Introductory Paragraph
Bacterial septicaemia is a major cause of mortality, but its pathogenesis remains poorly understood. In experimental pneumococcal murine intravenous infection, an initial reduction of bacteria in the blood is followed hours later by a fatal septicaemia. These events represent a population-bottleneck driven by efficient clearance of pneumococci by splenic macrophages and neutrophils, but as we show here, accompanied by occasional intracellular replication of bacteria that are taken up by a sub-set of CD169-positive splenic macrophages. In this model, proliferation of these sequestered bacteria provides a reservoir for dissemination of pneumococci into the bloodstream, as demonstrated by its prevention using an anti-CD169 mAb treatment. Intracellular replication of pneumococci within CD169+ splenic macrophages was also observed in an ex vivo porcine spleen, where the microanatomy is comparable to humans. We also showed that macrolides, that effectively penetrate macrophages, prevented septicaemia whereas beta-lactams, with inefficient intracellular penetration, failed to prevent dissemination to the blood. Our findings define a shift in our understanding of the pneumococcus from an exclusively extracellular pathogen to one with an intracellular phase. These findings open the door to the development of treatments that target this early, previously unrecognized intracellular phase of bacterial sepsis.
Introduction
Despite the existence of highly evolved mechanisms of immunity, humans may still develop life-threatening sepsis1. Clinical management of sepsis is challenging and the discouraging outcome of many anti-sepsis trials plus increasing incidence of antimicrobial resistance are among the compelling reasons for further research to better understand the pathogenesis of invasive infection (septicaemia) and sepsis2. The bacterium Streptococcus pneumoniae (pneumococcus) is one of the major causes of serious disease and death3. About half of patients with pneumococcal pneumonia are septicaemic and the presence of bacteria in the blood correlates to disease severity and outcome4 but it is not known why only a proportion of patients are septicaemic when there are no obvious co-morbidities or risk factors5. Notwithstanding decades of clinical and experimental research, there remain major gaps in our understanding of the early events in invasive pneumococcal infection6. Major unanswered questions are how pneumococci sustain a septicaemia that develops into clinical sepsis. Following intravenous inoculation of pneumococci in animals there is a consistent infection pattern in which a rapid reduction in the numbers of bacteria occurs such that within hours few, if any, organisms are detectable in the blood, a stage known as the ‘eclipse phase’7,8. In animals lacking adaptive immunity to the pneumococcus, this eclipse phase results from innate immune clearance of bacteria by splenic macrophages but, depending on the virulence of the pneumococcal strain, it may later be followed by the emergence of septicaemia and lethal sepsis9. Recently, time-lapse microscopy observations have added important data on the dynamic role of neutrophils in the containment of pneumococcal infection a few hours after infection10. However, the host and microbial determinants of the transition from the contained infection during the eclipse phase to overt septicaemia and sepsis remain poorly understood.
Previously we showed that the eclipse involves a single-cell population bottleneck8,11, which is succeeded by a ‘recovery phase’ with detection in the blood of pneumococci derived in most cases from clonal expansion of one founder bacterium. These events raised the critical question of how, within a few hours of virtual elimination of bacteria from the blood, a single founding pneumococcus could result in septicaemia. We hypothesised an extravascular site of pneumococcal replication, most likely in the spleen, because it is the major site of pneumococcal clearance12–14. Accordingly, we investigated the temporal pattern of localisation of bacteria in the spleen following intravenous inoculation. In particular, we investigated the involvement of different splenic macrophages. Here we show that the splenic metallophilic macrophages that are both CD169+ (Siglec-1, Sialoadhesin) and sulphated glycan positive (mannose receptor binding glycans; MRG+)15,16, henceforth referred to as CD169+act as a “sanctuary” during the first hours of pneumococcal infection. Our data reveal that septicaemia is initiated following uptake of pneumococci by CD169+ macrophages in mice and pigs. Within the CD169+ macrophages the internalised bacteria evade clearance, undergo replication, then cause macrophage lysis and disseminate to the blood. Further, we show that antimicrobial therapy, specifically targeted to abort this phase of intracellular replication, can prevent the occurrence of pneumococcal septicaemia.
Results
Live pneumococci are present in the spleen prior to septicaemia
To investigate the events leading to the eclipse phase and the subsequent occurrence of septicaemia, we hypothesised that there was an extravascular reservoir of pneumococci in the spleen. We induced pneumococcal infection in mice by intravenous inoculation of 1X106 pneumococci. Consistent with previous results8, bacteria were still present in the blood at 6 hours (h) after challenge, but were effectively cleared at 8h (eclipse phase). Following the virtual elimination of bacteria from the blood, bacteria re-appeared following the eclipse phase and by 24h mice showed signs of systemic infection (Fig. 1a). Using confocal microscopy, bacteria were visualised within the spleen during the eclipse phase (6 and 8h). Several staining combinations on tissue sections have been used to see the different splenic compartments (Supplementary Figure 1 a-b-c), showing the bacteria localised mainly in the marginal zone, the area surrounding the white pulp (Fig. 1c-d; Supplementary Figure 1 f-g). Higher magnification, revealed that bacteria were present as discrete clusters within the splenic tissue (Fig. 1e). In contrast, we did not detect bacteria in cultures of lung, cervical lymph node, liver and kidney homogenates at 8h.
Figure 1. The eclipse phase of pneumococcal bacteraemia.
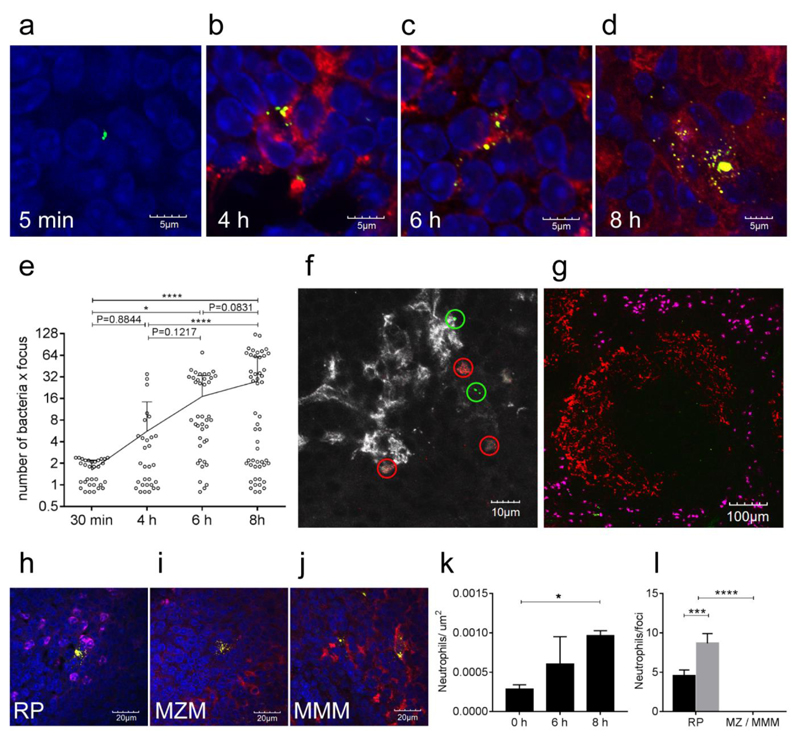
CD1 mice (n=15) were infected intravenously with 1x106 pneumococci (strain D39). (a) Shows the numbers of bacteria in the blood at 6h, 8h and 24h (filled circles, positive blood cultures; empty circles, negative blood cultures). In the spleen (b), bacteria can be found both at 6h and at 8 h after inoculation. Values are expressed as CFU/ml in blood or CFU/g of spleen [weight 100 to 200 mg]. The limit of detection is shown as a dotted line. Statistical differences (* P < 0.05, Fisher’s exact test, one tailed) are indicated by thin black lines. Panels c to e show representative immunofluorescence microscopy images of on infected spleen sections. Single optical sections (analysis of 5 sections from 3 different spleens for each staining combination) obtained from 6 h infected spleens show the presence of bacterial clusters as small green dots (c, 20x objective and d, 60x objective) which can be seen as clusters of single bacteria at a higher magnification (e, 60x objective). In panel c bacteria are shown in green (α-type2, AF488), bacterial foci are indicated by green circles, metallophilic macrophages in red (Cr-Fc, AF568b) and red pulp macrophages in magenta (α-F4/80, AF647). In panel d, bacterial clusters (green circles, α-type2, AF568) can be observed in the metallophilic macrophages area (Cr-Fc, AF647), in white marginal zone macrophages are also stained (aSIGN-R1b, AF488s). In panel e a big cluster of bacteria is shown in green (α-type2, AF488), nuclei in blue (DAPI) and actin in orange (pAF647 conjugated phalloidin). Antibody details in Supplementary Table 1.
Foci of infection are founded by a single bacterium
To further characterise the details of the persistence of pneumococci in the spleen, mice were sacrificed at 5 minutes (min), 30 min, 4 h, 6h and at 8h post-challenge with 1 X 106 pneumococci. Five and thirty min post-challenge, microscopy of spleen sections demonstrated only single bacterial cells or diplococci, localised mainly to the marginal zone macrophages in the marginal sinus (Fig. 2a; Supplementary figure 1 d). In contrast, at 4, 6 and 8h after challenge, pneumococci were predominantly in the marginal zone metallophilic macrophages (Fig. 1c-d; Fig. 2 b-d, Supplementary figure 1 e-g). The number of pneumococci in the clusters within the metallophilic macrophages increased over time (Fig. 2b-e). At 8h bacterial clusters appeared to extend to several adjacent host cells (Fig. 2d) and there were some single bacteria foci (Fig. 2e). These observations suggested lysis of infected cells, and release of bacteria to establish newly infected host cells.
Figure 2. Numbers of pneumococci within clusters, founded by single cells, increase over time in the infected spleens.
(a-d) Immunofluorescence microscopy (representative of 5 sections from 3 different samples) on spleen sections of CD1 mice infected by S. pneumoniae D39 shows an increase of the number of bacteria in the foci of infection over time: (a) 5m; (b) 4h; (c) 6h and (d) 8 h. Splenic architecture was identified by staining the red pulp magenta (α-F4/80, AF647), the metallophilic ring areas in red (Cr-Fc, AF568b) and nuclei in blue (DAPI). Bacteria (green, α-type2, AF488) are trapped in the marginal sinus already at 30 minutes (a) and after an initial clearance, the number and size of the foci increased (b-c-d). A deeper analysis of those images futher confirmed the increase of bacteria in time (e); for each time point, 25 microscope fields (60X magnification) were analysed to count the number of bacteria per focus (open circles) and number of foci. The trend line represents the arithmetic mean number of bacteria per focus, error bars show SD, statistical significance was determined using an ordinary one-way ANOVA with multiple comparisons. * P<0.05, **** p<0.0001. A representative spleen section of mice infected with GFP-D39 and RFP-D39 (n=4) labelled bacteria show how the pneumococci are present in clusters each of which is comprises either exclusively green or exclusively red fluorescent bacteria (green or red circles). Bacterial counts are shown in Supplementary Table 2. I In white metallophilic macrophages are also shown (Cr-Fc, AF568b) (f). In a representative spleen section 8 h after infection the neutrophil granulocytes in magenta (α-GR1, AF647) are seen in the red pulp (g) and do not colocalise with marginal zone macrophages (Cr-Fc, AF568b). At a higher magnification (60x, panels h-j), the neutrophils were found to localise next to all foci of infection in the red pulp (RP, α-F4/80, AF647) (h), but not in the marginal zone (MZM, nearby metallophilic macrophages are stained red, Cr-Fc, AF568b) (i) and in the metallophilic area (MMM, red, Cr-Fc, AF568b) (j). Five spleen sections from three mice each were analysed for this study, allowing for quantification of the number of neutrophils per square μm within the red pulp (k) and to quantify the number of neutrophils within a 25 μm radius of 5 or more discreet infectious foci in each zone of the spleen (l) (black bar 6h; grey bar 8 h). Error bars show SD, statistical significance was determined using a two tailed one-way ANOVA with Tukey’s post correction, * p<0.05, *** P<0.001, **** p<0.0001. Antibody details are in Supplementary Table 1.
To determine if the clusters of pneumococci originated as the result of the replication of single pneumococci, or through sequential phagocytic events by host cells, mice were inoculated intravenously with a 1:1 mixture of D39 strains expressing green fluorescent protein (GFP) or red fluorescent protein (RFP)17. The results showed that bacterial foci at 6h post-challenge consisted entirely of either green or red fluorescent bacteria (Fig. 2f). Multiple spleen samples showed complete absence of dual-labelled foci (2 spleens, 30 fields, Supplementary Table 2), clearly demonstrating that each infection focus was initiated by a single founder bacterium. Based on this finding and the bacterial counts within a focus (Fig. 2a-e), we estimated a mean generation time of approximately 60 min for pneumococcal replication within splenic tissue macrophages (5 generations of 60 min from 1 cell after 1h of infection to 32 cells at 8h). Comparable foci of infection were detected when mice were infected with a serotype 4 strain TIGR4 or a non-capsulated derivative of D39 (Supplementary Figure 2).
Previous work demonstrated the detrimental effect of neutrophil (PMN) depletion on bacterial clearance during the eclipse phase8. Therefore, we investigated the presence of neutrophils in spleen sections at 0, 6 and 8h post-challenge. Consistent with the recent observation showing involvement of mobile PMNs in early splenic clearance of pneumococci10, we detected PMNs at all times almost exclusively in the red pulp (Fig. 2 g) with an increase in number over time (Fig. 2 k). This increase was not uniform; PMNs localised next to pneumococcal cells and the number of PMNs per bacterial cluster also increased when comparing the samples at 6 and 8h (Fig. 2l). Clusters of pneumococci in red pulp macrophages were surrounded by PMNs (Fig. 2h), but neutrophils were not seen during the first 8 h in the proximity of the infectious foci in the marginal zone macrophages (Fig. 2i) or metallophilic macrophages (Fig. 2j).
CD169+ macrophages preferentially accumulate pneumococci
Soon after challenge, most bacteria were found next to the marginal sinus, the region where macrophages sample foreign antigens18. To investigate the fate of bacteria within the spleen, the distribution and number of infectious foci was determined 6h after intravenous challenge with 106 CFU/animal (Fig. 3 a). The compartments considered for this analysis were the white pulp, containing B and T cells (B220+ and KT3+ respectively), the marginal metallophilic macrophage area (CD169+ and MRG+)15,16, the marginal zone macrophage area (SIGN-R1+) and the red pulp (F4/80+) (Supplementary Figure 3 a-c). At this dose, the majority of infectious foci were in the red pulp (Supplementary Figure 3). After infection with a lower dose (105 CFU/animal) the clusters of pneumococci were predominantly observed next to the marginal sinus, the location of the marginal zone macrophages and the metallophilic macrophages, on the outside and inside of the sinus respectively (Supplementary Figure 3). Since the size of each compartment differs, multiple images of whole spleen sections were acquired and analysed to normalise the counts in the different areas of the spleen (Supplementary Figure 4). After normalisation, the data clearly showed that the proportion of infected CD169+ metallophilic macrophages, was significantly higher both at the high and at the low dose compared to any other compartment (Fig. 3a). Counting the number of bacteria present in the macrophage-associated clusters at 6h, showed marginal zone macrophages (MZM) harbouring mainly single bacteria, metallophilic macrophages (CD169+) with foci of about 4 bacteria and red pulp macrophages (F4/80+) with about 8 bacteria/focus (Fig. 3b).
Figure 3. Tissue localisation of infectious foci in the spleen.
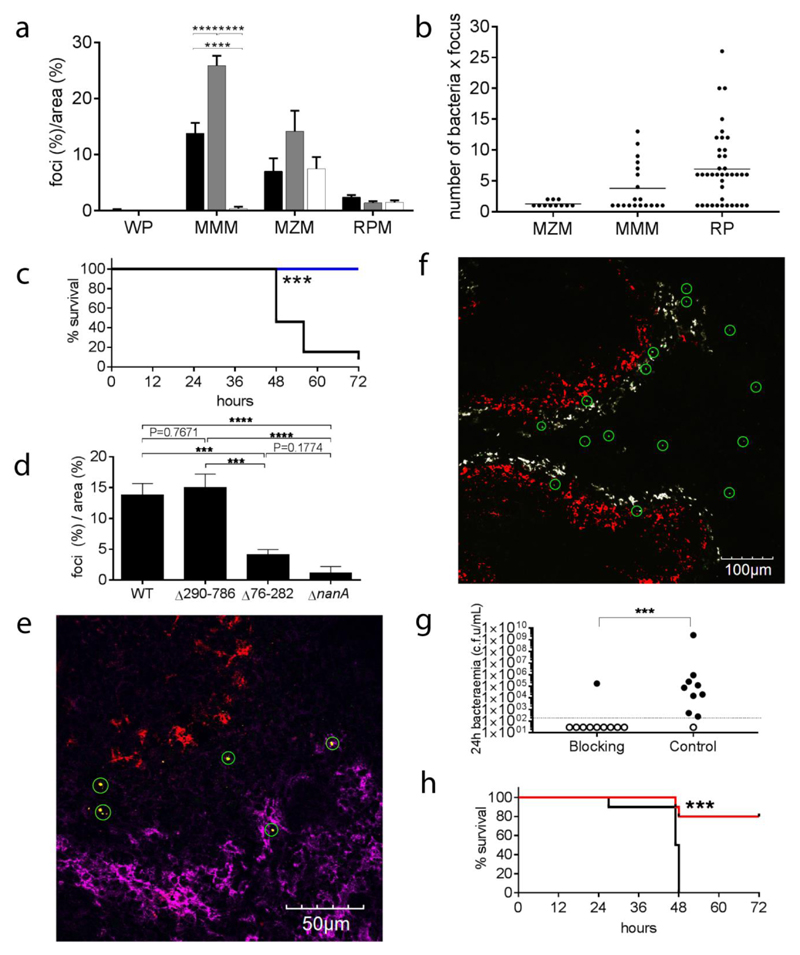
Panel a shows the distribution of the foci of pneumococci at 6 h after infection normalized for the area of each spleen compartment (Supplementary Figure 4). Compartment analysed were the white pulp (WP), the metallophilic macrophage area (MMM), the marginal zone macrophage area (MZM) and the red pulp area (RP) of the spleen. Three infected mice groups have been considered for this analysis: mice infected with high dose (1 x 106 CFU) of S. pneumoniae (black bars), mice infected with low dose (1 x 105 CFU) (grey bars) and mice infected with 1x106 CFU of the D39 after blocking with CD169 mAb (white bars). Counts were obtained from 30 random microscope fields from three independent infected spleens. Error bars show SD, Statistical differences in the percentages between the different infected groups are reported **** P< 0.0001 (two-tailed one-way ANOVA). Number of bacteria per focus for the each spleen compartment are shown in panel b. Mean (lines) and SD (error bars) are reported. Panels c to e report data on mouse infections with the nanA mutants (n=22). Panel c shows the survival graph (black wt n=5, blue ΔnanA n=5, *** P<0.001, kaplan-meier two-tailed test) and panel d the distribution of foci of infection in the metallophilic macrophages in mice infected with bacteria expressing wt NanA, no NanA (ΔnanA), NanAΔ290-786 (sialidase domain deletion), and NanAΔ76-282 (lectin domain deletion). Three independent spleens from mice infected with each nanA mutant were analysed to determine the localisation of foci. 20 fields were analysed from each spleen with a 40x objective. The data show the mean and standard deviation of 3 biological replicas. Statistical significance were determined using an ordinary one-way ANOVA with multiple comparisons, *** p<0.001, **** p<0.0001. Spleen section of D39ΔnanA infected mice (panel e, representative of 5 sections from 3 different samples) show bacterial foci (green circles) localising exclusively to the marginal zone (not stained) or the red pulp. Red pulp macrophages are shown in in magenta (α-F4/80, AF647), metallophilic macrophages in red (Cr-Fc, AF568b) an bacteria in green (α-type2, AF488). Panels from f to h show the data about blocking the CD169 receptor on metallophilic macrophages with a specific mAb (Rat IgG2a,k, Clone: 3D6.112) prior to infection. Panel f shows that in a representative spleen section (5 sections from 3 different samples), 4 hours after infection, bacterial clusters (green circles) do not localise anymore to the metallophilic macrophages (red) while they still are present in the marginal zone (white) or in the red pulp (black). Antibody details in Supplementary Table 1. Panel g shows the levels of bacteraemia 24 hours post-infection indicated by the number of CFU/mL of blood in mice treated with either anti-CD169 blocking antibody or with the isotype matched control (left, Isotype control, n=10, right, CD169 mAb, n=10). The dotted line indicates the limit of detection. Statistical significance were tested using a Mann-Whitney test. *** p=<0.001. In panel h the survival graph of the same experiment is reported (black, Isotype control, n=10, red, CD169 mAb, n=10, *** P<0.001, kaplan-meier two-tailed test).
The surface marker CD169, defining the metallophilic macrophages, previously was shown to be involved in the phagocytosis of sialylated bacteria19–21. While pneumococci are not sialylated, they possess sialidases, including NanA, that contains a sialic acid binding domain previously shown to be involved in adhesion and invasion of host cells22. To test if the surface protein NanA could be involved in macrophage uptake, we repeated the infection experiment with a nanA deletion mutant, that has reduced virulence (Fig. 3c), and two recombinant strains expressing either only the N-terminal lectin binding domain (NanAΔ290-786) or the C-terminal sialidase domain (NanAΔ76-282). Both the nanA deletion mutant and the lectin domain mutant, known to be involved in sialic-acid-mediated host cell binding and invasion22, showed reduction of the number of foci in the metallophilic macrophages (Fig. 3d). It should be noted that the nanA-recombinants are constructed on the background of the nanA deletion mutant and that the recombinant expressing the lectin-like domain (NanAΔ290-786) regains the capacity to localise to CD169+ cells.
CD169 macrophages are the source of bacteria causing invasive disease
Guided by reports showing that anti-CD169 antibodies prevented uptake of porcine reproductive and respiratory virus (PRRSV) by CD169+ macrophages23, we treated mice intravenously with an anti-CD169 mAb (Rat IgG2a,k, Clone: 3D6.112) 30 minutes prior to i.v. challenge with bacteria. At 4h post-infection, treatment with anti-CD169 prevented formation of foci in the metallophilic macrophages (Fig 3f). At 24h post-infection, the treatment with anti-CD169 prevented bacteraemia in almost all mice (Fig.3g) and this correlated with increased survival at 72h (Fig 3h). At 72h post-infection (or at the disease severity endpoint) almost all the mice treated with the CD169 mAb did not have detectable bacteria in blood and spleen (Supplementary Figure 5 a). It should be noted that the CD169 mAb, used as a single dose prior to challenge, was still detectable on the metallophilic macrophage surface at the conclusion of the experiment (Supplementary Figure 5 d).
Having established the importance of CD169+ macrophages in the pathogenesis of pneumococcal sepsis in our i.v. murine challenge model, we next sought to investigate the specific role of the CD169 surface expression marker in the interaction between bacteria and metallophilic macrophages. We compared infections of CD169 knock out mice and isogenic C57/BL6 controls 16. We found no difference in the bacterial counts in blood or spleen, nor in the size of infectious foci in the spleen (Supplementary Figure 5 f-g-h-i). These findings provided no evidence to support a direct role of CD169 itself in mediating interactions between pneumococci and metallophylic macrophages. However, C57/BL6 mice differ from CD1 mice in that splenic clearance is less efficient, blood counts are 100 fold higher and there is no eclipse phase (Fig 1a-b and Supplementary Figure 5 f-g), which limits the validity of the data by not allowing to evaluate the role of metallophylic macrophages in pneumococcal sepsis founded by a single cell bottleneck.
Intracellular localisation of pneumococci
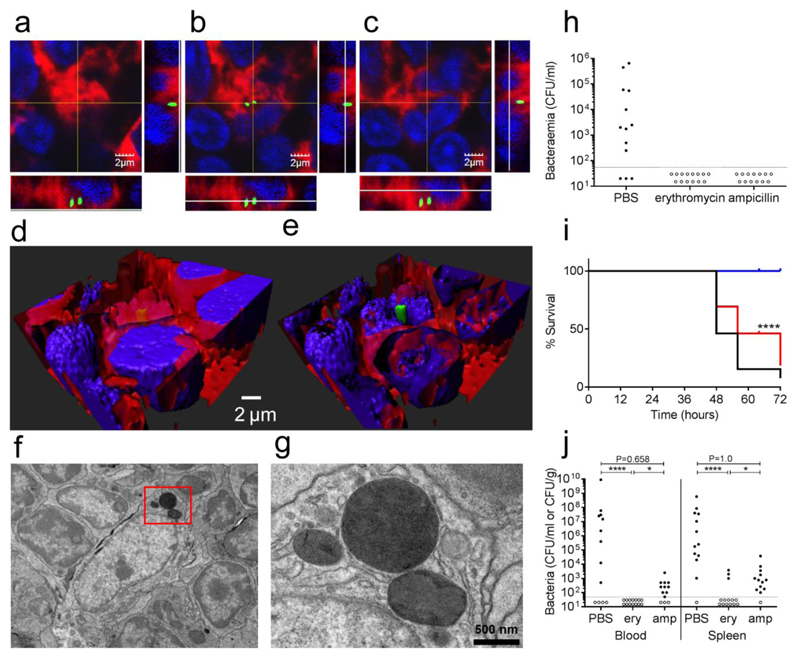
To investigate whether bacteria in a splenic focus were extracellular or intracellular we used a number of different approaches. To delineate host cell plasma membrane, splenic sections infected with GFP-expressing bacteria were labelled with WGA (wheat germ agglutinin). The infection foci at 4h post-infection were enclosed in single host cells. The use of GFP-expressing bacteria allowed differentiation of intact pneumococcal cells from lysed bacteria, as shown in the orthogonal view of a representative multi-stack image acquired using confocal microscopy (Fig. 4a-c). This image showed a cluster of pneumococci clearly identifiable between the cell nucleus (DAPI+) and the plasma membrane (WGA+). Its localisation was confirmed after deconvolution and 3D reconstruction of the tissue (Fig. 4d-e). Additionally, transmission electron microscopy showed groups of bacteria that localised within the host cell cytoplasm, without evident delimitation by a vacuolar membrane (Fig. 4f-g). The fine characterisation of the subcellular localisation of the bacteria within cells was impossible with these whole organ sections and must await the establishment of a validated model of primary cell culture of splenic CD169+ tissue macrophages.
Figure 4. The intracellular phase of S. pneumoniae in the early stages before overt septicaemia.
Panel a-c show three orthogonal views of intracellular pneumococci (green gfp), in this representative (5 sections from 3 different samples) multi-stack acquisition the green GFP-tagged bacteria are localised within the cytoplasm of the host cells 4 hours after the infection (60X magnification). The plasma membrane is shown in red (WGA 633 conjugated) and nuclei are stained in blue (DAPI). Three-dimensional reconstruction through deconvolution analysis clarifies the localisation of the pneumococci within the host cell (d-e). Further analysis with transmission electron microscopy identified groups of pneumococci in the cytoplasm of splenic macrophages. A representative image at 6000X magnification is shown in panel f (3 spleens analysed), while panel g is showing an enlarged insert of the same image (20000X magnification). The importance of the pneumococcal intracellular phase was assessed by treating CD1 mice after the infection (dose of 1 x 106 CFU of strain D39) with antibiotics that have different penetration rates into the host cells (h-j). Two doses of both erythromycin (high penetration rate) and ampicillin (low penetration rate) were administered intraperitoneally at 1 and 5 h post-infection to groups of mice (12-13 animal per group). The doses were chosen so that the predicted drug half-life would reduce the drug plasma levels in 11 h to the minimal inhibitory concentration (MIC). Blood counts at 24 h after infection (panel h) show antibiotics to be equally effective in preventing bacteraemia in comparison to control (p<0.001). The limit of detection is shown as a dotted line. Analysis of later time points indicate that survival is greater significantly higher in the erythromycin group (blue) both with respect to the ampicillin group (red) and control (panel i). **** P<0.0001 kaplan-meier two-tailed test. Survival correlates to the terminal bacterial blood and spleen counts which are lower in erythromycin (ery) treated mice with respect to control in blood and to both control and ampicillin (amp) in the spleen (panel j). (**** P<0.0001, * P<0.05, Fisher’s exact test, one tailed)
To provide further evidence of the intracellular location of pneumococci, we employed the excellent penetration of macrolide antibiotics into macrophages (10-40 mg/L cell: extracellular concentration ratio, C/E ratio), compared to beta-lactams (C/E ratio <0.2 mg/L)24. We hypothesised that we would detect differences in prevention of septicaemia in mice treated using an ultra-short course of each drug. Erythromycin (1.5 mg/animal; half-life 48 min; Minimum Inhibitory Concentration (MIC) 0.06 mg/L) and ampicillin (0.5 mg/animal, half-life 50 min, MIC 0.12 mg/L) were administered intraperitoneally at 1 and 5h post-infection. The doses were calculated so that the plasma concentration of both drugs would decrease below the MIC by 11h post-infection. Both antibiotics were effective in clearing pneumococci from the blood in the first 24h (Fig. 4h), but only erythromycin prevented later outgrowth of bacteria resulting in complete resolution of the infection and survival of mice (Fig. 4i-j). The ampicillin-treated animals showed a survival rate comparable to the control group treated with PBS (Fig. 4i). These data are in accordance with macrolides being effective in the sterilisation of an intracellular bacterial reservoir and thus prevention of late onset septicaemia.
A pig model confirming CD169+ replication-permissive splenic macrophages
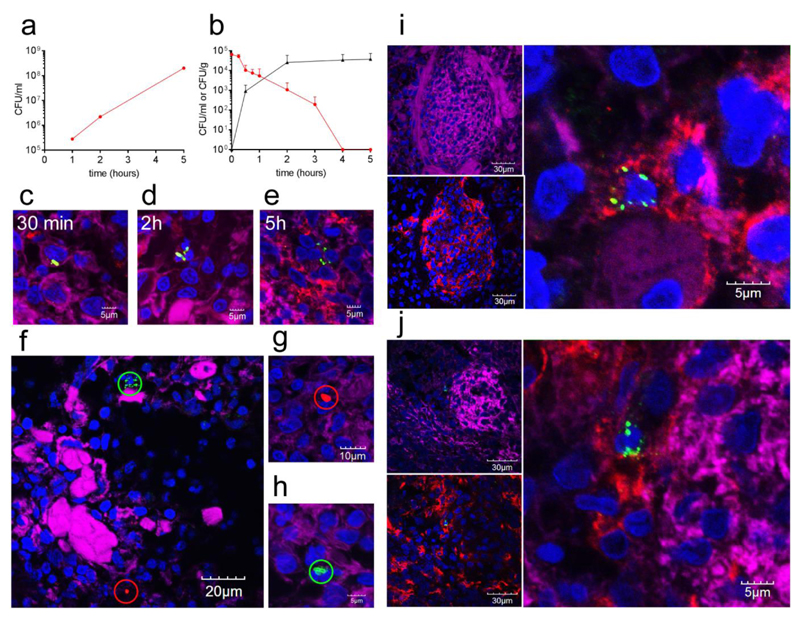
There are anatomical differences between human and mouse spleens. In particular, the human spleen has a relatively smaller white pulp, no marginal zone and importantly, the CD169+ macrophages localise to the perifollicular area, mostly associated to sheathed capillaries25,26. Human capillary sheaths are thus predicted to have a function similar to the marginal zone in mice and rats26. For these reasons, the use of a second model was of crucial importance to determine the potential for translation of our results to humans26. The porcine spleen represents a suitable model as it closely resembles its human counterpart, with a comparable microanatomy and subpopulations of splenic macrophages27,28 where CD169 is expressed by the perifollicular sheath macrophages29,30. Importantly the porcine experimental pneumococcal infection model also shows an eclipse phase following challenge31. Based on previous experience with porcine organ perfusion models32, we set up a normothermic, ex vivo porcine spleen perfusion model to test the dynamics of pneumococcal infection. The model invloves immediate perfusion of abattoir-sourced porcine spleens, followed by 5h perfusion with heparinised autologous blood. In this model it was possible to run a pneumococcal infection for 5h; long enough to study the critical early stages of infection. Thirty minutes after starting perfusion, the arterial circuit was injected with 6.5 x 107 CFU S. pneumoniae D39 (almost 105 CFU/ml). Subsequently, blood samples, spleen biopsies, and blood-gas parameters were taken at 30 min, 2, 4 and 5h. While pneumococci grew with a normal doubling time in heparinised pig blood (Fig 5a), the counts in the blood samples of the ex vivo model showed a steady decrease over time, while counts in the spleen steadily increased (Fig. 5b). This indicated that the spleen was performing its clearance role in this infection model. This porcine perfusion model of infection mirrored the data obtained in the mouse model. Bacterial clusters were found in increased numbers over time (Fig. 5c-e). In addition, independent foci derived from single bacterial cells were observed after infection with a mixed population of GFP- and RFP-expressing pneumococci (Fig. 5f-h). CD169+ macrophages in the porcine spleen localised in the peri-follicular zone (Fig. 5i-j). Analysis of spleen biopsies showed that, 5h post-infection, all foci of pneumococci were in CD169+ macrophages. The data from this whole organ pig model confirmed that early after infection single pneumococci are taken up by CD169+ macrophages and that these cells are permissive for rapid bacterial replication.
Figure 5. The pig spleen perfusion as a model of infection with S. pneumoniae.
Pneumococci grow in vitro with a doubling time of approximately 45 min in heparinised porcine blood (a). In the ex vivo spleen perfusion model D39 pneumococci injected into the arterial circuit are taken up by the spleen and cleared from the blood over time (b). The data show the average number of CFU recovered from blood samples (red) and spleen biopsies (black) of three independent experiments. Error bars represent standard deviation. Immunofluorescence microscopy of infected spleen sections showed an increase of the size of the foci of infection over time (c-e). Bacteria were stained in green (α-type2, AF488), CD169+ macrophages in red (α-CD169p, AF568c) and nuclei in blue (DAPI). Actin staining (in magenta) was used to identify the endothelial tissue of the arterioles (pAF647 conjugated phalloidin). In five-hour infected spleen sections GFP-D39 (green) and RFP-D39 (red) labelled bacteria are present in foci of infection (circles) containing either exclusively green or exclusively red fluorescent bacteria respectively (f-h) (actin in magenta and nuclei in blue). Sections of five hours infected porcine spleens are shown in panels i and j. Clusters of pneumococci were found localised in the CD169+ macrophages of porcine spleens in the perifollicular sheath (i-j). Inserts to the left in both panel i and j show the microarchitecture evidenced by actin staining (magenta) and the distribution of CD169+ macrophages (α-CD169p, AF568c), while the enlarged view is an enlarged view of the focus detected in the left upper panels. Staining was done as for Fig 5c. All the immunofluorescence images are representative of 5 sections from 3 different samples. Antibody details are in Supplementary Table 1.
Discussion
A conspicuous deficiency in our understanding of the pathophysiology of sepsis has been in detailing the host-microbial interactions occurring before the triggering of the catastrophic host inflammatory responses that are characteristic of overwhelming sepsis. Even when initially infected with many thousands of bacteria, our previous research showed that pneumococcal septicaemia is initiated by a single bacterial cell8,33. Recent data have reported the dynamics of S. pneumoniae clearance dependent on red pulp and marginal zone macrophages, aided by mobile neutrophils10. However, neither our work on the single cell bottleneck8,33, nor the time-lapse imaging of pneumococcal clearance in the spleen10, provided knowledge of how the founding organisms survive the highly efficient host innate immunity mechanisms. Now, here we have shown that a small number of bacteria occur within CD169-positive macrophages within the spleen and that these host cells represents an immune-priviledged sanctuary and a reservoir for the reseeding of bacteria into the blood to cause septicaemia and sepsis. Importantly, these findings are not exclusively based on the murine model, but are confirmed by data in the pig; a model of high functional relevance and predictive value for humans27,28. There is an obvious difference between the mouse and pig i.v. challenge models, which are designed to study events in sepsis in the absence of pneumonia, and the most frequent human invasive disease which is sepsis associated with pneumonia. Thus, other studies are required, including human clinical or autopsy samples, to establish the relative importance of the spleen, and intracellular replication therein, in the development of sepsis in the presence and absence of overt pneumonia. In addition to giving a revised perspective on early events leading to septicaemia, our observations have immediate therapeutic implications because they show that septicaemia and sepsis can be prevented by early short term administration of antimicrobials that target the early intracellular bacterial population. In the demonstration of this conclusion, macrolides were much superior to beta-lactams, presently the first choice therapy for pneumococcal pneumonia5, in preventing septicaemia. This is perfectly in line with the call for intracellularly active drugs to fight multi-drug resistant staphylococcal infections, another extracellular pathogens found to hide in intracellular sanctuaries34.
In a murine model of pneumococcal sepsis, we have documented the importance of replication within splenic CD169+ metallophilic macrophages, as an anti-CD169 antibody blocked bacterial uptake into these cells and protected against subsequent invasive disease. CD169 (Siglec-1 or Sialoadhesin) is a surface lectin receptor of macrophages reported to be involved in binding and uptake of a variety of sialylated viruses and bacteria19–21,35,36 but a role of CD169 in uptake of non-sialylated bacteria, such as pneumococci, has not been previously reported. It does appear, however, that sialic acid recognition by pneumococci is important because a mutant bacteria lacking the main pneumococcal sialidase, NanA, or the lectin domain of NanA showed reduced localisation to the metallophilic macrophages. Significantly, the lectin domain of NanA has previously shown to be required for sialic acid mediated invasion in other tissues 22, consistent with the suggestion that NanA mediates binding of pneumococci to macrophages, most likely to sialylated surface proteins. We conclude that while CD169 positive cells are critical, CD169 itself might not be directly involved.
To further examine the role of CD169 we infected CD169 KO mice, which still show the same ringlike distribution of MGR+ metallophilic macrophages around the white pulp (Fig. 5Si). As we saw that pneumococci localised to the marginal metallophilic macrophages with equal abundance in the knock out mice and the wt controls, this implies that CD169 is not directly involved in pneumococcal uptake into these macrophages and that the anti-CD169 antibodies are preventing infection of marginal metallophilic macrophages through indirect effects. For example, if CD169 exists in a molecular complex with other molecule(s) required for pneumococcal uptake by macrophages, antibodies directed to CD169 could block their function through steric effects. It has also been demonstrated that under some circumstances antibodies to CD169 can trigger endocytosis and non-specifically affect phagocytosis of latex beads and bacteria37. Nevertheless, the message of the studies reported here is that pneumococcal interaction with CD169+ macrophages in the spleen can be a critical precursor of septicaemia and sepsis. Invasive pneumococcal disease defined as bacteraemia without known primary focus or humans.
The functions of CD169+ macrophages are poorly defined but there are no reports that they represent a classical anti-microbial cell populations. Indeed pneumococcal survival and growth has not been reported in “typical” macrophages8,38–41, including splenic red pulp macrophages10. To date, CD169+ metallophilic macrophages have been implicated mainly in induction of adaptive immunity by enhancing antigen availability for CD8 T cell and B cell activation, as shown by their permissiveness for viral replication in mice and pigs42,43. Adaptations enabling productive viral infection in CD169+ cells include upregulation of Usp18, which inhibits signalling through the IFN-α and IFN-β receptors. CD169+ macrophages also have been shown to express higher levels of suppressors of cytokine signalling, which suggests unresponsiveness of these cells to selected stimuli42,43. CD169+ macrophages migrate to B cell follicles after LPS stimulation and this correlates with enhanced antibody responses to antigens targeted to these cells indicating that these macrophages can sense inflammatory stimuli44,45. Our data now show that they have a broader permissive state to infection than previously understood and that this permissive state includes intracellular bacterial replication. Combining this finding with the lack of neutrophil recruitment to the vicinity of infected CD169+ macrophages suggests that CD169+ macrophages are an Achilles heel of innate immunity to pneumococcal infections and this extracellular pathogen exploits their relatively ineffective bactericidal mechanisms.
For some bacterial pathogens an intracellular phase is well-known, for example Salmonella spp.46, and events leading to its invasive infection have been defined. However comparable data, especially insight from translational models, is not available for many pathogens, including the pneumococcus, whose within-host life style has been considered as extracellular. The intracellular phase described here documents a significant departure from accepted dogma. It alters perceptions of how pneumococcal bacteraemia occurs, despite efficient clearance mechanisms provided in the spleen, jointly by macrophages, complement and neutrophils10. It is remarkable that, despite life-threatening septicaemia being a relatively commonplace occurrence, so many gaps in our knowledge exist, including the tissue origins of bacteria and how they enter the vascular compartment and what leads to their replication to cause septicaemia47. Nevertheless a major lesson from the historical literature is that the sustained presence of bacteria in the blood cannot be plausibly explained by the one-off entry into the blood, but likely requires an extravascular immune-priviledged focus48,49, which once identified can alter strategies for intervention34. In the present studies, we identify such a crucial extravascular, intracellular site of replication in splenic macrophages that initiates dissemination of pneumococci. Importantly, the intracellular replication phase providing a jump start for single bacterial cells, also explains the generation of a monoclonal bacteraemia founded by a single-cell bottleneck that is observed at low multiplicities of infection8.
In summary, our findings define a crucial intracellular phase in the pathogenesis of invasive disease caused by the pneumococcus, classically considered to be the quintessential example of an extracellular bacterial pathogen. While the relevance of our findings for subacute bacteraemia or bacteraemia without a known primary focus appears evident, the applicability of our findings for pneumonia-associated bacteraemia will have to be defined. The findings open the door to the investigation of treatments that target this early, previously unrecognised yet critical intracellular phase of bacterial sepsis. Judicious use of anti-host therapeutic strategies or antimicrobials capable of efficient penetration of host cells may abort this crucial phase that is essential for initiating bacterial dissemination50. At the early intracellular stage in the pathogenesis, the numbers of organisms that need to be destroyed are relatively small, because this coincides with an extreme population bottleneck. Eliminating small numbers of intracellular bacteria is less challenging than contending with a much larger biomass, or even biofilm, consisting of millions of extracellular bacteria typical of the later stages of sepsis.
Methods
Pneumococcal strains and growth conditions
S. pneumoniae strain D39 (serotype 2), its non-encapsulated derivative R651,52, a sialidase/neuraminidase nanA (SPD_RS07935) deletion mutant53, the serotype 4 strain TIGR454, and GFP and RFP fluorescent D39 (kindly provided by Jan Willem Veening, Groningen)17, were used in this study. A capsule deletion mutant of D39 expressing GFP was constructed by replacing the capsule locus with a kanamycin cassette55. All pneumococcal strains were cultured in Tryptic Soy Broth (TSB, Becton Dickinson) which in the case of green fluorescent (GFP) and red fluorescent (RFP) D39 was supplemented with 5 mg/L chloramphenicol (Sigma-Aldrich). Strains were also grown on blood agar plates consisting of Tryptic Soy Agar (Becton Dickinson) supplemented with 3% v/v defibrinated horse blood. All strains were cultured at 37°C, 5% v/v CO2. For transformations, competence was induced by addition of competence stimulating peptide 1 (CSP-1) to a final concentration of 0.625 μg/mL.
Construction of nanA deletion mutants
Three nanA mutants were constructed in D39. One strain was constructed lacking the region encoding the sialidase domain (NanAΔ290-786), one lacking the lectin-like domain (NanAΔ290-786), and an inactivating insertion mutant. Unmarked mutants were constructed by transforming with PCR generated recombinant constructs (Supplementary Table 2) using a two step approach (Sung et al. 2001). Mutants showed absence of any SNP in the cloned portion by Sanger sequencing.
Ethics statement
All studies in Leicester utilising CD1 mice were performed in accordance with United Kingdom Home Office licence PPL60/4327 and P7B01C07A, and were approved by the University of Leicester Ethics Committee. All mice were scored for signs of disease56. Animals were culled at pre-determined time points, or at the point at which they showed moderate signs of disease in accordance with the Home Office Licence.
The animal protocols used for the KO mice in this study were approved by the Ethical Review Committee of the University of Dundee. All procedures were conducted according to the requirements of the United Kingdom Home Office Animals Scientific Procedures Act, 1986, under PPL PB232D3BA.
Mice
Eight-week old, female, outbred CD1 mice from Charles River (Margate, Kent CT9 4LT UK) were used in this study. Before use, mice were kept for at least 1 week under standard conditions, in the Central Research Facility animal facility at the University of Leicester, according to its guidelines for the maintenance of laboratory animals57–60. Blood samples from mice were collected by cardiac puncture under terminal anaesthesia, and treated with 100 U/ml of heparin (Sigma Aldrich, UK) to prevent blood coagulation. Mice were culled by cervical dislocation and death confirmed before the organs (spleen, lung, kidneys, cervical lymph nodes and liver) were collected post mortem. Mouse organs were either homogenised in 1 ml of TSB containing 10% v/v glycerol, for quantification of colony forming units (CFU), or fixed and embedded for sectioning.
The generation of Sialoadhesin KO (CD169 KO) mice has previously been described 16. All wild-type (WT) and CD169 KO mice used in experiments were derived from heterozygotes back-crossed for at least ten generations onto a C57BL/6J background. Animals were housed under specific pathogen-free conditions under standard housing conditions of 12h light cycle, 21°C and relative humidity of 55-65. Male WT and CD169 KO mice were used at 9-10 weeks for the confirmatory experiment and of 20 weeks for the pilot experiment.
Infection experiments
A total of 132 CD1 mice were used in this study. Sample size calculations were based on long standing experience with the infection models used57–59. Mice were randomised to cages and were challenged intravenously (i.v.) as previously described57–59 with 1.1x105or 1x106 CFU/mouse. At pre-set time points, pre-selected groups were sacrificed for the collection of blood and organ samples. Bacteria were enumerated by plating on selective medium. The bacterial doses for the experiments described herein were selected based on previous experience, in order to raise bacteraemia in at least 50% of the mice.
Experiments in C57BL/6J WT and CD169 KO mice included a confirmatory experiment using n=6/group and a pilot experiment of n=3/group. Mice were infected with 2 x 106 CFU/mouse with S. pneumoniae D39 via intravenous injection. Mice were monitored over a 6h period with no clinical signs observed. 6h post infection, mice were sacrificed by exposure to a rising concentration of carbon dioxide and blood taken by cardiac puncture. Whole blood was frozen in 1 ml RPMI/15% glycerol. Spleen was removed and half stored in 1 ml RPMI/15% glycerol. The remaining half was embedded and frozen in OCT. All samples were processed in Leicester.
The operator carrying out the sample and data analysis was blined for all experiemnts regarding the C57BL/6J mice, but not for work on CD1 mice.
CD169 blockade
Blockade experiments were performed by intravenously injecting 10 µg of anti-CD169 mAb (Rat IgG2a,k, Clone: 3D6.112), or isotype matched control (Rat IgG2a, clone RTK2758) 30 minutes prior to intravenous infection with 106 CFU S. pneumoniae D39 through the lateral tail vein. The distribution of the anti-CD169 mAb was assessed in the spleen through microscopy of sections (Supplementary Figure 5 b). Blood was taken by tail bleed, 24 hours after infection, to enumerate the number of CFU in the blood. Mice were culled at 72 hours post-infection, or when they showed moderate signs of disease in accordance with the home office licence. Blood were taken by cardiac puncture at death, and spleens were either frozen in OCT, or homogenised, serially diluted and plated to count viable bacteria. In all cases, death was confirmed by cervical dislocation. Signs of disease were monitored throughout the course of the experiments. Two independent receptor blockade experiments were performed, each with two groups of 5 mice (20 mice total).
Confocal Microscopy
Confocal imaging was performed on infected spleen sections and uninfected controls. The organs were frozen in dry ice, in Optimum Cutting Temperature (OCT) embedding matrix (Thermoscientific). A Leica CM1850UV cryostat was used to slice the tissues; 8 µm thick sections were cut and mounted on gelatin coated microscopy slides. Organ sections were then dried for 10 min at room temperature (RT) before fixing in phosphate-buffered saline (PBS) containing 4% v/v EM grade formaldehyde (Sigma) for 20 min at RT. Samples were washed three times in PBS, and incubated for 10 min with PBS containing 0.1% v/v Triton-x100 to permeabilise the tissues. Samples were then incubated for 30 min in blocking solution (PBS containing 5% v/v goat serum). Sections were incubated for 1 h with primary antibodies diluted in blocking solution, washed three times with PBS, and then incubated for 45 min with secondary antibody solution. Samples were then washed three times with PBS and once with H2O before adding the mounting medium containing DAPI (Thermoscientific ProLong Gold Antifade Mountant) and closing the slides with coverslips. Anti-serotype 2 and 4 capsule antibodies (Statens Serum Institute) were used to stain non-fluorescent bacteria. Different macrophages populations were labelled using a panel of antibodies (Supplementary Table 1). Metallophilic macrophages were labelled by anti-Mouse CD169 and the recombinant protein CR-Fc, which binds mannose receptor ligands expressed by the metallophilic CD169-positive macrophages61. Marginal zone macrophages were labelled with, anti-Mouse CD209b (SIGN-R1, eBioscience) and anti-Mouse F4/80 antibody (eBioscience). For other classes of immune cells, the following antibodies were used: anti-CD3 (clone Kt3 specific for T cells), anti-B220 (specific for B cells) and anti-FDC-M1 mAb (4C11 specific for follicular dendritic cells). For the studies of bacterial localisation Alexafluor 633-conjugated Wheat germ agglutinin and Alexafluor 647-conjugated phalloidin (Actin; Thermoscientific) were used. The porcine spleen samples were stained with anti-porcine CD169 and anti-porcine CD163 (both Biorad) (Supplementary Table 1). Combinations of Alexafluor conjugated antibodies from Thermoscientific were used as secondary antibodies (488, 568 or 647 with different host specificity) (Supplementary Table 1). An Olympus FV1000 confocal laser scanning microscope was used to acquire the images using 20X and60X objectives, and the free software ImageJ (http://imagej.nih.gov/ij/) was used for image processing. For visualization purpose some images were deconvolved using Huygens Essential deconvolution software version 16 (Scientific Volume Imaging, Netherlands) and viewed in Imaris 3D reconstruction software 9.4 (Bitplane, Switzerland).
Scanning of microscopy sections and spleen area measurements
Spleen sections were stained with different combinations of antibodies in order to identify the different spleen compartments. The sections were scanned using a fully motorised Nikon Eclipse Ti microscope equipped with a Plan Fluor 10X/0.3 objective and an Andor iXonEM+ EMCCD DU 885 camera. Automatic tissue scanning was performed using NIS-Elements software AR 4.51 (Nikon, Japan) and the images obtained were analysed using ImageJ software 1.51. Using the DAPI signal, the total area of each tissue section was calculated. Four different spleen compartments were identified and their areas calculated: white pulp, metallophilic macrophages, marginal zone and red pulp macrophages. The white pulp areas were defined as those areas that stained with a combination of B220 (B cells), CD3 (T cells) enclosed within the metallophilic rings stained withSiglec-1 (metallophilic macrophages marker). The areas of metallophilic macrophages (Siglec-1), marginal zone macrophages (SIGN-R1) and red pulp macrophages (F4/80) were defined as those areas stained for each cell marker. The proportion of the spleen compartments with respect to the total was calculated using the data obtained from at least 5 different images for each staining combination (Supplementary Figure 3). For neutrophils quantification cells were detected with an Ly-6G specific primary antibodies, and numbers of neutrophils were enumerated in at least 5 fields from 3 independent, biological replicates.
Transmission Electron Microscopy
Excised tissue was primarily fixed with 4% v/v Formaldehyde / 0.5% Glutaraldehyde in sodium cacodylate buffer with 2uM of calcium chloride overnight, followed by several washes in same buffer. The samples were post-fixed with aqueous 1% w/v osmium tetroxide/ 1.5% w/v potassium ferrocyanide for 60 min, washed with distilled, de-ionised water, followed by a further 60 minutes in 1% w/v aqueous osmium. After further washes with DDW, the tissue was en bloc stained in 2% w/v aqueous uranyl acetate for 60 min. Following dehydration through an ethanol series, the samples were treated with several short washes through ice cold acetone, before gradual infiltration with Durcopan resin (Sigma-Aldrich UK). Once fully infiltrated with Durcupan, the tissue was embedded in BEEM capsules, and polymerised at 60oC for 48 h.
Thin sections, of approximately 80 nm thickness, were cut from each sample using a Leica UC7 ultramicrotome, collected on copper mesh grids and counter stained Reynolds’ lead citrate. Sections were observed using a JEOL JEM-1400 transmission electron microscope, using an accelerating voltage of 100 kV. Digital Images were recorded using a SIS Megaview III Digital Camera (Olympus Soft Imaging Solutions, Germany) with iTEM Software V 5.1 (Olympus, Germany).
Porcine spleen perfusion model of infection
The methodology for perfusion of the porcine spleen was similar to that described previously for the perfusion of liver and kidney 32. Porcine spleens were collected from a local abattoir from domestic Large White pigs (40–50kg) immediately after slaughter. Following a midline laparotomy, a splenectomy was carried out close to its hilum. To perfuse the organ, the celiac trunk was isolated and cannulated, while the other arteries were carefully ligated and divided. The spleen was then perfused with saline solution containing heparin (LEO Laboratories Limited, UK) and human urokinase (Syner-Medica Ltd, UK) through the main splenic artery, and the blood excess was flushed out through the main splenic vein. The perfusion set-up consists of a SARNS 8000 extracorporeal roller pump (3M, St. Paul, MN, USA), Baby-RX venous reservoir and membrane oxygenator (Terumo, Ann Arbor, MI, USA), metal organ chamber, PVC tubings (Cellplex, Dandenong, VIC, Australia) and water bath temperature regulator.
The perfusate consisted of 1 litre of autologous whole blood collected via exsanguination, containing the two antimicrobials nalidixic acid (10 mg/L) and colistin (5 mg/L), glucose (5 ml/h), 500 µg epoprostenol sodium (vasodilatation), and 5000 IU heparin (microclots prevention; 1500units/hr). The temperature of the water bath was set at 39°C and oxygenation at 2 L/min. When the blood was warm and oxygenated, the organ was connected to the system. Initial flow rate was set at 0.2 L/min, adjusted to maintain a mean arterial pressure (MAP) at 70–80 mm Hg. As soon as the perfused spleen reached a stable flow (typically 30 min), the arterial circuit was injected with S. pneumoniae. Over the subsequent 5 h, serial blood samples and spleen biopsies were taken. Blood gas analysis was performed before and after the infection at predetermined time points to verify the functionality of the organ. Overall four infection perfusion experiments, and three negative controls using non-infected perfused organs were performed.
Statistical analysis
GraphPad Prism software version 6 was used to analyse all data. Fisher’s exact test was used to compare different cohorts of mice with the assumption that all samples containing more than 100 CFU/ml (or CFU/mg) were positive and the others negative. One-way Anova, with Tukey’s post-test, was used to compare the distribution of foci of bacteria in the spleen. Survival curves have been compared with a Log-rank (Mantel-Cox) test. Results were considered significant when P values were <0.05. Error bars in all figures show the Standard Deviation, unless otherwise stated.
Supplementary Material
Acknowledgements
GE was funded through an academic collaboration agreement between the Universities of Oxford and Leicester and in part by MRC grant MR/M003078/1. Authors would like to thank Jan Willem Veening for providing GFP and RFP pneumococci, Francesca Focarelli for construction of the non-encapsulated GFP expressing strain, Megan De Ste Croix for mutant construction, Sarah Glenn for help with the infection experiments, the Electron Microscopy Facility, University of Leicester for their technical support and Rohan Kumar and John Isherwood for help with the perfusion of the porcine organs at explant, the staff of Joseph Morris Butchers Ltd, Michael F Wood Butchers and the staff of Leicester Preclinical Research Facility for support.
Footnotes
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The recombinant protein CR-Fc, which binds mannose receptor ligands expressed by the metallophilic CD169-positive macrophages can be obtained for Luisa Martinez-Pomares and the CD169 knock out mice and their isogenic C57/BL6 controls can be obtained from Paul Crocker.
Statement on Data and Materials request
For any correspondence regarding data and results please contact Marco R Oggioni (mro5@leicester.ac.uk), for access to the recombinant protein CR-Fc please contact Luisa Martinez-Pomares (luisa.m@nottingham.ac.uk) and for access to the CD169 knock out mice and their isogenic C57/BL6 controls please contact Paul Crocker (p.r.crocker@dundee.ac.uk).
Author contributions
G.E. performed almost all experiments and wrote the manuscript. V.E.F. led and performed the animal infections. W.Y.C. led and performed the porcine spleen perfusion experiments. J.J.W. contributed to microscopy, animal infection work and contributed to the writing of the manuscript. C.D.B. contributed to the design of the experimental work. K.S. led the microscopy work. S.T. performed the infections of the CD169 ko mice. P.C. discussed the work and supervised the infections in the ko mice and contributed to the writing of the manuscript. A.D. designed and led the porcine perfusion work and contributed to the writing of the manuscript. L.M.P. designed the immunological work and overall setup of experimentation and contributed to the writing of the manuscript. P.W.A. participated in the overall design and setup of the experimentation and contributed to the writing of the manuscript. E.R.M. initiated and participated in the overall design and setup of the experimentation and contributed to the writing of the manuscript. M.R.O. led the design and setup of the project and contributed to the writing of the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Gratz N, Loh LN, Tuomanen E. In: Streptococcus Pneumoniae. Hammerschmidt Sven, Orihuela Carlos., editors. Academic Press; 2015. pp. 433–451. [Google Scholar]
- 2.van der Poll T. Future of sepsis therapies. Critical Care. 2016;20:106. doi: 10.1186/s13054-016-1274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Pneumococcal vaccines WHO position paper. WHO, Weekly Epidemiological Record (WER) 2012;87:129–144. http://www.who.int/wer/2012/wer8714/en/ [Google Scholar]
- 4.Musher DM, Thorner AR. Community-Acquired Pneumonia. New England Journal of Medicine. 2014;371:1619–1628. doi: 10.1056/NEJMra1312885. [DOI] [PubMed] [Google Scholar]
- 5.Lim WS, Smith DL, Wise MP, Welham SA. British Thoracic Society community acquired pneumonia guideline and the NICE pneumonia guideline: how they fit together. Thorax. 2015 doi: 10.1136/thoraxjnl-2015-206881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simell B, et al. The fundamental link between pneumococcal carriage and disease. Expert Review of Vaccines. 2012;11:841–855. doi: 10.1586/erv.12.53. [DOI] [PubMed] [Google Scholar]
- 7.Rogers DE. Host mechanisms which act to remove bacteria from the blood stream. Bacteriological Reviews. 1960;24:50–66. doi: 10.1128/br.24.1.50-66.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerlini A, et al. The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck. PLoS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1004026. e1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown EJ, Hosea SW, Frank MM. The role of the spleen in experimental pneumococcal bacteremia. Journal of Clinical Investigation. 1981;67:975–982. doi: 10.1172/JCI110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deniset JF, Surewaard BG, Lee W-Y, Kubes P. Splenic Ly6Ghigh mature and Ly6Gint immature neutrophils contribute to eradication of S. pneumoniae. The Journal of Experimental Medicine. 2017;214:1333–1350. doi: 10.1084/jem.20161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kono M, et al. Single Cell Bottlenecks in the Pathogenesis of Streptococcus pneumoniae. PLoS Pathogens. 2016;12 doi: 10.1371/journal.ppat.1005887. e1005887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horan M, Colebatch JH. Relation Between Splenectomy and Subsequent Infection: A Clinical Study. Archives of Disease in Childhood. 1962;37:398–414. doi: 10.1136/adc.37.194.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theilacker C, et al. Overwhelming Postsplenectomy Infection: A Prospective Multicenter Cohort Study. Clinical Infectious Diseases. 2016;62:871–878. doi: 10.1093/cid/civ1195. [DOI] [PubMed] [Google Scholar]
- 14.Shinefield HR, Steinberg CR, Kaye D. Effect of splenectomy on the susceptibility of mice inoculated with diplococcus pneumoniae. The Journal of Experimental Medicine. 1966;123:777–794. doi: 10.1084/jem.123.5.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Pomares L, et al. Fc chimeric protein containing the cysteine-rich domain of the murine mannose receptor binds to macrophages from splenic marginal zone and lymph node subcapsular sinus and to germinal centers. The Journal of experimental medicine. 1996;184:1927–1937. doi: 10.1084/jem.184.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oetke C, Vinson MC, Jones C, Crocker PR. Sialoadhesin-deficient mice exhibit subtle changes in B- and T-cell populations and reduced immunoglobulin M levels. Molecular and cellular biology. 2006;26:1549–1557. doi: 10.1128/mcb.26.4.1549-1557.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kjos M, et al. Bright Fluorescent Streptococcus pneumoniae for Live-Cell Imaging of Host-Pathogen Interactions. Journal of Bacteriology. 2015;197:807–818. doi: 10.1128/JB.02221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aichele P, et al. Macrophages of the Splenic Marginal Zone Are Essential for Trapping of Blood-Borne Particulate Antigen but Dispensable for Induction of Specific T Cell Responses. The Journal of Immunology. 2003;171:1148–1155. doi: 10.4049/jimmunol.171.3.1148. [DOI] [PubMed] [Google Scholar]
- 19.Jones C, Virji M, Crocker PR. Recognition of sialylated meningococcal lipopolysaccharide by siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Molecular Microbiology. 2003;49:1213–1225. doi: 10.1046/j.1365-2958.2003.03634.x. [DOI] [PubMed] [Google Scholar]
- 20.Heikema AP, et al. Enhanced, Sialoadhesin-Dependent Uptake of Guillain-Barré Syndrome-Associated Campylobacter jejuni Strains by Human Macrophages. Infection and Immunity. 2013;81:2095–2103. doi: 10.1128/IAI.01437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang Y-C, et al. Role of Macrophage Sialoadhesin in Host Defense Against the Sialylated Pathogen Group B Streptococcus. Journal of molecular medicine (Berlin, Germany) 2014;92:951–959. doi: 10.1007/s00109-014-1157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchiyama S, et al. The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. The Journal of Experimental Medicine. 2009;206:1845–1852. doi: 10.1084/jem.20090386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanderheijden N, et al. Involvement of sialoadhesin in entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages. Journal of virology. 2003;77:8207–8215. doi: 10.1128/JVI.77.15.8207-8215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maurin M, Raoult D. Antibiotic penetration within eukaryotic cells. Antimicrobial Agents and Intracellular Pathogens. 1993:21–37. [Google Scholar]
- 25.Steiniger B, Barth P, Hellinger A. The Perifollicular and Marginal Zones of the Human Splenic White Pulp: Do Fibroblasts Guide Lymphocyte Immigration? The American Journal of Pathology. 2001;159:501–512. doi: 10.1016/S0002-9440(10)61722-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steiniger BS. Human spleen microanatomy: why mice do not suffice. Immunology. 2015;145:334–346. doi: 10.1111/imm.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fairbairn L, Kapetanovic R, Sester DP, Hume DA. The mononuclear phagocyte system of the pig as a model for understanding human innate immunity and disease. Journal of Leukocyte Biology. 2011 doi: 10.1189/jlb.1110607. [DOI] [PubMed] [Google Scholar]
- 28.Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends in Microbiology. 2012;20:50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezquerra A, et al. Porcine myelomonocytic markers and cell populations. Developmental & Comparative Immunology. 2009;33:284–298. doi: 10.1016/j.dci.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Alvarez B, et al. Phenotypic and functional heterogeneity of CD169+ and CD163+ macrophages from porcine lymph nodes and spleen. Developmental & Comparative Immunology. 2014;44:44–49. doi: 10.1016/j.dci.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 31.de Greeff A, et al. Pneumococcal colonization and invasive disease studied in a porcine model. BMC Microbiology. 2016;16:102. doi: 10.1186/s12866-016-0718-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung WY, et al. Steps for the Autologous Ex vivo Perfused Porcine Liver-kidney Experiment. Journal of Visualized Experiments: JoVE. 2013:50567. doi: 10.3791/50567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moxon ER, Murphy PA. Haemophilus influenzae bacteremia and meningitis resulting from survival of a single organism. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:1534–1536. doi: 10.1073/pnas.75.3.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehar SM, et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature. 2015;527:323–328. doi: 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- 35.Heikema AP, et al. Characterization of the Specific Interaction between Sialoadhesin and Sialylated Campylobacter jejuni Lipooligosaccharides. Infection and Immunity. 2010;78:3237–3246. doi: 10.1128/IAI.01273-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klaas M, et al. Sialoadhesin Promotes Rapid Proinflammatory and Type I IFN Responses to a Sialylated Pathogen, Campylobacter jejuni. The Journal of Immunology Author Choice. 2012;189:2414–2422. doi: 10.4049/jimmunol.1200776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Schryver M, et al. Monoclonal antibody binding to the macrophage-specific receptor sialoadhesin alters the phagocytic properties of human and mouse macrophages. Cellular immunology. 2017;312:51–60. doi: 10.1016/j.cellimm.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Bewley MA, et al. Pneumolysin Activates Macrophage Lysosomal Membrane Permeabilization and Executes Apoptosis by Distinct Mechanisms without Membrane Pore Formation. mBio. 2014;5:e01710–01714. doi: 10.1128/mBio.01710-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dockrell DH, Lee M, Lynch DH, Read RC. Immune-Mediated Phagocytosis and Killing of Streptococcus pneumoniae Are Associated with Direct and Bystander Macrophage Apoptosis. The Journal of Infectious Diseases. 2001;184:713–722. doi: 10.1086/323084. [DOI] [PubMed] [Google Scholar]
- 40.Gordon SB, Irving GRB, Lawson RA, Lee ME, Read RC. Intracellular Trafficking and Killing of Streptococcus pneumoniae by Human Alveolar Macrophages Are Influenced by Opsonins. Infection and Immunity. 2000;68:2286–2293. doi: 10.1128/iai.68.4.2286-2293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis KM, Nakamura S, Weiser JN. Nod2 sensing of lysozyme-digested peptidoglycan promotes macrophage recruitment and clearance of S. pneumoniae colonization in mice. The Journal of Clinical Investigation. 2011;121:3666–3676. doi: 10.1172/JCI57761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honke N, et al. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat Immunol. 2012;13:51–57. doi: 10.1038/ni.2169. http://www.nature.com/ni/journal/v13/n1/abs/ni.2169.html#supplementary-information. [DOI] [PubMed] [Google Scholar]
- 43.Van Breedam W, Verbeeck M, Christiaens I, Van Gorp H, Nauwynck HJ. Porcine, murine and human sialoadhesin (Sn/Siglec-1/CD169): portals for porcine reproductive and respiratory syndrome virus entry into target cells. Journal of General Virology. 2013;94:1955–1960. doi: 10.1099/vir.0.053082-0. [DOI] [PubMed] [Google Scholar]
- 44.Backer R, et al. Effective collaboration between marginal metallophilic macrophages and CD8(+) dendritic cells in the generation of cytotoxic T cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:216–221. doi: 10.1073/pnas.0909541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veninga H, et al. Antigen targeting reveals splenic CD169(+) macrophages as promoters of germinal center B - cell responses. European Journal of Immunology. 2015;45:747–757. doi: 10.1002/eji.201444983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mastroeni P, Grant A, Restif O, Maskell D. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat Rev Micro. 2009;7:73–80. doi: 10.1038/nrmicro2034. [DOI] [PubMed] [Google Scholar]
- 47.Levin BR, Antia R. Why We Don't Get Sick: The Within-Host Population Dynamics of Bacterial Infections. Science. 2001;292:1112–1115. doi: 10.1126/science.1058879. [DOI] [PubMed] [Google Scholar]
- 48.Shaw S, Smith AL, Anderson P, Smith DH. The paradox of Hemophilus infuenzae type B bacteremia in the presence of serum bactericidal activity. Journal of Clinical Investigation. 1976;58:1019–1029. doi: 10.1172/JCI108525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grant AJ, et al. Modelling within-Host Spatiotemporal Dynamics of Invasive Bacterial Disease. PLoS Biology. 2008;6:e74. doi: 10.1371/journal.pbio.0060074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surewaard BGJ, et al. Identification and treatment of the Staphylococcus aureus reservoir in vivo. The Journal of Experimental Medicine. 2016 doi: 10.1084/jem.20160334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avery OT, MacLeod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types : Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. The Journal of Experimental Medicine. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iannelli F, Pearce BJ, Pozzi G. The Type 2 Capsule Locus of Streptococcus pneumoniae. Journal of Bacteriology. 1999;181:2652–2654. doi: 10.1128/jb.181.8.2652-2654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manco S, et al. Pneumococcal Neuraminidases A and B Both Have Essential Roles during Infection of the Respiratory Tract and Sepsis. Infection and Immunity. 2006;74:4014–4020. doi: 10.1128/IAI.01237-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tettelin H, et al. Complete Genome Sequence of a Virulent Isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 55.Pearce BJ, Iannelli F, Pozzi G. Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Research in Microbiology. 2002;153:243–247. doi: 10.1016/S0923-2508(02)01312-8. [DOI] [PubMed] [Google Scholar]
- 56.Morton D, Griffiths P. Guidelines on the recognition of pain, distress and discomfort in experimental animals and an hypothesis for assessment. Veterinary Record. 1985;116:431–436. doi: 10.1136/vr.116.16.431. [DOI] [PubMed] [Google Scholar]
- 57.Kadioglu A, et al. Sex-Based Differences in Susceptibility to Respiratory and Systemic Pneumococcal Disease in Mice. The Journal of Infectious Diseases. 2011;204:1971–1979. doi: 10.1093/infdis/jir657. [DOI] [PubMed] [Google Scholar]
- 58.Oggioni MR, et al. Antibacterial Activity of a Competence-Stimulating Peptide in Experimental Sepsis Caused by Streptococcus pneumoniae. Antimicrobial Agents and Chemotherapy. 2004;48:4725–4732. doi: 10.1128/AAC.48.12.4725-4732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oggioni MR, et al. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Molecular Microbiology. 2006;61:1196–1210. doi: 10.1111/j.1365-2958.2006.05310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee E-J, Pontes MH, Groisman EA. A Bacterial Virulence Protein Promotes Pathogenicity by Inhibiting the Bacterium's Own F(1)F(o) ATP Synthase. Cell. 2013;154:146–156. doi: 10.1016/j.cell.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor PR, et al. Development of a specific system for targeting protein to metallophilic macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1963–1968. doi: 10.1073/pnas.0308490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.