Abstract
All bacterial infections occur within a polymicrobial environment, from which a pathogen population emerges to establish disease within a host. Emphasis has been placed on prevention of pathogen dominance by competing microflora acting as probiotics1. Here we show that virulence of the human pathogen, Staphylococcus aureus is augmented by native, polymicrobial, commensal skin flora and individual species acting as “proinfectious agents”. The outcome is pathogen proliferation but not commensal. Pathogenesis augmentation can be mediated by particulate cell wall peptidoglycan (PGN), reducing the S. aureus infectious dose by over 1000-fold. This phenomenon occurs using a range of S. aureus strains, infection models and is not mediated by established receptor-mediated pathways including Nod1, Nod2, Myd88 and the NLPR3 inflammasome. During mouse sepsis, augmentation depends on liver resident macrophages (Kupffer cells, KC), that capture and internalise both pathogen and ‘proinfectious agent’, leading to reduced production of reactive oxygen species, pathogen survival and subsequent multiple liver abscess formation. The augmented infection model more closely resembles the natural situation and establishes the role of resident environmental microflora in initiation of disease by an invading pathogen. As human microflora is ubiquitous2 its role in increasing susceptibility to infection S. aureus highlights potential strategies for disease prevention.
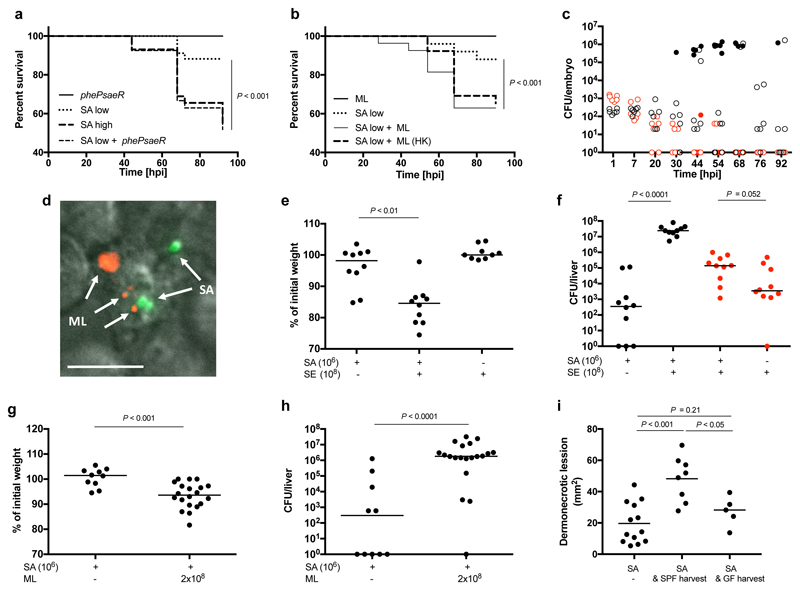
Whilst S. aureus exists as part of a heterogeneous resident microflora3, it often emerges as an invasive human pathogen, capable of in vivo persistence and dissemination4. The surrounding commensal community is protective in some contexts5,6, while in others coinfection can be mutually beneficial for pathogen and commensal7. During pathogenesis, the population of S. aureus expands clonally, as individual organisms within the original infecting cohort found the characteristic abscesses8,9. As animal host mortality is dose dependent10, most of the inoculum does not directly contribute to disease. We hypothesised that virulence might be enhanced by co-inoculation with non-infectious organisms. To test this, we first used the established zebrafish embryo infection model10 and demonstrated that a virulence attenuated mutant (pheP saeR; deficient in an amino acid permease and a global regulator of virulence factors10,11) of S. aureus SH1000 is able to augment infection caused by low dose of a virulent strain (Fig. 1a). Only the low dose virulent organism substantially benefits (Supplementary Data Fig. 1a). To test if unrelated non-pathogenic organisms can similarly augment infection, the skin commensal M. luteus was co-injected with S. aureus (Fig. 1b). M. luteus alone does not cause disease and is swiftly eliminated from the host when injected alone or in combination with S. aureus (Fig. 1c, Supplementary Data Fig. 1b). However, its presence significantly enhances S. aureus virulence leading to host mortality and pathogen proliferation. Both pathogen and commensal can be found co-localised in phagocytes in vivo (Fig. 1d), characteristic of S. aureus infection dynamics in this model.
Figure 1. S. aureus virulence is augmented by live commensal flora.
a, Survival curves of fish injected with low dose S. aureus SH1000 (150 CFU, SA low) and/or S. aureus SH1000 phePsaeR (1350 CFU). S. aureus SH1000 high dose (1500 CFU, SA high) was injected as a positive control. Data are representative of three independent experiments; n ≥ 28, log-rank (Mantel-Cox) test. b, Survival of fish injected with low dose S. aureus SH1000 (150 CFU, SA low) with or without live or heat killed (HK) M. luteus (2000 CFU, ML). Data are representative of three independent experiments; n ≥ 28, log-rank (Mantel-Cox) test. c, Growth of bacteria within embryos after co-injection with M. luteus (2000 CFU) and S. aureus SH1000 (150 CFU). Open circles, live and filled circles, dead embryos, M. luteus (red), S. aureus (black) CFU in each fish. n ≥ 60. d, In vivo imaging of pHrodo (red) labelled M. luteus (2000 CFU, ML indicated by arrows) and S. aureus SH1000-GFP (150 CFU, SA indicated by arrows) 2 hpi. Within the zebrafish circulation valley, phagocytes were viewed at x 60 magnification). Images are representative of 5 embryos from two independent experiments. Scale bar 10 μm. e,f, Co-injection of live 1x108 CFU S. epidermidis (SE) and low dose (1x106 CFU) S. aureus NEWHG into mice (SA) with weight loss (e) and liver CFU (f) recorded (S. aureus, black; S. epidermidis, red). n = 10 per group; median value shown, Mann-Whitney two-sided test. g,h, Co-injection of live M. luteus (ML, 2x108 CFU) and low dose S. aureus NEWHG (SA, 1x106 CFU) into mice with weight loss (g) and liver CFU (S. aureus) (h) recorded. n = 10-20 per group; median value shown, Mann-Whitney two-sided test. i, Dermonecrotic lesion size for C57BL/6J mice injected (on the left flank) with S. aureus NewHG (SA, 107 CFU, n= 13) or co-injected with S. aureus NewHG 107 CFU and either isolated skin commensals from SPF mice (SA & SPF harvest, n= 8) or skin commensals from GF mice (SA & GF harvest, n=5). Median value shown, one-way ANOVA with Tukey post-test.
S. aureus infection of humans is often iatrogenic, resulting in co-inoculation of skin (or other) microflora. The ability of human skin commensal organisms to augment S. aureus mammalian infection was next tested. 1-2 x 108 CFU S. epidermidis or M. luteus led to augmentation (Fig. 1e-h). Survival of S. epidermidis was not enhanced by S. aureus, and M. luteus was completely cleared (Fig. 1f). 1 x 108 CFU M. luteus could augment as low as 1 x 105 CFU S. aureus (Supplementary data Fig. 1c). Combining 1 x 106 CFU of both S. aureus and M. luteus gave a significant increase in S. aureus liver CFU (Supplementary data Fig. 1d). Live commensal flora, whilst able to augment infection are cleared by the host, likely because they do not have the multiple mechanisms that enable S. aureus to avoid killing by the innate immune system12,13. The number of commensal bacteria necessary to augment S. aureus infection is comparable to that found on the skin, where punch biopsies have demonstrated at least 106 CFU/cm2 14. Also in a study of vascular catheters, a range of bacteria were found with numbers up to 107 CFU15. However, it was important to demonstrate the ability of the natural mix of mammalian skin microflora to augment pathogenesis. Thus, skin-associated, microbiota containing material from either naturally colonised or GF mice was harvested and used directly to augment S. aureus infection. Pathogenesis of S. aureus could be augmented by material from mice colonised with native microflora, whereas material from GF mice could not (Fig. 1i, P<0.05). This demonstrates that native flora has the capacity to augment S. aureus infection. Given the varied molecular moieties that can augment infection, we have named them “proinfectious agents”.
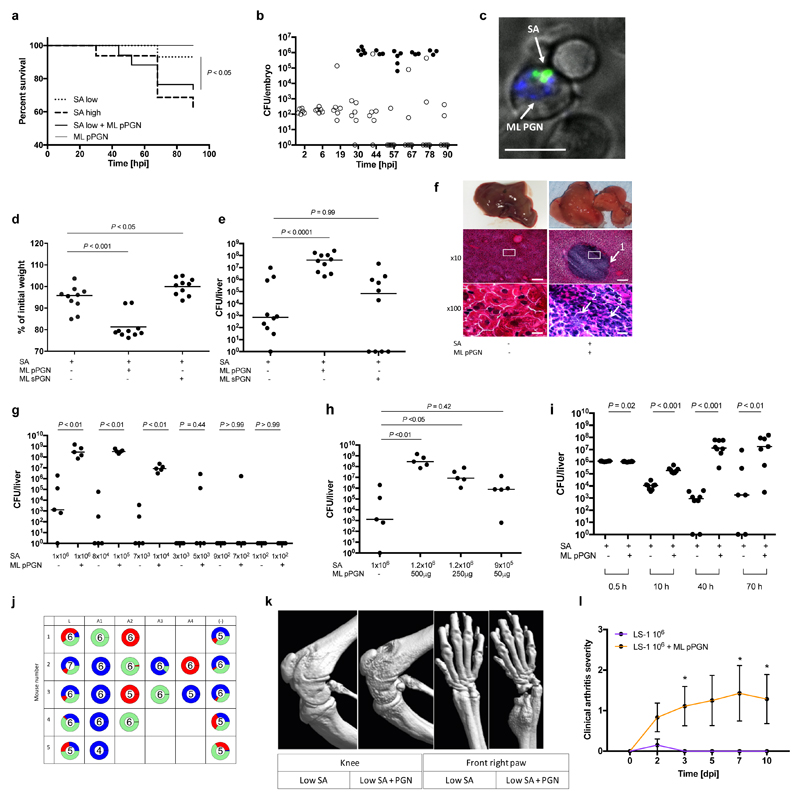
To determine the molecular basis of “proinfectious agents” we first established that heat killed M. luteus can augment zebrafish infection, strongly suggesting a bacterial cellular component may be responsible (Fig. 1b). Peptidoglycan (PGN) is a bacterial cell wall polymer, known to have many host immune system interactions16,17. Particulate, but not soluble, M. luteus PGN can augment infection and is co-localised within phagocytes with S. aureus (Fig. 2a, b, c, Supplementary Data Fig. 1e). Latex beads are also co-phagocytosed but do not alter infection dynamics or outcome (Supplementary Data Fig. 1f-i, Supplementary video 5), demonstrating this is not a simple niche-filling phenomenon. PGN as a proinfectious agent was then tested in the murine sepsis model of infection, where a mixed inoculum consisting of S. aureus NewHG (low dose, 1x106 CFU) and M. luteus PGN (500 μg) were injected intravenously, compared to each component alone. PGN alone had no effect on animal weight or health status (Supplementary Fig. 2a). Mice receiving the mixed inoculum lost significantly more weight than low dose controls (P<0.001), with exceedingly high S. aureus numbers (around 108 CFU) recovered from livers (Fig. 2d, e). Only mixed inocula caused severe structural deterioration of liver parenchyma including multiple, small abscesses (Fig. 2f). At 72 hours post infection (hpi), in the presence of PGN, classical abscesses were formed where a central extracellular nidus of S. aureus is surrounded by a dense neutrophilic infiltrate. Solubilised PGN and latex beads did not augment infection (Fig. 2d, e, Supplementary Data Fig. 2b-d). PGN augmented infection with the community acquired MRSA strain JE2 (Supplementary Data Fig. 2e-g), leading to increased kidney CFU and weight loss. As the augmentation phenomenon has been demonstrated with three distinct strains, ranging from laboratory to emergent clinical epidemic strains18, and including both methicillin sensitive (MSSA) and methicillin resistant strains (MRSA), it is inferred that strain specificity does not play a significant role.
Figure 2. Gram-positive PGN augments S. aureus pathogenesis in animal models.
a, Survival curves of fish injected with low dose S. aureus SH1000 (150 CFU, SA low) and 5 ng polymeric M. luteus PGN (ML pPGN). S. aureus SH1000 high dose (1500 CFU, SA high) was injected as a positive control. Data are representative of three independent experiments; n ≥ 28, log-rank (Mantel-Cox) test. b, Growth of bacteria within embryos after co-injection with low dose S. aureus SH1000 (150 CFU) and 5 ng M. luteus PGN. Open circles, live and filled circles, dead embryos. n ≥ 60. c, In vivo imaging of Alexafluor 647 (blue) labelled M. luteus PGN (5 ng, ML PGN indicated by arrow) and S. aureus SH1000-GFP (150 CFU, SA indicated by arrow) 2 hpi. Within the zebrafish circulation valley, phagocytes were viewed at x 60 magnification). Images are representative of 5 embryos from two independent experiments. Scale bar 10 μm. d, e, BALB/c mice were injected i.v. with low dose (1x106 CFU) S. aureus NEWHGkan with or without 500 μg M. luteus particulate PGN (pPGN) or soluble PGN (sPGN). Weight loss (d) and liver (e) CFU were measured. n = 10 per group; median value shown, Mann-Whitney two-sided test. f, Representative images of histopathological changes during infection. Arrows show 1, large abscess within liver parenchyma; 2, accumulation of extracellular S. aureus; 3, dense infiltrate of polymorphonuclear leukocytes (PMNs). Inset box at x10 magnification (scale bar 100 μm) is displayed at x100 (scale bar 10 μm) in bottom panels. n = 5 per group. g, Liver CFU recovered from BALB/c mice injected i.v. with a decreasing dose of S. aureus NEWHGkan with or without 500 μg M. luteus pPGN. n = 5 per group; median value shown, Mann-Whitney two-sided test. h, Liver CFU recovered from BALB/c mice injected i.v. with low dose (1x106 CFU) S. aureus NEWHGkan with or without a decreasing dose of M. luteus pPGN. n = 5 per group; median value shown, Mann-Whitney two-sided test. i, Liver CFU at various time points after co-injection of low dose (1x106 CFU) S. aureus with or without 500 μg M. luteus pPGN. n = 8 per group; median value shown, Mann-Whitney two-sided test. j, Livers from mice injected with low dose (1x106 CFU; 1:1:1 mixture of NewHG EryR, TetR or KanR, n = 5 per group S. aureus NEWHGkan plus 500 μg M. luteus PGN were harvested. Individual abscesses were dissected and bacterial CFU enumeration from each abscess was determined (A1-A4). Bacterial CFUs from residual liver tissue post dissection (-) was also enumerated and added to the abscess CFUs to provide a total CFU count for each liver (L). k, Micro-CT imaging of knee and front right paw of an NMRI mouse injected i.v. with low dose (1x106 CFU) S. aureus LS-1 with or without 1 mg M. luteus pPGN. Images are representative of 10 animals. l, Clinical arthritis severity of NMRI mice injected i.v. with S. aureus LS-1 low dose (1x106 CFU) and 1 mg M. luteus pPGN. dpi, days post infection. n = 10 per group; error bars, mean and s.e.m, Mann-Whitney two-sided test. * P < 0.05.
Large inocula are required to reliably establish infection in S. aureus murine models, with 107-8 CFU injected as standard19–21. It is improbable that such large doses are mirrored in human infection and early work notes that ‘a nasal droplet of 100 μm diameter could not accommodate this number, even if it consisted entirely of staphylococci22. However, the S. aureus infectious dose can be drastically reduced when augmented with PGN. Significant weight loss occurred with a dose of S. aureus as low as 1x105 CFU in the co-inoculum (Supplementary Data Fig. 2h) and strikingly, high liver bacterial numbers were recovered from all mice receiving 1x104 CFU (Fig. 2g). Astonishingly, one mouse exhibited a liver burden of 106 CFU with an inoculum of only 700 CFU. A lower PGN dose of 250 μg also augmented (Fig 2h, Supplementary Data Fig. 2a).
To determine how augmentation enhances disease outcome, S. aureus population dynamics during infection were evaluated. We have previously identified a phagocyte-dependent immunological bottleneck, from which clonal expansion of a small number of bacteria results in characteristic kidney abscesses8. Mice were injected with three marked but otherwise isogenic, S. aureus strains in a 1:1:1 ratio totaling 1 x 106 CFU. 30 minutes post-infection, regardless of PGN addition, the majority of the CFU were in the liver and without PGN, bacterial numbers subsequently declined (Fig. 2i). As infection progressed there were significantly more S. aureus in the liver, kidneys and spleens of mice receiving mixed inocula. To understand clonal expansion in this context, we assessed contribution to the final bacterial load of the three marked strains, in each organ. By 70 hpi, dominance by individual or pairs of strains indicated clonal expansion in kidneys, but less so in the liver (Fig. 2j, Supplementary Data Fig. 2k,l). However, careful dissection and bacterial enumeration of individual liver abscesses showed these were clonal (Fig. 2j, P<0.001).
To determine the molecular basis for infection augmentation, PGNs from a range of species including Staphylococcus epidermidis, Curtobacterium flaccumfaciens, Bacillus subtilis and S. aureus strains were used. Infection could be augmented in the murine sepsis model by PGN from all species tested (Supplementary Data Fig. 3a-l), having a diversity of amino acids in the peptide side chain, suggesting the conserved glycan moiety is important. C. flaccumfaciens has unusual PGN for a Gram-positive organism as it contains glycine as the first peptide in the side chain as opposed to L-alanine. Bacillus subtilis PGN contains meso-diaminopimelic acid (m-DAP) at stem peptide position 3, an amino acid commonly found in the PGN of Gram-negative bacteria23. PGN from S. epidermidis however is similar to that of S. aureus, the only difference being altered composition of the crosslinking side chains. The presence of wall teichoic acids on PGN (i.e. the PGN was not HF treated) did not alter augmentation (Supplementary Data Fig. 3m-o). S. aureus lipoproteins are immunostimulatory via TLR224, however PGN from a lipoprotein deficient mutant (lipoprotein diacylglyceryl transferase, lgt) could still augment pathogenesis (Supplementary Data Fig. 3p–r). Furthermore, solubilisation of PGN abrogates augmentation eliminating a contaminating moiety within the preparations as the mechanism of augmentation. PGN can also augment infection in other murine models. Using S. aureus LS-1 and NMRI mice, M. luteus PGN caused increased severity in both septic arthritis (Fig. 2k, l) and subcutaneous abscess models (Supplementary Data Fig. 4a,b) in which PGN alone had no effect.
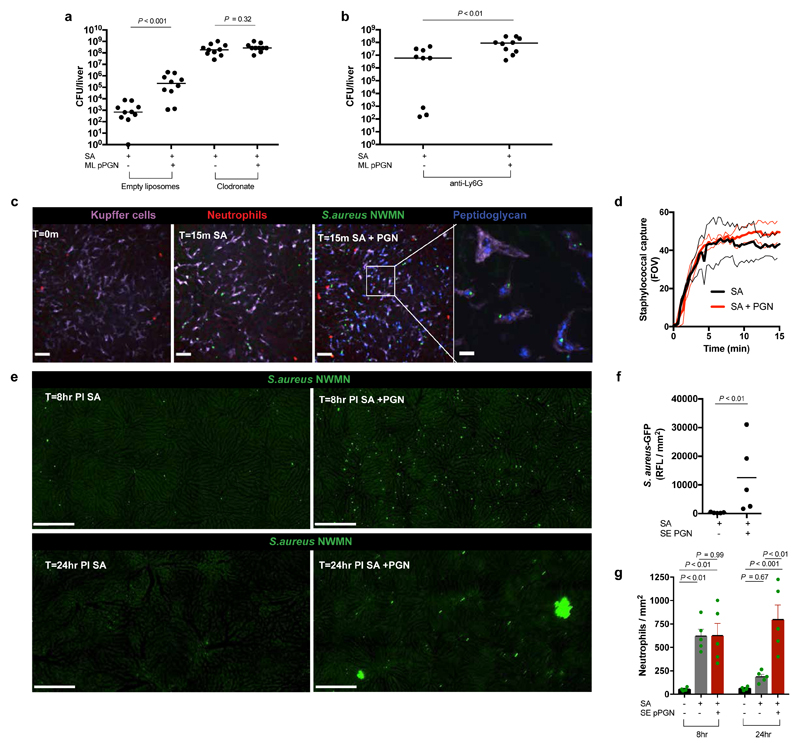
Augmentation circumvents the immune bottleneck during pathogenesis which we have hypothesised occurs inside phagocytes8,9 (Fig. 2c). Therefore, we depleted either neutrophils or macrophages (and macrophage-like cells) prior to challenge in the murine sepsis model. Depletions led to an expected increased susceptibility to S. aureus, thus requiring a reduced inoculum of 1x105 CFU. Macrophage depletion using clodronate liposomes resulted in multiple, small liver abscesses reminiscent of PGN augmentation. However, addition of PGN to the inoculum did not lead to augmentation suggesting a critical role for macrophages (or related cells) in augmentation of pathogenesis (Fig. 3a). Conversely, infection of neutropenic mice was still PGN augmented (Fig. 3b) and this is in agreement with in vitro data where co-incubation of S. aureus and PGN with human derived neutrophils did not promote survival of S. aureus compared to bacteria only controls (Supplementary Data Fig. 4c). However, survival of intracellular S. aureus in human monocyte derived macrophages (MDMs) was increased in the presence of PGN (Supplementary Data Fig. 4d; P<0.01), demonstrating a potential human relevance for our findings.
Figure 3. Kupffer cells are key mediators of augmentation.
a, Liver CFUs of BALB/c mice injected i.v. with low dose (1x105 CFU) S. aureus NEWHG with or without 500 μg M. luteus PGN post treatment with empty liposomes or clodronate. n = 10 per group; median value shown, Mann-Whitney two-sided test. b, Liver CFUs of BALB/c mice injected i.v. with low dose (5x105 CFU) S. aureus NEWHG with or without 500 μg M. luteus PGN post treatment with anti-Ly6G. n = 9-10 per group; median value shown, Mann-Whitney two-sided test. c, Representative SD-IVM images of liver neutrophils (Ly6g; red) and Kupffer cells (KC) (F4/80; purple) at baseline or after i.v. injection of S. aureus (5x107 CFU, BSG1; green) or S. aureus plus S. epidermidis PGN (PGN-AF647; blue) at 15 min in female C57BL/6J mice; scale bars 50 μm. Insert shows higher magnification image of KCs with internalized S. epidermidis PGN and S. aureus. Scale bar 10 μm. n = 5 per group. d, Quantification of SD-IVM images of S. aureus (5x107 CFU, BSG1) catching by KC in the livers of female C57BL/6J mice with (red) and without (black) co-injection of 500 μg S. epidermidis PGN (FOV – field of view); n = 4; thin lines, mean and s.e.m. e, Representative stitched SD-IVM images of mouse livers at 8 h (infected with S. aureus, 107 CFU, BSG1) or 24 h (infected with S. aureus 106 CFU, BSG1) with and without co-injection of 500 μg S. epidermidis PGN in male C57BL/6J; scale bar 250 μm; n = 5. f, Quantification of 2 mm2 stitched SD-IVM images for GFP-fluorescence (S. aureus, BSG1) in murine livers, assessed at 24 h post i.v. injection of S. aureus with or without S. epidermidis PGN. n = 5 per group; mean value shown, Mann-Whitney two-sided test. g, Quantification of 2mm2 stitched SD-IVM images for TdTomato fluorescence (Neutrophils) assessed at 8 h post i.v. injection with S. aureus (107 CFU, BSG1) or at 24 h post i.v. injection with S. aureus (106 CFU, BSG1) with and without co-injection of S epidermidis PGN in Catchup mice. n = 4-5 per group. Error bars, mean with s.e.m. Tukey’s multiple comparisons test applied.
Augmentation of infection by PGN leads to liver abscesses, so to decipher organ and cellular level mechanisms, we employed spinning-disk intravital microscopy (SD-IVM) to visualise Kupffer cells (KC; liver-resident macrophages), and other innate immune cells. Polymorphonuclear neutrophils (PMNs) are crucial for S. aureus control by the host in both animal models25,26 and humans27. Additionally, an important role for KC in capturing and eliminating S. aureus during bacteremia has been recently described19,28. Fluorescently labelled PGN and S. aureus-GFP were co-injected into C57BL/6J mice and both were rapidly engulfed by KC (purple), but not neutrophils (red) (Fig. 3c, Video 1 and 2). The rate of S. aureus capture by KC was not perturbed by PGN augmentation (Fig. 3d). However, at 8 hpi mice receiving mixed inocula had significantly more S. aureus-GFP within the liver than controls and by 24 hpi they contained small, multi-lobar focal abscesses (Fig. 3e), concomitant with an elevated S. aureus burden (Fig. 3f). Augmentation with PGN did not affect neutrophil recruitment at 8 hpi (Fig. 3g), but by 24 hpi there was a significant increase (Fig. 3g) consistent with abscess formation (Figs. 2f and 3e). These data suggest that a mixed inoculum leads to inadequate control of S. aureus inside KC.
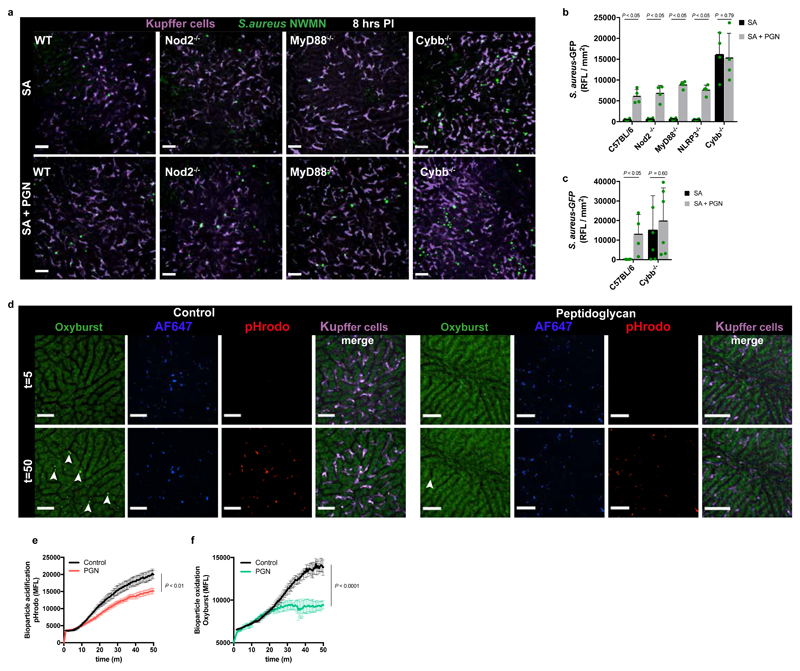
It has been previously shown that during natural colonisation of mice an immune tolerance to S. aureus infection occurs, mediated by Nod129. To test whether prior exposure to commensal organisms affects augmentation, we tested the phenomenon in germ-free (GF) mice, which are incidentally more susceptible to S. aureus30. Using SD-IVM it was observed that the rate of staphylococcal capture by KC is comparable to that seen in wildtype mice and importantly, S. aureus pathogenesis is augmented by particulate PGN (Supplementary Data Fig. 4e-g).
Host PGN recognition has been attributed to TLR2 receptors, however this is now known to be due to lipoprotein contamination31. We demonstrated no role for MyD88 dependent signaling in augmentation and also ruled out the cytosolic PGN receptor Nod2 and the NLRP3 inflammasome (Fig. 4a,b). Furthermore, the range of PGN structures able to augment precluded a role for Nod132 and the NLRP3 inflammasome33 (Supplementary Data Fig. 3a-l). However, Cybb-/- mice, missing the NADPH oxidase34, required by phagocytes to produce reactive oxygen species (ROS), showed lack of augmentation at 8 hpi (Fig. 4b). Cybb-/- mice are highly susceptible to S. aureus infection (Fig. 4b), not surviving until 24 hpi. At a lower inoculum of 105 CFU, 8 hpi imaging was not possible due to low fluorescence levels, but at 24 hpi, augmentation was still not observed (Fig. 4c) confirming the likely mechanistic involvement of ROS. Augmentation with PGN led to greatly diminished oxidation (P<0.0001) and to less acidification (P<0.01) of the phagolysosomes containing S. aureus in KC (Fig. 4d-f, Video 3 and 4), highlighting the critical role of ROS.
Figure 4. Reduced oxidative burst in KCs permits augmentation of S. aureus virulence.
a,b, Representative SD-IVM image of mouse livers (a) or quantification of 2 mm2 stitched SD-IVM images for GFP-fluorescence, scale bar 50 μm (b) at 8 h after i.v. infection with S. aureus BSG1 (SA, 107 CFU) with and without co-injection of 500 μg PGN in male C57BL/6J, Nod2-/-, MyD88-/-, NLRP3-/- or Cybb-/- mice, n = 4 per group; mean shown, error bar s.d., unpaired t-test two-tailed. c, Quantification of 2 mm2 stitched SD-IVM images for GFP-fluorescence at 24 h after i.v. infection with S. aureus BSG1 (SA, 105 CFU) with and without co-injection of 500 μg PGN in male C57BL/6J or Cybb-/- mice, n = 4 per group, mean shown, error bar s.d., unpaired t-test two-tailed. d, SD-IVM image of mouse livers injected with pH-rodo S. aureus bioparticles (red) additionally labelled with AF647 (blue) as a reference fluorophore and OxyBURST (green) with and without (control) co-injection of 500 μg PGN at 5 and 50 min post infection, scale bar 50 μm. Arrows point to oxidized bioparticles. n = 3 per group. e,f, Quantification of intracellular acidification of pH-rodo (e) or oxidation of OxyBURST labelled S. aureus bioparticles (f) in KC over time in C57BL/6J mice with and without (control) co-injection of 500 μg PGN. Data represent the mean fluorescence of S. aureus bioparticles compiled from five separate FOV per time point, n = 3 per group, error bars, s.e.m, two-way ANOVA.
A characteristic feature of many S. aureus infection models is a high inoculum. Here we have established that the majority of the infecting material can be commensal bacteria or even cell wall peptidoglycan. This has important implications for infection prevention where both the pathogen and other organisms or material, previously thought innocuous, need to be considered. Deciphering the cellular and molecular mechanisms involved will allow exploitation for development of novel interventions17,35,36. Potentially S. aureus responds to augmenting material resulting in an increased capability of the pathogen to initiate infection. P. aeruginosa is known to respond to PGN to enhance its virulence37. As well as at the initiation of infection, augmentation could occur during the action of antibiotics, where death of a proportion of the bacterial population may give rise to cell wall fragments. Also, indwelling medical devices, such as intravascular catheters, reside in-situ for several days where the prosthetic material can become colonised by commensal flora15. As catheters are regularly accessed, both commensal flora (e.g. M. luteus and S. epidermidis) and pathogen (S. aureus) could be flushed into the bloodstream simultaneously. Our work establishes a precedent for how a human pathogen can initiate disease using proinfectious agents, as microbial crowdsourcing to circumvent immune system control. In clinical practice, infection by all bacterial pathogens occurs from within a microflora and is therefore initially polymicrobial. This raises the likelihood of a more general role for proinfectious agents, requiring revision of existing models of bacterial pathogenesis and highlighting the involvement of commensal organisms as unwitting accomplices in infection initiation.
Methods
Bacterial strains and culture conditions
S. aureus, M. luteus, C. flaccumfaciens and S. epidermidis strains (Table 1) were grown using brain heart infusion (BHI) liquid or solid medium (Oxoid) at 37°C with the exception of M. luteus which was grown at 30°C. Bacillus sp. strains were grown using nutrient agar liquid or solid medium (Oxoid) at 37°C. Supplementation with the following antibiotics was added where appropriate: kanamycin 50 μg/ml, tetracycline 5 μg/ml or erythromycin 5 μg/ml plus lincomycin 25 μg/ml (Sigma-Aldrich). To distinguish bacterial populations in mixed inocula experiments, M. luteus was serially passaged on BHI media with or without rifampicin (0.03 μg ml-1) and incubated at 30°C until a rifampicin resistant derivative was identified. The same was conducted for S. epidermidis. For all murine experiments, pre-grown batches of bacteria were thawed, washed and diluted to the desired concentration in endotoxin-free PBS (Sigma). Staining of bacterial cells for microscopy was carried out as described for PGN below. SH1000 mCherry and SH1000 GFP were constructed as follows. The pMV158mCherry plasmid was constructed by introducing a gene encoding mCherry (Uniprot: X5DSL3-1) into pMV158GFP48 and replacing the existing GFP gene. The plasmids, pMV158GFP and pMV158mCherry, were then introduced into S. aureus RN4220 by electroporation, resulting in transformants expressing GFP and mCherry, respectively. The plasmids were subsequently transferred into S. aureus SH1000 by Φ11 transduction and the obtained transductants were verified by fluorescence microscopy. To create an avirulent S. aureus mutant, SH1000pheP11 (deficient in amino acid permease) was transduced into SH1000saeR49 (mutation affecting the two component system required for innate immune evasion) using ϕ 11 phage. The same phage was used to transduce lgt::ermB from S. aureus SA113 background into SH1000.
Table 1. Bacterial species and strains and plasmids used in this study.
| Species | Strain | Description | Reference |
|---|---|---|---|
| Staphylococcus aureus | SH1000 | rsbU+ derivative of S. aureus 8325-4 | 38 |
| Staphylococcus aureus | NewHG | Newman with saeSL allele from strain RN1 | 39 |
| Staphylococcus aureus | Newman | NCTC 8178 | 40 |
| Staphylococcus aureus | BSG 1 | NewHG carrying pCM29-GFP | 41,42 |
| Staphylococcus aureus | BSG 2 | MW2 carrying pCM29-GFP | 42 |
| Staphylococcus aureus | JE2 | USA300 LAC cured of p01 and p03 | 43 |
| Staphylococcus aureus | NewHGery | NewHG lysA::pGM068 (EryR) lysA+ | 8 |
| Staphylococcus aureus | NewHGkan | NewHG lysA::pGM072 (KanR) lysA+ | 8 |
| Staphylococcus aureus | NewHGtet | NewHG lysA::pGM070 (TetR) lysA+ | 8 |
| Staphylococcus aureus | TJ1 | LS1 | 44 |
| Staphylococcus aureus | SH1000 GFP | SH1000 carrying pMV158-GFP | This study |
| Staphylococcus aureus | SH1000 mCherry | SH1000 carrying pMV158-mCherry | This study |
| Staphylococcus aureus | phePsaeR | SH1000 sae::EryR pheP::TetR | This study |
| Staphylococcus aureus | SA113 | lgt::ermB | 31 |
| Staphylococcus aureus | SJF4591 | SH1000lgt::ermB | This study |
| Micrococcus luteus | SJF 256 | ATCC 4698 | Sigma |
| Micrococcus luteus | SJF4393 | ATCC 4698 (RifR) | This study |
| Staphylococcus epidermidis | SJF229 | 138 | 45 |
| Staphylococcus epidermidis | SJF4381 | 138 (RifR) | This study |
| Bacillus subtilis | SJF 1 | 168 | Lab stock |
| Bacillus cereus | SJF 1657 | ATCC 14579 | Lab stock |
| Curtobacterium flaccumfaciens | SJF 449 | Wildtype | Lab stock |
Preparation of latex beads, peptidoglycan and bioparticles
For in vivo latex bead experiments, polystyrene latex beads (1.1 μm, Sigma) were washed in endotoxin free PBS, diluted to the desired concentrations and sonicated for 3 x 30 sec (Soniprep 150, MSE, UK) prior to injection into infection models. To prepare PGN, a mid-exponential phase bacterial culture was prepared in appropriate medium and purification was carried out as previously described50. Endotoxin assay was carried out using Pyrochrome (Associates of Cape Cod Inc.) as per manufacturer’s instructions. No significant difference in endotoxin concentration was detected between enzyme only control and the various solubilised Gram-positive PGN preparations. To solubilise PGN, 250 μg ml-1 mutanolysin (50 mM sodium phosphate buffer, pH 5.5) was added and incubated at 37°C overnight on a rotary shaker (100 rpm). Thereafter, the mixture was heated to 95°C for 5 mins before being centrifuged (13 000 rpm, 8 min) to remove any remaining insoluble material. Succinimidyl esters (Life Technologies) were used to stain PGN. pHrodo (2.5 mM), Fluorescein-5- EX (16.95 mM) or Alexa-Fluor 647 were used as per manufacturer’s protocols. In brief, 5 mg PGN was recovered by centrifugation (13 000 rpm, 2 min) and resuspended in an appropriate volume of PBS pH 9 and 200 μl of the suspension mixed in a microcentrifuge tube with 1 μl of S-ester. After 30 min incubation at 37°C on a rotary shaker (100 rpm), excess dye was removed by sequential washing in PBS pH 8, Tris pH 8.5 and again in PBS pH 8 each followed by mixing, centrifugation and gentle removal of the supernatant. Stained PGN was finally re-suspended in 200 μl PBS pH 7.4. Staphylococcal pHrodo bioparticles (Life Technologies) were additionally labelled with Alexa-Fluor 647 Succinimidyl ester (Life Technologies) and OxyBURST green (H2DCFDA SE) according to manufacturer’s protocols to generate reporter bioparticles for acidification and oxidation. pHrodo red S. aureus BioParticles were labeled at 2 mg ml−1 with 50 μg ml−1 AlexaFluor 647 NHS ester and 100 μg ml−1 OxyBURST in 100 mM bicarbonate, pH 8.3, buffered saline for 30 min at room temperature under vigorous agitation. Activation of OxyBURST was accomplished by adding 250 μl 1.5 M hydroxylamine, pH 8.5, and incubating for 30 min on ice. Labelled BioParticles were washed twice with PBS and checked for labeling efficiency by flow cytometry or injected i.v. into mice.
Phagocyte bacterial challenge and quantification of viable intracellular bacteria
Human blood was obtained from healthy volunteers, with informed consent, in compliance with the guidelines of the South Sheffield Research Ethics Committee (07/Q2305/7).
Neutrophils were purified from anti-coagulated human blood as previously described51. In triplicate and with two biological repeats, approximately 2.5x106 cells/ml were co-incubated at 37°C with S. aureus NewHG bacteria to produce a multiplicity of infection (MOI) of 5 in the presence or absence of M. luteus PGN. At 30 and 90 min post co-culture, extracellular and intracellular CFU was calculated. To measure intracellular CFU, the co-culture was transferred to a microcentrifuge tube, centrifuged at 2000 rpm for 2 mins before the supernatant was removed. The neutrophils were then lysed with 1% (w/v) saponin for 10 mins at RT. Bacterial enumeration was calculated from serial dilutions. At 90 mins, lysostaphin (20 μg ml-1) was added to the remaining co-culture wells for 30 mins to lyse extracellular bacteria for the final 120 minute timepoint.
Monocyte derived macrophages (MDMs) were isolated from peripheral blood mononuclear cells (PBMCs) from healthy donors, as previously described52. PBMCs were isolated by Ficoll Plaque (GE Healthcare) density centrifugation, seeded at 2x106 cells/ml in RPMI 1640 medium with 2 mmol/l L-glutamine (Lonza) supplemented with 10% v/v newborn foetal calf serum (Gibco) in 24 well plates (Corning) with 1 ml/well to achieve approximately 2x105 MDM/ml. After 24 hours, non-adherent cells were removed and adherent cells were cultured in RPMI 1640 medium with 2 mmol/l L-glutamine supplemented with 10% v/v low endotoxin heat inactivated foetal calf serum (Biosera) and used at 14 days. Differentiated MDMs were challenged with S. aureus NewHG at a MOI of 0.5. The bacteria were thawed, washed in PBS and added to the MDMs in fresh media. M. luteus PGN (sonicated for 3 x 30 s bursts) was added to the bacteria-containing media at a concentration of 100 μg/ml. The MDMs were incubated on ice for 1 hour then at 37°C, 5% v/v CO2. Following a 4-h total challenge, infected media was removed and the MDMs washed with ice cold PBS. Residual extracellular bacteria were killed by addition of 100 μg/ml gentamicin in fresh media and incubated for 30 mins, then maintained in media containing 20 μg/ml of gentamicin (Sanofi-Aventis) until the desired time point. MDMs were washed with PBS and incubated with 2% v/v saponin (Sigma) at 37°C for 12 min. PBS was added and cells lysed by scraping and pipetting. Estimation of viable intracellular bacteria was determined by surface viable count6. To confirm complete killing of extracellular bacteria, some wells were fixed with 2% v/v paraformaldehyde before bacterial challenge, then exposed to gentamicin and lysed as described, demonstrating absence of bacteria in lysates. Intracellular killing after initial bacterial challenge was estimated by lysing cells maintained in 20 μg/ml of gentamicin for 0.5h – 3.5h.
Animal experiments
Murine work was carried out according to UK law in the Animals (Scientific Procedures) Act 1986, under Project License PPL 40/3699; approved by the Animal Research Ethical Committee of Gothenburg or approved by the University of Calgary Animal Care Committee (AC12 0162) in compliance with the Canadian Council for Animal Care Guidelines.
Murine models
All mice (Table 2) were housed in designated animal facilities in standard environmental conditions of temperature and light and fed laboratory chow and water ad libitum. For the haematogenous septic arthritis model, S. aureus LS-144 was inoculated i.v. into the tail vein female of NMRI mice (n=10), 6-8 weeks old (Charles River Laboratories, Germany) with 0.2 ml of low dose LS-1 (1x106 CFU), with or without 1 mg M. luteus PGN. Mice were regularly weighed and examined for clinical arthritis by observers blinded to the groups as previously described53. In brief, a clinical scoring system ranging from 0–3 was used for each paw (0- no inflammation; 1- mild visible swelling and/or erythema; 2- moderate swelling and/or erythema; 3- marked swelling and/or erythema). The clinical arthritis severity overall score was constructed by adding the scores from all 4 limbs for each animal. On day 10, mice were sacrificed and limbs were resected for microcomputed tomography (micro-CT) radiological examination of bone erosion.
Table 2. Mouse strains used in this study.
| Strain | Description | Source |
|---|---|---|
| BALB/c | Wildtype | Charles River Laboratories |
| C57BL/6J | Wildtype | The Jackson Laboratory |
| Cybb-/- | Cybb-deficient | The Jackson Laboratory |
| MyD88-/- | MyD88 deficient | The Jackson Laboratory |
| Nod2-/- | Nod2 deficient | The Jackson Laboratory |
| NLRP3-/- | NLRP3 deficient | The Jackson Laboratory |
| Catchup | TdTomato driven from Ly6G-cre | University of Duisburg-Essen46 |
| NMRI | Wildtype | Charles River Laboratories |
| SPF | Wildtype C57BL/6J | Taconic or the Jackson Laboratory |
| Germ-free C57BL/6J | Germ-free wildtype C57BL/6J | Taconic, or The Jackson Laboratory and rederived under Germ-free conditions47 |
For the staphylococcal skin infection model, NMRI mice (n=18) were anaesthetised with ketamine/medetomidine and the dorsum was shaved before subcutaneous (s.c.) injection of one flank with 0.05 ml of S. aureus SH1000 (1x106 CFU/spot) and a mixture of SH1000 (1x106 CFU/spot) with PGN (250 μg/spot) in the other flank. PGN alone did not cause any inflammation or skin lesions. Two observers blinded to the treatment groups measured the lesion size of each mouse with a caliper on day 4. The skin lesion was calculated using the mathematical formula for the area of an ellipse. After sacrificing the mice on day 4, skin was disinfected with 70% v/v ethanol and skin biopsies encompassing the entire infected area were taken with a sterile 8 mm biopsy puncher (Kai Medical, Seki, Japan). Biopsy samples were homogenised (Ultra Turrax T25 homogeniser, Germany) and viable counts of bacteria were assessed54.
For the mouse sepsis model, female BALB/c mice (Charles River Laboratories, UK) or male and female C57BL/6 at 7-8 weeks old (The Jackson Laboratory) were inoculated in the tail vein with 0.1 ml S. aureus (NEWHG or JE2), M. luteus or S. epidermidis either alone (dose range 102 – 107 CFU as indicated) or with PGN (50 μg - 1 mg as indicated) or with PGN only. Mice were monitored and sacrificed at 72 hpi unless otherwise stated or according to experimental design. Mouse organs were individually homogenised in PBS and after serial dilution, plated onto BHI agar supplemented with antibiotics as needed for bacterial number enumeration. Germ-free (GF) and C57BL/6(J) mice were bred and maintained in flexible-film isolators or in individually ventilated cages (IVC) at the Clean Mouse Facility, University of Bern, Switzerland. At the age of 6 weeks these mice were shipped under germ-free conditions to the University of Calgary. Alternatively, GF mice were obtained from Taconic. GF status was routinely monitored by culture-dependent and -independent methods and all mice were independently confirmed to be pathogen-free.
For the dermonecrosis model, mice were left in their cage without changing the bedding material for at least 7 days. Mice were euthanized and skin samples (2cm2) were surgically removed from the abdominal region of specific pathogen free (SPF) C57BL/6J or GF mice. Isolation of skin microorganisms was performed by incubation of the skin samples in PBS containing 0.1% Triton X-100 for 1hr at 37°C at high agitation. Skins were gently massaged against a 100 μm filter with additional PBS-Triton. Isolated microorganisms were centrifuged and washed 3x with PBS and stored on ice until the infection experiment. Skin microbes were enumerated by culturing on TSA blood agar plates for 48 hours at 25°C. 24 hours prior to infection experiments hair from the dorsal region of C57BL/6J mice was removed by shaving and hair removal cream (Nair). The left dorsal flank of each mouse was subcutaneously injected with 107 CFU S. aureus (injection volume 50 μL) and the right dorsal flank received a co-injection of the isolated skin microbes and 107 CFU S. aureus. At 48 hpi mice were euthanized and skin lesions were photographed. Dermonecrotic lesion area was measured using ImageJ.
For SD-IVM experiments, a tail vein catheter was inserted into mice after anesthetization with 200 mg kg−1 ketamine (Bayer Animal Health) and 10 mg kg−1 xylazine (Bimeda-MTC). Surgical preparation of the liver for intravital imaging was performed as previously described55. Mouse body temperature was maintained at 37°C with a heated stage. Image acquisition was performed using an Olympus IX81 inverted microscope, equipped with an Olympus focus drive and a motorized stage (Applied Scientific Instrumentation) and fitted with a motorized objective turret equipped with 4×/0.16 UPLANSAPO, 10×/0.40 UPLANSAPO, and 20×/0.70 UPLANSAPO objective lenses and coupled to a confocal light path (WaveFx; Quorum Technologies) based on a modified Yokogawa CSU-10 head (Yokogawa Electric Corporation). Target cells were visualized using fluorescently stained antibodies or fluorescent reporter bacteria. Typically, KCs and neutrophils were stained by i.v. injection of 2.5 μg anti–F4-80 or 3.5 μg anti-ly6G fluorescent conjugated mAbs. Laser excitation wavelengths 491, 561, 642, and 730 nm (Cobolt) were used in rapid succession, together with the appropriate band-pass filters (Semrock). A back-thinned EMCCD 512 × 512 pixel camera was used for fluorescence detection (Hamamatsu). Volocity software (Perkin Elmer) was used to drive the confocal microscope and for 3D rendering, acquisition, and analysis of images. For quantification of staphylococcal catching or measurements of Bioparticle oxidation and acidification in the liver, five random fields of view (FOV) with 10× objective were selected before injection of bacteria. Fluorescent reporter bacteria or reporter Bioparticles were injected i.v. into mice 1 min after initiation of acquiring background images. Find objects function in Volocity software was used to identify individual captured bacteria or Bioparticles by Kupffer cells (F4/80+ cells in liver) and when appropriate autofluorescent spots were subtracted from the final quantification. For Bioparticles the particles were selected in the AF647 reference channel and an increase in pHrodo (acidification) and OxyBURST (oxidation) fluorescence was quantified in the first hour after infection. Quantification of SD-IVM images; 4 hpi, computer generated stitched images of 2mm2 were generated using the stitched image function in Volocity. Volocity software was also used to quantify relative GFP fluorescence as a measurement of the presence of S. aureus-GFP or TD-tomato for the quantification of neutrophils in the liver after staphylococcal infection. SD-IVM images of uninfected mice were used to determine the background fluorescence and S. aureus-GFP was quantified with the same settings for all mutants and treatments.
Macrophage and Neutrophil depletion
Macrophages were depleted using clodronate liposomes (NvR). The mice were injected i.v. with 1 ml of liposomes per 100 g on day 1 as per manufacturer’s instructions. The mice were then injected with 1x105 CFU of S. aureus NewHG (1:1:1 mixture of NewHG EryR, TetR or KanR) on day 2. Blank liposomes were used as a control. Macrophage depletion was confirmed using histology sections of the liver stained with anti-macrophage antibody (rat anti mouse F4/80, AbD Serotec, catalog number MCA497R). For antibody based neutrophil depletion, in vivo anti-ly6G mouse antibody (1A8, BioXcell, catalog number BE0075-1) was used as per the previously published protocol56. The mice were injected with 1.5 mg/mouse of antibody (200 μl per mouse) on day 1 with the mice being injected with 5x105 CFU S. aureus NewHG (1:1:1 mixture of NewHG EryR, TetR or KanR) on day 2. 100 μl of blood was collected via tail bleeding at the time of S. aureus injection and at the end of the experiment via terminal anaesthesia and heart puncture. The blood samples were mixed with 20 μl of Heparin each and then stained with APC Rat Anti-Mouse Ly-6G antibody (BD biosciences, catalog number 560599) according to the BD bioscience protocol. The samples were then processed using the BD LSRII flow cytometer to confirm neutrophil depletion.
Histological analysis of mouse organs
Carried out within a liquid nitrogen dewar, individual organs were placed in an embedding cube, partially pre-filled with optimal cutting temperature (OCT) medium. Ensuring correct positioning for optimal sectioning, the remaining cube was filled with medium and stored at -80°C. Pre-sectioning, organs were placed at -20°C and 200 μm slices were taken before staining with hematoxylin and eosin (H&E) stain as previously established49.
Zebrafish model
Zebrafish embryos less than 5 days post fertilisation (dpf) are not protected under the Animals (Scientific Procedures) Act 1986 but all zebrafish work was carried out according to the details set out in Project License PPL 40/3574.
For zebrafish experiments, London wild-type (LWT) embryos were incubated in E3 medium at 28°C according to standard protocols57. Embryos were microinjected at 30 hours post fertilization (hpf) into the circulation valley as previously described10. Following injection, embryos were kept individually in 100 μl E3 medium and survival recorded for up to 90 hpi. For bacterial growth experiments in vivo, at various time points, embryos were collected and bacterial numbers enumerated as described for mouse organs above.
For microscopy, live anaesthetised zebrafish were mounted onto 15 mm petri dishes in 1% (w/v) low melting point agarose and E3 solution. Images were acquired using either the TE-2000U microscope (Nikon) with a Hamamatsu Orca-AG camera (objectives used: 4× Nikon Plan Fluor objective NA 0.13 and 60× Nikon Plan Apo oil objective NA 1.4; fluorophores excited with either 488 nm (GFP) or 543 nm (mCherry)) or the UltraVIEW VoX spinning disk confocal microscope (Perkin Elmer) (GFP, mCherry and Alexa Fluor® 647 were excited by the 457-51 nm argon laser, 561 nm sapphire laser and 642 nm diode laser, respectively). Image acquisition and processing were performed with Volocity™ software.
Statistical analysis
Sample sizes were predetermined for mouse and zebrafish experiments based on previous experimental data8,10. The refined murine sepsis model, augmenting S. aureus sepsis with PG, increased reproducibility and decreased the spread of results. Revised power calculations (80% power; 95% confidence) permitted comparisons with 5 animals per group (n=5) for both weight loss (10% difference) and liver CFU (2 log difference). Animal experiments were not blinded but to reduce bias, selected experiments were performed by different team researchers. Mice were randomly selected for experimental or control groups and kept in separate cages throughout the experiment. In instances of unexpected death in animals which had otherwise been showing normal health, animals were excluded from analysis as per pre-established criteria. All statistical tests were appropriate for the type of data obtained. For zebrafish embryo survival experiments, the Kaplan-Meier method was employed. Comparison between survival curves was made using the log-rank (Mantel Cox) test. For bacterial count comparison, skin lesion size or clinical arthritis severity in murine experiments, the Mann-Whitney U and Wilcoxson signed rank tests were used for comparison of unpaired and paired data, respectively. For comparison of two or more independent samples (parametric) a one-way ANOVA was used with Tukey’s multiple comparison test. A two-way repeated measurements ANOVA compared pHrodo and OxyBURST data as shown and the CFU quantification in neutrophils. For macrophage assays, a 2-way ANOVA with Tukey’s multiple comparison post-test was used between the first two time-points. To compare the number of intracellular bacteria without and with PGN at all other time-points in the macrophage assays, a paired t-test was used. Statistical analysis was performed using Prism version 6.0 (GraphPad) and P < 0.05 was considered significant. Individual P values are reported. Multinomial probability was calculated to compare the expected frequency of outcomes with observed outcomes for strain ratios within individual abscesses in murine liver.
Supplementary Material
Acknowledgements
This work was funded by the Wellcome Trust (099957/Z/12/Z, 089981), Innovate UK (27486-188210), the Swedish Research Council (2013-09302), an MRC Programme Grant to SAR (MR/M004864/1), an MRC Grant to SJF (MR/R001111/1) and the University of Sheffield 2022 Futures programme via the Florey Institute. Imaging used the Wolfson Light Microscopy Facility (supported by MRC grant MR/K015753/1). P. Kubes is supported by Alberta Innovates Health Solutions (AIHS), the Canadian Institutes of Health Research (CIHR) and the Canada Research Chairs Program. B.G.J. Surewaard is partially funded by a postdoctoral fellowship of CIHR. We are grateful for the use of the Bateson Centre aquarium, Biological Services Unit, Core Histology Service and the Flow Cytometry Facility at the University of Sheffield. We thank the International Microbiome Centre from the UofC for their assistance. We thank the Bateson Centre aquaria staff for their assistance with zebrafish husbandry; Lynne Prince, Dingyi Yang, Josh Hooker, Fiona Wright and Astrid Hendriks for advice and assistance; Professor Friedrich Götz for kindly sending SA113lgt::ermB and M. Gunzer and A. Hasenberg for providing Ly6G-tdTomato reporter mice.
Footnotes
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information.
The authors declare no competing interests.
Author Contributions
E.B., B.G.J.S., D.S., M.N., Y.F., A.A., A.W., E.J.G.P., P.S., P.M., and T.K.P. performed and analysed the experiments. K.D.M., T.J., D.H.D., J.A.G.S., P.K., S.A.R and S.J.F. contributed to study design and data analysis. E.B. and S.J.F. wrote the manuscript. All authors discussed the results and commented on the manuscript.
References
- 1.Isolauri E, Kirjavainen PV, Salminen S. Probiotics: a role in the treatment of intestinal infection and inflammation? Gut. 2002;50:iii54–iii59. doi: 10.1136/gut.50.suppl_3.iii54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice EA, et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naber CK. Staphylococcus aureus Bacteremia: Epidemiology, Pathophysiology, and Management Strategies. Clin Infect Dis. 2009;48:S231–S237. doi: 10.1086/598189. [DOI] [PubMed] [Google Scholar]
- 5.Naik S, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01230. 1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo C-W, Lai Y-K, Liu Y-T, Gallo RL, Huang C-M. Staphylococcus aureus Hijacks a Skin Commensal to Intensify Its Virulence: Immunization Targeting β-Hemolysin and CAMP Factor. J Invest Dermatol. 2011;131:401–409. doi: 10.1038/jid.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McVicker G, et al. Clonal Expansion during Staphylococcus aureus Infection Dynamics Reveals the Effect of Antibiotic Intervention. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003959. e1003959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prajsnar TK, et al. A privileged intraphagocyte niche is responsible for disseminated infection of Staphylococcus aureus in a zebrafish model. Cell Microbiol. 2012;14:1600–1619. doi: 10.1111/j.1462-5822.2012.01826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prajsnar TK, Cunliffe VT, Foster SJ, Renshaw SA. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell Microbiol. 2008;10:2312–2325. doi: 10.1111/j.1462-5822.2008.01213.x. [DOI] [PubMed] [Google Scholar]
- 11.Horsburgh MJ, Wiltshire MD, Crossley H, Ingham E, Foster SJ. PheP, a Putative Amino Acid Permease of Staphylococcus aureus, Contributes to Survival In Vivo and during Starvation. Infect Immun. 2004;72:3073–3076. doi: 10.1128/IAI.72.5.3073-3076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bera A, Biswas R, Herbert S, Götz F. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect Immun. 2006;74:4598–4604. doi: 10.1128/IAI.00301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng AG, et al. Contribution of Coagulases towards Staphylococcus aureus Disease and Protective Immunity. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grice EA, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherertz RJ, et al. Three-year experience with sonicated vascular catheter cultures in a clinical microbiology laboratory. J Clin Microbiol. 1990;28:76–82. doi: 10.1128/jcm.28.1.76-82.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorbara MT, Philpott DJ. Peptidoglycan: a critical activator of the mammalian immune system during infection and homeostasis. Immunol Rev. 2011;243:40–60. doi: 10.1111/j.1600-065X.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- 17.Wheeler R, Chevalier G, Eberl G, Gomperts Boneca I. The biology of bacterial peptidoglycans and their impact on host immunity and physiology. Cell Microbiol. 2014;16:1014–1023. doi: 10.1111/cmi.12304. [DOI] [PubMed] [Google Scholar]
- 18.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Surewaard BGJ, et al. Identification and treatment of the Staphylococcus aureus reservoir in vivo. J Exp Med. 2016;213:1141–1151. doi: 10.1084/jem.20160334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thammavongsa V, Missiakas DM, Schneewind O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science. 2013;342:863–866. doi: 10.1126/science.1242255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 22.Elek SD, Conen PE. The virulence of Staphylococcus pyogenes for man; a study of the problems of wound infection. Br J Exp Pathol. 1957;38:573–586. [PMC free article] [PubMed] [Google Scholar]
- 23.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto M, et al. Lipoprotein is a predominant Toll-like receptor 2 ligand in Staphylococcus aureus cell wall components. Int Immunol. 2006;18:355–362. doi: 10.1093/intimm/dxh374. [DOI] [PubMed] [Google Scholar]
- 25.Verdrengh M, Tarkowski A. Role of neutrophils in experimental septicemia and septic arthritis induced by Staphylococcus aureus. Infect Immun. 1997;65:2517–2521. doi: 10.1128/iai.65.7.2517-2521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol. 2012;34:237–259. doi: 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heyworth PG, Cross AR, Curnutte JT. Chronic granulomatous disease. Curr Opin Immunol. 2003;15:578–584. doi: 10.1016/s0952-7915(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Z, et al. CRIg Functions as a Macrophage Pattern Recognition Receptor to Directly Bind and Capture Blood-Borne Gram-Positive Bacteria. Cell Host Microbe. 2016;20:99–106. doi: 10.1016/j.chom.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gauguet S, et al. Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect Immun. 2015 doi: 10.1128/IAI.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoll H, Dengjel J, Nerz C, Götz F. Staphylococcus aureus Deficient in Lipidation of Prelipoproteins Is Attenuated in Growth and Immune Activation. Infect Immun. 2005;73:2411–2423. doi: 10.1128/IAI.73.4.2411-2423.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chamaillard M, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 33.Wolf AJ, et al. Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan. Cell. 2016;166:624–636. doi: 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollock JD, et al. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet. 1995;9:202–209. doi: 10.1038/ng0295-202. [DOI] [PubMed] [Google Scholar]
- 35.Boyle JP, Parkhouse R, Monie TP. Insights into the molecular basis of the NOD2 signalling pathway. Open Biol. 2014;4 doi: 10.1098/rsob.140178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 37.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horsburgh MJ, et al. SigmaB Modulates Virulence Determinant Expression and Stress Resistance: Characterization of a Functional rsbU Strain Derived from Staphylococcus aureus 8325-4. J Bacteriol. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mainiero M, et al. Differential Target Gene Activation by the Staphylococcus aureus Two-Component System saeRS. J Bacteriol. 2010;192:613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duthie ES, Lorenz LL. Staphylococcal Coagulase: Mode of Action and Antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 41.Pang YY, et al. agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Surewaard BGJ, et al. Inactivation of Staphylococcal Phenol Soluble Modulins by Serum Lipoprotein Particles. PLOS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002606. e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fey PD, et al. A Genetic Resource for Rapid and Comprehensive Phenotype Screening of Nonessential Staphylococcus aureus Genes. mBio. 2013;4:e00537–12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bremell T, et al. Outbreak of spontaneous staphylococcal arthritis and osteitis in mice. Arthritis Rheum. 1990;33:1739–1744. doi: 10.1002/art.1780331120. [DOI] [PubMed] [Google Scholar]
- 45.Modun BJ, Cockayne A, Finch R, Williams P. The Staphylococcus aureus and Staphylococcus epidermidis transferrin-binding proteins are expressed in vivo during infection. Microbiology. 1998;144:1005–1012. doi: 10.1099/00221287-144-4-1005. [DOI] [PubMed] [Google Scholar]
- 46.Hasenberg A, et al. Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat Methods. 2015;12:445–452. doi: 10.1038/nmeth.3322. [DOI] [PubMed] [Google Scholar]
- 47.Gomez de Agüero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 48.Nieto C, Espinosa M. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the green fluorescent protein. Plasmid. 2003;49:281–285. doi: 10.1016/s0147-619x(03)00020-9. [DOI] [PubMed] [Google Scholar]
- 49.Cheng AG, et al. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner RD, et al. Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nat Commun. 2010;1:26. doi: 10.1038/ncomms1025. [DOI] [PubMed] [Google Scholar]
- 51.Sabroe I, Williams TJ, Hébert CA, Collins PD. Chemoattractant cross-desensitization of the human neutrophil IL-8 receptor involves receptor internalization and differential receptor subtype regulation. J Immunol. 1997;158:1361–1369. [PubMed] [Google Scholar]
- 52.Dockrell DH, Lee M, Lynch DH, Read RC. Immune-mediated phagocytosis and killing of Streptococcus pneumoniae are associated with direct and bystander macrophage apoptosis. J Infect Dis. 2001;184:713–722. doi: 10.1086/323084. [DOI] [PubMed] [Google Scholar]
- 53.Ali A, et al. CTLA4 Immunoglobulin but Not Anti-Tumor Necrosis Factor Therapy Promotes Staphylococcal Septic Arthritis in Mice. J Infect Dis. 2015;212:1308–1316. doi: 10.1093/infdis/jiv212. [DOI] [PubMed] [Google Scholar]
- 54.Kwiecinski J, Jin T, Josefsson E. Surface proteins of Staphylococcus aureus play an important role in experimental skin infection. APMIS. 2014;122:1240–1250. doi: 10.1111/apm.12295. [DOI] [PubMed] [Google Scholar]
- 55.Wong CHY, Jenne CN, Lee W-Y, Léger C, Kubes P. Functional Innervation of Hepatic iNKT Cells Is Immunosuppressive Following Stroke. Science. 2011;334:101–105. doi: 10.1126/science.1210301. [DOI] [PubMed] [Google Scholar]
- 56.Wilson R, et al. Protection against Streptococcus pneumoniae lung infection after nasopharyngeal colonization requires both humoral and cellular immune responses. Mucosal Immunol. 2015;8:627–639. doi: 10.1038/mi.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nusslein-Volhard C, Dahm R. Zebrafish. A Practical Approach. New York: Oxford University Press; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.