Abstract
1. Failure to predict human pharmacokinetics of aldehyde oxidase (AO) substrates using traditional allometry has been attributed to species differences in AO metabolism.
2. To identify appropriate species for predicting human in vivo clearance by single species scaling (SSS) or multispecies allometry (MA), we scaled in vitro intrinsic clearance (CLint) of five AO substrates obtained from hepatic S9 of mouse, rat, guinea pig, monkey, and minipig to human in vitro CLint.
3. When predicting human in vitro CLint, average absolute fold-error was ≤ 2.0 by SSS with monkey, minipig, and guinea pig (rat/mouse >3.0), and was <3.0 by most MA species combinations (including rat/mouse combinations).
4. Interspecies variables, including fraction metabolized by AO (Fm,AO) and hepatic extraction ratios (E) were estimated in vitro. SSS prediction fold-errors correlated with the animal:human ratio of E (r2 =0.6488), but not Fm,AO (r2 =0.0051).
5. Using plasma clearance (CLp) from the literature, SSS with monkey was superior to rat or mouse at predicting human CLp of BIBX1382 and zoniporide, consistent with in vitro SSS assessments.
6. Evaluation of in vitro allometry, Fm,AO, and E, may prove useful to guide selection of suitable species for traditional allometry and prediction of human pharmacokinetics of AO substrates.
Keywords: aldehyde oxidase, clearance, S9, molybdenum hydroxylase, pharmacokinetics, non-cytochrome P450, drug metabolism, species differences
Introduction
Aldehyde oxidase (AO) is a molybdo-flavoenzyme known to oxidize aldehydes and aromatic azaheterocycles, as well as reduce N- or S-oxides, nitro groups and some heterocycles. Interest in AO-mediated drug metabolism has increased in recent years as new generations of drug candidates increasingly contain AO-susceptible aromatic azaheterocyles (Pryde et al., 2010). Moreover, the termination of several promising development programs during clinical trial assessment due to unrecognized or underestimated AO metabolism highlights the necessity for innovative approaches to predict human pharmacokinetics (PK) and disposition where AO is the primary clearance mechanism. For example, discontinuation of BIBX1382 (Dittrich et al., 2002), FK3453 (Akabane et al., 2011), and RO1 (Zhang et al., 2011) during clinical trials resulted from unexpectedly poor oral bioavailability attributed to AO-mediated clearance that went unidentified in preclinical and in vitro studies. The use of microsomes (lacking cytosol, thus lacking AO) and preclinical species with decreased AO activity (e.g. rat), or in species altogether missing an AO metabolism gene (e.g. dog), ultimately resulted in a lack of clinical translation in PK and anticipated exposure of these candidate drugs (and their metabolites).
A challenge in predicting human drug disposition where AO-mediated metabolism predominates has been attributed to species differences in AO expression and activity. While humans express only one functional gene, AOX1, AO expression in other species commonly used to model human PK ranges anywhere from 2–4 isoforms (e.g. rat) to a complete absence of the gene in dog (dog does express non-hepatic, non-drug metabolizing isoforms) (Garattini and Terao, 2012). Human liver S9, cytosolic fractions, and hepatocytes have proven useful in identifying AO metabolism, although at times these systems have resulted in under-prediction or variable activity, proposed to be associated with possible single nucleotide polymorphisms (SNPs) in the AOX1 gene, instability of the dimer, or deficiency of the essential molybdenum cofactor (Hartmann et al., 2012, Hutzler et al., 2012, Fu et al., 2013, Hutzler et al., 2014b), as well as extra-hepatic expression of AO (Kurosaki et al., 1999, Moriwaki et al., 2001, Nishimura and Naito, 2006, Terao et al., 2016), although the contribution of these mechanisms are poorly understood. Consequently, promising drug candidates are routinely discarded or structurally modified to eliminate AO metabolism, often at the expense of pharmacological potency and/or selectivity.
While research has been conducted to evaluate human in vitro methods for direct scaling of AO-mediated clearance (Zientek et al., 2010, Hutzler et al., 2012, Hutzler et al., 2014b), studies investigating allometric scaling approaches are limited. Allometric scaling of in vivo plasma clearance in nonclinical species is commonly used to predict human total body clearance of drugs eliminated renally and/or via hepatic metabolism (Mahmood, 2007). However, the aforementioned differences observed in AO expression and activity of species commonly employed in allometric scaling (particularly dog) have resulted in decreased confidence in the utility of this method to predict AO-mediated clearance. Interestingly, similar to AO, metabolism mediated by uridine diphosphate-glucuronosyltransferases (UGTs) has been found to vary with species, yet success in predicting human clearance of UGT-metabolized drugs with multispecies allometry (MA) and single-species scaling (SSS) has been demonstrated (Deguchi et al., 2011). Furthermore, while rat underestimated the human plasma clearance of the AO substrate BIBX1382, a report by Hutzler et al. demonstrated comparable BIBX1382 plasma clearance (as a percentage of liver blood flow) between cynomolgus monkey and human, indicating SSS with monkey may be useful in predicting the clearance of drugs subject to AO metabolism (Hutzler et al., 2014a). Choughule et al., however, reported that relative hepatic intrinsic clearance (CLint) mediated by AO in human, rhesus monkey, and guinea pig cytosol was substrate-dependent, suggesting that no single species could be reliably employed to consistently predict human clearance (Choughule et al., 2013b). Given that the ability to successfully predict human clearance of AO substrates by allometry using a particular species (or combination of species) is likely to be substrate-dependent in nature, we sought to investigate the utility of scaling in vitro CLint of AO substrates with allometry to human in vitro CLint as a tool to determine which species (or combination of species) may be suitable for conducting traditional allometric scaling of in vivo clearance. Specifically, the in vitro CLint of five compounds (Figure 1) known to be cleared either predominantly or partially by AO in human was determined in hepatic S9 from multiple nonclinical species known to express hepatic AO (mouse, rat, guinea pig, cynomolgus monkey, rhesus monkey, and minipig). Subsequently, CLint obtained from multispecies hepatic S9 incubations was subjected to MA or SSS to predict human in vitro hepatic CLint, and a folderror analysis was used to evaluate which species may be most suitable for estimating human in vivo clearance. In addition, we estimated Fm,AO (fraction of metabolism mediated by AO) and hepatic extraction ratio (E) in each species (in vitro) to understand the potential influence of these variables on the accuracy of prediction by the scaling methods presented herein.
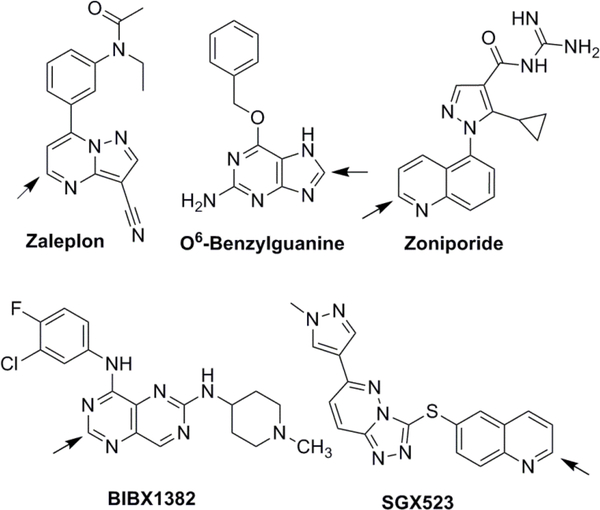
Figure 1.
Structures of AO substrates subjected to in vitro allometric scaling. Arrows indicate site of AO oxidation.
Materials and Methods
Materials
Potassium phosphate, formic acid, NADPH, MgCl2, zaleplon, O6-benzylguanine, and hydralazine hydrochloride were purchased from Sigma-Aldrich (St. Louis, MO). Zoniporide dihydrochloride, BIBX1382 dihydrochloride, and SGX523 were purchased from Tocris Bioscience (R&D Systems, Minneapolis, MN). Pooled human hepatic S9 (150-donor, mixed gender pool) was obtained from BD Biosciences (San Diego, CA), and male Sprague-Dawley rat (n=36 pool), cynomolgus monkey (n=2, pool), and CD-1 mouse (n=170 pool) hepatic S9 were obtained from Corning Inc. (Tewksbury, MA). Male rhesus monkey (n= 6 pool) and Hartley guinea pig (n=50 pool) hepatic S9 were purchased from XenoTech (Lenexa, KS), and male Gottingen minipig (n= 7 pool) hepatic S9 was purchased from BioreclamationIVT (Baltimore, MD). All solvents used for bioanalysis were purchased from Sigma-Aldrich or Fisher Scientific (Waltham, MA) and were of high-performance liquid chromatography (HPLC) grade.
Incubations with Multispecies Hepatic S9 Fractions
Substrates (1 μM) were incubated at 37°C for 60 minutes in a potassium phosphatebuffered reaction (100 mM, pH 7.4) containing hepatic S9 from human (mixed gender) or male mouse, rat, guinea pig, cynomolgus monkey, rhesus monkey, or minipig (2.5 mg/mL; +/− NADPH, 1 mM) and MgCl2 (3 mM), with preincubation in the presence or absence of hydralazine (50 μM) for estimation of fraction metabolized by AO (Fm,AO). Reactions were initiated with addition of substrate, and at designated times (t = 0, 7, 15, 30, 45, and 60 min), aliquots were removed and precipitated with ice-cold acetonitrile containing an internal standard (carbamazepine, 50 nM). The mixture was centrifuged at 3500 rcf for 5 min and resulting supernatants diluted with water in preparation for LC/MS/MS analysis of substrate depletion, by monitoring the analyte/internal standard peak area ratio.
Liquid Chromatography-Mass Spectrometry Analysis
The extent of substrate depletion in S9 fractions was determined employing LC/MS/MS analysis with an electrospray ionization enabled Sciex API-4000 triple quadrupole instrument (Sciex, Foster City, CA) that was coupled to LC-10AD pumps (Shimadzu, Columbia, MD) and a CTC PAL autosampler (Leap Technologies, Carrboro, NC). Analytes were separated by gradient elution using a Fortis C18 column (3 × 50 mm, 3 μm; Fortis Technologies Ltd., Cheshire, UK) warmed to 40°C. Mobile phase A was 0.1% formic acid in water (pH unadjusted); mobile phase B was 0.1% formic acid in acetonitrile, and the flow rate was 0.5 mL/min. Mass spectral analyses were performed using multiple reaction monitoring (MRM), with transitions and voltages specific for each analyte using a Turbo Ion Spray source (source temp 500°C) in positive ionization mode (5.0 kV spray voltage). MRM transitions were the following: zaleplon (m/z 306→236), O6-benzylguanine (m/z 242→91), zoniporide (m/z 321→262), BIBX1382 (m/z 388→98), SGX523 (m/z 360→160), and carbamazepine (m/z 237 → 194). Data were analyzed using Sciex Analyst 1.5.1 software.
Data Analysis
Intrinsic Clearance (CLint)
In vitro hepatic intrinsic clearance (CLint, mL/min/kg) for each species was extrapolated from hepatic S9 using the substrate depletion method and eq. 1 (Zientek et al., 2010):
| (1) |
The scaling factor, A, for each species is listed in Supplemental Table 1. CLint was not calculated if the mean ln[C] versus time slope was not significantly different from zero (determined using GraphPad Prism version 5.04 by an F test with a significance level of p < 0.05) or if the slope was exclusively dependent on the terminal time point in order to be considered different from zero.
Hepatic Clearance (CLHEP)
Hepatic clearance (CLHEP, mL/min/kg) was estimated using eq. 2, according to the wellstirred model, uncorrected for fraction unbound in plasma (Obach, 1999):
| (2) |
Where QH is species specific hepatic blood flow (Supplemental Table 1) and CLint is the intrinsic clearance calculated from eq. 1. Protein binding was purposely not incorporated into CLHEP estimations, as the intent of the CLHEP estimations was not to scale in vitro CLint to in vivo clearance, but rather to compare the relative extent of metabolism-mediated clearance across species (via estimation of the hepatic extraction ratio, eq. 3 below).
Hepatic Extraction Ratio (E)
The estimated hepatic extraction (E) was calculated using eq 3:
| (3) |
Estimation of fraction metabolized by AO (Fm,AO)
The Fm,AO of the five compounds was estimated for each species. Hydralazine has been reported as a specific AO inhibitor suitable for use in determination of Fm,AO (Strelevitz et al., 2012). Utilizing this method in hepatic S9 fractions, incubations were fortified with NADPH in the presence or absence of hydralazine, and eq. 4 (method A) was applied to estimate the Fm,AO:
| (4) |
where CLint is the intrinsic clearance in S9 fortified with NADPH and CLint (+Hyd) is the intrinsic clearance in S9 containing both NADPH and hydralazine. Alternatively, the Fm,AO can also be estimated with eq. 5 (method B), utilizing S9 in the absence of NADPH (AO catalytic activity is NADPH-independent):
| (5) |
where CLint (no NADPH) is the intrinsic clearance in S9 without NADPH and CLint (no NADPH+Hyd) is the intrinsic clearance in S9 containing hydralazine without NADPH. In some cases, low turnover prevented an estimation of CLint in S9 incubations containing NADPH and hydralazine (CLint (+Hyd) = 0) or in S9 incubations absent NADPH (CLint (no NADPH) = 0), which results in an Fm,AO estimation equal to 1 by method A or 0 by method B, respectively. Due to this limitation, both methods A and B were used to approximate Fm,AO. In addition, agreement between the two methods provides more confidence in the estimate. In some cases low turnover prevented a calculation of Fm,AO by both methods A and B.
Multi-species Allometry (MA) and Single-Species Scaling (SSS)
For allometric scaling of in vitro hepatic intrinsic clearance (CLint), the in vitro values obtained from incubations of hepatic S9 from preclinical species were allometrically scaled to predict human hepatic S9 CLint. The simple allometric equation (eq. 6) was applied for predictions using 3 or 4 species:
| (6) |
where CLint is the predicted intrinsic clearance, W is body weight, and a and b are the allometric coefficient and exponent, respectively. The coefficient and exponents a and b were obtained from a plot of CLint versus body weight (W) (Supplemental Figure 1). A standard body weight for each species was used in the analyses (Supplemental Table 1). Following the same principle, eq. 7 was applied in single-species scaling (SSS) analyses, using a fixed exponent of 0.75, which has been proposed for use in SSS of PK parameters based on the understanding that many physiological factors including basal metabolic rate and passive renal clearance may be scaled using this exponent (Hosea et al., 2009):
| (7) |
Success Criteria
The success of each prediction method was assessed by calculation of the average absolute fold-error (AAFE) and the average fold-error (AFE), described by eq. 8 and 9, respectively (Obach et al., 1997, Tang et al., 2007),
| (8) |
| (9) |
where N equals the total number of compounds, and fold-error (eq. 10) is the ratio of the predicted clearance (CLpred) to the observed clearance (CLobs),
| (10) |
where CLobs represents the human CLint (obtained using human hepatic S9, eq. 1) and CLpred equals the predicted human hepatic S9 CLint scaled from either MA or SSS of CLint obtained using hepatic S9 of preclinical species. AFE is equal to the geometric mean of the fold-error and represents a measurement of the overall bias in both directions (above or below the reference value of 1), whereas the AAFE gives both over-predictions and under-predictions equal value. Therefore, the overall bias of the prediction method towards under- or over-prediction is represented by the AFE, and the AAFE is an unbiased representation of the fold-error. An AAFE of ≤ 3 was considered successful; this criteria is in line with a convention reported in a recent PhRMA consensus paper by Ring et.al., identifying a < 2-, < 3- and <10-fold criteria in the identification of successful scaling methods (>10-fold criteria considered unsuccessful) (Ring et al., 2011). The percentage of compounds within 2-fold-error (fold-error = 0.5–2.0) and 3-folderror (fold-error = 0.33–3.0) was also considered when assessing each method.
SSS Correlation with Fm or E
The fold-error for in vitro SSS CLint predictions was plotted against the animal:human ratio of either Fm,AO or E, with correlation coefficients (r2) determined using GraphPad Prism version 5.04. Fm,AO estimates calculated by method B were used for the analysis (except Fm,AO of zaleplon in mouse and minipig because method B resulted in Fm,AO = 0 due to low turnover; estimates calculated from method A were instead used in these two instances). Fm,AO of zaleplon in rat, O6-benzylguanine in mouse, rat, and minipig, and BIBX1382 in mouse could not be determined by either method and were thus excluded from the analysis. Fm,AO estimations > 1, were assumed to be equal to 1. No data were excluded from the analysis involving E.
For comparison to another non-cytochrome P450 pathway, similar correlation analyses were performed using in vivo data derived from a report by Deguchi et al. of SSS to predict human CLp of uridine diphosphate-glucuronosltransferase (UGT) substrates (Deguchi et al., 2011). For twelve UGT substrates, Deguchi et al. reported CLp in each species (mouse, rat, monkey, and dog), human predictions of CLp obtained from SSS, and Fm,UGT in each species (estimated from in vivo production of glucuronide metabolites excreted into the bile and urine). For our correlation analyses using UGT substrate data from Deguchi et al., fold-errors of the SSS predictions were calculated as described above in eq. 10 from the observed human CLp values and SSS CLp predictions reported by Deguchi et al. Fm,UGT was not reported by Deguchi et al for imipramine in mouse, nor was it reported for levofloxacin and telmisartan in human, resulting in exclusion of levofloxacin and telmisartan from the Fm,UGT analysis. Similar to the correlation analysis involving E, an analysis was conducted using CLp as a percentage of QH obtained from Deguchi et al’s report. Eq. 3 was adapted to calculate CLp as a percentage of QH using CLp values reported by Deguchi et al. and species-specific hepatic blood flow, resulting in eq. 11:
| (11) |
When CLp is exclusively mediated by hepatic elimination, this value (CLp as a percentage of QH) will be equal to E. If extra-hepatic elimination is present, this value will be greater than E. Therefore, levofloxacin and furosemide, which are predominantly excreted unchanged in the urine, were excluded from this analysis.
Results
Intrinsic clearance in hepatic S9 fractions and relationship to hepatic extraction ratio
Considering the test articles employed in the present investigation are metabolized by multiple drug metabolizing enzymes, namely P450 enzymes and aldehyde oxidase (AO), we limited the sub cellular fraction employed in the in vitro pharmacokinetic assays to S9 fractions (containing both cytosol/AO and microsomes/P450). Importantly, the ability to attenuate the contribution(s) of P450 enzymes by limiting co-factor fortification (i.e., NADPH) was of utmost importance in the determination of the total contribution of these individual enzymes in the clearance of the test articles in vitro, and towards the determination of a fraction-metabolized by AO (Fm,AO; vide infra). The intrinsic clearance (CLint) estimates measured in hepatic S9 fractions of human and nonclinical species are summarized in Table 1. These data represent estimates from incubations in the presence of NADPH, thus encompassing clearance mediated by NADPH-independent (e.g. AO) as well as any NADPH-dependent (e.g. P450) pathways. CLint was converted to hepatic clearance (CLHEP) according to equation 2 for the purpose of estimating the hepatic extraction ratio (E), according to equation 3 (data summarized in Table 2). CLint estimates for zaleplon were low for all species, with E estimated to be ≤ 0.32 in each species. O6-benzylguanine CLint estimates were moderate in human, monkey, and guinea pig and lower in rat, mouse and minipig, resulting in E estimates that ranged from approximately 0.1 (rat, mouse, minipig) to approximately 0.5 (human, cynomolgus, and guinea pig). Conversely, zoniporide was moderately cleared in human, monkey, guinea pig, and minipig S9 incubations (E = 0.41 – 0.59), while it was rapidly cleared in incubations with rat and mouse S9 (E = 0.87 and 0.79, respectively). Estimated CLint of BIBX1382 was high in human, monkey, and minipig (E = 0.72 – 0.83), moderate in guinea pig (E = 0.54), and low in rat and mouse (E = 0.30 and 0.27, respectively). SGX523 exhibited low-moderate clearance in all species, with E ranging from 0.25 – 0.50. Consistent with the observations of others (Choughule et al., 2013a), no single species was fully representative of human when considering E for each compound, but rather substratedependence was observed. However, E estimated in cynomolgus and rhesus monkey was most similar to human overall, followed by guinea pig and minipig; rat and mouse generally provided a poorer representation of human E. These data highlight the need to conduct an interspecies evaluation for selection of appropriate species for human PK prediction
Table 1.
Multispecies intrinsic clearance (CLint, mL/min/kg) of zaleplon, O6-benzylguanine, zoniporide, BIBX1382, and SGX523 in incubations with hepatic S9 (in the presence of NADPH).
| Zaleplon | O6-benzylguanine | Zoniporide | BIBX1382 | SGX523 | |
|---|---|---|---|---|---|
| Human | 5.8 ± 1.02 | 17.8 ± 1.94 | 17.9 ± 1.77 | 80.2 ± 9.61 | 7.1 ± 2.28 |
| Mouse | 24.4 ± 5.28 | 20.7 ± 7.10 | 332 ± 11.3 | 33.2 ± 7.10 | 39.4 ± 7.19 |
| Rat | 15.6 ± 3.05 | 8.3 ± 2.03 | 466 ± 27.0 | 30.7 ± 3.56 | 35.3 ± 6.39 |
| Guinea Pig | 14.2 ± 1.89 | 60.3 ± 4.29 | 79.9 ± 7.47 | 72.7 ± 4.05 | 42.4 ± 4.71 |
| Cynomolgus Monkey | 20.3 ± 2.45 | 42.5 ± 8.40 | 62.8 ± 8.34 | 181 ± 11.5 | 44.4 ± 5.60 |
| Rhesus Monkey | 15.0 ± 2.06 | 22.9 ± 3.98 | 30.2 ± 3.29 | 114 ± 7.46 | 29.6 ± 7.22 |
| Minipig | 5.55 ± 1.34 | 4.43 ± 1.44 | 23.4 ± 2.07 | 134 ± 5.69 | 25.4 ± 1.46 |
Data represent means of triplicate determinations from 2–3 experiments (± SD).
Table 2.
Multispecies hepatic clearance (CLHEP, mL/min/kg) of zaleplon, O6-benzylguanine, zoniporide, BIBX1382, and SGX523 in incubations with hepatic S9 (in the presence of NADPH) and the estimated hepatic extraction ratio (E).
| Zaleplon | O6-benzylguanine | Zoniporide | BIBX1382 | SGX523 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CLHEP | E | CLHEP | E | CLHEP | E | CLHEP | E | CLHEP | E | |
| Human | 4.55 ± 0.625 | 0.22 | 9.62 ± 0.593 | 0.46 | 9.64 ± 0.503 | 0.46 | 16.6 ± 0.417 | 0.79 | 5.22 ± 1.15 | 0.25 |
| Mouse | 19.0 ± 3.35 | 0.21 | 16.6 ± 4.51 | 0.18 | 70.8 ± 0.517 | 0.79 | 24.1 ± 3.85 | 0.27 | 27.2 ± 3.66 | 0.30 |
| Rat | 12.7 ± 2.10 | 0.18 | 7.40 ± 1.61 | 0.11 | 60.8 ± 0.485 | 0.87 | 21.3 ± 1.73 | 0.30 | 23.3 ± 2.85 | 0.33 |
| Guinea Pig | 11.5 ± 1.25 | 0.19 | 30.3 ± 1.11 | 0.50 | 34.5 ± 1.40 | 0.57 | 33.1 ± 0.838 | 0.54 | 24.9 ± 1.64 | 0.41 |
| Cynomolgus Monkey | 13.9 ± 1.16 | 0.32 | 21.4 ± 2.28 | 0.49 | 25.8 ± 1.43 | 0.59 | 35.4 ± 0.434 | 0.80 | 22.0 ± 1.39 | 0.50 |
| Rhesus Monkey | 11.2 ± 1.14 | 0.25 | 15.0 ± 1.75 | 0.34 | 17.9 ± 1.16 | 0.41 | 31.7 ± 0.593 | 0.72 | 17.5 ± 2.69 | 0.40 |
| Minipig | 4.60 ± 0.934 | 0.16 | 3.79 ± 1.04 | 0.14 | 12.7 ± 0.620 | 0.45 | 23.1 ± 0.170 | 0.83 | 13.3 ± 0.401 | 0.48 |
Data, calculated using CLint values from Table 1, represent means of triplicate determinations from 2–3 experiments (± SD).
Estimation of Fm,AO in hepatic S9 fractions
Table 3 summarizes Fm,AO estimated for each compound in each species using methods A and B (eq. 4 and 5, respectively). Fm,AO obtained for O6-benzylguanine, zaleplon, zoniporide, and BIBX1382 in human is consistent with previous reports (Hutzler et al., 2012, Strelevitz et al., 2012). In addition, with a few exceptions, the two methods used to estimate human Fm,AO generally produced similar values, providing more confidence in these estimates. It is important to note that the lower the rate of depletion, the less accurate the estimation of CLint, (and thus, Fm,AO) using the substrate depletion method due to difficulty in distinguishing legitimate metabolism-mediated depletion from biological or bioanalytical variability in detection (Di and Obach, 2015, Hutzler et al., 2015). For example, in isolated incidences, low turnover resulted in an Fm,AO estimation of 1 or 0, by method A or B, respectively (e.g. estimation of zaleplon Fm,AO in rat), in which case, Fm,AO was not reported. Ideally, under circumstances of low turnover, CLint could be estimated via metabolite formation and determination of Michaelis-Menten parameters (CLint = Vmax /Km); however, this method requires authentic standards of each major metabolite (e.g. AO and P450-mediated metabolites) contributing to the overall clearance.
Table 3.
Multispecies intrinsic clearance (CLint) estimated from S9 fractions containing NADPH, containing NADPH and hydralazine, without NADPH, or containing hydralazine without NADPH, and the estimated fraction metabolized by AO (Fm,AO) calculated from the CLint data using methods A and B.
| Species | CLint (mL/min/kg) | Fm,AO | ||||
|---|---|---|---|---|---|---|
| +NADPH | +NADPH +Hyd | −NADPH | −NADPH +Hyd | Method A | Method B | |
| Zaleplon | ||||||
| Human | 5.57 ± 1.01 | n/c | 3.96 ± 1.35 | n/c | 1 | 0.71 |
| Mouse | 28.0 ± 2.81 | 22.0 ± 6.14 | n/c | n/c | 0.22 | 0 |
| Rat | 18.0 ± 0.698 | n/c | n/c | n/c | n/a | n/a |
| Guinea Pig | 13.5 ± 1.90 | n/c | 6.73 ± 3.63 | n/c | 1 | 0.50 |
| Cynomolgus Monkey | 18.3 ± 1.41 | n/c | 12.8 ± 1.34 | n/c | 1 | 0.70 |
| Rhesus Monkey | 16.7 ± 1.31 | 9.61 ± 0.964 | 10.8 ± 0.743 | n/c | 0.42 | 0.65 |
| Minipig | 6.70 ±0.621 | 5.54* | n/c | n/c | 0.17 | 0 |
| O6-benzylguanine | ||||||
| Human | 17.8 ± 3.05 | 5.37 ± 1.16 | 14.8 ± 1.98 | n/c | 0.70 | 0.83 |
| Mouse | 15.5 ± 2.48 | n/c | n/c | n/c | n/a | n/a |
| Rat | 6.59 ± 0.664 | n/c | n/c | n/c | n/a | n/a |
| Guinea Pig | 58.7 ± 4.65 | 15.0 ± 2.14 | 48.7 ± 5.15 | n/c | 0.74 | 0.83 |
| Cynomolgus Monkey | 35.6 ± 5.10 | n/c | 33.5 ± 3.84 | n/c | 1 | 0.94 |
| Rhesus Monkey | 19.5 ± 2.05 | n/c | 21.3 ± 1.52 | n/c | 1 | 1# |
| Minipig | 4.68 ± 2.20 | n/c | n/c | n/c | n/a | n/a |
| Zoniporide | ||||||
| Human | 17.0 ± 1.49 | 4.00 ± 0.442 | 17.1 ± 1.53 | 4.80 ±0.685 | 0.77 | 0.72 |
| Mouse | 331 ± 17.5 | 31.7 ± 1.12 | 297 ± 16.0 | n/c | 0.90 | 0.90 |
| Rat | 482 ± 13.9 | 125 ± 11.0 | 357 ± 25.1 | 9.70 ± 1.90 | 0.74 | 0.72 |
| Guinea Pig | 73.6 ± 2.61 | 11.6 ± 2.06 | 84.4 ± 2.75 | 12.9 ± 2.54 | 0.84 | 0.97 |
| Cynomolgus Monkey | 55.2 ± 1.73 | 5.74 ± 1.16 | 55.3 ± 1.63 | n/c | 0.90 | 1 |
| Rhesus Monkey | 27.8 ± 2.03 | 6.47 ± 3.97 | 31.2 ± 2.60 | n/c | 0.77 | 1# |
| Minipig | 22.1 ± 1.87 | 3.67 ± 1.55 | 21.5 ± 1.01 | n/c | 0.83 | 0.97 |
| BIBX1382 | ||||||
| Human | 71.6 ± 1.92 | 5.83 ± 1.22 | 69.6 ± 2.10 | 3.93 ± 1.75 | 0.92 | 0.92 |
| Mouse | 21.4 ± 10.9 | n/c | n/c | n/c | n/a | n/a |
| Rat | 27.6 ± 0.995 | 25.1 ± 6.29 | 19.0 ± 3.17 | n/c | 0.09 | 0.69 |
| Guinea Pig | 69.4 ± 1.66 | 24.5 ± 2.92 | 50.4 ± 1.13 | n/c | 0.65 | 0.73 |
| Cynomolgus Monkey | 172 ±1.26 | n/c | 209 ± 19.6 | n/c | 1 | 1# |
| Rhesus Monkey | 108 ± 5.43 | 6.03 ± 3.34 | 92.7 ± 1.21 | n/c | 0.94 | 0.86 |
| Minipig | 133 ± 6.98 | 20.0 ± 2.92 | 127 ± 7.08 | n/c | 0.85 | 0.96 |
| SGX523 | ||||||
| Human | 7.34 ± 3.34 | 4.70 ± 2.40 | 4.51 ± 0.310 | n/c | 0.36 | 0.61 |
| Mouse | 35.9 ± 10.3 | 24.2 ± 5.04 | 24.4 ± 9.31 | 16.5 ± 4.03 | 0.33 | 0.22 |
| Rat | 36.9 ± 4.23 | 21.4 ± 0.664 | 10.4 ± 3.20 | n/c | 0.42 | 0.28 |
| Guinea Pig | 38.2 ± 1.13 | 29.4 ± 3.05 | 15.8 ± 6.15 | 14.6 ± 6.05 | 0.23 | 0.03 |
| Cynomolgus Monkey | 39.4 ± 1.25 | 16.6 ± 3.42 | 26.9 ± 4.35 | n/c | 0.58 | 0.68 |
| Rhesus Monkey | 23.6 ± 4.13 | 15.6 ± 2.43 | 11.3 ± 0.942 | 7.97 ± 3.59 | 0.34 | 0.14 |
| Minipig | 26.0 ± 1.20 | 21.4 ± 1.98 | 3.20 ± 0.574 | n/c | 0.18 | 0.12 |
CLint data represent means of triplicate determinations (±SD); Fm,AO calculated using mean CLint values
n/c = CLint not calculated; mean ln[C] versus time slope not significantly different from zero
n/a = insufficient CLint data to calculate Fm,AO
mean of duplicate determinations
calculations resulting in an Fm,AO > 1 were assumed to be equal to 1
Zaleplon Fm,AO.
CLint of zaleplon in human S9 incubations with hydralazine could not be measured; however, a value of 0.71 was determined using method B, consistent with previous reports (Strelevitz et al., 2012). Cynomolgus monkey, rhesus monkey, and guinea pig demonstrated similar Fm,AO to human, with estimates ranging between 0.42–0.70, while mouse and minipig estimates were ≤ 0.22. Turnover of zaleplon in rat S9 was only measurable in incubations with NADPH absent hydralazine, preventing an Fm,AO estimate from being obtained.
O6-benzylguanine Fm,AO.
A high Fm,AO (≥ 0.70) was estimated for O6-benzylguanine in all species except rat, mouse, and minipig, which could not be determined due to low turnover of the compound. As was the case for zaleplon in rat S9 incubations, a lack of measurable turnover of O6benzylguanine in rat, mouse, and minipig S9 incubations in the presence of both NADPH and hydralazine as well as incubations absent NADPH prevented estimation of Fm,AO.
Zoniporide Fm,AO.
A high Fm,AO (> 0.70) for zoniporide was estimated in all species. In human, rat, and guinea pig, NADPH-independent clearance of zoniporide was not completely inhibited by hydralazine. Literature reports indicate hydrolysis as a secondary metabolism pathway of zoniporide (Dalvie et al., 2010, Strelevitz et al., 2012), which may account for this observation.
BIBX1382 Fm,AO.
Fm,AO for BIBX1382 was estimated to be ≥ 0.70 by both methods in all species except rat and mouse. BIBX1382 reportedly undergoes some degree of P450 2D6-mediated metabolism (Dittrich et al., 2002), and it has been reported that hydralazine may exert mild human 2D6 inhibition (Strelevitz et al., 2012, Zientek and Youdim, 2015), in which case method A could potentially estimate an inflated Fm,AO; however, both methods A and B resulted in the same Fm,AO estimation in human, suggesting that either P450 2D6-mediated clearance was insignificant, or hydralazine did not inhibit this pathway. This result is in agreement with a previous report phenotyping BIBX1382 clearance in human hepatocytes, where substrate depletion was predominantly mediated by AO (Hutzler et al., 2012). Interestingly, CLint in rat S9 incubations fortified with NADPH was only slightly inhibited by hydralazine (decreased from 29 to 25 mL/min/kg), suggestive of predominantly NADPH-dependent clearance; however, in incubations absent NADPH, a CLint of 19 mL/min/kg still remained. This finding indicates the possibility that the cytosolic enzyme xanthine oxidase (XO) may mediate part of the NADPH-independent clearance in rat S9 since XO is not inhibited by hydralazine; however, no measurable substrate depletion was observed in incubations containing hydralazine without NADPH. Minor depletion of BIBX1382 CLint was observed in human S9 incubations containing hydralazine without NADPH, but no other species exhibited measurable depletion under these conditions.
SGX523 Fm,AO.
Considerable variability between the two methods was observed in the Fm,AO of SGX523 in some species (e.g. guinea pig), but overall a low-moderate Fm,AO was estimated in all species (range of 0.03 – 0.68). In some species (mouse, rhesus, and guinea pig), not all NADPHindependent activity was inhibited by hydralazine, indicating potential involvement of XO; Diamond et al. reported an absence of XO metabolism in formation of the SGX523 oxidative metabolite M11 in human and cynomolgus S9, but rather that the metabolite was solely attributed to AO (Diamond et al., 2010). It is possible that the challenges associated with low substrate turnover, as described previously, contributed to our observations.
In general, the Fm,AO calculated by the two different methods were in better agreement for compounds/species exhibiting more rapid clearance, and thus, more confidence can be placed in the accuracy of these estimations. Monkey and guinea pig demonstrated Fm,AO values most similar to human. However, because low turnover prevented an Fm,AO calculation for some compounds in mouse, rat, and minipig, it is unclear how closely these species replicate human Fm,AO.
Prediction of human hepatic S9 clearance by multi-or single species allometry
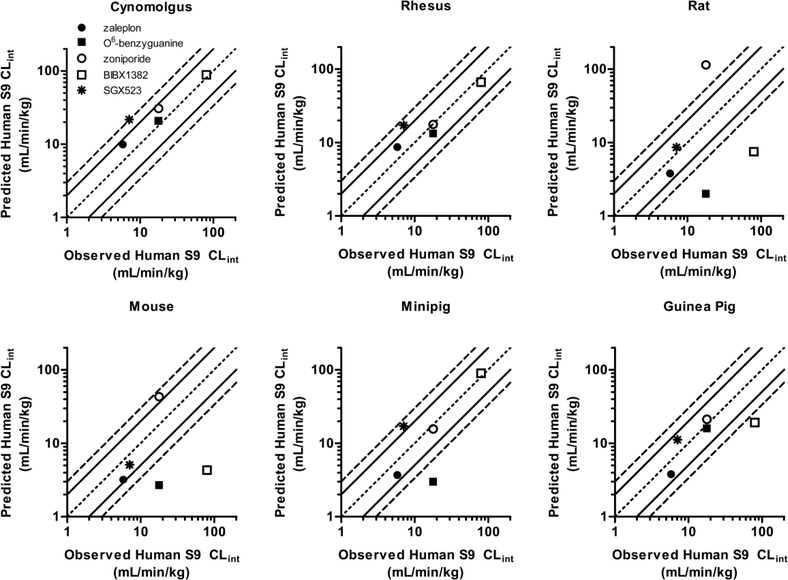
Intrinsic clearance estimates from hepatic S9 incubations from mouse, rat, guinea pig, cynomolgus, rhesus, and minipig were employed in various combinations of 3 or 4 species for allometric scaling (MA) as well as individually for direct extrapolation from a single-species (SSS). Human CLint values predicted from MA and SSS were compared to CLint measured in incubations with human hepatic S9. The overall performance of each method is summarized in Supplemental Table 2 (MA) and Table 4 (SSS), and plots of the observed CLint for each compound versus the human CLint predicted by each method are displayed in Supplemental Figure 2 (MA) and Figure 2 (SSS) (see also tabulated data in Supplemental Tables 3 and 4, along with the allometric exponents obtained for each MA method).
MA of CLint.
An AAFE of ≤ 2.0 was obtained from all of the 4-species combinations and from the rhesus/rat/mouse combination, with rhesus/rat/mouse yielding the lowest AAFE (1.7) and 80% of the five compounds predicted within 2-fold of the experimentally measured human S9 CLint (Supplemental Table 2). Only two combinations, cynomolgus/rat/mouse and cynomolgus/guinea pig/mouse, resulted in AAFE of > 3.0 (3.3-fold and 3.5-fold respectively). SGX523 is the only compound of the five for which a fold-error of < 3 could not be obtained by at least one of the species combinations; however, the minipig/rhesus/guinea pig combination yielded a fold-error of 3.0 (Supplemental Figure 2, Supplemental Table 3). Interestingly, all but one of the species combinations (minipig/rat/mouse) had an AFE of ≥ 1.0, indicating that the predictions were biased towards over-prediction rather than under-prediction. This clearly was not the case in every instance, however, particularly for O6-benzylguanine and zoniporide (Supplemental Figure 2, Supplemental Table 3).
SSS of CLint.
Single-species scaling (SSS) has previously been reported to be as accurate or better than MA in predicting human pharmacokinetics, regardless of clearance mechanism, including P450 and non-P450 metabolism (Hosea et al., 2009). Therefore, we chose to investigate this method in addition to MA for prediction of human S9 CLint. SSS predictions yielded AAFEs of ≤ 2.0 when scaling from cynomolgus and rhesus monkey, guinea pig, and minipig S9 CLint, with 80% of compounds predicted within 3-fold-error for cynomolgus, guinea pig, and minipig, and 100% for rhesus (Table 4; Figure 2). The compound falling outside of 3-fold-error differed for each species—SGX523 for cynomolgus, BIBX1382 for guinea pig, and O6-benzylguanine for minipig (Figure 2, Supplemental Table 4). AAFEs for rat and mouse were 4.1 and 3.8, respectively, with only 40% and 60% of compounds predicted within 3-fold-error, respectively. A tendency towards under-prediction via SSS was exhibited in all species except monkey, which had AFEs of 1.6 (cynomolgus) and 1.2 (rhesus).
Table 4.
Average absolute fold-error (AAFE), average fold-error (AFE), and percentage of compounds predicted within 2 or 3 fold-error of observed CLint measured in human S9, as predicted by single-species scaling.
| Single species scaling | AAFE | AFE | % within 2 fold | % within 3 fold |
|---|---|---|---|---|
| Cynomolgus Monkey | 1.6 | 1.6 | 80% | 80% |
| Rhesus Monkey | 1.4 | 1.2 | 80% | 100% |
| Rat | 4.1 | 0.56 | 40% | 40% |
| Mouse | 3.8 | 0.38 | 40% | 60% |
| Minipig | 2.0 | 0.76 | 60% | 80% |
| Guinea Pig | 1.7 | 0.76 | 80% | 80% |
Figure 2.
Plots of observed human S9 CLint vs that predicted from single-species scaling. Inner dotted line represents unity, solid line represents 2-fold-error, and outer dashed line represents 3fold-error. Zaleplon (•), O6-benzylguanine (■), zoniporide (○), BIBX1382 (□), and SGX523 (*).
Relationship of Fm or E to SSS prediction accuracy
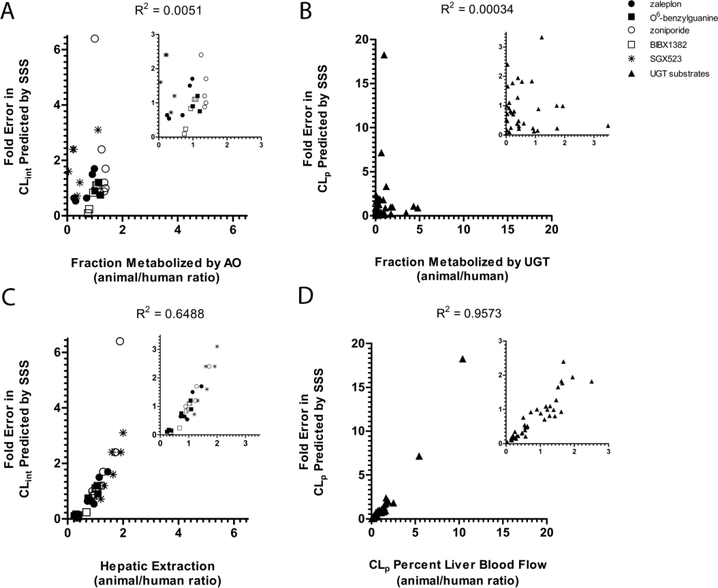
Based on their studies examining allometric scaling of UGT substrates, Deguchi et al. reported that overall Fm,UGT values in monkey were more similar to human than other species evaluated, concluding that this likely contributed to a higher rate of prediction accuracy from monkey SSS relative to the other species investigated (mouse, rat, and dog) (Deguchi et al., 2011). Likewise, we observed similar Fm,AO values between monkey and human, as well as better overall success with predictions from monkey SSS. However, a similar Fm,AO between animals and human did not always translate to a more accurate CLint prediction. Furthermore, in Deguchi’s report it can also be observed when comparing individual Fm,UGT of each compound for each species with predicted CLp by SSS, that a species exhibiting a similar Fm,UGT to human did not always yield a more accurate prediction versus another species displaying a Fm,UGT substantially different from human (Deguchi et al., 2011). In some cases however we observed a similar E between human and a given species despite a divergence in Fm,AO, suggesting that other metabolism pathways are contributing to compensate for the lacking AO pathway, resulting in a reasonable human CLint prediction. To evaluate the relationship between Fm,AO or E and the accuracy of prediction by SSS, the CLint prediction fold-error by SSS was plotted against the animal:human ratio of either Fm,AO or E (Figure 3A and C) A correlation was not observed between the animal:human ratio of Fm,AO and the fold-error in the CLint predicted from SSS (r2 = 0.0051). However, a positive correlation was observed between SSS CLint prediction fold-error and E (r2 = 0.6488). To determine if similar trends existed among the 12 UGT substrates evaluated by Deguchi et al., data were obtained from this report (Deguchi et al., 2011) to plot the CLp prediction fold-error by SSS against the animal:human ratio of Fm,UGT (Figure 3B) or against the animal:human ratio of CLp as a percentage of QH (Figure 3D). Because CLp as a percentage of QH will be equal to E when clearance is mediated by hepatic elimination, we excluded UGT substrates that are predominantly cleared renally. Once again, correlation with Fm,UGT was poor (r2 = 0.00034), but was strong with CLp as a percentage of QH (r2 = 0.9573). Overall, these data suggest that the fold-error in clearance prediction by SSS is more closely associated with the overall hepatic extraction efficiency than the Fm between human and a given species.
Figure 3.
Correlation of SSS fold-error and animal/human ratios of Fm E, or CLp as a percentage of liver blood flow for in vitro data with AO substrates (A and C) or in vivo data with UGT substrates reported by Deguchi et al. (B and D). (A) fold-error in SSS of CLint vs animal/human ratio of Fm,AO (inset, axes magnified), (B) fold-error in SSS of CLp vs animal/human ratio of Fm,UGT (inset, axes magnified), (C) fold-error in SSS of CLint vs animal/human ratio of E (inset, axes magnified), and (D) fold-error in SSS of CLp vs animal/human ratio of CLp as a percentage of liver blood flow (inset, axes magnified). Zaleplon (•), O6-benzylguanine (■), zoniporide (○), BIBX1382 (□), and SGX523 (*). UGT substrates are collectively represented by (▲).
SSS of zoniporide and BIBX1382 CLp
In vivo plasma clearance (CLp) reported in the literature for zoniporide and BIBX1382 in mouse, rat, and cynomolgus monkey (Dalvie et al., 2013, Hutzler et al., 2014a) were subjected to SSS to predict human CLp. These data are displayed in Table 5, alongside in vitro data for comparison. When comparing zoniporide in vitro data (E, Fm,AO, and SSS human CLint prediction) of mouse, rat, cynomolgus monkey, and human, the species exhibiting the most similarity to human across all data is cynomolgus monkey. Accordingly, SSS of CLp using cynomolgus monkey resulted in a more accurate human zoniporide CLp prediction (15.2 mL/min/kg) versus mouse and rat (39.2 and 62.7, respectively) relative to the observed human CLp (21 mL/min/kg). Comparison of in vitro data for BIBX1382 also demonstrated greater similarities between human and cynomolgus monkey relative to mouse and rat. Likewise, SSS of CLp using cynomolgus monkey yielded a better human BIBX1382 CLp prediction (56 mL/min/kg) versus mouse and rat (7.2 and 13.4, respectively) relative to the observed human CLp (25–55 mL/min/kg).
Table 5.
Interspecies comparison of hepatic extraction ratio (E), fraction metabolized by AO (Fm,AO), and SSS predictions using in vitro CLint (mL/min/kg) or in vivo CLp (mL/min/kg) for zoniporide (top) and BIBX1382 (bottom).
| Species | E* | Fm,AO# | S9 CLint | SSS Human Predicted S9 CLint | CLp | SSS Human Predicted CLp |
|---|---|---|---|---|---|---|
| Zoniporide | ||||||
| Human | 0.46 | 0.72–0.77 | 17.9 | --- | 21§ | --- |
| Mouse | 0.79 | 0.90 | 332 | 43.1 | 298§ | 39.2 |
| Rat | 0.87 | 0.72–0.74 | 466 | 114 | 237§ | 62.7 |
| Cynomolgus Monkey | 0.59 | ≥ 0.90 | 62.8 | 30.7 | 31§ | 15.2 |
| BIBX1382 | ||||||
| Human | 0.79 | 0.92 | 80.2 | --- | 25–55¶ | --- |
| Mouse | 0.27 | n/a | 33.2 | 4.3 | 55¶ | 7.2 |
| Rat | 0.30 | ≤ 0.69 | 30.7 | 7.5 | 55¶ | 13.4 |
| Cynomolgus Monkey | 0.80 | 1.0 | 181 | 88.7 | 118¶ | 56.0 |
Data from Table 2
Data from Table 3
Obtained from: Dalvie D, Zhang C, Chen W, et al. (2010). Drug Metab Dispos 38:641–654.
Obtained from: Hutzler JM, Cerny MA, Yang YS et al. (2014). Drug Metab Dispos 42:1751–1760.
Discussion
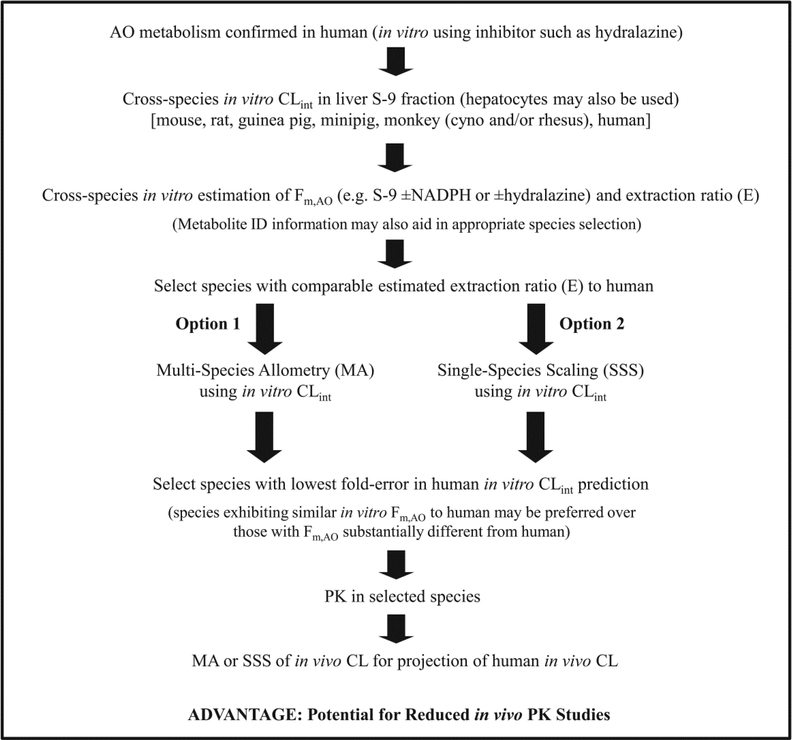
The general assumption in the utility of traditional allometric scaling to predict human clearance is that it requires conserved drug elimination mechanisms across species. Accordingly, where AO-mediated clearance exists, confidence in this approach is lacking due to differences in AO expression and activity between human and preclinical species traditionally employed (e.g. mouse, rat, dog), resulting in limited studies examining allometry to predict human clearance of AO substrates. In the present investigation, human in vitro hepatic CLint of five AO substrates was successfully predicted by MA and SSS with certain species, indicating the possibility that allometry may be useful for predicting human in vivo clearance of AO substrates when the appropriate species are utilized. Others have proposed that species expressing only the AOX1 isoenzyme in the liver (e.g. guinea pig, monkey) may serve as better species to estimate human AO-mediated metabolism versus species expressing both the AOX1 and AOX3 isoenzymes (e.g. rat, mouse) (Garattini and Terao, 2012). Consistent with this proposal, our in vitro data indicate that the hepatic CLint of AO substrates may be scaled from preclinical species to human by SSS with monkey, guinea pig, and minipig with reasonable accuracy and precision, while this relationship does not appear to be consistent (highly substrate-dependent) when directly scaling from rat or mouse. However, even SSS with guinea pig, monkey, and minipig exhibited some degree of substrate-dependency, which was similarly reported by Choughule et al. with regard to guinea pig and rhesus monkey cytosolic CLint of AO substrates DACA and phthalazine (Choughule et al., 2013b). Given these observations, we propose that allometric scaling of in vitro hepatic CLint may be beneficial for guiding selection of suitable species to be utilized for prediction of human in vivo clearance via allometry. In addition, we propose that species selection may be further aided by interspecies comparison of Fm,AO and E estimated from in vitro CLint. Table 5 illustrates an example of this approach, where comparison of in vitro data (E, Fm,AO, and human CLint prediction by SSS) indicates that SSS with cynomolgus monkey would be more appropriate than SSS with mouse or rat to estimate human CLp of zoniporide and BIBX1382. Accordingly, when CLp obtained from the literature was scaled to human CLp via SSS with cynomolgus monkey, rat, or mouse (Table 5), SSS with cynomolgus monkey did in fact yield the most accurate estimate of human CLp for both zoniporide and BIBX1382. These examples together support the proposed in vitro allometry approach as a potentially useful method to identify suitable nonclinical species for estimating human in vivo clearance via traditional allometry. Figure 4 depicts a flow chart summarizing this approach, which may prevent unnecessary PK studies in species that are not likely to reflect human PK. Furthermore, though beyond the scope of the present investigation, this approach may also benefit (in combination with in vitro biotransformation experiments) selection of appropriate species for toxicity testing.
Figure 4.
Flow chart integrating current strategies to identify and predict human AO-mediated metabolism with a novel in vitro approach to guide selection of appropriate species to be employed in traditional (in vivo) allometric scaling for projection of human in vivo clearance.
A potential limitation to the utility of this in vitro allometry approach is the use of hepatic S9, which only represents clearance mediated by hepatic metabolism. Though the contribution of extrahepatic metabolism to total body clearance of AO-metabolized compounds is currently poorly understood, extrahepatic expression of AO has been demonstrated in human as well as nonclinical species (Kurosaki et al., 1999, Moriwaki et al., 2001, Nishimura and Naito, 2006, Terao et al., 2016). Furthermore, differences in the AOX1 mRNA levels observed in various tissues between humans and mice (Terao et al., 2016) indicate tissue-specific expression patterns may not parallel across species. For example, Hutzler et al. demonstrated metabolism of BIBX1382 in lung and kidney S9 fractions of human and cynomolgus monkey (Hutzler et al., 2014a), although species differences in the relative rates of elimination of BIBX1382 from these two tissues were noted. Indeed, human CLp for four of the five substrates we evaluated are reported in the literature and were each under-represented by our CLHEP estimates from human S9 incubations, including BIBX1382 (Table 6). This is not an uncommon observation, which may be attributed to extrahepatic metabolism, among other possibilities, such as SNPs or other sources of donor variability, ex vivo protein instability, or procedural differences in tissue procurement that may yield lot-to-lot variability in AO activity from commercial sources of S9 (Hartmann et al., 2012, Hutzler et al., 2012, Fu et al., 2013, Hutzler et al., 2014b). While our in vitro studies do not account for potential extrahepatic clearance which may occur in vivo, they were conducted under the assumption that hepatic AO-mediated metabolism is largely responsible for drug clearance, such that hepatic CLint measurements will permit suitable species selection, even though the in vitro CLint may underrepresent the total body clearance occurring in vivo. Accordingly, despite differences observed in elimination from extrahepatic S9 fractions of cynomolgus and human (Hutzler et al., 2014a), SSS of cynomolgus CLp of BIBX1382 still predicted the rapid CLp observed in clinical pharmacokinetic studies (Table 5), and our in vitro assessments with hepatic S9 successfully identified cynomolgus as an appropriate species. Future research to establish species-specific tissue expression patters, mechanisms regulating AO expression, and importantly, to develop standardized in vitro scaling factors that can be used to estimate total organ clearance from in vitro CLint in extra-hepatic tissues will all be critical steps towards understanding the potential contribution of extra-hepatic metabolism.
Table 6.
Human i.v. plasma clearance of zoniporide, O6-benzylguanine, zaleplon, and BIBX1382 reported in the literature.
| Compound | Human Plasma Clearance (mL/min/kg) |
|---|---|
| Zoniporide | 21* |
| O6-Benzylguanine | 14.5# |
| Zaleplon | 16§ |
| BIBX1382 | 22–55¶ |
| SGX523 | N/A |
Dalvie D, Zhang C, Chen W, et al. (2010). Drug Metab Dispos 38:641–654.
Dolan ME, Roy SK, Fasanmade AA, et al. (1998). J Clin Oncol 16:1803–1810.
Rosen AS, Fournie P, Darwish M, et al. (1999). Biopharm Drug Dispos 20:171–175.
Dittrich C, Greim G, Borner M, et al. (2002). Eur J Cancer 38:1072–1080.
In addition to our in vitro allometry assessments, we evaluated the relationship between interspecies Fm,AO or E and the fold-error of human CLint prediction by SSS. Interestingly, comparison of prediction fold-errors by SSS with the animal:human ratio of either Fm,AO or E revealed little correlation with Fm,AO, but a positive correlation with E; similar trends were also observed for UGT substrates when plotting SSS prediction fold-error of CLp versus the animal:human ratio of Fm,UGT or CLp as a percentage of QH. We observed some examples where a discrepancy in Fm,AO between human and animal did not preclude a similar E, which suggests a non-AO metabolism pathway may compensate for the lacking AO pathway to result in an overall similar hepatic extraction efficiency. For example, the Fm,AO obtained for zaleplon was substantially higher in human (≥ 0.71) versus minipig (≤ 0.17), while the E between the two species was similar (human = 0.22, minipig = 0.16). Consequently, these data may indicate that compounds containing a mixed AO/P450 metabolism phenotype, or possessing P450 favorable sites in addition to the AO metabolism site, could help to enable allometric scaling approaches if alternate metabolism pathway(s) in certain species compensate for reduced AO-mediated clearance. Nonetheless, these observations suggest that there is not a strong relationship between Fm and the prediction fold-error by SSS or that methods to obtain Fm are not sufficiently accurate to observe this relationship. With regard to the present study, it should be noted that our Fm,AO calculations assume that 50 μM hydralazine is adequate to selectively and completely inhibit AO metabolism, whereas the potency and selectivity of hydralazine is not known for all species studied. In addition, our Fm,AO calculations are dependent on the ability to measure substrate depletion when turnover may be low, which presents an additional challenge in obtaining an accurate Fm,AO estimate. Importantly, the challenge of low substrate turnover is not unique to the present AO investigation, as the P450 literature is replete with similar data generated for the purposes of scaling a particular PK parameter (such as clearance), and subsequently resulting in a successful model or simulation. Current research efforts among the drug metabolism community are focused on the development of novel in vitro models towards the resolution of this issue (Di and Obach, 2015, Hutzler et al., 2015). Finally, four of the five compounds exhibited an Fm,AO in human of ≥ 0.70. A larger data set consisting of compounds exhibiting a broader range of Fm,AO would help to better understand the importance of this value to obtaining an accurate human hepatic clearance prediction by SSS.
For our in vitro allometry assessment, we included data obtained from minipig, which is gaining popularity in preclinical development (particularly safety testing) due to similarities in anatomy and physiology to humans, as well as advantages related to regulatory acceptability and animal welfare (Bode et al., 2010, Ellegaard et al., 2010, van der Laan et al., 2010). With regard to drug metabolism, minipigs hold an advantage over dogs (concerning nonrodent/large animals) when AO is involved, as dogs are essentially devoid of AO activity in the liver (Dalgaard, 2015, Terao et al., 2016). In addition, a recent report found minipig to be useful in allometric scaling of drugs mostly cleared by P450 or glucuronidation (Yoshimatsu et al., 2016). For the present investigation, we chose to replace dog, which is commonly used for allometric scaling, with minipig, a species of similar body weight. Indeed, the use of minipig for MA enabled human S9 CLint to be predicted with an AAFE of < 3, and SSS with minipig predicted CLint within 3-fold for four out of five substrates. However, as noted previously, the Fm,AO of zaleplon was higher in human versus minipig, indicating that a non-AO metabolism pathway enabled the accurate human CLint prediction (unlike monkey and guinea pig which demonstrated similar Fm,AO to human). Along with substantial underprediction of O6-benzylguanine human CLint by minipig SSS, this observation highlights the potential substrate-dependency associated with this species. Interestingly, based on the AAFE and individual evaluation of fold-error for each of the five compounds, MA methods utilizing four species (including minipig) may prove to be more reliable than 3-species combinations, even though rat and mouse (which yielded poorer predictions by SSS) were included in the 4-species combinations (Supplemental Tables 2 and 3; Supplemental Figure 2). Evaluation of these species combinations in vivo will be important to fully understand this approach.
Conclusions
In summary, our data support prior postulations that guinea pig and monkey represent better models of AO-mediated drug clearance in human versus commonly employed nonclinical models such as rat or mouse (Garattini and Terao, 2012, Hutzler et al., 2013, Hutzler et al., 2014a), while reiterating that no single species should be expected to reflect human clearance of all AO substrates (Choughule et al., 2013b, Hutzler et al., 2013). Moreover, the minipig represents a large animal species to consider when investigating AO metabolism, particularly when employed in multispecies allometry and drug safety assessments of NCEs (Bode et al., 2010, Ellegaard et al., 2010, van der Laan et al., 2010) Collectively, our data support the need for a multiple species assessment when gauging the intrinsic lability of new chemical entities (NCEs) towards AO metabolism and the projection of drug clearance in human, and we have offered a potentially useful in vitro approach to aid in selecting nonclinical species that may provide the best estimates of human clearance when evaluated in vivo (potentially reducing the number of unnecessary in vivo PK studies conducted in inappropriate species). While mechanisms behind variable AO activity and contributions of extra-hepatic metabolism remain important unanswered questions towards the implementation of standardized methods pertaining to AO-mediated drug disposition, application of the methodology presented herein would likely reduce the risk of encountering unexpected rapid AO-mediated clearance in clinical trials due to the use of inappropriate species for preclinical assessments.
Supplementary Material
Acknowledgements
This work was supported by the Pharmaceutical Research and Manufacturers of America Foundation (PhRMA Foundation Pre-Doctoral Fellowship); the National Institutes of Health under Grant T32GM07628.
Footnotes
Declaration of Interest
The authors report no conflicts of interest. The authors are responsible for the content and writing of the article.
References
- Akabane T, Tanaka K, Irie M, Terashita S & Teramura T, 2011. Case report of extensive metabolism by aldehyde oxidase in humans: Pharmacokinetics and metabolite profile of FK3453 in rats, dogs, and humans. Xenobiotica, 41, 372–384. [DOI] [PubMed] [Google Scholar]
- Bode G, Clausing P, Gervais F, Loegsted J, Luft J, Nogues V & Sims J, 2010. The utility of the minipig as an animal model in regulatory toxicology. J Pharmacol Toxicol Methods, 62, 196–220. [DOI] [PubMed] [Google Scholar]
- Choughule KV, Barr JT & Jones JP, 2013a. Evaluation of Rhesus Monkey and Guinea Pig Hepatic Cytosol Fractions as Models for Human Aldehyde Oxidase. Drug Metabolism and Disposition, 41, 1852–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choughule KV, Barr JT & Jones JP, 2013b. Evaluation of rhesus monkey and guinea pig hepatic cytosol fractions as models for human aldehyde oxidase. Drug Metab Dispos, 41, 1852–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard L, 2015. Comparison of minipig, dog, monkey and human drug metabolism and disposition. J Pharmacol Toxicol Methods, 74, 80–92. [DOI] [PubMed] [Google Scholar]
- Dalvie D, Xiang C, Kang P & Zhou S, 2013. Interspecies variation in the metabolism of zoniporide by aldehyde oxidase. Xenobiotica, 43, 399–408. [DOI] [PubMed] [Google Scholar]
- Dalvie D, Zhang C, Chen W, Smolarek T, Obach RS & Loi CM, 2010. Cross-species comparison of the metabolism and excretion of zoniporide: contribution of aldehyde oxidase to interspecies differences. Drug Metab Dispos, 38, 641–54. [DOI] [PubMed] [Google Scholar]
- Deguchi T, Watanabe N, Kurihara A, Igeta K, Ikenaga H, Fusegawa K, Suzuki N, Murata S, Hirouchi M, Furuta Y, Iwasaki M, Okazaki O & Izumi T, 2011. Human Pharmacokinetic Prediction of UDP-Glucuronosyltransferase Substrates with an Animal Scale-Up Approach. Drug Metabolism and Disposition, 39, 820–829. [DOI] [PubMed] [Google Scholar]
- Di L & Obach RS, 2015. Addressing the challenges of low clearance in drug research. AAPS J, 17, 352–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond S, Boer J, Maduskuie TP, Falahatpisheh N, Li Y & Yeleswaram S, 2010. Species-Specific Metabolism of SGX523 by Aldehyde Oxidase and the Toxicological Implications. Drug Metabolism and Disposition, 38, 1277–1285. [DOI] [PubMed] [Google Scholar]
- Dittrich C, Greim G, Borner M, Weigang-Köhler K, Huisman H, Amelsberg A, Ehret A, Wanders J, Hanauske A & Fumoleau P, 2002. Phase I and pharmacokinetic study of BIBX 1382 BS, an epidermal growth factor receptor (EGFR) inhibitor, given in a continuous daily oral administration. European Journal of Cancer, 38, 1072–1080. [DOI] [PubMed] [Google Scholar]
- Ellegaard L, Cunningham A, Edwards S, Grand N, Nevalainen T, Prescott M & Schuurman T, 2010. Welfare of the minipig with special reference to use in regulatory toxicology studies. J Pharmacol Toxicol Methods, 62, 167–83. [DOI] [PubMed] [Google Scholar]
- Fu C, Di L, Han X, Soderstrom C, Snyder M, Troutman MD, Obach RS & Zhang H, 2013. Aldehyde oxidase 1 (AOX1) in human liver cytosols: quantitative characterization of AOX1 expression level and activity relationship. Drug Metab Dispos, 41, 1797–804. [DOI] [PubMed] [Google Scholar]
- Garattini E & Terao M, 2012. The role of aldehyde oxidase in drug metabolism. Expert Opinion on Drug Metabolism & Toxicology, 8, 487–503. [DOI] [PubMed] [Google Scholar]
- Hartmann T, Terao M, Garattini E, Teutloff C, Alfaro JF, Jones JP & Leimkuhler S, 2012. The impact of single nucleotide polymorphisms on human aldehyde oxidase. Drug Metab Dispos, 40, 856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosea NA, Collard WT, Cole S, Maurer TS, Fang RX, Jones H, Kakar SM, Nakai Y, Smith BJ, Webster R & Beaumont K, 2009. Prediction of Human Pharmacokinetics From Preclinical Information: Comparative Accuracy of Quantitative Prediction Approaches. The Journal of Clinical Pharmacology, 49, 513–533. [DOI] [PubMed] [Google Scholar]
- Hutzler JM, Cerny MA, Yang YS, Asher C, Wong D, Frederick K & Gilpin K, 2014a. Cynomolgus monkey as a surrogate for human aldehyde oxidase metabolism of the EGFR inhibitor BIBX1382. Drug Metab Dispos, 42, 1751–60. [DOI] [PubMed] [Google Scholar]
- Hutzler JM, Obach RS, Dalvie D & Zientek MA, 2013. Strategies for a comprehensive understanding of metabolism by aldehyde oxidase. Expert Opinion on Drug Metabolism & Toxicology, 9, 153–168. [DOI] [PubMed] [Google Scholar]
- Hutzler JM, Ring BJ & Anderson SR, 2015. Low-Turnover Drug Molecules: A Current Challenge for Drug Metabolism Scientists. Drug Metab Dispos, 43, 1917–28. [DOI] [PubMed] [Google Scholar]
- Hutzler JM, Yang Y-S, Albaugh D, Fullenwider CL, Schmenk J & Fisher MB, 2012. Characterization of Aldehyde Oxidase Enzyme Activity in Cryopreserved Human Hepatocytes. Drug Metabolism and Disposition, 40, 267–275. [DOI] [PubMed] [Google Scholar]
- Hutzler JM, Yang Y-S, Brown C, Heyward S & Moeller T, 2014b. Aldehyde Oxidase Activity in Donor-Matched Fresh and Cryopreserved Human Hepatocytes and Assessment of Variability in 75 Donors. Drug Metabolism and Disposition, 42, 10901097. [DOI] [PubMed] [Google Scholar]
- Kurosaki M, Demontis S, Barzago MM, Garattini E & Terao M, 1999. Molecular cloning of the cDNA coding for mouse aldehyde oxidase: tissue distribution and regulation in vivo by testosterone. Biochem J, 341 ( Pt 1), 71–80. [PMC free article] [PubMed] [Google Scholar]
- Mahmood I, 2007. Application of allometric principles for the prediction of pharmacokinetics in human and veterinary drug development. Adv Drug Deliv Rev, 59, 1177–92. [DOI] [PubMed] [Google Scholar]
- Moriwaki Y, Yamamoto T, Takahashi S, Tsutsumi Z & Hada T, 2001. Widespread cellular distribution of aldehyde oxidase in human tissues found by immunohistochemistry staining. Histol Histopathol, 16, 745–53. [DOI] [PubMed] [Google Scholar]
- Nishimura M & Naito S, 2006. Tissue-specific mRNA expression profiles of human phase I metabolizing enzymes except for cytochrome P450 and phase II metabolizing enzymes. Drug Metab Pharmacokinet, 21, 357–74. [DOI] [PubMed] [Google Scholar]
- Obach RS, 1999. Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: An examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos, 27, 1350–9. [PubMed] [Google Scholar]
- Obach RS, Baxter JG, Liston TE, Silber BM, Jones BC, Macintyre F, Rance DJ & Wastall P, 1997. The Prediction of Human Pharmacokinetic Parameters from Preclinical and In Vitro Metabolism Data. Journal of Pharmacology and Experimental Therapeutics, 283, 46–58. [PubMed] [Google Scholar]
- Pryde DC, Dalvie D, Hu Q, Jones P, Obach RS & Tran T-D, 2010. Aldehyde Oxidase: An Enzyme of Emerging Importance in Drug Discovery. Journal of Medicinal Chemistry, 53, 8441–8460. [DOI] [PubMed] [Google Scholar]
- Ring BJ, Chien JY, Adkison KK, Jones HM, Rowland M, Jones RD, Yates JW, Ku MS, Gibson CR, He H, Vuppugalla R, Marathe P, Fischer V, Dutta S, Sinha VK, Bjornsson T, Lave T & Poulin P, 2011. PhRMA CPCDC initiative on predictive models of human pharmacokinetics, part 3: comparative assessement of prediction methods of human clearance. J Pharm Sci, 100, 4090–110. [DOI] [PubMed] [Google Scholar]
- Strelevitz TJ, Orozco CC & Obach RS, 2012. Hydralazine As a Selective Probe Inactivator of Aldehyde Oxidase in Human Hepatocytes: Estimation of the Contribution of Aldehyde Oxidase to Metabolic Clearance. Drug Metabolism and Disposition, 40, 1441–1448. [DOI] [PubMed] [Google Scholar]
- Tang H, Hussain A, Leal M, Mayersohn M & Fluhler E, 2007. Interspecies prediction of human drug clearance based on scaling data from one or two animal species. Drug Metab Dispos, 35, 1886–93. [DOI] [PubMed] [Google Scholar]
- Terao M, Romao MJ, Leimkuhler S, Bolis M, Fratelli M, Coelho C, Santos-Silva T & Garattini E, 2016. Structure and function of mammalian aldehyde oxidases. Arch Toxicol, 90, 753–80. [DOI] [PubMed] [Google Scholar]
- Van Der Laan JW, Brightwell J, Mcanulty P, Ratky J & Stark C, 2010. Regulatory acceptability of the minipig in the development of pharmaceuticals, chemicals and other products. J Pharmacol Toxicol Methods, 62, 184–95. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu H, Konno Y, Ishii K, Satsukawa M & Yamashita S, 2016. Usefulness of minipigs for predicting human pharmacokinetics: Prediction of distribution volume and plasma clearance. Drug Metab Pharmacokinet, 31, 73–81. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu HH, Weller P, Zheng M, Tao W, Wang J, Liao G, Monshouwer M & Peltz G, 2011. In silico and in vitro pharmacogenetics: aldehyde oxidase rapidly metabolizes a p38 kinase inhibitor. Pharmacogenomics J, 11, 15–24. [DOI] [PubMed] [Google Scholar]
- Zientek M, Jiang Y, Youdim K & Obach RS, 2010. In Vitro-In Vivo Correlation for Intrinsic Clearance for Drugs Metabolized by Human Aldehyde Oxidase. Drug Metabolism and Disposition, 38, 1322–1327. [DOI] [PubMed] [Google Scholar]
- Zientek MA & Youdim K, 2015. Reaction phenotyping: advances in the experimental strategies used to characterize the contribution of drug-metabolizing enzymes. Drug Metab Dispos, 43, 163–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.