Abstract
Background:
Idiopathic scoliosis is a form of spinal deformity that affects 2–3% of children and results in curvature of the spine without structural defects of the vertebral units. The pathogenesis of idiopathic scoliosis remains poorly understood, in part due to the lack of a relevant animal model.
Results:
We performed a forward mutagenesis screen in zebrafish to identify new models for idiopathic scoliosis. We isolated a recessive zebrafish mutant, called skolios, which develops isolated spinal curvature that arises independent of vertebral malformations. Using meiotic mapping and whole genome sequencing, we identified a nonsense mutation in kinesin family member 6 (kif6gw326) unique to skolios mutants. Three additional kif6 frameshift alleles (gw327, gw328, gw329) were generated with transcription activator-like effector nucleases (TALENs). Zebrafish homozygous or compound heterozygous for kif6 frameshift mutations developed a scoliosis phenotype indistinguishable from skolios mutants, confirming that skolios is caused by the loss of kif6. Although kif6 may play a role in cilia, no evidence for cilia dysfunction was seen in kif6gw326 mutants.
Conclusions:
Overall, these findings demonstrate a novel role for kif6 in spinal development and identify a new candidate gene for human idiopathic scoliosis.
Keywords: kinesin, scoliosis, Danio rerio
Introduction
Idiopathic scoliosis is a multifaceted genetic disorder that causes curvature of the spine in 2–3% of humans, with approximately 10% of patients progressing to deformity warranting bracing or surgical treatment (Miller, 1999). Although idiopathic scoliosis is defined by a deviation of the spine from the midline in the medial–lateral plane, curvature is typically complex and causes a three dimensional deformity affecting all three planes (medial– lateral, dorsal–ventral, and transverse). Unlike secondary scoliosis that arises from congenital vertebral malformations or scoliosis arising from other syndromic conditions, idiopathic scoliosis is poorly understood and no obvious structural vertebral abnormalities are present. A major challenge in determining the pathogenesis of idiopathic scoliosis has been the lack of a relevant animal model. Animal models of idiopathic scoliosis have been primarily limited to experimental forms of scoliosis, where scoliosis is induced by invasive surgical procedures, immobilization or the use of the systemic agents (reviewed in Janssen et al., 2011). Because spinal deformity develops secondary to harsh interventions, experimentally induced forms of scoliosis are unlikely to recapitulate many features of human idiopathic scoliosis.
With the exception of humans, naturally occurring scoliosis has been described only rarely in the animal kingdom (Ouellet and Odent, 2013). Humans ambulate in an upright position with their center of gravity over the pelvis, a characteristic that is unique even among other primates (D’Aout et al., 2002). This erect, bipedal posture significantly alters the dorsal shear loads of the spine, which has been proposed as an important prerequisite for scoliosis development (Castelein et al., 2005). In support of this model, pinealectomized rats develop scoliosis when also made bipedal, whereas similarly treated quadrupedal rats do not (Machida et al., 2005). Although pinealectomy and bipedalism may be important prerequisites for scoliosis in some species, pinealectomy in bipedal nonhuman primates does not cause spinal deformity (Cheung et al., 2005). Moreover, pinealectomy in guppy (Gorman et al., 2007) and salmon (Fjelldal et al., 2004). induces spinal curvature, demonstrating that scoliosis induced by pinealectomy does not require bipedalism.
A mutant guppy strain, curveback, is one of the few examples of an animal model with noninduced spinal deformity that parallels human idiopathic scoliosis (Gorman et al., 2007). The curveback mutants develop a spinal curvature sporadically in the dorsal–ventral and medial–lateral plane that is not due to vertebral malformations. Sporadic spinal deformity has also been noted in other teleosts, including medaka and swordtail, suggesting that teleosts may be ideal animal models to identify and characterize naturally occurring scoliosis (reviewed in Gorman and Breden, 2007). Of the teleosts, zebrafish (Danio rerio) are particularly well-suited model organisms, as zebrafish are highly tractable and have extensive genomic resources. The zebrafish mutant leviathan is caused by recessive mutations in collagen type VIII alpha1a (col8a1a) and develops scoliosis (Gray et al., 2014). However, leviathan mutants also develop notochord defects and congenital vertebral malformations and it remains unknown whether zebrafish are susceptible to naturally occurring scoliosis without vertebral malformations. Therefore, we sought to identify novel zebrafish scoliosis mutants that could serve as a model for human idiopathic scoliosis. Using an N-ethyl-N-nitrosourea (ENU) mutagenesis forward genetic screen, we identify and describe skolios, a zebrafish mutant with recessively inherited scoliosis that shares several characteristics with human idiopathic scoliosis.
Results
Identification of skolios Mutant
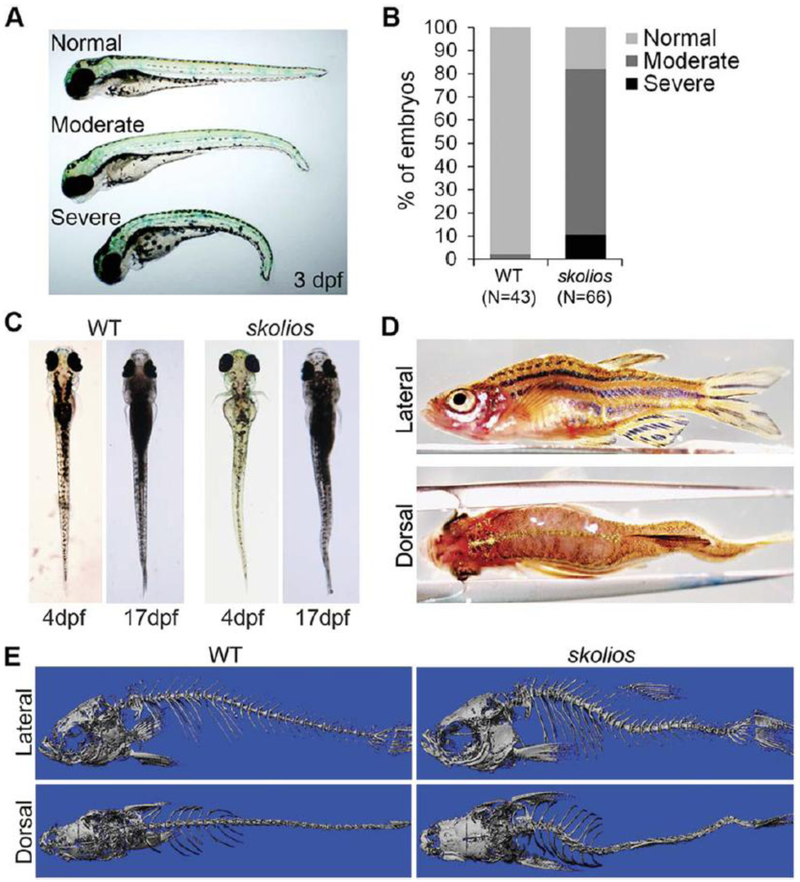
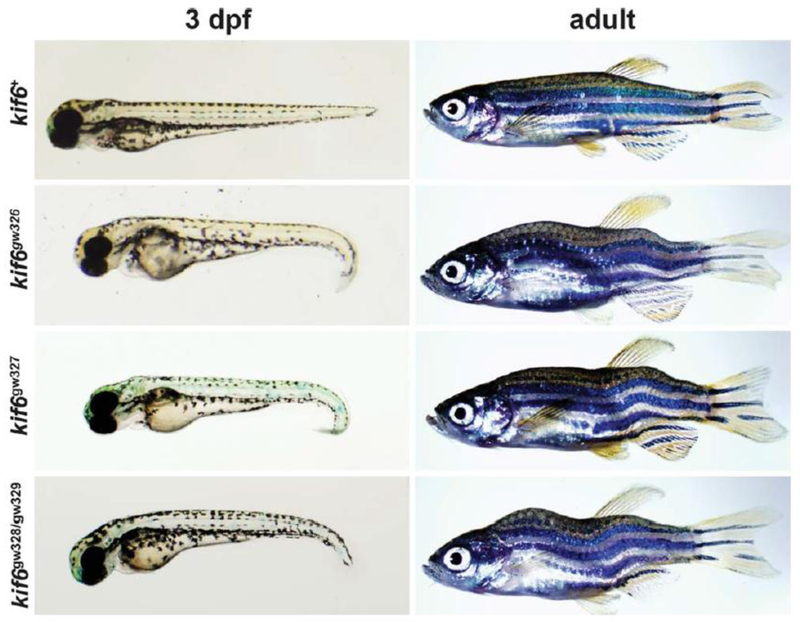
To identify zebrafish mutants with scoliosis, we performed an ENU mutagenesis screen on wild-type (WT) AB zebrafish. From this screen, we identified a new zebrafish mutant, skolios, which develops recessively inherited curvature of the body axis. Body curvature first develops in skolios mutants during embryonic hatching stages (2–3 days postfertilization [dpf]), causing a moderate ventral curvature of the body axis that is frequently limited to the distal end of the tail (Fig. 1A). This early curvature arises without observable defects in the notochord (e.g., kinking) and is incompletely penetrant with variable expressivity. Approximately 20% of homozygous skolios embryos appear normal at 3 dpf, while the majority exhibit either moderate (_70%) or severe (10%) ventral body curvature (Fig. 1A,B). Unlike the incompletely penetrant ventral body curvature first seen during hatching stages, curvature in the medial–lateral plane becomes fully penetrant during early larval stages (4–5 dpf) and progresses through later larval and juvenile stages (5–89 dpf) (Fig. 1C). Although there is large phenotypic variability in the appearance and severity of the curve, body curvature defects are easily observed in both the dorsal–ventral and medial–lateral planes of all adult skolios mutants (Fig. 1D). Despite the severity of the deformity, adult skolios mutants are both viable and fertile. Heterozygous mutants exhibit no overt embryonic or postembryonic phenotype.
Fig. 1.
Skolios mutants develop recessively inherited curvature of the body axis. A: Ventral curvature phenotypes observed in skolios mutant embryos at 3 dpf. B: Relative frequencies of ventral curvature phenotypes in WT and skolios embryos (3 dpf). C: Dorsal view of embryonic (4 dpf) and juvenile (17 dpf) zebrafish showing development of sagittal body curvature in skolios mutants. D: Severe body curvature in both the medial– lateral and dorsal–ventral planes of an adult skolios mutant. E: MicroCT imaging at approximately 50 dpf shows marked curvature of the abdominal
Characterization of Spinal Curvature in skolios Mutants
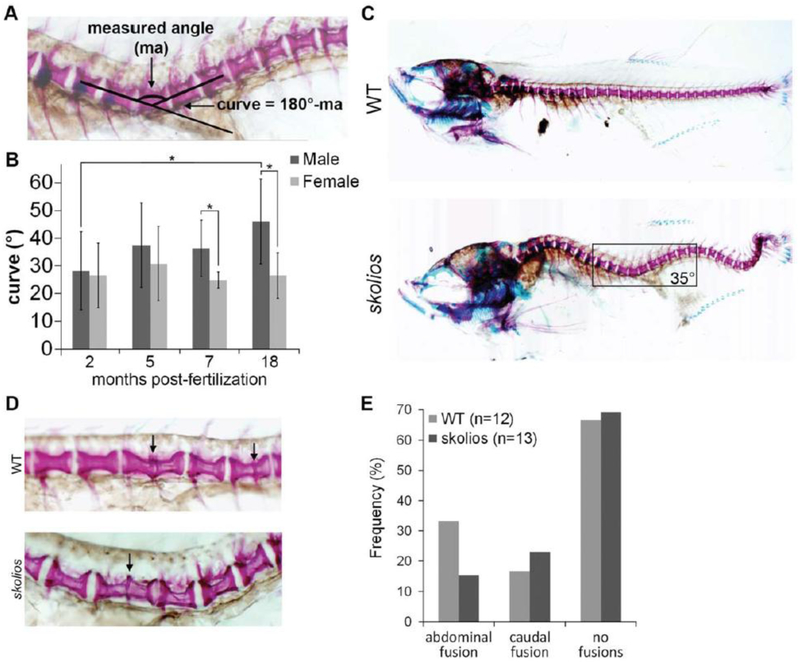
MicroCT imaging of a skolios mutant and WT zebrafish was performed at 50 dpf to determine if vertebral abnormalities underlie the curved body axis in skolios. Compared with WT, the skolios mutant developed a marked curvature of the abdominal and caudal spine in both the dorsal–ventral and medial–lateral planes (Fig. 1E). To evaluate the temporal onset of curvature and to screen for vertebral defects, we performed a two-staining protocol using alcian blue, a positively charged dye that is thought to react with the acidic mucopolysaccharides in cartilage, and alizarin red, a dye that binds to mineralized bone (Walker and Kimmel, 2007). Vertebral morphology was evaluated at several developmental time points (2–18 months postfertilization [mpf]) for WT and skolios mutants. To track the progression of the spinal curve, curves were measured in the dorsal–ventral plane as shown in Figure 2A. Although human idiopathic scoliosis is measured in the medial–lateral plane, individual zebrafish vertebrae were more easily observed when viewed laterally, which allowed for more accurate curve measurements. All curves were measured at the curve apex near the transition from abdominal to caudal vertebrae. At 2 mpf, the average spinal curve was similar in both male and female skolios mutants (Fig. 2B). In males, spinal curvature was progressive and by 18 mpf, the average curve was 46_ (SD.15.3_, N.5), compared with an average of 28_ at 2 mpf (SD.14.1_, N.10) (P.0.043, Student’s t-test). Although the curves were similar at 2 mpf, males developed more severe spinal curves than females at both 7 mpf (36_ vs. 25_, P.0.042, Student’s t-test) and 18 mpf (46_ vs. 27_, P.0.037, Student’s t-test). Female curves were stable and did not change between 2 and 18 mpf. In contrast, WT zebrafish rarely had spinal curvature and if present, were consistently small but variable. WT zebrafish measured at 2 and 7 mpf had an average spinal curve of 5.3_ (SD.6.7_, N.8) in males and 3.4_ (SD.2.1_, N.9) in females (not shown).
Fig. 2.
Spinal curvature in skolios mutants occurs independent of major vertebral abnormalities and is progressive through adult stages in males. A: Method of measuring spinal curve in zebrafish using skeletal histomorphology. Lines were drawn through the middle of the two vertebrae adjacent to the apex of the curve. The measured angle from the intersection was subtracted from 180_ to calculate the spinal curve. B: Spinal curves measured for female skolios mutants at 2 (N.11), 5 (N.9), 7 (N.5) and 18 (N.5) mpf and male skolios mutants at 2 (N.10), 5 (N.8), 7 (N.5) and 18 (N.5) mpf. The average spinal curve is shown for each time point. At 2 mpf, males and females are similarly affected; however, by later stages, males continue to progress in severity. *P<0.05. C: Skeletal histomorphology of a 2 mpf female WT and a skolios mutant with a 35_ spinal curve near the transition from abdominal to caudal vertebrae that developed in the absence of vertebral fusions or other major vertebral abnormalities. Box identifies region shown in A. D: Zebrafish between 2 and 7 mpf were stained with Alizarin red to visualize individual vertebrae. Staining shows vertebral fusions (arrows) identified in WT and skolios mutants. E: The overall frequency of vertebral fusions in skolios mutants is similar to WT. (rib vertebrae) and caudal (vertebrae posterior to ribs) spine in skolios mutants.
Spinal curvature in skolios mutants occurred without an increased frequency of vertebral fusions or malformations (Fig. 2C). Although vertebral fusions and malformations were noted in approximately 30% of all skolios mutants (Fig. 2D), this was comparable to frequencies observed in WT zebrafish (Fig. 2E).
Identification of kinesin family member 6 Nonsense Mutation (kif6gw326) in skolios Mutants
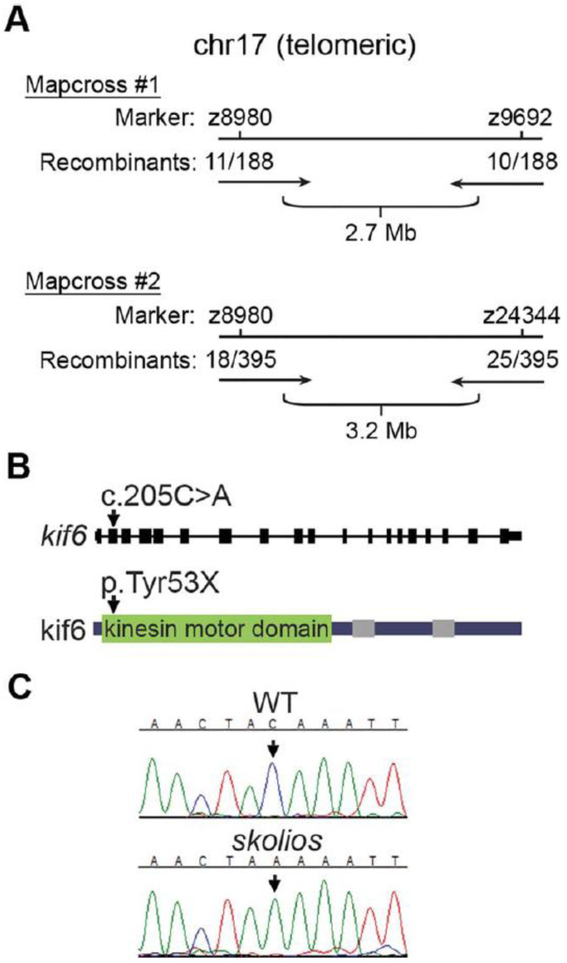
To determine the genetic basis of the skolios mutant phenotype, we performed meiotic mapping, which localized the mutation to a region bounded by markers z8980 on one side and either z9692 or z24344 on the other (z9692 but not z24344 was polymorphic in one WIK fish used for a mapcross, while z24344 but not z9692 was polymorphic in a second WIK fish used for a mapcross) (Fig. 3A). The marker z24344 is not currently listed in Ensembl Zv9 but the sequence can be found in GenBank (G47401.1) and can subsequently be located in Zv9. The smaller region, between z8980 and z9692, mapped the phenotype to a 2.7 Mb locus at chr17:47,500,198–50,216,501 (Zv9).
Fig. 3.
Meiotic mapping and whole genome sequencing identifies a kif6 nonsense mutation (kif6gw326) as candidate for skolios. A: Summary of mapping experiments. Two map-crosses identified a _3 Mb region on chromosome 17 harboring the causative genetic lesion for skolios. B: Gene and protein schematic with corresponding location of the kif6 nonsense mutation (kif6gw326). The C>A mutation in the second exon of kif6 causes a premature stop early in the kif6 protein sequence. Kif6 protein domains are drawn according to the simple modular architecture research tool (SMART) (Schultz et al., 1998). The kinesin motor domain is shown, with grey bars indicating coiled-coil regions. C: Sanger sequencing validation of kif6gw326 (arrow) in skolios.
Although mapping limited the region harboring the mutation responsible for skolios to a 2.7 Mb region on chromosome 17, approximately 20 genes remained within the locus. To simultaneously investigate all genes within the interval, whole genome sequencing was performed on a single skolios mutant. Next generation sequencing reads were aligned to the Zv9 T€ubingen reference genome (Table 1). Nearly three-quarters of the reference genome was covered at _3X, which we deemed sufficient to identify a homozygous base change. In the mapped interval, 18,941 total variants were identified. To eliminate strain-specific polymorphisms resulting from the alignment of skolios (generated on an AB background) to the Zv9 sequence (T€ubingen), variants present in sequence data from an unrelated zebrafish with a similar genetic background (AB) were removed. Of 20 homozygous, nonsynonymous variants, only one mutant allele (gw326) produced a nonsense mutation, caused by a G>T transversion at chr17:48958292 (Zv9) in the second exon of kif6 (encoding kinesin family member 6). The kif6gw326 mutant allele, corresponding to a C>A base change at position 205 in the kif6 mRNA (NM_001077431, kif6 is encoded on the minus strand), is predicted to truncate kif6 in the kinesin motor domain (Tyr53X) (Fig. 3B). This variant was validated in additional skolios mutants using Sanger sequencing (Fig. 3C).
TABLE 1.
Identification of Candidate Gene for skolios Phenotype Using Whole Genome Sequencing
| Sequence and variant identification | |
| 101 bp paired-end reads | 98,289.069 |
| Aligned paired-end reads | 65,99,507 |
| >3X coveragve | 74% |
| Total genomic variants | 8,043,651 |
| Variant filtering | |
| Total variants in 2.7 MB mapped locus | 18.943 |
| And absent in related zebrafish | 4166 |
| With similar genetic background And homozygous |
3156 |
| And causing a nonsynonymous amino Acid change |
20 |
| And causing a nonsense mutation (gene) | 1 (kif6) |
TALEN-induced Mutations in kif6
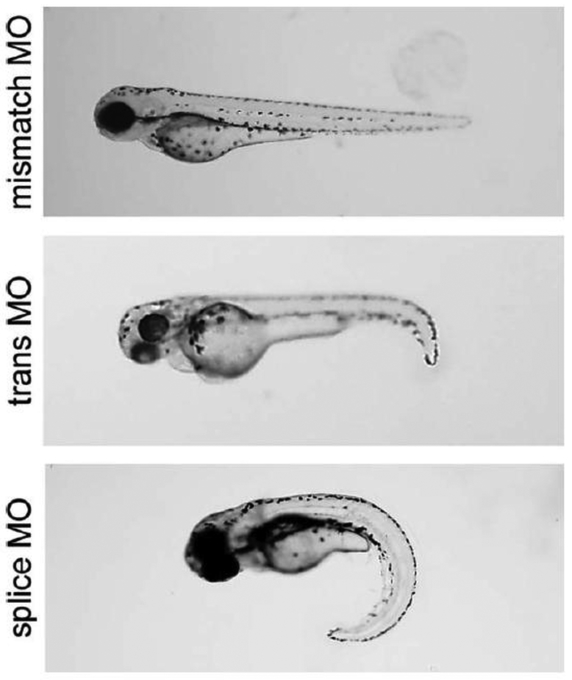
Transient knock-down of kif6 with morpholinos targeting the translation start site or the first exon–intron boundary recapitulated the embryonic curvature defects (Fig. 4); however, kif6 morphants developed into normal adults (not shown). Because the transient nature of morpholinos limits our ability to assess adult phenotypes, we generated stable germline mutations in kif6 using TALENs (transcription activator-like effector nucleases), which have been shown to effectively induce new mutations in zebrafish (Sander et al., 2011).
Fig. 4.
Transient knockdown of kif6 using morpholinos. WT zebrafish were injected with morpholinos targeting either the translation start site or a splice site, at the 1–4 cell stage. A morpholino targeting the kif6 translation start site but containing a 5 base mismatch (mismatch MO) served as a negative control. Both translation start site and splice site morphants recapitulated the early embryonic curvature of skolios.
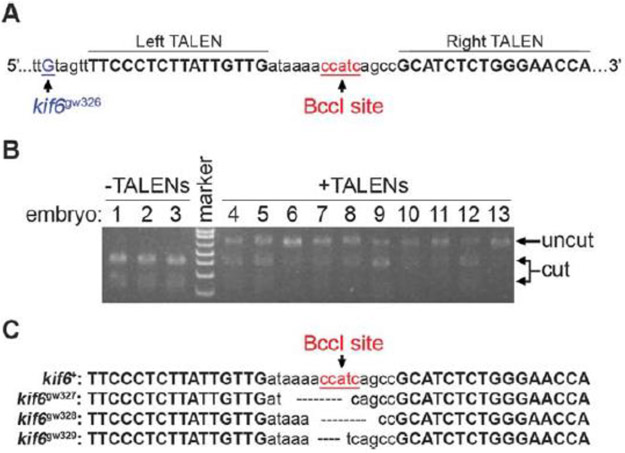
We designed a TALEN pair targeting a region in the second exon of kif6, which flanked a BccI restriction site and was adjacent to kif6gw326 (Fig. 5A). TALENs were combined at equal concentrations and injected into one- to four-cell stage WT embryos. Polymerase chain reaction (PCR) amplification and subsequent digestion with BccI showed that all embryos injected with TALENs had some degree of undigested product, indicating that the BccI restriction site was disrupted by an induced mutation (Fig. 5B). Because the F0 (injected) zebrafish are mosaic, zebrafish were outcrossed with WT adults to create stable lines. Three new mutant alleles were identified in the F1 generation: two 8 bp deletions (gw327 and gw328) and a 4 bp deletion (gw329) (Fig. 5C). Sibling crosses generated F2 zebrafish that were either homozygous or compound heterozygous for TALEN-induced kif6 mutant alleles. All combinations produced zebrafish with curvature of the body axis in both embryonic and adult stages that were indistinguishable from kif6gw326 mutants, providing strong support for kif6 as the genetic cause of skolios (Fig. 6). The curvature phenotype was fully penetrant by larval and adult stages for all allelic combinations.
Fig. 5.
Design and efficiency of kif6 TALENs. A: Genomic sequence showing left and right TALENs that were designed in the second exon of kif6 near the gw326 mutant allele. The TALEN pair flank a BccI restriction site. B: BccI restriction digest of PCR amplified DNA from WT embryos. Embryos 1–3 were not injected with kif6 TALENs and BccI cleaves the PCR product completely. Embryos 4–13 were injected with kif6 TALENs at the 1–4 cell stage and all embryos had varying levels of uncut product, indicating a mutation was induced at the BccI restriction site. C: Three mutations identified in TALEN-injected embryos.
Fig. 6.
TALEN-induced mutations in kif6 recapitulate the skolios phenotype. Zebrafish with TALEN-induced mutations in kif6 (gw327, gw328, gw329) develop both embryonic and adult body axis curvature, similar to kif6gw326 mutants.
Characterization of kif6 Expression in Zebrafish
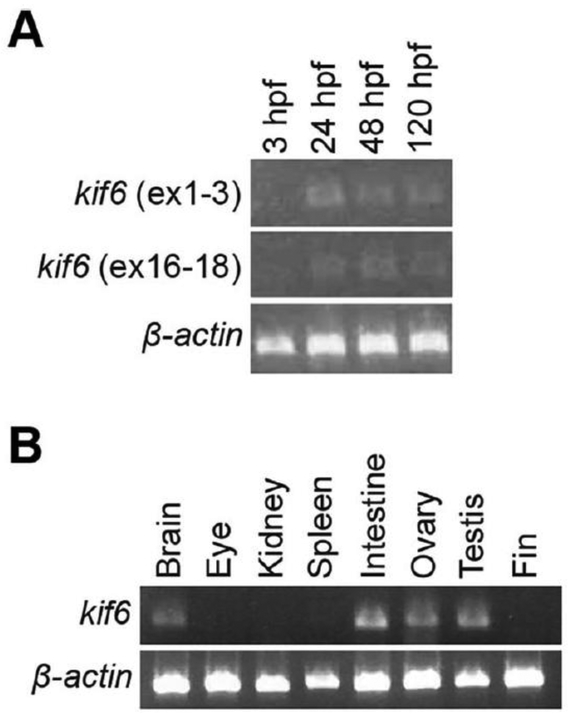
We analyzed kif6 expression at various embryonic time points and in adult tissues using RT-PCR (Fig. 7A). In WT embryos, kif6 was expressed at extremely low levels by 24 hours postfertilization (hpf) and expression was maintained through 120 hpf, a finding corroborated by qPCR experiments demonstrating that kif6 is expressed several orders of magnitude below common housekeeping genes (not shown). Maternal expression was not detected at 3 hpf. In WT adult tissues (Fig. 7B), kif6 was expressed at low levels in brain, intestine, ovary and testis but was not detectible in eye, kidney, spleen, or fin (dermal bone). Whole-mount in situ hybridization using a DIG-labeled antisense RNA probe showed very low level expression of kif6 in WT zebrafish embryos at 24, 48, and 72 hpf, which was diffuse and nonspecific (data not shown). In the Eurexpress Transcriptome Atlas Database for Mouse Embryo (www.eurexpress.org), in situ hybridization similarly showed diffuse, low level kif6 expression, with weak expression in the choroid plexus in the mouse embryo (embryonic day 14.5). In the Allen Brain Atlas (www.brain-map.org), kif6 localizes to the central canal of the spinal cord in both juvenile (postnatal day 4) and adult (postnatal day 56) mice.
Fig. 7.
RT-PCR expression of kif6 in WT embryos and WT adult tissues. A: RT-PCR in WT zebrafish embryos shows low levels of kif6 between 24 and 120 hpf. B: RT-PCR of zebrafish tissues shows kif6 expression primarily in brain, intestine and reproductive organs.
Assessment of Cilia-related Phenotypes in kif6 Mutants
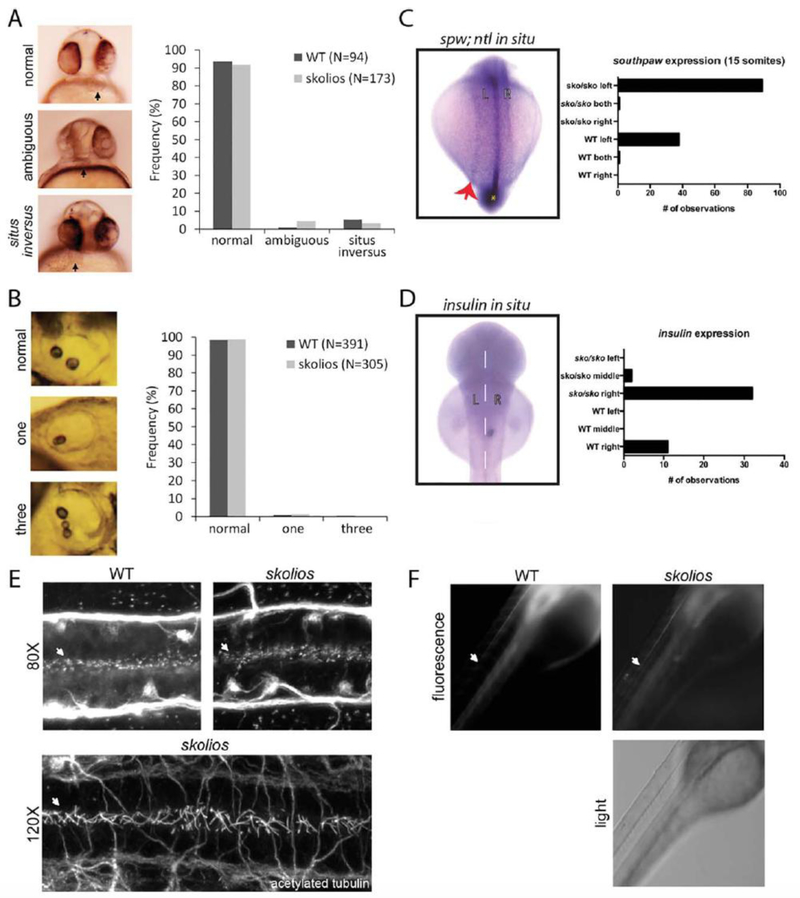
The function of kif6 is unknown, but it has been predicted to be involved in cilia/flagellum assembly or function (Miki et al., 2005; Ha et al., 2013; Hameed et al., 2013), particularly in ciliated ependymal cells (Jacquet et al., 2009). Several characteristic phenotypes are common in zebrafish with defects of motile cilia, including ventral curvature of the embryo, left– right asymmetry defects, kidney cysts, hydrocephalus, and otolith abnormalities (reviewed in Jaffe et al., 2010; Malicki et al., 2011). Because kif6 mutants develop embryonic body curvature typical of other zebrafish mutants with cilia defects, we considered if additional ciliopathy-related phenotypes were present in kif6 mutants. Normal heart positioning was identified in 93.6% of WT embryos (N.94) and 91.9% of kif6gw326 embryos (N.173) at 30 hpf (Fig. 8A), suggesting that the loss of kif6 is not associated with an increased incidence of left–right asymmetry defects. Similarly, otoliths appeared normal at 48–72 hpf in 98.5% and 98.7% of WT (N.391) and kif6gw326 mutants (N.305), respectively (Fig. 8B). In addition, in situ hybridization showed that two additional markers of symmetry, southpaw (Fig. 8C) and insulin (Fig. 8D), were correctly expressed on the left and right sides of skolios embryos, respectively. Other easily observed phenotypes that are characteristic of cilia defects, such as hydrocephalus and kidney cysts, were never observed in kif6gw326 mutants, suggesting that kif6 is not globally important for cilia formation or function.
Fig. 8.
Cilia structure and function are normal in kif6gw326 mutants. A: Defects in left–right asymmetry were evaluated by visualizing the position of the heart in 30 hpf embryos for WT (N.94) and kif6gw326 mutants (N.173). Normally, zebrafish hearts are located to the left of the midline (right side when viewed ventrally) at 30 hpf. In zebrafish with ambiguous heart positioning, hearts appear at the midline and to the right side (left side when viewed ventrally) in zebrafish with situs inversus. Ambiguous and inversed orientations of the heart were observed at similar frequencies in WT and kif6gw326 embryos. B: Otoliths were observed laterally in 48–72 hpf zebrafish embryos. Normal otolith numbers (two) and abnormal otolith numbers (one or three) were quantified WT (N.371) and kif6gw326 mutants (N.305). C: In situ hybridization for southpaw (spw) to highlight LR patterning and no tail (ntl) to highlight the midline shows no defect in left–right patterning in kif6 sko/sko mutants (n.39 WT and 90 kif6 sko/sko mutants). D: In situ hybridization for insulin (ins) to highlight LR patterning in the pancreas shows no defect in the normally rightward place of the pancreas in kif6 sko/sko mutants (n.11 WT and 34 kif6 sko/sko mutants). E: Whole-mount confocal immunostaining of 28 hpf WT and kif6gw326 mutant embryos shows normal appearance of cilial in the central canal (arrows). F: Live 48 hpf WT and kif6gw326 embryos were injected with 5% tetramethylrhodamine conjugated to 70,000 molecular weight dextran. Sixty minutes after injection, the dye has migrated similar distances in skolios mutants compared with WT embryos.
Because kif6 is thought to have a role in ciliated ependymal cells (Jacquet et al., 2009) and is expressed in the central canal of the rodent spinal cord (www.brain-map.org), we evaluated the structure and function of cilia in the central canal in kif6gw326 mutants using whole-mount confocal immunostaining with an antibody that labels acetylated tubulin. No gross abnormalities in ciliary number or morphology were detected in 28 hpf embryos compared with WT (Fig. 8E). To evaluate ciliary function, we assessed the movement of injected dye in the central canal of WT and kif6gw326 embryos at 48 hpf using a previously described assay (Kramer-Zucker et al., 2005). No differences in dye movement were observed in kif6gw326 mutants compared with WT (Fig. 8F).
Discussion
Idiopathic scoliosis affects 2–3% of the population. Despite its frequency, the pathogenesis of idiopathic scoliosis remains elusive. Here, we describe a new zebrafish mutant, called skolios, which develops spinal deformity that parallels many aspects of human idiopathic scoliosis.
First, skolios mutants develop a complex, three-dimensional curvature of the spine without an increased frequency of structural vertebral abnormalities. In humans, idiopathic scoliosis is diagnosed only after other causes of scoliosis have been ruled out, including congenital vertebral defects or other syndromic diagnoses that can cause scoliosis (e.g., cerebral palsy, Marfan syndrome). While vertebral defects were absent in approximately 70% skolios mutants, mild vertebral defects (vertebral fusions) were noted occasionally, but were present at the same frequency in WT, suggesting curvature is unlinked to these vertebral abnormalities in skolios mutants. Moreover, spinal curvature is the only phenotype observed in skolios mutants, suggesting that spinal curvature is not occurring secondary to a broader, systemic condition. Second, like human idiopathic scoliosis, we noted sexual dimorphism in skolios for the most severe curves. In humans, mild idiopathic scoliosis affects males and females equally, but females develop severe deformity 10 times more frequently than males (Lenke, 2004). In skolios, both genders were affected, but over time, adult male mutants developed a more severe spinal deformity compared with adult females. Although this is opposite to the female bias observed in humans, it nevertheless demonstrates a role for gender in the progression of spinal deformity but not for overall susceptibility.
Using meiotic mapping and whole genome sequencing, we identified a nonsense mutation (gw326) in kif6, a poorly characterized member of the kinesin family, as the genetic basis of skolios. Our results, along with others (Bowen et al., 2012; Leshchiner et al., 2012; Obholzer et al., 2012; Voz et al., 2012), demonstrate that whole genome sequencing is an effective approach to identify mutations in zebrafish. Kif6 TALEN mutants (kif6gw327 and kif6gw328/gw329) recapitulated both the embryonic and postembryonic skolios phenotypes, confirming that skolios is caused by the loss of kif6.
Kinesins are motor proteins that have diverse functions, but can be identified by their highly conserved motor domain that binds to microtubules and powers movement through the hydrolysis of ATP. Using degenerate primers designed to this conserved motor domain, kif6 was first identified in cDNA from mouse hippocampus (Nakagawa et al., 1997). In a large mouse cDNA library, kif6 was identified in spinal ganglion and cerebellum (Miki et al., 2003) and Western blotting of mouse tissues has shown kif6 expression in various tissues, including brain and spinal cord (Hameed et al., 2013). Kif6 is expressed in the choroid plexus and central canal of the rodent spinal cord in the Eurexpress Transcriptome database (www.eurexpress.org) and Allen Brain Atlas (www.brain-map.org), respectively. Our expression analysis in zebrafish demonstrated complimentary results, with kif6 expressed in the brain, but also in the intestine, ovary and testis. To our knowledge, this is the first demonstration of kif6 expression in zebrafish and the first demonstration of a mutant phenotype resulting from a mutation in kif6.
Kif6 is poorly characterized and its function is unknown. Most investigations of kif6 have been limited to a common single nucleotide polymorphism (rs20455) in humans, which was associated with coronary artery disease (Bare et al., 2007; Iakoubova et al., 2008a,b; Shiffman et al., 2008a,b), although a large metaanalysis failed to replicate the association (Assimes et al., 2010) and the importance of the common polymorphism in kif6 is currently being debated. A kif6 mutant mouse was found to have normal cardiac function, while additional phenotypes were not described (Hameed et al., 2013). Kif6 was originally identified as an orphan kinesin (Miki et al., 2001, 2003), but has also been included with kinesin-family member 9 (kif9) in the Kinesin-9 superfamily. Although kif9 and kif6 share a region of conserved sequence near the kinesin motor domain that is specific to the Kinesin-9 superfamily (Miki et al., 2005), the proteins are phylogenetically distant (Miki et al., 2001). Members of the Kinesin-9 superfamily have only been identified in vertebrates and protozoa, which is consistent with a role in cilia and flagella (Miki et al., 2005).
Further evidence for a role of kif6 in cilia was suggested by its down regulation in forebrain of forkhead box J1 (FoxJ1)-null mice (Jacquet et al., 2009), a transcription factor that is necessary and sufficient for motile cilia generation (Stubbs et al., 2008; Yuet al., 2008). FoxJ1 is also required for the differentiation of ependymalcells (Li et al., 2009), which are ciliated cells that line theventricles in the brain and the central canal in the spinal cord. Ependymal cells are responsible for circulating cerebral spinal fluid, although they may have additional functions. In the AllenBrain Atlas (www.mouse.brain-map.org/), kif6 expression is limited to the ependymal layer of the ventricle (Li et al., 2009) and central canal of the spinal cord. Gross abnormalities in cilia structure or function, as assessed by movement of central spinal fluid in the central canal, were absent, as were additional cilia related phenotypes in kif6 mutants (defects in left–right asymmetry, otolith abnormalities, hydrocephalus, or kidney cysts). Although we did not assess ciliary function by means of a direct analysis of Kupffer’s vesicle, disruption of ciliary function in Kupffer’s vesicle invariably leads to downstream defects in left–right symmetry, which we failed to observe. However, these results do not rule out other cilia-related functions for kif6. For example, cells in the developing neural tube must interpret sonic hedgehog (shh) signals to specify cell fates and correctly pattern the neural tube (Su et al., 2012). In vertebrates, the primary cilium is a shh signaling center, requiring cilia-related proteins to correctly localize members of the hedgehog pathway to the cilium (Goetz and Anderson, 2010; Larkins et al., 2011). Curvature of the body axis is a common phenotype in zebrafish embryos with cilia defects (Malicki et al., 2011) and has also been observed in adult cilia mutants (Bachmann-Gagescu et al., 2011), which is consistent with a cilia-related defect in kif6 mutants. Cilia defects have been proposed as a mechanism for human idiopathic scoliosis (Burwell et al., 2006), although scoliosis is not common in human ciliopathies. Future studies are needed to clarify the function of kif6. Identifying the predominant cell types expressing kif6 may be particularly useful to identify its molecular role and elucidate the mechanism by which it leads to spinal curvature in skolios mutants. Given the absence of kif6 expression in bone (fin rays), a skeletal phenotype autonomous to bone-forming cells (osteoblasts/scleroblasts) seems unlikely. Because in situ hybridization yielded nonspecific, low-level expression of kif6, and qPCR experiments demonstrate that kif6 is expressed several orders of magnitude below common housekeeping genes, more robust methods such as knocking in a GFP-tagged kif6 into WT zebrafish are likely needed. It is unclear whether kif6 mutations are important in the genesis of human idiopathic scoliosis. Efforts aimed at identifying kif6 mutations in a DNA database of pediatric patients with musculoskeletal disorders, including scoliosis, are currently under way.
In summary, this study identifies skolios, a zebrafish mutant with spinal deformity. The spinal curvature phenotype occurs isolated from other features and is not secondary to vertebral malformations, which suggests that skolios may serve as a model for human idiopathic scoliosis and could be a valuable resource to better understand this complex human condition. Moreover, we have established zebrafish as a viable model for spinal deformity that may be of use to confirm genetic findings in human studies of idiopathic scoliosis. Using meiotic mapping and whole genome sequencing, we identify kif6 as genetic basis of skolios. These finding highlight a new role for kif6 in spinal development and stability, expanding upon the already diverse cellular functions of kinesins.
Experimental Procedures
Zebrafish Maintenance
Zebrafish were maintained under standard conditions (Westerfield, 1993). Zebrafish were bred by in vitro fertilization using adults anesthetized in 0.16% tricaine methanesulfonate (3-amino benzoic acidethylester, Sigma-Aldrich) diluted in E3 embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4 in H2O) or by natural pairwise mating. With the exception of WIK strains used for meiotic mapping, all wild-type (WT) zebrafish were from an AB genetic background. Zebrafish embryos were maintained in E3 embryo medium in Petri dishes (50 embryos/dish) until 5 dpf, when they were transferred to 100- ml beakers containing 50 ml E3 medium (25 embryos/beaker). Larvae were fed twice daily from 6 to 14 dpf with _1 ml of rotifer (Brachionis plicatilis) culture maintained on Nannochloropsis algae (Reed Mariculture, California). After 14 dpf, larvae were fed 4 times daily with either rotifer culture or brine shrimp (yourfishstuff.com). After 4 weeks juvenile zebrafish were placed on flowing water and fed a combination of brine shrimp and Hatchfry Encapsulon (Argent Labs) until 8 weeks of age. After 8 weeks, adult zebrafish were fed a combination of brine shrimp and Hikari Micro Pellets (Aquatic Ecosystems). Experiments were carried out in accordance with the animal protocol guidelines at Washington University.
Chemical Mutagenesis and Forward Genetic Screening
The generation of mutagenized F1 female fish was carried out with ENU as previously described (Gansner et al., 2008). Gynogenetic diploids were subsequently derived by the early pressure method (Obholzer et al., 2012) and screened for spinal curvature.
Analysis of Spinal and Vertebral Morphology
MicroCT imaging was performed using a VivaCt40 machine (ScanCo) as previously described (Gray et al., 2014). Zebrafish observations were imaged using an Olympus DP70 Digital Microscope Camera fixed to an Olympus MVX10 MacroView fluorescence microscope with MicroSuite Basic Edition software (Olympus). For bone and cartilage staining, zebrafish at various time points were euthanized with tricaine methanesulfonate and fixed in 4% paraformaldehyde/phosphate buffered saline (PFA/ PBS). Fixed zebrafish were immersed in 0.1% alcian blue in 7:3 EtOH:glacial acetic acid for 24–48 hr and destained for 0.5–2 hr in 7:3 EtOH/glacial acetic acid to visualize cartilage. Bones were then stained with alizarin red S in 0.5% KOH for 6–24 hr and destained in 0.5% KOH. Stained zebrafish were stored in glycerol. The spinal curvature was quantified by imaging zebrafish positioned laterally. For consistency, spinal curvature was only quantified near the transition from abdominal (rib vertebrae) to caudal vertebrae (vertebrae posterior of ribs), which excluded the most rostral and caudal vertebrae. The apex of the curve was identified within the defined region and lines were drawn through the center of the two vertebrae adjacent to the apex. The angle of the two intersecting lines was measured using Photoshop CS3 (Adobe). Measured angles were subtracted from 180_ to calculate the spinal curve.
Meiotic Mapping
The skolios mutation was localized to chromosome 17 using centromeric linkage analysis (Johnson et al., 1995, 1996). Fine mapping was achieved by mapcrossing to the polymorphic WIK strain and assessing for recombination along chromosome 17 by sequence length polymorphism analysis (Shimoda et al., 1999).
Whole Genome Sequencing
Genomic DNA from a skolios mutant was extracted using Proteinase K (Roche) and 25:24:1 phenol:chloroform:isoamyl (Roche) according to the manufacturer’s instructions. Whole genome sequencing was performed by the Genome Technology Access Center at Washington University using 3 mg DNA from a single skolios mutant. One lane of a HiSeq 2000 sequencer (Illumina) was used to generate 101 bp paired-end reads. Raw sequencing reads were aligned to the Zv9 genome assembly using Novoalign (Novocraft Technologies). Variants were identified using SAMtools software (Li et al., 2009) and annotated using SnpEff software (Cingolani et al., 2012). Variants were validated by Sanger sequencing using an ABI 3730 Sequencer (Life Technologies) and visualized using Sequencher DNA Sequencing software (Gene Codes).
RT-PCR of Adult Zebrafish Tissues
Tissue samples were harvested from adult zebrafish killed in tricaine methanesulfonate. Tissue samples were dissected and pooled from 5 to 10 zebrafish and total RNA was extracted using Trizol Reagent (Invitrogen). RNA concentrations were measured using a NanoDrop 2000 UV-Vis spectrophotometer. cDNA synthesis was performed using SuperScript II Reverse Transcriptase (Invitrogen) using 1 mg of RNA according to the manufacturer’s instructions. As a negative control, cDNA synthesis reactions were also performed with the reverse transcriptase omitted. PCR reactions were performed with Bullseye Taq Polymerase (Midsci) using 1 ml of cDNA and 35–40 amplification cycles with a 58_ annealing temperature and 30-s extension. The following primers were used: kif6.exon 1–3. 5’-CGTGCAGGTAAACAGTAAAAGC-3’, 5’-CAGCAACAGGTTTTGCAATG-3’; kif6.exon 16–18. 5’-GCGGAGAGAACACAGGAAAC-3’, 5’-GTGCTGTTGAGCAGGTTGTG-3’; beta-actin 5’-TACAATGAGCTCCGTGTTGC-3’, 5’-AAGGAAGGCTGGAAGAGAGC-3’.
In Situ Hybridization
Embryos were manually dechorionated at the indicated developmental stages, fixed in 4% PFA/PBS overnight at 4_C, and dehydrated by methanol series. DIG-labeled antisense RNA probes were synthesized using a DIG-labeling kit (Roche), and wholemount in situ hybridization was performed as previously described (Thisse and Thisse, 2008).
Microinjection of Morpholinos
Morpholinos (MOs) were purchased from Gene Tools, LLC to transiently knockdown zebrafish kif6. MOs were designed to target the translation start site (trans-block) and the boundary between exon 1 and intron 1 (splice-block). An additional MO targeting the translation start site but containing a 5 base mismatch served as a negative control (mismatch). The 30 end of the splice blocking and mismatch MOs were modified with a fluorescein tag. The sequences of the MOs are as follows: trans-block 5’-TTGCTTTTACTGTTTACCTGCACGT-3’, splice-block 5’-ACAAAAGCAAAACACTCACCGAGGT-3, mismatch 5’-TGCTTAACGATCTTAATCATCGCTT-3’. MOs were prepared at concentrations ranging from 0.05 mM to 3 mM and approximately 500 pL were injected in one- to fourcell stage embryos.
Transcription Activator-like Effector Nuclease (TALEN) Design and Assembly
Two TALENs (left and right) were designed to target the second exon of kif6. These sites flanked a BccI restriction site that was used for mutation screening. TALENs were generated using the Golden Gate method as previously described (Cermak et al., 2011). Plasmids were acquired from the Golden Gate TALEN and TAL Effector Kit (AddGene) and RVD repeat arrays were cloned into pCS2TAL3DD and pCS2TAL3RR (AddGene). Plasmids were transformed into DH5a competent cells (Invitrogen) and isolated using the QIAprep Spin Miniprep Kit (Qiagen). The following sequence targets and RVD sites were used to generate kif6 TALENs: Left target DNA TCCCTCTTATTGTTG, LeftRVDsequence NGHDHDHDNGHD NGNGNINGNGNNNGNGNN Right target DNA GCATCTCTGGGAACC RightRVDsequence NNNNNGNGHDHDHDNINN NINNNINGNNHD
Injection of TALEN RNA in Zebrafish Embryos
Complete plasmids (containing the scaffold and RVD arrays) were linearized using XhoI and 50-capped mRNA was generated by in vitro transcription using the mMESSAGE mMACHINE SP6 Transcription Kit (Life Technologies). Capped mRNA was purified using the RNeasy Mini Kit (Qiagen). Left and right TALENs were combined at equal concentrations. WT embryos were collected and 40–50 pg of pooled TALENs were injected into the yolk of one- to four-cell stage embryos. Injected embryos and progeny were screened for TALEN-induced mutations by digesting PCR amplified DNA with BccI (New England Biolabs). The following primers were used for screening the BccI restriction site: BccI 5’-TGCAATAAATGGAAACAAGAACC-3’, 5’-TTCGCACATTTGTTCAGTGAC-3’
Evaluation of Cilia and Cilia-related Phenotypes
Defects in left–right asymmetry were determined by visualizing the position of the heart in 30 hpf embryos. Abnormal left–right asymmetry was classified as ambiguous when the heart was positioned at the midline and as situs inversus when the heart was positioned left of the midline when viewed ventrally (the right side of the embryo). Otoliths were evaluated by visualizing zebrafish laterally between 48 and 72 hpf. Immunofluorescence staining was performed as previously described (Ha et al., 2013) using embryos collected at 28 hr postfertilization (hpf). Embryos were dechorionated in protease type XIV (Sigma) and fixed in 4% paraformaldehyde for immunostaining with anti-acetylated tubulin monoclonal antibody 6–11-B1 (Sigma). Fluid flow in the central canal was measured as previously stated (Kramer-Zucker et al., 2005). Live 48 hpf zebrafish were anesthetized in tricaine methanesulfonate and imaged at various time points after injection of 5% tetramethylrhodamine conjugated to 70,000 molecular weight dextran into the brain ventricle.
Statistical Analysis
A one-tailed Student’s t-test was used for all statistical analyses.
Acknowledgment
We thank Douglas Oppedal for assistance with zebrafish husbandry.
References
- Assimes TL, Holm H, Kathiresan S, Reilly MP, Thorleifsson G, Voight BF, Erdmann J, Willenborg C, Vaidya D, Xie C, Patterson CC, Morgan TM, Burnett MS, Li M, Hlatky MA, Knowles JW, Thompson JR, Absher D, Iribarren C, Go A, Fortmann SP, Sidney S, Risch N, Tang H, Myers RM, Berger K, Stoll M, Shah SH, Thorgeirsson G, Andersen K, Havulinna AS, Herrera JE, Faraday N, Kim Y, Kral BG, Mathias RA, Ruczinski I, Suktitipat B, Wilson AF, Yanek LR, Becker LC, Linsel-Nitschke P, Lieb W, Konig IR, Hengstenberg C, Fischer M, Stark K, Reinhard W, Winogradow J, Grassl M, Grosshennig A, Preuss M, Schreiber S, Wichmann HE, Meisinger C, Yee J, Friedlander Y, Do R, Meigs JB, Williams G, Nathan DM, MacRae CA, Qu L, Wilensky RL, Matthai WH Jr., Qasim AN, Hakonarson H, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser VE, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Martinelli N, Olivieri O, Trabetti E, Malerba G, Pignatti PF, Guiducci C, Mirel D, Parkin M, Hirschhorn JN, Asselta R, Duga S, Musunuru K, Daly MJ, Purcell S, Eifert S, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Myocardial Infarction Genetics C, Wellcome Trust Case Control C, Cardiogenics, Ouwehand WH, Deloukas P, Scholz M, Cambien F, Huge A, Scheffold T, Salomaa V, Girelli D, Granger CB, Peltonen L, McKeown PP, Altshuler D, Melander O, Devaney JM, Epstein SE, Rader DJ, Elosua R, Engert JC, Anand SS, Hall AS, Ziegler A, O’Donnell CJ, Spertus JA, Siscovick D, Schwartz SM, Becker D, Thorsteinsdottir U, Stefansson K, Schunkert H, Samani NJ, Quertermous T. 2010. Lack of association between the Trp719Arg polymorphism in kinesin-like protein-6 and coronary artery disease in 19 case-control studies. J Am Coll Cardiol 56: 1552–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann-Gagescu R, Phelps IG, Stearns G, Link BA, Brockerhoff SE, Moens CB, Doherty D. 2011. The ciliopathy gene cc2d2a controls zebrafish photoreceptor outer segment development through a role in Rab8-dependent vesicle trafficking. Hum Mol Genet 20:4041–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bare LA, Morrison AC, Rowland CM, Shiffman D, Luke MM, Iakoubova OA, Kane JP, Malloy MJ, Ellis SG, Pankow JS, Willerson JT, Devlin JJ, Boerwinkle E. 2007. Five common gene variants identify elevated genetic risk for coronary heart disease. Genet Med 9:682–689. [DOI] [PubMed] [Google Scholar]
- Bowen ME, Henke K, Siegfried KR, Warman ML, Harris MP. 2012. Efficient mapping and cloning of mutations in zebrafish by lowcoverage whole-genome sequencing. Genetics 190:1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwell RG, Dangerfield PH, Freeman BJ, Aujla RK, Cole AA, Kirby AS, Pratt RK, Webb JK, Moulton A. 2006. Etiologic theories of idiopathic scoliosis: the breaking of bilateral symmetry in relation to left-right asymmetry of internal organs, right thoracic adolescent idiopathic scoliosis (AIS) and vertebrate evolution. Stud Health Technol Inform 123:385–390. [PubMed] [Google Scholar]
- Castelein RM, van Dieen JH, Smit TH. 2005. The role of dorsal shear forces in the pathogenesis of adolescent idiopathic scoliosis–a hypothesis. Med Hypotheses 65:501–508. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. 2011. Efficient design and assembly of custom TALEN and other TAL effectorbased constructs for DNA targeting. Nucleic Acids Res 39:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KM, Wang T, Poon AM, Carl A, Tranmer B, Hu Y, Luk KD, Leong JC. 2005. The effect of pinealectomy on scoliosis development in young nonhuman primates. Spine (Phila Pa 1976) 30: 2009–2013. [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aout K, Aerts P, De Clercq D, De Meester K, Van Elsacker L. 2002. Segment and joint angles of hind limb during bipedal and quadrupedal walking of the bonobo (Pan paniscus). Am J Phys Anthropol 119:37–51. [DOI] [PubMed] [Google Scholar]
- Fjelldal PG, Grotmol S, Kryvi H, Gjerdet NR, Taranger GL, Hansen T, Porter MJ, Totland GK. 2004. Pinealectomy induces malformation of the spine and reduces the mechanical strength of the vertebrae in Atlantic salmon, Salmo salar. J Pineal Res 36:132–139. [DOI] [PubMed] [Google Scholar]
- Gansner JM, Madsen EC, Mecham RP, Gitlin JD. 2008. Essential role for fibrillin-2 in zebrafish notochord and vascular morphogenesis. Dev Dyn 237:2844–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. 2010. The primary cilium: a signaling centre during vertebrate development. Nat Rev Genet 11:331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman KF, Breden F. 2007. Teleosts as models for human vertebral stability and deformity. Comp Biochem Physiol C Toxicol Pharmacol 145:28–38. [DOI] [PubMed] [Google Scholar]
- Gorman KF, Tredwell SJ, Breden F. 2007. The mutant guppy syndrome curveback as a model for human heritable spinal curvature. Spine (Phila Pa 1976) 32:735–741. [DOI] [PubMed] [Google Scholar]
- Gray RS, Wilm TP, Smith J, Bagnat M, Dale RM, Topczewski J, Johnson SL, Solnica-Krezel L. 2014. Loss of col8a1a function during zebrafish embryogenesis results in congenital vertebral malformations. Dev Biol 386:72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha K, Buchan JG, Alvarado DM, McCall K, Vydyanath A, Luther PK, Goldsmith MI, Dobbs MB, Gurnett CA. 2013. MYBPC1 mutations impair skeletal muscle function in zebrafish models of arthrogryposis. Hum Mol Genet 22:4967–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed A, Bennett E, Ciani B, Hoebers LP, Milner R, Lawrie A, Francis SE, Grierson AJ. 2013. No evidence for cardiac dysfunction in Kif6 mutant mice. PLoS One 8:e54636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoubova OA, Sabatine MS, Rowland CM, Tong CH, Catanese JJ, Ranade K, Simonsen KL, Kirchgessner TG, Cannon CP, Devlin JJ, Braunwald E. 2008a. Polymorphism in KIF6 gene and benefit from statins after acute coronary syndromes: results from the PROVE IT-TIMI 22 study. J Am Coll Cardiol 51:449–455. [DOI] [PubMed] [Google Scholar]
- Iakoubova OA, Tong CH, Rowland CM, Kirchgessner TG, Young BA, Arellano AR, Shiffman D, Sabatine MS, Campos H, Packard CJ, Pfeffer MA, White TJ, Braunwald E, Shepherd J, Devlin JJ, Sacks FM. 2008b. Association of the Trp719Arg polymorphism in kinesin-like protein 6 with myocardial infarction and coronary heart disease in 2 prospective trials: the CARE and WOSCOPS trials. J Am Coll Cardiol 51:435–443. [DOI] [PubMed] [Google Scholar]
- Jacquet BV, Salinas-Mondragon R, Liang H, Therit B, Buie JD, Dykstra M, Campbell K, Ostrowski LE, Brody SL, Ghashghaei HT. 2009. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development 136:4021–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe KM, Thiberge SY, Bisher ME, Burdine RD. 2010. Imaging cilia in zebrafish. Methods Cell Biol 97:415–435. [DOI] [PubMed] [Google Scholar]
- Janssen MM, de Wilde RF, Kouwenhoven JW, Castelein RM. 2011. Experimental animal models in scoliosis research: a review of the literature. Spine J 11:347–358. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Africa D, Horne S, Postlethwait JH. 1995. Half-tetrad analysis in zebrafish: mapping the ros mutation and the centromere of linkage group I. Genetics 139:1727–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Gates MA, Johnson M, Talbot WS, Horne S, Baik K, Rude S, Wong JR, Postlethwait JH. 1996. Centromere-linkage analysis and consolidation of the zebrafish genetic map. Genetics 142:1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, Drummond IA. 2005. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development 132:1907–1921. [DOI] [PubMed] [Google Scholar]
- Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T. 2011. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell 22:4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenke L 2004. Idiopathic scoliosis. Philadelphia: Lippincot, Williams & Wilkins 3. [Google Scholar]
- Leshchiner I, Alexa K, Kelsey P, Adzhubei I, Austin-Tse CA, Cooney JD, Anderson H, King MJ, Stottmann RW, Garnaas MK, Ha S, Drummond IA, Paw BH, North TE, Beier DR, Goessling W, Sunyaev SR. 2012. Mutation mapping and identification by whole-genome sequencing. Genome Res 22:1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida M, Saito M, Dubousset J, Yamada T, Kimura J, Shibasaki K. 2005. Pathological mechanism of idiopathic scoliosis: experimental scoliosis in pinealectomized rats. Eur Spine J 14:843–848. [DOI] [PubMed] [Google Scholar]
- Malicki J, Avanesov A, Li J, Yuan S, Sun Z. 2011. Analysis of cilia structure and function in zebrafish. Methods Cell Biol 101:39–74. [DOI] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. 2005. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol 15: 467–476. [DOI] [PubMed] [Google Scholar]
- Miki H, Setou M, Hirokawa N, Group RG, Members GSL. 2003. Kinesin superfamily proteins (KIFs) in the mouse transcriptome. Genome Res 13:1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N. 2001. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A 98:7004–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NH. 1999. Cause and natural history of adolescent idiopathic scoliosis. Orthop Clin North Am 30:343–352, vii. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Tanaka Y, Matsuoka E, Kondo S, Okada Y, Noda Y, Kanai Y, Hirokawa N. 1997. Identification and classification of 16 new kinesin superfamily (KIF) proteins in mouse genome. Proc Natl Acad Sci U S A 94:9654–9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obholzer N, Swinburne IA, Schwab E, Nechiporuk AV, Nicolson T, Megason SG. 2012. Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using wholegenome sequencing. Development 139:4280–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet J, Odent T. 2013. Animal models for scoliosis research: state of the art, current concepts and future perspective applications. Eur Spine J 22(suppl 2):S81–S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. 2011. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 29:697–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 95:5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiffman D, Chasman DI, Zee RY, Iakoubova OA, Louie JZ, Devlin JJ, Ridker PM. 2008a. A kinesin family member 6 variant is associated with coronary heart disease in the Women’s Health Study. J Am Coll Cardiol 51:444–448. [DOI] [PubMed] [Google Scholar]
- Shiffman D, O’Meara ES, Bare LA, Rowland CM, Louie JZ, Arellano AR, Lumley T, Rice K, Iakoubova O, Luke MM, Young BA, Malloy MJ, Kane JP, Ellis SG, Tracy RP, Devlin JJ, Psaty BM. 2008b. Association of gene variants with incident myocardial infarction in the Cardiovascular Health Study. Arterioscler Thromb Vasc Biol 28:173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, de Sauvage F, Jacob H, Fishman MC. 1999. Zebrafish genetic map with 2000 microsatellite markers. Genomics 58: 219–232. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C. 2008. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet 40:1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ‘Su CY, Bay SN, Mariani LE, Hillman MJ, Caspary T. 2012. Temporal deletion of Arl13b reveals that a mispatterned neural tube corrects cell fate over time. Development 139:4062–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69. [DOI] [PubMed] [Google Scholar]
- Voz ML, Coppieters W, Manfroid I, Baudhuin A, Von Berg V, Charlier C, Meyer D, Driever W, Martial JA, Peers B. 2012. Fast homozygosity mapping and identification of a zebrafish ENUinduced mutation by whole-genome sequencing. PLoS One 7: e34671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. 2007. A two-color acid-free cartilage and bone stain for zebrafish larvae Biotech Histochem 82:23–28. [DOI] [PubMed] [Google Scholar]
- Westerfield M 1993. The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio). Eugene, OR: M. Westerfield. [Google Scholar]
- Yu X, Ng CP, Habacher H, Roy S. 2008. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet [DOI] [PubMed] [Google Scholar]