independent Research DFF – 4183-00384 (N.J.) and the Novo Nordisk Foundation NNF13OC0003882 (N.J.). E.Z. was supported by a Sir Henry Wellcome postdoctoral fellowship. C.C.H. was supported by a Canadian Diabetes Association postdoctoral fellowship.
Footnotes
Reporting summary. Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability. Uncropped Western blot images are available in Supplementary section. A Life Sciences Reporting Summary is available (linked to this article).
Author Contributions
R.W.H. and K.S. designed the study. R.W.H. performed all the biochemical assays, and the majority of in vivo experiments assisted by K.S. Analysis of FBP1 structure and design of the mutants were performed by E.Z. and F.S. M.P. performed molecular cloning and mutagenesis of FBP1. N.J. and E.I.S. performed [11C]-metformin uptake kinetics study and analyzed the data. C.C.H. and L.L. performed metformin-euglycemic clamp and analyzed the data. D.H.W. supervised C.C.H. and L.L., and contributed to data interpretation of the clamp study. R.W.H and K.S. wrote the manuscript. All the authors reviewed, edited and approved the manuscript.
Competing Financial Interests Statements
K.S. is a full-time employee of the Nestlé Institute of Health Sciences S.A., Switzerland.
References
- 1.Rena G, Pearson ER, Sakamoto K. Molecular mechanism of action of metformin: old or new insights? Diabetologia. 2013;56:1898–1906. doi: 10.1007/s00125-013-2991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348(Pt 3):607–614. [PMC free article] [PubMed] [Google Scholar]
- 3.El-Mir MY, et al. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 4.Bridges HR, Jones AJ, Pollak MN, Hirst J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. Biochem J. 2014;462:475–487. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou G, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogt J, Traynor R, Sapkota GP. The specificities of small molecule inhibitors of the TGFss and BMP pathways. Cell Signal. 2011;23:1831–1842. doi: 10.1016/j.cellsig.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Fullerton MD, et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med. 2013;19:1649–1654. doi: 10.1038/nm.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foretz M, et al. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest. 2010;120:2355–2369. doi: 10.1172/JCI40671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller RA, et al. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madiraju AK, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasenour CM, et al. 5-Aminoimidazole-4-carboxamide-1-beta-D-ribofuranoside (AICAR) effect on glucose production, but not energy metabolism, is independent of hepatic AMPK in vivo. J Biol Chem. 2014;289:5950–5959. doi: 10.1074/jbc.M113.528232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent MF, Marangos PJ, Gruber HE, Van den Berghe G. Inhibition by AICA riboside of gluconeogenesis in isolated rat hepatocytes. Diabetes. 1991;40:1259–1266. doi: 10.2337/diab.40.10.1259. [DOI] [PubMed] [Google Scholar]
- 13.Pagliara AS, Karl IE, Keating JP, Brown BI, Kipnis DM. Hepatic fructose-1,6-diphosphatase deficiency. A cause of lactic acidosis and hypoglycemia in infancy. J Clin Invest. 1972;51:2115–2123. doi: 10.1172/JCI107018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouskila M, et al. Allosteric regulation of glycogen synthase controls glycogen synthesis in muscle. Cell Metab. 2010;12:456–466. doi: 10.1016/j.cmet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Ouyang J, Parakhia RA, Ochs RS. Metformin activates AMP kinase through inhibition of AMP deaminase. J Biol Chem. 2011;286:1–11. doi: 10.1074/jbc.M110.121806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gidh-Jain M, et al. The allosteric site of human liver fructose-1,6-bisphosphatase. Analysis of six AMP site mutants based on the crystal structure. J Biol Chem. 1994;269:27732–27738. [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Fructose-1,6-bisphosphatase regulates glucose-stimulated insulin secretion of mouse pancreatic beta-cells. Endocrinology. 2010;151:4688–4695. doi: 10.1210/en.2009-1185. [DOI] [PubMed] [Google Scholar]
- 18.Faupel RP, Seitz HJ, Tarnowski W, Thiemann V, Weiss C. The problem of tissue sampling from experimental animals with respect to freezing technique, anoxia, stress and narcosis. A new method for sampling rat liver tissue and the physiological values of glycolytic intermediates and related compounds. Arch Biochem Biophys. 1972;148:509–522. doi: 10.1016/0003-9861(72)90170-1. [DOI] [PubMed] [Google Scholar]
- 19.Erion MD, et al. MB06322 (CS-917): A potent and selective inhibitor of fructose 1,6-bisphosphatase for controlling gluconeogenesis in type 2 diabetes. Proc Natl Acad Sci U S A. 2005;102:7970–7975. doi: 10.1073/pnas.0502983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincent MF, Erion MD, Gruber HE, Van den Berghe G. Hypoglycaemic effect of AICAriboside in mice. Diabetologia. 1996;39:1148–1155. doi: 10.1007/BF02658500. [DOI] [PubMed] [Google Scholar]
- 21.Guigas B, et al. 5-Aminoimidazole-4-carboxamide-1-beta-D-ribofuranoside and metformin inhibit hepatic glucose phosphorylation by an AMP-activated protein kinase-independent effect on glucokinase translocation. Diabetes. 2006;55:865–874. doi: 10.2337/diabetes.55.04.06.db05-1178. [DOI] [PubMed] [Google Scholar]
- 22.Vincent MF, Bontemps F, Van den Berghe G. Substrate cycling between 5-amino-4-imidazolecarboxamide riboside and its monophosphate in isolated rat hepatocytes. Biochem Pharmacol. 1996;52:999–1006. doi: 10.1016/0006-2952(96)00413-3. [DOI] [PubMed] [Google Scholar]
- 23.Hunter RW, et al. Mechanism of action of compound-13: an alpha1-selective small molecule activator of AMPK. Chem Biol. 2014;21:866–879. doi: 10.1016/j.chembiol.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey CJ, Wilcock C, Scarpello JH. Metformin and the intestine. Diabetologia. 2008;51:1552–1553. doi: 10.1007/s00125-008-1053-5. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida T, et al. Metformin primarily decreases plasma glucose not by gluconeogenesis suppression but by activating glucose utilization in a non-obese type 2 diabetes Goto-Kakizaki rats. Eur J Pharmacol. 2009;623:141–147. doi: 10.1016/j.ejphar.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Takashima M, et al. Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action. Diabetes. 2010;59:1608–1615. doi: 10.2337/db09-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stepensky D, Friedman M, Raz I, Hoffman A. Pharmacokinetic-pharmacodynamic analysis of the glucose-lowering effect of metformin in diabetic rats reveals first-pass pharmacodynamic effect. Drug Metab Dispos. 2002;30:861–868. doi: 10.1124/dmd.30.8.861. [DOI] [PubMed] [Google Scholar]
- 28.Duca FA, et al. Metformin activates a duodenal Ampk-dependent pathway to lower hepatic glucose production in rats. Nat Med. 2015;21:506–511. doi: 10.1038/nm.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu H, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 30.Grundemann D, Gorboulev V, Gambaryan S, Veyhl M, Koepsell H. Drug excretion mediated by a new prototype of polyspecific transporter. Nature. 1994;372:549–552. doi: 10.1038/372549a0. [DOI] [PubMed] [Google Scholar]
- 31.Kjobsted R, et al. Prior AICAR stimulation increases insulin sensitivity in mouse skeletal muscle in an AMPK-dependent manner. Diabetes. 2015;64:2042–2055. doi: 10.2337/db14-1402. [DOI] [PubMed] [Google Scholar]
- 32.Shaw RJ, et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel K, et al. The LKB1-salt-inducible kinase pathway functions as a key gluconeogenic suppressor in the liver. Nat Commun. 2014;5 doi: 10.1038/ncomms5535. 4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuel VT, et al. Fasting hyperglycemia is not associated with increased expression of PEPCK or G6Pc in patients with Type 2 Diabetes. Proc Natl Acad Sci U S A. 2009;106:12121–12126. doi: 10.1073/pnas.0812547106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cool B, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3:403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Graham GG, et al. Clinical pharmacokinetics of metformin. Clin Pharmacokinet. 2011;50:81–98. doi: 10.2165/11534750-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 37.Christensen MM, et al. The pharmacogenetics of metformin and its impact on plasma metformin steady-state levels and glycosylated hemoglobin A1c. Pharmacogenet Genomics. 2011;21:837–850. doi: 10.1097/FPC.0b013e32834c0010. [DOI] [PubMed] [Google Scholar]
- 38.Lalau JD, Lemaire-Hurtel AS, Lacroix C. Establishment of a database of metformin plasma concentrations and erythrocyte levels in normal and emergency situations. Clin Drug Investig. 2011;31:435–438. doi: 10.2165/11588310-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 39.Bleasby K, et al. Expression profiles of 50 xenobiotic transporter genes in humans and pre-clinical species: a resource for investigations into drug disposition. Xenobiotica; the fate of foreign compounds in biological systems. 2006;36:963–988. doi: 10.1080/00498250600861751. [DOI] [PubMed] [Google Scholar]
- 40.Terada T, et al. Molecular cloning, functional characterization and tissue distribution of rat H+/organic cation antiporter MATE1. Pharm Res. 2006;23:1696–1701. doi: 10.1007/s11095-006-9016-3. [DOI] [PubMed] [Google Scholar]
- 41.Gormsen LC, et al. In Vivo Imaging of Human 11C-Metformin in Peripheral Organs: Dosimetry, Biodistribution, and Kinetic Analyses. J Nucl Med. 2016;57:1920–1926. doi: 10.2967/jnumed.116.177774. [DOI] [PubMed] [Google Scholar]
- 42.Jensen JB, et al. [11C]-Labeled Metformin Distribution in the Liver and Small Intestine Using Dynamic Positron Emission Tomography in Mice Demonstrates Tissue-Specific Transporter Dependency. Diabetes. 2016;65:1724–1730. doi: 10.2337/db16-0032. [DOI] [PubMed] [Google Scholar]
- 43.Chen L, et al. OCT1 is a high-capacity thiamine transporter that regulates hepatic steatosis and is a target of metformin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9983–9988. doi: 10.1073/pnas.1314939111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawley SA, Gadalla AE, Olsen GS, Hardie DG. The antidiabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism. Diabetes. 2002;51:2420–2425. doi: 10.2337/diabetes.51.8.2420. [DOI] [PubMed] [Google Scholar]
- 45.Hawley SA, et al. Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 2010;11:554–565. doi: 10.1016/j.cmet.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Argaud D, Roth H, Wiernsperger N, Leverve XM. Metformin decreases gluconeogenesis by enhancing the pyruvate kinase flux in isolated rat hepatocytes. European journal of biochemistry / FEBS. 1993;213:1341–1348. doi: 10.1111/j.1432-1033.1993.tb17886.x. [DOI] [PubMed] [Google Scholar]
- 47.McCarty MF. A proposal for the locus of metformin's clinical action: potentiation of the activation of pyruvate kinase by fructose-1,6-diphosphate. Med Hypotheses. 1999;52:89–93. doi: 10.1054/mehy.1997.0597. [DOI] [PubMed] [Google Scholar]
- 48.van Poelje PD, Dang Q, Erion MD. Discovery of fructose-1,6-bisphosphatase inhibitors for the treatment of type 2 diabetes. Curr Opin Drug Discov Devel. 2007;10:430–437. [PubMed] [Google Scholar]
- 49.Tao H, Zhang Y, Zeng X, Shulman GI, Jin S. Niclosamide ethanolamine-induced mild mitochondrial uncoupling improves diabetic symptoms in mice. Nat Med. 2014;20:1263–1269. doi: 10.1038/nm.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.Dang Q, et al. Discovery of potent and specific fructose-1,6-bisphosphatase inhibitors and a series of orally-bioavailable phosphoramidase-sensitive prodrugs for the treatment of type 2 diabetes. J Am Chem Soc. 2007;129:15491–15502. doi: 10.1021/ja074871l. [DOI] [PubMed] [Google Scholar]
- 2.Giroux E, Williams MK, Kantrowitz ER. Shared active sites of fructose-1,6-bisphosphatase. Arginine 243 mediates substrate binding and fructose 2,6-bisphosphate inhibition. J Biol Chem. 1994;269:31404–31409. [PubMed] [Google Scholar]
- 3.Tashima Y, Mizunuma H, Hasegawa M. Purification and properties of mouse liver fructose 1,6-bisphosphatase. J Biochem. 1979;86:1089–1099. doi: 10.1093/oxfordjournals.jbchem.a132603. [DOI] [PubMed] [Google Scholar]
- 4.Smiley KL, Jr, Berry AJ, Suelter CH. An improved purification, crystallization, and some properties of rabbit muscle 5'-adenylic acid deaminase. J Biol Chem. 1967;242:2502–2506. [PubMed] [Google Scholar]
- 5.Han P, Han G, McBay H, Johnson J. Adenosine 5'-monophosphate-removing system in fructose-1,6-bisphosphatase assay mixture: a new approach. Anal Biochem. 1982;122:269–273. doi: 10.1016/0003-2697(82)90280-9. [DOI] [PubMed] [Google Scholar]
- 6.Nelson SW, Choe JY, Honzatko RB, Fromm HJ. Mutations in the hinge of a dynamic loop broadly influence functional properties of fructose-1,6-bisphosphatase. J Biol Chem. 2000;275:29986–29992. doi: 10.1074/jbc.M000473200. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski P, Rymaszewski M. Alkaline polyethylene glycol-based method for direct PCR from bacteria, eukaryotic tissue samples, and whole blood. Biotechniques. 2006;40:454. doi: 10.2144/000112149. 456,458. [DOI] [PubMed] [Google Scholar]
- 8.Zarghi A, Foroutan SM, Shafaati A, Khoddam A. Rapid determination of metformin in human plasma using ion-pair HPLC. J Pharm Biomed Anal. 2003;31:197–200. doi: 10.1016/s0731-7085(02)00608-8. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura K, Maeda H, Kawaguchi H. Enzymatic assay of hemoglobin in tissue homogenates with chlorpromazine. Anal Biochem. 1987;165:28–32. doi: 10.1016/0003-2697(87)90196-5. [DOI] [PubMed] [Google Scholar]
- 10.Bosselaar M, Smits P, van Loon LJ, Tack CJ. Intravenous AICAR during hyperinsulinemia induces systemic hemodynamic changes but has no local metabolic effect. J Clin Pharmacol. 2011;51:1449–1458. doi: 10.1177/0091270010382912. [DOI] [PubMed] [Google Scholar]
- 11.Ayala JE, et al. Hyperinsulinemic-euglycemic clamps in conscious, unrestrained mice. J Vis Exp. 2011 doi: 10.3791/3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasenour CM, et al. Mass spectrometry-based microassay of (2)H and (13)C plasma glucose labeling to quantify liver metabolic fluxes in vivo. Am J Physiol Endocrinol Metab. 2015;309:E191–203. doi: 10.1152/ajpendo.00003.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal Chem. 2011;83:3211–3216. doi: 10.1021/ac200012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young JD. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014;30:1333–1335. doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Landau BR, et al. Contributions of gluconeogenesis to glucose production in the fasted state. J Clin Invest. 1996;98:378–385. doi: 10.1172/JCI118803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satapati S, et al. Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. J Lipid Res. 2012;53:1080–1092. doi: 10.1194/jlr.M023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakobsen S, et al. A PET Tracer For Renal Organic Cation Transporters, 11C-metformin: Radiosynthesis and Preclinical Proof-of-Concept Studies. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2016 doi: 10.2967/jnumed.115.169292. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Faupel RP, Seitz HJ, Tarnowski W, Thiemann V, Weiss C. The problem of tissue sampling from experimental animals with respect to freezing technique, anoxia, stress and narcosis. A new method for sampling rat liver tissue and the physiological values of glycolytic intermediates and related compounds. Arch Biochem Biophys. 1972;148:509–522. doi: 10.1016/0003-9861(72)90170-1. [DOI] [PubMed] [Google Scholar]
- 21.Keppler D, Decker K. Glycogen determination with amyloglucosidase. In: Bergmeyer HU, editor. Methods of Enzymatic Analysis. Vol. 3. 1974. pp. 1127–1131. [Google Scholar]
- 22.Ryll T, Wagner R. Improved ion-pair high-performance liquid chromatographic method for the quantification of a wide variety of nucleotides and sugar-nucleotides in animal cells. J Chromatogr. 1991;570:77–88. doi: 10.1016/0378-4347(91)80202-n. [DOI] [PubMed] [Google Scholar]
- 23.Noll F. Methods of Enzymatic Analysis. 1984 [Google Scholar]
- 24.Passonneau JV, Lowry OH. Enzymatic analysis. A practical guide. 1993 [Google Scholar]
- 25.Racker E. In: Methods of Enzymatic Analysis. Bergmeyer HU, editor. 1965. pp. 160–163. [Google Scholar]
- 26.Van Schaftingen E, Lederer B, Bartrons R, Hers HG. A kinetic study of pyrophosphate: fructose-6-phosphate phosphotransferase from potato tubers. Application to a microassay of fructose 2,6-bisphosphate. Eur J Biochem. 1982;129:191–195. doi: 10.1111/j.1432-1033.1982.tb07039.x. [DOI] [PubMed] [Google Scholar]
- 27.Davidson AL, Arion WJ. Factors underlying significant underestimations of glucokinase activity in crude liver extracts: physiological implications of higher cellular activity. Arch Biochem Biophys. 1987;253:156–167. doi: 10.1016/0003-9861(87)90648-5. [DOI] [PubMed] [Google Scholar]
- 28.Castano JG, Nieto A, Feliu JE. Inactivation of phosphofructokinase by glucagon in rat hepatocytes. J Biol Chem. 1979;254:5576–5579. [PubMed] [Google Scholar]
- 29.Blair JB, Cimbala MA, Foster JL, Morgan RA. Hepatic pyruvate kinase. Regulation by glucagon, cyclic adenosine 3'-5'-monophosphate, and insulin in the perfused rat liver. J Biol Chem. 1976;251:3756–3762. [PubMed] [Google Scholar]
- 30.Petrescu I, et al. Determination of phosphoenolpyruvate carboxykinase activity with deoxyguanosine 5'-diphosphate as nucleotide substrate. Anal Biochem. 1979;96:279–281. doi: 10.1016/0003-2697(79)90582-7. [DOI] [PubMed] [Google Scholar]
- 31.Saheki S, Takeda A, Shimazu T. Assay of inorganic phosphate in the mild pH range, suitable for measurement of glycogen phosphorylase activity. Anal Biochem. 1985;148:277–281. doi: 10.1016/0003-2697(85)90229-5. [DOI] [PubMed] [Google Scholar]
- 32.Srere PA. Citrate synthase. Methods in Enzymology. 1969;13:3–11. [Google Scholar]
- 33.Thomas JA, Schlender KK, Larner J. A rapid filter paper assay for UDPglucose-glycogen glucosyltransferase, including an improved biosynthesis of UDP-14C-glucose. Anal Biochem. 1968;25:486–499. doi: 10.1016/0003-2697(68)90127-9. [DOI] [PubMed] [Google Scholar]
- 34.Gilboe DP, Larson KL, Nuttall FQ. Radioactive method for the assay of glycogen phosphorylases. Anal Biochem. 1972;47:20–27. doi: 10.1016/0003-2697(72)90274-6. [DOI] [PubMed] [Google Scholar]
- 35.Stalmans W, Hers HG. The stimulation of liver phosphorylase b by AMP, fluoride and sulfate. A technical note on the specific determination of the a and b forms of liver glycogen phosphorylase. Eur J Biochem. 1975;54:341–350. doi: 10.1111/j.1432-1033.1975.tb04144.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.