Abstract
Background
Perinatal asphyxia interferes with neonatal development, resulting in long-term deficits associated with systemic and neurological diseases. Despite the important role of poly (ADP-ribose) polymerase 1 (PARP-1) in the regulation of gene expression and DNA repair, overactivation of PARP-1 in asphyxia-exposed animals worsens the ATP-dependent energetic crisis. Inhibition of PARP-1 offers a therapeutic strategy for diminishing the effects of perinatal asphyxia.
Methods
We designed a nanosystem that incorporates a specific siRNA for PARP-1 knockdown. The siRNA was complexed with gold nanorods (AuNR) conjugated to the peptide CLPFFD for brain targeting.
Results
The siRNA was efficiently delivered into PC12 cells, resulting in gene silencing. The complex was administered intraperitoneally in vivo to asphyxia-exposed rat pups, and the ability of the AuNR-CLPFFD/siRNA complex to reach the brain was demonstrated.
Conclusion
The combination of a nanosystem for delivery and a specific siRNA for gene silencing resulted in effective inhibition of PARP-1 in vivo.
Keywords: neonatal hypoxia, siRNA delivery, PARP-1 knockdown, gold nanorods, in vivo administration, PC12, rats
Background
Perinatal asphyxia (PA) is defined as oxygen deprivation occurring around the time of birth. Survivors may show early or delayed systemic and neurological deficits.1 To ensure cell survival after a metabolic insult, several sentinel proteins are activated, including poly (ADP-ribose) polymerase 1 (PARP-1).2 PARP-1 is the principal member of the PARP superfamily and is responsible for most of the poly(ADP-ribosyl)ation that occurs in living cells.3 PARP-1 uses NAD+ to synthesize polymers of ADP-ribose that then bind to protein acceptors that are mainly associated with chromatin.4 PARP-1 participates in multiple cellular processes and plays a prominent role in DNA repair.5 Activation of PARP-1 results in certain molecular events such as 1) rearrangement of chromatin organization and transcription,6 2) gene regulation,7 and 3) double-strand break repair and replication.8,9 PARP-1 also regulates inflammatory pathways, increasing the expression of pro-inflammatory cytokines.10,11
The fact that overactivation of PARP-1 can be deleterious to stressed cells leads to the idea that PARP-1 inhibition could be beneficial; such inhibition has been described as a possible protective intervention against neuronal damage, reducing ischemic injury and inflammation affecting the neonatal brain and preventing the effects of PA on the dopaminergic system.12
siRNA provides an endogenous regulatory mechanism of gene expression by promoting mRNA degradation in a highly sequence-specific manner.13 The application of siRNAs as potential therapeutic agents in vivo requires specialized delivery approaches to enhance their pharmacodynamics and pharmacokinetics.14 Additionally, the presence of the blood– brain barrier (BBB) is a major challenge to the delivery of molecules into the brain, limiting the penetration of most central nervous system (CNS) drug candidates.15
Based on the physicochemical similarities of DNA and siRNA, carriers developed for DNA have also been applied to siRNA. These carriers can be broadly divided into two categories: 1) viral vectors and 2) nonviral vectors.16–18 Due to the potential toxicity associated with viral vectors, the use of nonviral vectors is a potential alternative.19
A powerful approach to drug delivery is based on the use of gold nanoparticles (AuNPs).20–23 AuNPs can be conjugated to peptides, nucleic acids, and a wide range of small molecules; they provide a high surface-to-volume ratio, thereby maximizing the payload/carrier ratio and the coupling to other loading systems and making it possible to adjust the charge and hydrophobicity of the complex to maximize transfection efficiency while minimizing toxicity.24,25 Several studies have demonstrated the biomedical potential of AuNPs.26–29
The purpose of this study was to develop a nanoparticle delivery system for systemic administration that possesses the capacity to cross the BBB and to transport siRNA molecules that inhibit the expression of PARP-1 in vitro and in vivo. We prepared a gold nanorod (AuNR)-siRNA complex and evaluated the efficiency of its delivery and its ability to knockdown the expression of PARP-1 in vitro using PC12 cells.30,31 This cell line was selected based on the extensive number of publications in which it has been shown to mimic dopaminergic cells.32–35
A previous report showed that treatment with AuNR-siRNA efficiently induced gene silencing in the rat hippocampus in vivo following stereotaxic administration to adult rats.36 However, to date, there have been no studies involving the systemic administration of AuNPs to neonatal rats. Previous results from our group showed that the amphipathic peptide CLPFFD, derived from the sequence of beta-amyloid, can be used to stabilize and improve brain delivery of spherical AuNPs (12 nm diameter) administered intraperitoneally (ip) to rats.37,38 Furthermore, Li et al demonstrated that AuNRs conjugated to the peptide QKLVFF, which is also derived from the beta-amyloid sequence, cross the BBB.39
In this work, AuNRs were conjugated with CLPFFD to obtain an AuNR-CLPFFD conjugate that was associated with siRNA-PARP-1 in an AuNR-CLPFFD/siRNA complex. We show here the effects of treatment with the AuNR-CLPFFD/siRNA complex on cell viability and demonstrate for the first time the capacity of this complex to cross the BBB of neonatal rats and reach the brain following systemic administration and to successfully knockdown the expression of PARP-1 in specific brain regions of asphyxia-exposed animals.
Methods
Synthesis of gold nanorods (AuNR) and conjugation with the CLPFFD peptide
AuNRs were synthesized using a seed-mediated approach.40–42 For the conjugation of CLPFFD to AuNRs, a centrifugation step was performed, and the pellets were resuspended in 4 mL of an aqueous solution containing the peptide at a concentration of 0.25 mg/mL. The mixture was allowed to react at 27°C for 72 hours, yielding AuNR-CLPFFD40 conjugate (Supplementary materials).
Quantification of AuNR-CLPFFD
The method described by Adura et al was used to determine the concentration of AuNR-CLPFFD in the solution.40 The AuNR-CLPFFD concentration in the solution, which had an OD of 1 in the longitudinal peak at 771 nm, was found to be 0.9 nM (Supplementary materials).
siRNA sequence and modifications
The siRNA PARP-1 sequence was previously described and its specificity and efficiency tested by Huang et al.43 The siRNA-PARP-1 sequence was modified to increase its stability44,45 and conjugated to the fluorescent dye Alexa 647, which emits a red fluorescent signal that makes it possible to localize the siRNA molecules. The modified siRNA sequence “siSTABLE” was purchased from Dharmacon, Inc. (Lafayette, CO, USA) (Supplementary materials).
Synthesis of AuNR-CLPFFD/siRNA complex
The previously conjugated AuNR-CLPFFD (0.9 nM) was concentrated by centrifugation (1 hour at 1,500 rcf) and resuspended in 100 µL of nuclease-free H2O at a final concentration of 9 nM. The siRNA (0.6 µg) was added, and the sample was gently mixed and incubated at room temperature for 5 minutes. The resulting complex was diluted to the desired concentration with nuclease-free H2O.
Previously, to find the optimal AuNR-CLPFFD:siRNA ratio, we prepared the complex using various AuNR-CLPFFD:siRNA ratios and evaluated the retardation of their migration in an agarose gel. Various AuNR-CLPFFD:siRNA ratios were assayed to determine the maximum loading of the siRNA (Figure S1).
AuNR-CLPFFD and AuNR-CLPFFD/siRNA complex characterization
Each new batch of synthesized AuNR-CLPFFD and AuNR-CLPFFD/siRNA complex was characterized by: 1) UV-vis-NIR spectra, measured with a 2501PC UV-vis recording spectrophotometer (Shimadzu Corporation, Kyoto, Japan); 2) hydrodynamic diameter and zeta potential, measured with a Zetasizer 3000 (Malvern Instruments, Malvern, UK) (determined in triplicate in aqueous solution); and 3) transmission electron microscopy (TEM), in which 20 µL of the colloidal AuNR-CLPFFD suspension was air-dried onto an FCF300-Cu Formvar carbon film on a 300-mesh copper grid and recorded using a Phillips TECNAI 12 instrument and an Olympus G2 camera.
PC12 cell cultures
PC12 cells (PC12; ATCC® CRL-1721™; Manassas, VA, USA) were cultured in complete RPMI 1640 medium (11875–093; Thermo Fisher Scientific Waltham, MA, USA) containing 1% penicillin/streptomycin (15140–122; Thermo Fisher Scientific), 10% heat-inactivated horse serum (26050– 088; Thermo Fisher Scientific), and 5% FBS (16000–044; Thermo Fisher Scientific). PC12 cell lines were purchased from ATCC (ATCC CRL-1721).
MTS cell viability assay
PC12 cells were seeded in pretreated wells and allowed to attach at 37°C for 24 hours. The medium was then removed, the cells were washed with PBS, fresh serum-free medium containing the AuNR-CLPFFD/siRNA complex was added, and the cells were incubated for 6 hours. Thereafter, serum was added, and the complex was incubated for 24 hours. On the third day, the treatment medium was removed, and colorless medium containing the (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)- 2H-tetrazolium inner salt) (MTS)/phenazine methosulfate (PMS) solution was added; after incubation for 1 hour, the absorbance of the culture medium at 490/655 nm was recorded using a microplate reader (Synergy HT; BioTek, Winooski, VT, USA).
Cell internalization of the AuNR-CLPFFD/siRNA complex
CellLight™ Early Endosomes-GFP, BacMam 2.0, (10586, Thermo Fisher Scientific) reagent was used to detect early endosomes, and cell nuclei were stained with Hoechst 33342. The medium was removed, the cells were washed with PBS, and the medium was replaced with fresh serum-free medium. The cells were then treated with either 1) nuclease-free H2O, 2) 0.1 nM naked siRNA (0.6 µg), 3) FuGENE® HD Transfection Reagent (3:1, 0.6 µg siRNA) (E2311; Promega Corporation, Fitchburg, WI, USA), or 4) 0.1 nM AuNR-CLPFFD/siRNA complex. After 1 hour, the cells were washed twice with PBS, fixed in 4% formalin solution (4% paraformaldehyde), coverslipped with hydrophilic resin (Fluoromount) and examined by confocal microscopy (Olympus-fv10i).
To investigate the mechanism of uptake of the AuNR-CLPFFD/siRNA complex by PC12 cells, the cells were treated with 0.1 nM AuNR-CLPFFD/siRNA complex at different temperatures (37°C and 4°C) for 1 hour. The samples were washed with PBS, and the fluorescent cells were analyzed by flow cytometry on a BD FACSCanto A (Becton Dickinson, San José, CA, USA) (Figure S2).
Cesarean section, hysterectomy, and induction of PA
On the last day of gestation (G22), pregnant female rats were anesthetized, cervically dislocated, and hysterectomized (n=12). One or two pups were removed immediately and used as nonasphyxiated cesarean-delivered control rats (CS) (n=36). The uterine horns containing the remaining fetuses were immersed in a water bath at 37°C for 21 minutes to produce asphyxia-exposed rats (AS) (n=36). Following asphyxia, the uterine horns were incised, and the fetuses were delivered and stimulated to breathe. All procedures were conducted in accordance with the animal care and use protocol established by the Local Ethics Committee for Experimentation with Laboratory Animals at the Medical Faculty of Universidad de Chile. The protocol was approved by the Ethics Committee (Protocol CBA 0838 FMUCH).
Determination of the gold content of neonatal rat brain
One hour after birth, AS animals were injected ip with a single dose of 2.2 nM of the AuNR-CLPFFD/siRNA complex in a volume of 100 µL. The rats were euthanized 30 minutes, 1 hour, and 2 hours after the treatment, their brains were removed, and selected tissues were dissected. The tissue was immersed in PBS and stored in liquid nitrogen. Thereafter, the tissue was freeze-dried and pulverized to generate samples for neutronic activation analysis (NAA) to quantify the gold in the neonatal brain.
Western blots
Sampling and treatment
PC12 cell line: After removal of the medium, the cells were washed with PBS, and the medium was replaced with fresh serum-free medium for 6 hours; then, serum and one of the treatments listed below were added, and the cells were incubated for 24 hours. The cells were then washed with PBS, and total protein was extracted. The treatments were: 1) nuclease-free H2O, 2) 0.1 nM siRNA (0.6 µg), 3) FuGENE HD Transfection Reagent (3:1) (E2311; Promega Corporation), and 4) 0.1 nM AuNR-CLPFFD/siRNA complex.
Neonatal rats: One hour after birth, AS and CS animals were injected ip with a single dose of a solution of either 1) saline (100 µL), 2) 2.2 nM naked siRNA, or 3) the AuNR-CLPFFD/siRNA complex in a volume of 100 µL using a 0.5 mL insulin syringe. The neonates were nursed by a surrogate dam and euthanized 24 hours after birth; selected brain tissues were removed and dissected immediately to be assayed for PARP-1 levels by Western blotting.
Western blot analysis
Total protein extracts were mixed with sample buffer (125 mM Tris-HCl, pH 6.8, 2% SDS w/v, 20% glycerol v/v, 20 µg/µL bromophenol blue, and 5% β-mercaptoethanol), boiled for 10 minutes, separated by electrophoresis on a 10% SDS-PAGE gel, and blotted onto a BioTrace™ nitrocellulose membrane (Pall Corporation, Port Washington, NY, USA). The membranes were blocked with Tris-buffered saline (TBS) containing 0.1% Tween-20% and 5% nonfat dry milk at room temperature for 1 hour and incubated with antibodies to PARP-1 (46D11; rabbit mAb #9532; Cell Signaling Technology, Danvers, MA, USA and mouse β-actin (A5316; Sigma-Aldrich Co., St. Louis, MO, USA). The membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibody in blocking solution at 4°C. The immune complex was visualized using Pierce™ ECL Western Blotting Substrate, enhanced chemiluminescent luminol substrate (#32106; Thermo Fisher Scientific) according to the manufacturer’s instructions. The chemiluminescent signal was captured by a ChemiScope 34|00 (Clinx; Science Instruments Co., Ltd., Shanghai, China). Reactive bands were quantified by densitometry using ImageJ software.
Statistical analysis
All data were expressed as the mean ± standard error of the mean (SEM). Pairwise comparisons were evaluated by Student’s t-test or F-ANOVA followed by Dunnett’s post hoc test using GraphPad Prism® software, La Jolla, CA, USA. The significance level was set at P<0.05.
Results
Synthesis and characterization of AuNR-CLPFFD and AuNR-CLPFFD/siRNA
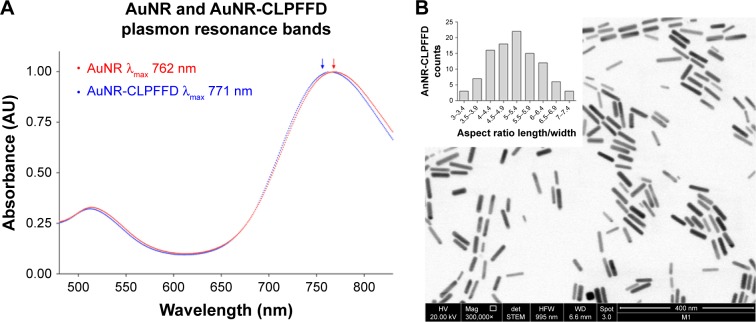
The formation of the AuNR and their conjugation with the CLPFFD peptide were monitored by following their optical properties. Figure 1A shows the UV-vis spectra of the AuNR and AuNR-CLPFFD. For AuNR, characteristic spectral bands corresponding to the transverse and longitudinal plasmon resonance bands were seen at 512 nm and 762 nm, respectively. After conjugation with CLPFFD, shifts of the longitudinal and transverse peaks to 514 nm and 771 nm, respectively, were observed.
Figure 1.
UV-visible and TEM characterization.
Notes: (A) UV-visible spectra characterization. AuNR (blue line) produces a longitudinal 512 nm peak and a transverse 762 nm peak (blue arrow). After conjugation with the CLPFFD peptide, the AuNR-CLPFFD (red line) presented a shift to a longitudinal 514 nm peak and a transverse 771 nm peak (red arrow). The graphic representation shows the peak displacement, indicating the conjugation of the CLPFFD peptide to the surface of the AuNR. (B) Characterization of the shape and aspect ratio of AuNR-CLPFFD. A representative TEM image of AuNR-CLPFFD showing the cylindrical shape of the nanoparticles is presented. The inset shows the aspect ratio histogram of the observed nanoparticles. The average aspect ratio, which was 5±0.9 (length/width), was determined by measuring 150 nanoparticles in different images.
Abbreviations: TEM, transmission electron microscopy; AuNR, gold nanorod.
TEM was used to determine the conformation, size, length, and width of the AuNR-CLPFFD particles. The particles were 50±0.7 nm in length and 10±0.1 nm in width. Figure 1B shows a TEM image of AuNR-CLPFFD. The mean aspect ratio of the AuNR-CLPFFD was 5±0.9 (length/width) as determined by measurement of 150 particles from different images.
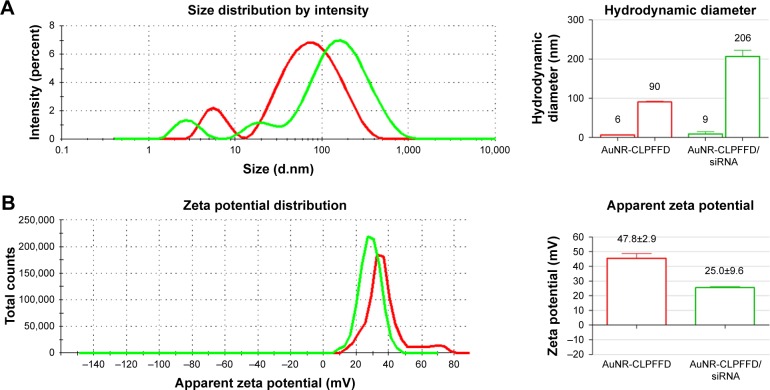
The previously synthesized and characterized AuNR-CLPFFD was incubated with siRNA against the rat PARP-1 mRNA sequence under conditions that generate electrostatic interactions. The AuNR-CLPFFD/siRNA complex formed under these conditions was characterized by dynamic light scattering and zeta potential measurements. Figure 2A shows the size distributions of the AuNR-CLPFFD (red line) and the AuNR-CLPFFD/siRNA complex (green line). The characteristic two-peak pattern of AuNR indicates the corresponding shapes of the particles. The observed hydrodynamic diameters of the AuNR-CLPFFD were 90±0.1 nm and 6±0.8 nm, with a polydispersity index (PDI) of 0.5. The hydrodynamic diameters of the AuNR-CLPFFD/siRNA complex increased to 206 nm and 9 nm (column graphic), yielding a PDI of 0.7. Figure 2B shows the zeta potential values for AuNR-CLPFFD determined at +48 mV (red line) and the shift for the AuNR-CLPFFD/siRNA complex at +25 mV (green line).
Figure 2.
Hydrodynamic diameter and zeta potential characterization.
Notes: (A) Hydrodynamic diameter of AuNR-CLPFFD and the AuNR-CLPFFD/siRNA complex. The average hydrodynamic diameter of AuNR-CLPFFD was 90 nm in length and 6 nm in width with a polydispersity index (PDI) of 0.5. The hydrodynamic diameter of the AuNR-CLPFFD/siRNA complex increased to 206 nm in length and 9 nm in width with a PDI of 0.7 (column graphic), suggesting the association of multiple AuNR-CLPFFD with siRNA molecules and their molecular rearrangement. (B) Zeta potential changes from 47.8 mV in AuNR-CLPFFD (red line) to 25 mV in AuNR-CLPFFD/siRNA complex (green line) (quantified in the column graphic) were observed, suggesting that electrostatic interactions occur between positively charged AuNR-CLPFFD and negatively charged siRNA.
Abbreviation: AuNR, gold nanorod.
The formation of the AuNR-CLPFFD/siRNA complex was confirmed by measuring the change in the electrophoretic mobility of the siRNA molecules. The interaction between AuNR-CLPFFD and the siRNA increased the diameter of the nanoparticles. The retained siRNA forming the complex retarded the migration of the complex on the agarose gel, resulting in a mobility pattern that differed from that of the free siRNA molecules. These results suggest that AuNR-CLPFFD was able to interact with anionic siRNA molecules via electrostatic interactions and that this interaction changed the migration pattern of the siRNA molecules (Figure S1).
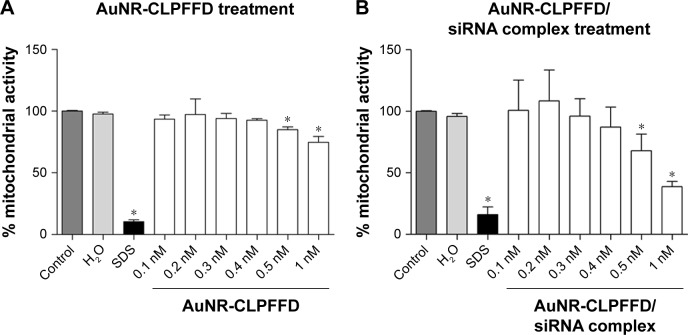
Evaluation of the effects of AuNR-CLPFFD and AuNR-CLPFFD/siRNA complexes on the viability of PC12 cells
An MTS assay was performed to evaluate the effects of AuNR-CLPFFD and AuNR-CLPFFD/siRNA complexes on cell viability. This assay allowed for the determination of the effect of the treatment on mitochondrial activity, which is directly involved in cell viability.46 Cells were exposed to 0.1–1 nM AuNR-CLPFFD or AuNR-CLPFFD/siRNA complex for 24 hours. Two additional treatments were performed as controls: the cells were treated with 1) 10% SDS solution or 2) nuclease-free H2O. Figure 3 shows that cell viability decreased after 24-hour treatment with AuNR-CLPFFD or the AuNR-CLPFFD/siRNA complex only at the higher assayed concentrations and only after sustained treatment. This result indicates that AuNR-CLPFFD and the AuNR-CLPFFD/siRNA complex led to cellular toxic effects only when applied at very high concentrations for a sustained period.
Figure 3.
Effect of AuNR-CLPFFD and AuNR-CLPFFD/siRNA complexes on PC12 cell viability.
Notes: PC12 cells were treated with increasing concentrations of (A) AuNR-CLPFFD and (B) AuNR-CLPFFD/siRNA complexes for 24 hours. The MTS assay shows decreased mitochondrial activity only in the cells that were treated with the highest concentrations (0.5–1 nM) of the compounds. Comparisons were analyzed using Student’s t-test (*P<0.05 compared to the control; n=6 for the respective experimental groups).
Abbreviations: AuNR, gold nanorod; MTS, 3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium.
Internalization of the AuNR-CLPFFD/siRNA complex into PC12 cells
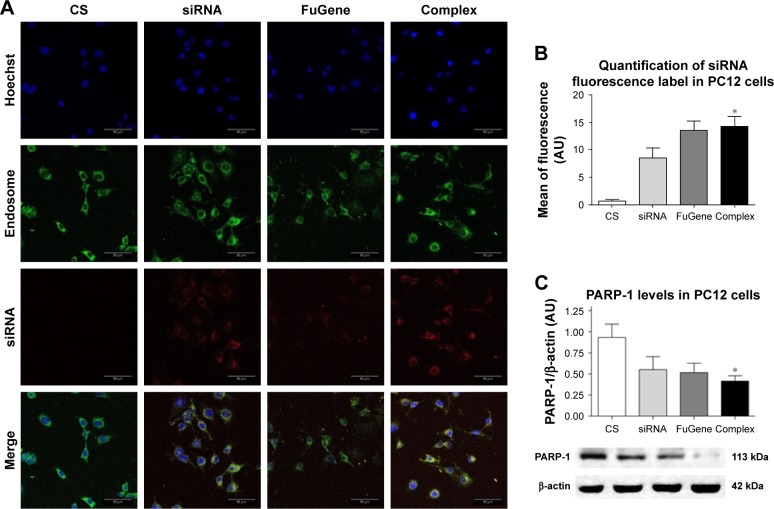
The delivery of siRNA into PC12 cells by the AuNR-CLPFFD/siRNA complex was evaluated by confocal microscopy. PC12 cells were first exposed to an early endosome fluorescent marker and were then treated for 1 hour with the AuNR-CLPFFD/siRNA complex at a concentration of 0.1 nM. Two additional controls were performed. First, a commercial transfection reagent FuGENE that had been previously loaded with 0.1 nM siRNA (3:1) was used as a positive transfection control. Second, cells were exposed to 0.1 nM naked PARP-1 siRNA molecules in the absence of vector. Figure 4A shows the cellular uptake of the AuNR-CLPFFD/siRNA complex. The nuclei, which are stained with Hoechst 33342, are seen in blue. Early endosomes (green) showed a dotted pattern that was localized to the cytoplasmic compartment; such a pattern is a characteristic feature of endosomal vesicles. The siRNA fluorescent labeling is shown in red (Alexa 647). The merged view of the three panels makes it possible to observe the apparent colocalization of the AuNR-CLPFFD/siRNA complex and early endosomes in the cytoplasm, supporting the above observation that PC12 cells can take up the complex. Quantification of the labeled siRNA by measurement of the emitted fluorescence intensity confirmed that the siRNA molecules were taken up by the cells, suggesting the existence of a more efficient cell entry mechanism for the AuNR-CLPFFD/siRNA complex than for the commercial reagent and the naked siRNA molecules (Figure 4B). Figure S3 show a micrograph of a single cell that was treated with 0.1 nM AuNR-CLPFFD/siRNA for 1 hour. The set of images shows the colocalization of early endosome vesicles and the AuNR-CLPFFD/siRNA complex (Supplementary materials).
Figure 4.
Cellular uptake of the AuNR-CLPFFD/siRNA complex and knockdown of PARP-1 protein.
Notes: (A) PC12 cells treated with 1) nuclease-free H2O (CS), 2) naked PARP-1 siRNA (siRNA, 0.1 nM), 3) FuGENE® loaded with siRNA (3:1) (FuGene), or 4) AuNR-CLPFFD/siRNA complex (Complex, 0.1 nM) 1 hour after the treatment. Images were obtained by confocal microscopy: Hoechst 33342-stained nuclei (blue), early endosomes (green), fluorescent siRNA label dye 647 (red). The images shown represent the merging of three pictures taken at 1 µm intervals. Scale bar: 50 µm; magnification, 60×. The image suggests the colocalization of siRNA with the dotted pattern of early endosomes. (B) Relative quantification of siRNA fluorescence. (C) Quantification of PARP-1 protein levels in PC12 cells. The results show that the complex was able to transport the siRNA into the cells at 1 hour post-treatment. The siRNA inhibited the expression of PARP-1 protein 24 hours post-treatment (*P<0.03 compared to the saline control; F-ANOVA followed by Dunnett’s post hoc test [n=4 for each group]).
Abbreviations: AuNR, gold nanorod; CS, cesarean-delivered control rats.
We also compared the internalization of the complexes at different temperatures and observed a decrease in cell inter-nalization at 4°C. Figure S2 show that the mean detected fluorescence per cell after treatment with the AuNR-CLPFFD/siRNA complex significantly decreased in cells incubated at 4°C compared with cells incubated at 37°C. This result could be attributed to uptake of the AuNR-CLPFFD/siRNA complex in PC12 cells by an energy-dependent endocytosis mechanism (Supplementary materials).
Efficiency of the siRNA-PARP-1 molecular knockdown
The siRNA sequence used in this study was previously described by Huang et al.43 To evaluate the efficiency of the siRNA-PARP-1 knockdown of PARP-1 mRNA, PARP-1 protein levels were quantified by Western blotting (Figure 4C). For this purpose, a monoclonal antibody against PARP-1 (~113 kDa) was used together with a monoclonal antibody against β-actin (~42 kDa), a housekeeping and loading control protein. PARP-1 protein levels were evaluated in whole protein extracts obtained from PC12 cells. The cells were exposed to the previously described treatments for 24 hours to allow the siRNA molecules to inhibit the translation of PARP-1 mRNA. As shown in Figure 4C, the levels of PARP-1 were decreased by the treatments, reaching a statistically significant level in cells treated with the AuNR-CLPFFD/siRNA complex.
Determination of the gold content of neonatal rat brain
To evaluate the capacity of the AuNR-CLPFFD/siRNA complex to reach the brains of neonatal rats, 60-minute-old neonates were injected ip with a single dose (100 µL) of 2.2 nM AuNR-CLPFFD/siRNA. The presence of gold in the brains of AS and CS animals was quantified at 30 minutes, 1 hour, and 2 hours after the treatment. The samples were analyzed by NAA. Figure 5, which shows the quantification of gold levels in the neonatal rat brains, reveals the presence of gold in the brains of asphyxia-exposed and control pups. No significant differences in gold content between control and asphyxia-exposed pups were observed, and similar levels were observed at all of the analyzed times.
Figure 5.
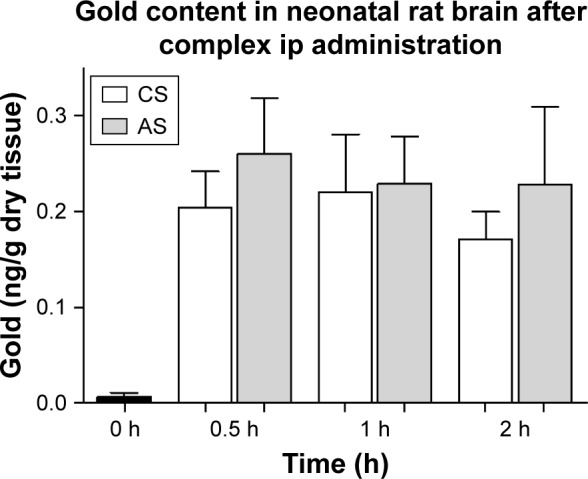
Gold content of neonatal rat brain.
Notes: Distribution of gold (ng/g dry tissue) in brain samples from AuNR-CLPFFD/siRNA complex treated neonatal rats after a single dose (2.2 nM, ip). Samples were obtained at 30 minutes, 1 hour, and 2 hours after treatment. The result shows that gold accumulates in the brains of the neonatal rats after passing through the BBB. No differences between CS and AS animals were observed. ANOVA followed by Dunnett’s post hoc test (n=6 for each group).
Abbreviations: AuNR, gold nanorod; ip, intraperitoneal; CS, cesarean-delivered control rats; AS, asphyxia-exposed rats; BBB, blood–brain barrier.
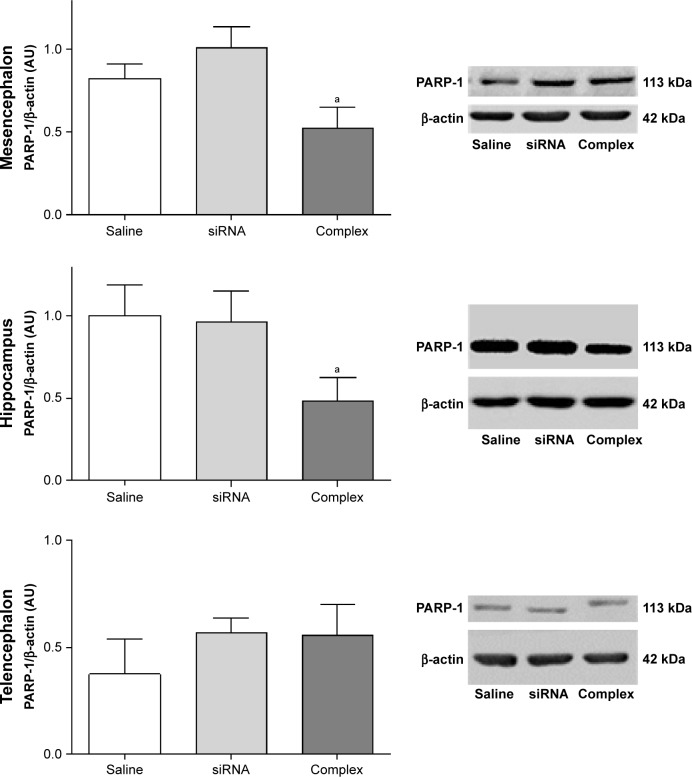
Knockdown of the expression of PARP-1 in vivo
The effect of treatment with the AuNR-CLPFFD/siRNA complex on PARP-1 expression in AS animals was analyzed by Western blotting of brain tissue obtained from three areas: the mesencephalon, the hippocampus, and the telencephalon (Figure 6). The animals were treated with a solution containing either saline, naked siRNA, or the AuNR-CLPFFD/siRNA complex. Samples were collected 24 hours post-treatment and processed to obtain whole-cell protein extracts.
Figure 6.
In vivo knockdown of PARP-1 in AS-exposed animals.
Notes: Administration of a single dose of AuNR-CLPFFD/siRNA complex (ip) to AS-exposed animals. The animals were treated with 1) saline solution (Saline), 1) naked siRNA (siRNA), or 3) AuNR-CLPFFD/siRNA complex (Complex) 1 hour after birth. Total protein extracts were obtained 24 hours after treatment. PARP-1 protein levels in the mesencephalon, hippocampus, and telencephalon were quantified by Western blotting. The quantification shows that treatment with the AuNR-CLPFFD/siRNA complex decreased PARP-1 levels in the mesencephalon and hippocampus (aP<0.05 compared to the saline treatment, Student’s t-test; n=6 for each condition).
Abbreviations: AuNR, gold nanorod; ip, intraperitoneal; AS, asphyxia-exposed rats.
Treatment with the AuNR-CLPFFD/siRNA complex decreased PARP-1 levels in the mesencephalon and hippocampus of AS rats, demonstrating that the complex is able to knockdown the expression of PARP-1, as proposed. However, treatment with the AuNR-CLPFFD/siRNA complex did not result in any change in PARP-1 levels in the telencephalon.
The effects of treatment of control animals with the AuNR-CLPFFD/siRNA complex on PARP-1 expression in the mesencephalon, the telencephalon, and the hippocampus were also analyzed. Figure S4 show the differential response in each analyzed region. Treatment with the AuNR-CLPFFD/siRNA complex decreased the expression of PARP-1 in the mesencephalon and telencephalon but not in the hippocampus (Supplementary materials).
Discussion
PARP-1 knockdown has been proposed as a target for therapeutic strategies designed to alleviate the consequences of hypoxia and oxidative stress, thereby improving survival, minimizing secondary effects, and re-establishing normal development.47,48 This proposal is supported by experimental evidence showing that nicotinamide decreases the cellular deficit of NAD+, lessening the damage produced by PARP-1 overactivation.10,49 To further test the idea of PARP-1 over-activation, we used siRNA to knockdown the expression of PARP-1, expecting to decrease PARP-1 expression and overactivation.50–52
The physicochemical properties of siRNAs (including their size, hydrophilicity, negative charge, and sensitivity to nuclease degradation) hinder transfection. Thus, in this study, we used structurally modified siRNA molecules (siSTABLE RNAs from Dharmaco, Inc.). These modifications increase the nuclease resistance and stability of the siRNAs, causing them to be more than four times more resistant and stable than unmodified siRNAs.51,53
Although modification of the siRNAs increased their stability, the increase in stability was not sufficient to allow them to be used as naked siRNAs for systemic administration. For in vitro purposes, the use of a transfection reagent such as FuGENE that can penetrate into the cell was required. However, these reagents still carry toxicity risks and exhibit instability in blood, nonspecific distribution, and low transfection efficiency after systemic administration.54 Thus, significant challenges for in vivo delivery,14,55 as well as obstacles to generating a vector for use as a therapeutic agent in the CNS, remain.
AuNPs, nontoxic carriers that can be used in drug and gene delivery applications,56 bind to nucleic acid molecules by electrostatic interactions and protect the molecules from enzymatic digestion.57,58 In this work, we used peptide-functionalized AuNRs as carriers to improve the efficiency of siRNA in the treatment of PA.
We synthesized AuNRs using a method that was previously described by Adura et al.40 The AuNRs, which were initially stabilized with hexadecyltrimethylammonium bromide (CTAB), were functionalized with the CLPFFD peptide (AuNR-CLPFFD), decreasing their z-potential and displacing the CTAB. The light absorption of the complexes showed a strong band associated with the surface plasmon resonance (SPR) effect of the nanoparticles.27 Characterization of the nanoparticles by spectrophotometry (Figure 1A) showed the effect of conjugation of the CLPFFD peptide and AuNR on the SPR; the peaks changed due to chemisorption of the peptide to the surface of the nanoparticle. The spectral change, which can be attributed to the coating of the AuNR by the peptide, is generated by the stably conjugated coated nanoparticles and by a partial displacement of the CTAB.40 TEM allowed the real size of a dense electronic area (corresponding to the gold atoms) to be determined, revealing the actual diameter of the gold nanorods and their length– width proportions and confirming their cylindrical shape (Figure 1B). Hence, the size of each AuNR-CLPFFD was determined. The quantification of the AuNR concentration could then be calculated according to Adura et al40 assuming a cylindrical shape for the AuNR-CLPFFD (Supplementary materials). The absolute zeta potential value of the AuNR-CLPFFD was 48 mV (Figure 2B), indicating that electrostatic interactions with siRNA molecules occurred during the generation of an AuNR-CLPFFD/siRNA complex. Thus, the AuNR-CLPFFD/siRNA complex was created as a result of electrostatic interactions between the negatively charged siRNA molecules (due to the presence of phosphate in the bond structure of the nucleic acid) and the positively charged AuNR-CLPFFD. In addition, we report here an increase in the hydrodynamic diameter of the nanoparticles from 90 nm to 206 nm that is attributable to multiple interactions of the AuNR-CLPFFD with the siRNA molecules (Figure 2A). We observed a PDI of 0.7. The high PDI value can be attributed to the presence of two peaks in the gold nanorods, attributed to the effect of the shape, the particle diffusion, and the interaction with the light. The diffusion coefficient of a particle depends not only on the mass of the particle but also on its shape and surface chemistry because these parameters affect the particle–solvent interactions and therefore the Brownian motion of the particles. Additionally, it has been shown that the intensity distribution increases significantly with increased aspect ratio.59 The interaction of siRNA with the complex was demonstrated by the change in the electrophoretic mobility of the siRNA on agarose gels when the complex was prepared at a concentration ratio of 9 nM (100 µL)/0.2 nmol (Figure S1).
The effects on cell viability and the delivery capacity of the synthesized AuNR-CLPFFD/siRNA complex were studied in a PC12 rat cell line. A decrease in cell viability was only observed at the highest tested concentrations (0.5–1 nM) (Figure 3). We would expect that under in vivo conditions the concentration of the nanoparticles should rapidly decrease, making it unlikely that the nanoparticles will reach toxic concentrations in vivo.
A fluorescent label (Alexa647) (λ max abs 645 nm, λ max em 662 nm) was attached to the 3′ end of the antisense strand to make it possible to visualize and monitor the siRNA molecule and to track the internalization of the AuNR-CLPFFD/siRNA complex (0.1 nM) into the cell. Evaluation of the internalization by confocal microscopy demonstrated that the AuNR-CLPFFD/siRNA complex was able to cross the cell membrane and be taken up by PC12 cells, likely colocalizing with early endosomes. This result strongly suggests that the complex entered the cell by an endocytic mechanism (Figure 4A). Previously, Adura et al37 observed the internalization and accumulation of AuNR-CLPFFD in vesicles in SH-SY5Y neuroblastoma cells40 as well as in an in vitro blood–brain barrier model.60 We also assayed the uptake of the complex at low temperature (4°C) and found it to be decreased, indicating that the mechanism of internalization was energy-dependent endocytosis61,62 (Figure S2).
The uptake of the complex decreased PARP-1 protein levels in PC12 cells by 50% compared to the corresponding controls (Figure 4C), demonstrating the usefulness of the complex as a carrier and showing that it protected the integrity of the siRNA molecule while favoring its interaction with PARP-1 mRNA. As a whole, the present in vivo experiments support the hypothesis that 1) the AuNR-CLPFFD/siRNA complex safely transports siRNA molecules into cells; 2) the complex maintains the integrity of the siRNA molecules; and 3) the complex is able to knockdown the expression of a particular protein (in the present study, PARP-1). These results confirm previous observations by confocal microscopy. Tatiparti et al63 recently described some strategies for nanoparticle delivery of siRNA molecules.
It is important to note that to achieve inhibition of the PARP-1 produced by the siRNA, it is necessary for the siRNA to escape from the endosomes to the cytoplasm.64 The endosomal escape of the siRNA to the cytoplasm could occur as a result of dissociation of the complex within the endosome due to its acidic pH,65 considering that the peptide CLPFFD acquires a positive charge (isoelectric point 5.6).66 The positive charge triggers an interaction between the carrier and the endosomal membrane for endosomal escape.67–69 However, the exact mechanism of endosomal escape of many siRNA delivery carriers is still poorly understood.70
For in vivo characterization, we used a model of global PA,2 administering the complex in a single dose (i.p.) one hour after birth. The dose was calculated and adjusted for body weight based on a previous report in adult rats.37 None of the treated pups died or showed any evidence of physical impairment for 24 hours, when the pups were sacrificed for tissue sampling.
In this study, we measured gold concentrations in specific brain regions 30 minutes, 1 hour and 2 hours after systemic administration and compared the levels achieved in the brains of asphyxia-exposed and control rats. The results demonstrate that following a single dose of the AuNR-CLPFFD/siRNA complex (2.2 nM in 100 µL, i.p.), the complex reached the targeted tissue. No differences in brain gold levels were observed between asphyxia-exposed and control animals (Figure 5). The amount of gold detected in the brain was ~0.04% of the injected dose. This result is consistent with previous work37,71 showing that a single dose (i.p.) of spherical AuNPs conjugated to the CLPFFD peptide crossed the BBB of adult rats. It can be suggested that the CLPFFD peptide, which has an amino acidic sequence that is identical to that of the βA peptide, enters the brain via the receptor for advanced glycation end-products (RAGE) in a process that is critical for the regulation of βA homeostasis. RAGE, a βA receptor found on the BBB and on the surfaces of neurons and microglia, is also the major endothelial receptor for βAinflux into the brain.72 These experiments show for the first time that AuNPs ip administered to neonatal rats cross the BBB and successfully reach the brain without affecting the survival of the animals.
The capacity of the complex to knockdown the in vivo expression of PARP-1 in the mesencephalon, hippocampus and telencephalon was evaluated by Western blotting. Treatment with the AuNR/CLPFFD-siRNA complex decreased PARP-1 levels in the mesencephalon and hippocampus of asphyxia-exposed animals, whereas treatment with the naked siRNA had no effect, in agreement with the results of in vitro experiments in PC12 cells.
The present study suggests that there is a region-dependent vulnerability of the brain to the effects of PA that can be targeted by treatment with the AuNR/CLPFFD-siRNA complex; individual brain regions respond differently to the treatment, probably depending upon their PARP-1 levels. The specific history of brain tissues with respect to hypoxic and/or reoxygenation insults, as well as different postnatal maturational timing, primes PARP-1 expression and/or PARP-1 activity.73
We also observed a decrease in PARP-1 expression in the mesencephalon and telencephalon of control animals treated with the complex. Furthermore, decreased PARP-1 expression was observed after treatment with naked siRNA. However, these effects were not detected in asphyxia-exposed animals. Further investigation will be necessary to determine the basis for this observation (Figure S4).
Conclusion
A nanocarrier (AuNR-CLPFFD) that interacts with siRNA molecules was prepared. This nanocarrier forms a stable complex (AuNR-CLPFFD/siRNA) and facilitates endosomal release. The capacity of the complex to transport siRNA molecules was demonstrated in PC12 cells, and it was shown to knockdown PARP-1 expression.
The in vivo experiments demonstrated that AuNR-CLPFFD/siRNA complex administered ip into neonatal rats is able to cross the BBB, successfully reach the brain, and decrease PARP-1 expression in the mesencephalon and hippocampus of asphyxia-exposed animals. The effect of the treatment upon the reported vulnerability of brain regions, ie, the mesencephalon and hippocampus, was region dependent. The use of functionalized gold nanorods represents a promising platform for delivery of siRNA into the brain.
Supplementary materials
Methods
Synthesis of gold nanorods (AuNR) and conjugation with CLPFFD peptide
In a first step, a seed solution was prepared by reduction of 250 µL of HAuCl4 in a solution of 9.75 mL of 0.1 M hexadecyltrimethylammonium bromide (CTAB) with a cold-prepared sodium borohydride (600 µL, 0.01 M). Seeds were kept at 27°C for 2 hours before use. Next, 55 µL of 0.1 M ascorbic acid was added to a growth solution, containing 75 µL of 0.01 M AgNO3, 9.5 mL of 0.1 M CTAB, and 500 µL of HAuCl4 0.01 M. Then, 250 µL of 0.1 M HCl and 12 µL of the previously prepared seed solution were added. The solution was allowed to react at 27°C for 10 minutes before centrifugation at 7,030× g for 15 minutes. After centrifugation, the supernatant was removed, and the pellet was resuspended in milli-Q water.
Quantification of AuNR-CLPFFD
An aliquot of a known volume of AuNR solution was lyophilized and gold concentration (CAu) was determined by neutron activation analysis (NAA), at the Chilean Nuclear Energy Commission (CCHEN). The samples were sealed by friction welding and exposed for 17 hours to a neutron flux of 0.25–1.3 n/cm2·s with a power of 5 mW using a RECH-1 reactor at the CCHEN, thereby triggering the conversion of 197Au to 198Au. After 7–12 days of neutron bombardment, the γ-rays emitted by the samples were counted and sorted by energies using a germanium detector coupled to a PC-based multichannel γ-ray spectrometer. The γ-ray spectra were analyzed with the software GENIE 2000 Canberra. Gold standards were run with the experimental samples to standardize a library of gold element data for calculating the amount of gold. Considering that the detection limit by neutron activation can be influenced by the elemental composition of the sample, background levels were determined by irradiating untreated tissue samples (control) of similar size and composition to that shown by the experimental samples.
The final uncertainty is evaluated by combining the individual components with that contributed by neutron activation: µ-weight of the sample, µ-weight of the standard reference material, µ-measure of the analyte in the sample, µ-measure of the analyte in the standard, µ-neutron flux of the standard reference material, µ-neutron flux of the sample, and µ-correction of the dead time in the sample and in the standard referent material. The informed uncertainty corresponds to the expanded uncertainty (U), which was calculated by a cover factor equal to 2. This factor is equivalent to a level of confidence of 95%.
With the quantity of gold (CAu) data and the known length and width averages of the AuNR-CLPFFD (both determined by TEM), the concentration (nanomolar) of AuNR-CLPFFD was determined as follows:
ρAu: density of gold atoms (59 át/nm3); CAu: gold concentration determined by NAA (CCHEN); L: length of AuNR previously determinate by TEM (50 nm); W: width of AuNR previously determined by TEM (10 nm).
siRNA sequence
The used sequence was:
Sense sequence: DY647-5′-GGA UGA UCU UCG ACG UGG AUU-3′.
Antisense sequence: 5′-P UCC ACG UCG AAG AUC AUC CUU-3′.
Custom siRNAs synthesized with the siSTABLE modification are sequences specially modified to provide nuclease resistance. The final product and modification were described by the manufacturer (Dharmacon, Inc., Lafayette, CO, USA) as siSTABLE. For more information: http://dharmacon.horizondiscovery.com/rnai/controls/sirna/reporter-sistable-other/#overview.
Dynamic light scattering and z-potential measurement
Smolochowski’s approximation was used to calculate the zeta potential from the measured electrophoretic mobility. Each sample was measured in triplicate using a disposable polycarbonate capillary cell (DTS 1061; Malvern Instruments) under precise temperature control (25°C).
Determination of the electrophoretic mobility
The formation of the complex AuNR-CLPFFD/siRNA was evaluated by the delay in the electrophoretic mobility of the siRNA. The interaction between AuNR-CLPFFD and the siRNA enlarged the diameter of the nanoparticles and retained the siRNA molecules in the gel. The AuNR-CLPFFD was gently mixed with the siRNA at various weight ratios, and incubated for 5 minutes at room temperature. The complexes were then characterized by 0.8% agarose gel electrophoresis for 10 minutes at 100 V in Tris base, acetic acid and ethylenediaminetetraacetic acid buffer solution. The gel was stained with ethidium bromide and the image was taken under UV illumination.
PC12 cells treatment for confocal microscopy
More information about the cell line can be obtained at https://www.atcc.org/products/all/CRL-1721.aspx#documentation. About 200,000 cells/well were seeded in a six-well plate pretreated with poly-l-lysine and allowed to get attached for 24 hours at 37°C.
PC12 cells treatment for flow cytometry
About 1×106 PC12 cells were washed with PBS, removed the media, replaced with fresh serum-free media, and then treated with 1) nuclease-free H2O at 37°C for 1 hour, 2) 0.1 nM AuNR-CLPFFD/siRNA complex at 37°C for 1 hour, and 3) 0.1 nM AuNR-CLPFFD/siRNA complex at 4°C for 1 hour. Samples were washed with PBS and the fluorescent cells were analyzed by flow cytometry using BD FACSCanto A (Becton Dickinson, San José, CA, USA).
Animal model
Wistar albino rats from the animal station of the Molecular & Clinical Pharmacology Program, Instituto de Ciencias Biomédicas (ICBM), Faculty of Medicine, University of Chile, Santiago, Chile, were used. The animals were kept on a temperature and humidity-controlled environment with a 12/12-hour light/dark cycle and fed ad libitum, when not used in the experiments. Neonate rat treatment was performed with a 0.5 mL insulin syringe.
Results
Change in the electrophoretic mobility
Cellular uptake and endocytosis mechanism
The aim for incubating the cells at 4°C was to test whether the uptake mechanism was by energy-dependent endocytosis. The method consisted of increasing intracellular energy consumption by reducing the temperature to cut down ATP levels inside the cells, which can indirectly inhibit endocytosis. The uptake efficacy was analyzed by flow cytometry. Figure S2 shows that the fluorescent uptake was decreased when cells were incubated al 4°C, demonstrating that the uptake mechanism was by endocytosis.
AuNR-CLPFFD/siRNA complex formation evaluated by changes of the electrophoretic mobility pattern.
Notes: (A) Agarose electrophoresis under non-denaturing conditions. Lane 1 shows the migration pattern of 0.6 µg of free siRNA (0.2 nmol). Lane 2 shows the conjugation of 9 nM AuNR-CLPFFD with 0.6 µg of siRNA. Lane 3 shows 0.9 nM of AuNR-CLPFFD with 0.6 µg of siRNA and lane 4 shows 0.2 nM of AuNR-CLPFFFD with 0.6 µg of siRNA. These results show that at 9 nM of AuNR-CLPFFD, the nanoparticles can form electrostatic interaction with siRNA molecules, retaining the siRNA molecules forming the AuNR-CLPFFD/siRNA complex, allowing to observe a difference in the electrophoretic mobility of free siRNA molecules. In contrast, lanes 3 and 4 show that nanoparticles were not concentrated enough, leaving high amount of free siRNA molecules in solution and conserving the migration pattern of free siRNA. (B) A schematic representation of the siRNA molecules with AuNR-CLPFFD.
Abbreviation: AuNR, gold nanorod.
Cellular uptake is by endocytosis.
Notes: PC12 cells were treated with 1) nuclease-free H2O at 37°C (CS), 2) 0.1 nM AuNR-CLPFFD/siRNA complex at 37°C (Complex), or 3) 0.1 nM AuNR-CLPFFD/siRNA complex at 4°C (Complex) for 1 hour. The mean of the quantified fluorescence was measured by flow cytometry expressed in arbitrary units. The result shows that the fluorescent uptake was decreased when cells were incubated al 4°C, demonstrating that the uptake mechanism was by endocytosis. Inset shows a representative population of analyzed cells (*P<0.05 compared to the saline control); F-ANOVA followed by Bonferroni’s multiple comparison test (n=3 for each group).
Abbreviations: AuNR, gold nanorod; CS, cesarean-delivered control rats.
Single cell image of cellular uptake of the AuNR-CLPFFD/siRNA complex.
Notes: PC12 cells were treated with 0.1 nM AuNR-CLPFFD/siRNA complex for 1 hour. Images were obtained by confocal microscopy: (A) transmitted light microphotograph, (B) Hoechst 33342 stained nuclei (blue), (C) early endosomes (green), (D) fluorescent siRNA label dye647 (red), and (E) merge of (B, C, and D) images. The result shows the colocalization of early endosome vesicles and the AuNR-CLPFFD/siRNA complex. Images are the merge of three pictures taken every 1 µm. Scale bar: 20 µm; 60× optic, 3.1 optic zoom.
Abbreviation: AuNR, gold nanorod.
In vivo knockdown of PARP-1 in control animals.
Notes: Administration of a single dose of AuNR-CLPFFD/siRNA complex (ip) to control animals. Treatment: 1) saline solution (Saline); 2) naked siRNA (siRNA), or 3) AuNR-CLPFFD/siRNA complex (Complex), 1 hour after birth. Total protein extracts were obtained 24 hours after the treatment. PARP-1 protein levels were quantified by Western blots, in mesencephalon, hippocampus, and telencephalon. The quantification showed that in mesencephalon and telencephalon, the AuNR-CLPFFD/siRNA complex treatment decreased PARP-1 levels. We also showed the decreasing levels of PARP-1 in rats treated with naked siRNA administration. Further study will be needed to explain the nature of this effect (aP<0.05 compared to the saline control; Student’s t-test; n=6 for each condition).
Abbreviations: AuNR, gold nanorod; ip, intraperitoneal.
Acknowledgments
The contract grant sponsors: FONDECYT-Chile (contracts: 1080447 and 11070192 to MHM and PM, and 1170929 to MJK); Fondap (15130011 to MJK); and MECESUP-Chile fellows (UCH0704 to VV). EA acknowledges Proyecto Nucleo DI-36-18/N. The excellent technical and secretarial help from Mr Juan Santibanez and Ms Carmen Almeyda is kindly acknowledged. We thank Cecilia Zúñiga for valuable help with the cytometer measurements.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Fattuoni C, Palmas F, Noto A, Fanos V, Barberini L. Perinatal asphyxia: a review from a metabolomics perspective. Molecules. 2015;20(4):7000–7016. doi: 10.3390/molecules20047000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrera-Marschitz M, Morales P, Leyton L, et al. Perinatal asphyxia: current status and approaches towards neuroprotective strategies, with focus on sentinel proteins. Neurotox Res. 2011;19(4):603–627. doi: 10.1007/s12640-010-9208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26(8):882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 4.Miwa M, Ida C, Yamashita S, Tanaka M, Fujisawa J. Poly(ADP-ribose): Structure, Physicochemical Properties and Quantification in vivo, with special reference to poly(ADP-ribose) binding protein modules. Curr Protein Pept Sci. 2016;17(7):683–692. doi: 10.2174/1389203717666160419145246. [DOI] [PubMed] [Google Scholar]
- 5.Hegedűs C, Virág L. Inputs and outputs of poly(ADP-ribosyl)ation: Relevance to oxidative stress. Redox Biol. 2014;2(2c):978–982. doi: 10.1016/j.redox.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim MY. Regulation of chromatin structure by PARP-1. Methods Mol Biol. 2011;780:227–236. doi: 10.1007/978-1-61779-270-0_14. [DOI] [PubMed] [Google Scholar]
- 7.Kraus WL, Hottiger MO. PARP-1 and gene regulation: progress and puzzles. Mol Aspects Med. 2013;34(6):1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp Cell Res. 2014;329(1):18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 9.de Murcia G, Ménissier de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci. 1994;19(4):172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 10.Neira-Peña T, Rojas-Mancilla E, Munoz-Vio V, et al. Perinatal asphyxia leads to PARP-1 overactivity, p65 translocation, IL-1β and TNF-α over-expression, and apoptotic-like cell death in mesencephalon of neonatal rats: prevention by systemic neonatal nicotinamide administration. Neurotox Res. 2015;27(4):453–465. doi: 10.1007/s12640-015-9517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haskó G, Mabley JG, Németh ZH, Pacher P, Deitch EA, Szabó C. Poly(ADP-ribose) polymerase is a regulator of chemokine production: relevance for the pathogenesis of shock and inflammation. Mol Med. 2002;8(5):283–289. [PMC free article] [PubMed] [Google Scholar]
- 12.Allende-Castro C, Espina-Marchant P, Bustamante D, et al. Further studies on the hypothesis of PARP-1 inhibition as a strategy for lessening the long-term effects produced by perinatal asphyxia: effects of nicotinamide and theophylline on PARP-1 activity in brain and peripheral tissue: nicotinamide and theophylline on PARP-1 activity. Neurotox Res. 2012;22(1):79–90. doi: 10.1007/s12640-012-9310-2. [DOI] [PubMed] [Google Scholar]
- 13.Dykxhoorn DM, Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu Rev Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- 14.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123(Pt 8):1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 15.Pardridge WM. The blood-brain barrier: bottleneck in brain drug development. NeuroRx. 2005;2(1):3–14. doi: 10.1602/neurorx.2.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durand S, Cimarelli A. The inside out of lentiviral vectors. Viruses. 2011;3(2):132–159. doi: 10.3390/v3020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srikanth M, Kessler JA. Nanotechnology-novel therapeutics for CNS disorders. Nat Rev Neurol. 2012;8(6):307–318. doi: 10.1038/nrneurol.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109(2):259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 19.Martin SE, Caplen NJ. Applications of RNA interference in mammalian systems. Annu Rev Genomics Hum Genet. 2007;8:81–108. doi: 10.1146/annurev.genom.8.080706.092424. [DOI] [PubMed] [Google Scholar]
- 20.Kim BY, Rutka JT, Chan WC. Nanomedicine. N Engl J Med. 2010;363(25):2434–2443. doi: 10.1056/NEJMra0912273. [DOI] [PubMed] [Google Scholar]
- 21.Velasco-Aguirre C, Morales F, Gallardo-Toledo E, et al. Peptides and proteins used to enhance gold nanoparticle delivery to the brain: preclinical approaches. Int J Nanomedicine. 2015;10:4919–4936. doi: 10.2147/IJN.S82310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velasco-Aguirre C, Morales-Zavala F, Salas-Huenuleo E, et al. Improving gold nanorod delivery to the central nervous system by conjugation to the shuttle Angiopep-2. Nanomedicine. 2017;12(20):2503–2517. doi: 10.2217/nnm-2017-0181. [DOI] [PubMed] [Google Scholar]
- 23.Mccully M, Sanchez-Navarro M, Teixido M, Giralt E. Peptide Mediated Brain Delivery of Nano- and Submicroparticles: a Synergistic Approach. Curr Pharm Des. 2018;24(13):1366–1376. doi: 10.2174/1381612824666171201115126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawasaki ES, Player A. Nanotechnology, nanomedicine, and the development of new, effective therapies for cancer. Nanomedicine. 2005;1(2):101–109. doi: 10.1016/j.nano.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Doane TL, Burda C. The unique role of nanoparticles in nanomedicine: imaging, drug delivery and therapy. Chem Soc Rev. 2012;41(7):2885–2911. doi: 10.1039/c2cs15260f. [DOI] [PubMed] [Google Scholar]
- 26.Cabuzu D, Cirja A, Puiu R, Grumezescu AM. Biomedical applications of gold nanoparticles. Curr Top Med Chem. 2015;15(16):1605–1613. doi: 10.2174/1568026615666150414144750. [DOI] [PubMed] [Google Scholar]
- 27.Daniel MC, Astruc D. Gold nanoparticles: assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem Rev. 2004;104(1):293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 28.Dreaden EC, Austin LA, Mackey MA, El-Sayed MA. Size matters: gold nanoparticles in targeted cancer drug delivery. Ther Deliv. 2012;3(4):457–478. doi: 10.4155/tde.12.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai M, Ingle AP, Birla S, Yadav A, Santos CA. Strategic role of selected noble metal nanoparticles in medicine. Crit Rev Microbiol. 2015;3:1–24. doi: 10.3109/1040841X.2015.1018131. [DOI] [PubMed] [Google Scholar]
- 30.Roda LG, Nolan JA, Kim SU, Hogue-Angeletti RA. Isolation and characterization of chromaffin granules from a pheochromocytoma (PC 12) cell line. Exp Cell Res. 1980;128(1):103–109. doi: 10.1016/0014-4827(80)90392-4. [DOI] [PubMed] [Google Scholar]
- 31.Fornai F, Lenzi P, Lazzeri G, et al. Fine ultrastructure and biochemistry of PC12 cells: a comparative approach to understand neurotoxicity. Brain Res. 2007;1129(1):174–190. doi: 10.1016/j.brainres.2006.10.071. [DOI] [PubMed] [Google Scholar]
- 32.Jung JY, Kim WJ. Involvement of mitochondrial- and Fas-mediated dual mechanism in CoCl2-induced apoptosis of rat PC12 cells. Neurosci Lett. 2004;371(2–3):85–90. doi: 10.1016/j.neulet.2004.06.069. [DOI] [PubMed] [Google Scholar]
- 33.Seta KA, Spicer Z, Yuan Y, Lu G, Millhorn DE. Responding to hypoxia: lessons from a model cell line. Sci STKE. 2002;2002(146):re11. doi: 10.1126/stke.2002.146.re11. [DOI] [PubMed] [Google Scholar]
- 34.Vavilis T, Delivanoglou N, Aggelidou E, et al. Oxygen-Glucose Deprivation (OGD) Modulates the Unfolded Protein Response (UPR) and Inflicts Autophagy in a PC12 Hypoxia Cell Line Model. Cell Mol Neurobiol. 2016;36(5):701–712. doi: 10.1007/s10571-015-0250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee M, Kang H, Jang SW. CoCl2 induces PC12 cells apoptosis through p53 stability and regulating UNC5B. Brain Res Bull. 2013;96:19–27. doi: 10.1016/j.brainresbull.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Bonoiu AC, Bergey EJ, Ding H, et al. Gold nanorod – siRNA induces efficient in vivo gene silencing in the rat hippocampus. Nanomedicine. 2011;6(4):617–630. doi: 10.2217/nnm.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerrero S, Araya E, Fiedler JL, et al. Improving the brain delivery of gold nanoparticles by conjugation with an amphipathic peptide. Nanomedicine. 2010;5(6):897–913. doi: 10.2217/nnm.10.74. [DOI] [PubMed] [Google Scholar]
- 38.Guerrero S, Herance JR, Rojas S, et al. Synthesis and in vivo evaluation of the biodistribution of a 18F-labeled conjugate gold-nanoparticle-peptide with potential biomedical application. Bioconjug Chem. 2012;23(3):399–408. doi: 10.1021/bc200362a. [DOI] [PubMed] [Google Scholar]
- 39.Li M, Guan Y, Zhao A, Ren J, Qu X. Using Multifunctional Peptide Conjugated Au Nanorods for Monitoring β-amyloid Aggregation and Chemo-Photothermal Treatment of Alzheimer’s Disease. Theranostics. 2017;7(12):2996–3006. doi: 10.7150/thno.18459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adura C, Guerrero S, Salas E, et al. Stable conjugates of peptides with gold nanorods for biomedical applications with reduced effects on cell viability. ACS Appl Mater Interfaces. 2013;5(10):4076–4085. doi: 10.1021/am3028537. [DOI] [PubMed] [Google Scholar]
- 41.Nikoobakht B, El-Sayed MA. Preparation and Growth Mechanism of Gold Nanorods (NRs) Using Seed-Mediated Growth Method. Chem Mater. 2003;15(10):1957–1962. [Google Scholar]
- 42.Jana NR, Gearheart L, Murphy CJ. Wet Chemical Synthesis of High Aspect Ratio Cylindrical Gold Nanorods. J Phys Chem B. 2001;105(19):4065–4067. [Google Scholar]
- 43.Huang D, Yang C, Wang Y, Liao Y, Huang K. PARP-1 suppresses adiponectin expression through poly(ADP-ribosyl)ation of PPAR gamma in cardiac fibroblasts. Cardiovasc Res. 2009;81(1):98–107. doi: 10.1093/cvr/cvn264. [DOI] [PubMed] [Google Scholar]
- 44.Wang S, Kool ET. Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2′-hydroxyl effects. Biochemistry. 1995;34(12):4125–4132. doi: 10.1021/bi00012a031. [DOI] [PubMed] [Google Scholar]
- 45.Sioud M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: a central role for 2′-hydroxyl uridines in immune responses. Eur J Immunol. 2006;36(5):1222–1230. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]
- 46.Bahadar H, Maqbool F, Niaz K, Abdollahi M. Toxicity of Nanoparticles and an Overview of Current Experimental Models. Iran Biomed J. 2016;20(1):1–11. doi: 10.7508/ibj.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Virág L, Szabó C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54(3):375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 48.Azzopardi DV, Strohm B, Edwards AD, et al. TOBY Study Group Moderate hypothermia to treat perinatal asphyxial encephalopathy. N Engl J Med. 2009;361(14):1349–1358. doi: 10.1056/NEJMoa0900854. [DOI] [PubMed] [Google Scholar]
- 49.Raina A, Pandita A, Harish R, Yachha M, Jamwal A. Treating perinatal asphyxia with theophylline at birth helps to reduce the severity of renal dysfunction in term neonates. Acta Paediatr. 2016;105(10):e448–e451. doi: 10.1111/apa.13469. [DOI] [PubMed] [Google Scholar]
- 50.Lee SJ, Son S, Yhee JY, et al. Structural modification of siRNA for efficient gene silencing. Biotechnol Adv. 2013;31(5):491–503. doi: 10.1016/j.biotechadv.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 51.Lam JK, Chow MY, Zhang Y, Leung SW. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol Ther Nucleic Acids. 2015;4(9):e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mcmanus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3(10):737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 53.Scaringe SA, Kitchen D, Kaiser RJ, Marshall WS. Preparation of 5′-silyl-2′-orthoester ribonucleosides for use in oligoribonucleotide synthesis. Curr Protoc Nucleic Acid Chem. 2004:10. doi: 10.1002/0471142700.nc0210s16. Chapter 2:Unit 2. [DOI] [PubMed] [Google Scholar]
- 54.Vhora I, Patil S, Amrutiya J, Misra A. Liposomes and Lipid Envelope-Type Systems for Systemic siRNA Delivery. Curr Pharm Des. 2015;21(31):4541–4555. doi: 10.2174/138161282131151013185850. [DOI] [PubMed] [Google Scholar]
- 55.Leng Q, Woodle MC, Lu PY, Mixson AJ. Advances in Systemic siRNA Delivery. Drugs Future. 2009;34(9):721. doi: 10.1358/dof.2009.034.09.1413267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghosh P, Han G, de M, Kim CK, Rotello VM. Gold nanoparticles in delivery applications. Adv Drug Deliv Rev. 2008;60(11):1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 57.Guo S, Huang Y, Jiang Q, et al. Enhanced gene delivery and siRNA silencing by gold nanoparticles coated with charge-reversal polyelectrolyte. ACS Nano. 2010;4(9):5505–5511. doi: 10.1021/nn101638u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett. 2009;9(5):2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 59.Liu H, Pierre-Pierre N, Huo Q. Dynamic light scattering for gold nanorod size characterization and study of nanorod–protein interactions. Gold Bull. 2012;45(4):187–195. [Google Scholar]
- 60.Ruff J, Hüwel S, Kogan MJ, Simon U, Galla HJ. The effects of gold nanoparticles functionalized with β-amyloid specific peptides on an in vitro model of blood-brain barrier. Nanomedicine. 2017;13(5):1645–1652. doi: 10.1016/j.nano.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 61.Mukherjee S, Ghosh RN, Maxfield FR. Endocytosis. Physiol Rev. 1997;77(3):759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- 62.Huotari J, Helenius A. Endosome maturation. Embo J. 2011;30(17):3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tatiparti K, Sau S, Kashaw SK, Iyer AK. siRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials. 2017;7(4):pii: E77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Li J, Liu N, et al. In vitro studies of phospholipid-modified PAMAM-siMDR1 complexes for the reversal of multidrug resistance in human breast cancer cells. Int J Pharm. 2017;530(1–2):291–299. doi: 10.1016/j.ijpharm.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 65.Hu YB, Dammer EB, Ren RJ, Wang G. The endosomallysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener. 2015;4:18. doi: 10.1186/s40035-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olmedo I, Araya E, Sanz F, et al. How changes in the sequence of the peptide CLPFFD-NH2 can modify the conjugation and stability of gold nanoparticles and their affinity for beta-amyloid fibrils. Bioconjug Chem. 2008;19(6):1154–1163. doi: 10.1021/bc800016y. [DOI] [PubMed] [Google Scholar]
- 67.Shete HK, Prabhu RH, Patravale VB. Endosomal escape: a bottleneck in intracellular delivery. J Nanosci Nanotechnol. 2014;14(1):460–474. doi: 10.1166/jnn.2014.9082. [DOI] [PubMed] [Google Scholar]
- 68.Tanaka H, Nakatani T, Furihata T, et al. In Vivo Introduction of mRNA Encapsulated in Lipid Nanoparticles to Brain Neuronal Cells and Astrocytes via Intracerebroventricular Administration. Mol Pharm. 2018;15(5):2060–2067. doi: 10.1021/acs.molpharmaceut.7b01084. [DOI] [PubMed] [Google Scholar]
- 69.Ma D. Enhancing endosomal escape for nanoparticle mediated siRNA delivery. Nanoscale. 2014;6(12):6415–6425. doi: 10.1039/c4nr00018h. [DOI] [PubMed] [Google Scholar]
- 70.Li J, Liang H, Liu J, Wang Z. Poly (amidoamine) (PAMAM) dendrimer mediated delivery of drug and pDNA/siRNA for cancer therapy. Int J Pharm. 2018;546(1–2):215–225. doi: 10.1016/j.ijpharm.2018.05.045. [DOI] [PubMed] [Google Scholar]
- 71.Prades R, Guerrero S, Araya E, et al. Delivery of gold nanoparticles to the brain by conjugation with a peptide that recognizes the transferrin receptor. Biomaterials. 2012;33(29):7194–7205. doi: 10.1016/j.biomaterials.2012.06.063. [DOI] [PubMed] [Google Scholar]
- 72.Keaney J, Campbell M. The dynamic blood-brain barrier. Febs J. 2015;282(21):4067–4079. doi: 10.1111/febs.13412. [DOI] [PubMed] [Google Scholar]
- 73.Morales P, Fiedler JL, Andrés S, et al. Plasticity of hippocampus following perinatal asphyxia: effects on postnatal apoptosis and neurogenesis. J Neurosci Res. 2008;86(12):2650–2662. doi: 10.1002/jnr.21715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AuNR-CLPFFD/siRNA complex formation evaluated by changes of the electrophoretic mobility pattern.
Notes: (A) Agarose electrophoresis under non-denaturing conditions. Lane 1 shows the migration pattern of 0.6 µg of free siRNA (0.2 nmol). Lane 2 shows the conjugation of 9 nM AuNR-CLPFFD with 0.6 µg of siRNA. Lane 3 shows 0.9 nM of AuNR-CLPFFD with 0.6 µg of siRNA and lane 4 shows 0.2 nM of AuNR-CLPFFFD with 0.6 µg of siRNA. These results show that at 9 nM of AuNR-CLPFFD, the nanoparticles can form electrostatic interaction with siRNA molecules, retaining the siRNA molecules forming the AuNR-CLPFFD/siRNA complex, allowing to observe a difference in the electrophoretic mobility of free siRNA molecules. In contrast, lanes 3 and 4 show that nanoparticles were not concentrated enough, leaving high amount of free siRNA molecules in solution and conserving the migration pattern of free siRNA. (B) A schematic representation of the siRNA molecules with AuNR-CLPFFD.
Abbreviation: AuNR, gold nanorod.
Cellular uptake is by endocytosis.
Notes: PC12 cells were treated with 1) nuclease-free H2O at 37°C (CS), 2) 0.1 nM AuNR-CLPFFD/siRNA complex at 37°C (Complex), or 3) 0.1 nM AuNR-CLPFFD/siRNA complex at 4°C (Complex) for 1 hour. The mean of the quantified fluorescence was measured by flow cytometry expressed in arbitrary units. The result shows that the fluorescent uptake was decreased when cells were incubated al 4°C, demonstrating that the uptake mechanism was by endocytosis. Inset shows a representative population of analyzed cells (*P<0.05 compared to the saline control); F-ANOVA followed by Bonferroni’s multiple comparison test (n=3 for each group).
Abbreviations: AuNR, gold nanorod; CS, cesarean-delivered control rats.
Single cell image of cellular uptake of the AuNR-CLPFFD/siRNA complex.
Notes: PC12 cells were treated with 0.1 nM AuNR-CLPFFD/siRNA complex for 1 hour. Images were obtained by confocal microscopy: (A) transmitted light microphotograph, (B) Hoechst 33342 stained nuclei (blue), (C) early endosomes (green), (D) fluorescent siRNA label dye647 (red), and (E) merge of (B, C, and D) images. The result shows the colocalization of early endosome vesicles and the AuNR-CLPFFD/siRNA complex. Images are the merge of three pictures taken every 1 µm. Scale bar: 20 µm; 60× optic, 3.1 optic zoom.
Abbreviation: AuNR, gold nanorod.
In vivo knockdown of PARP-1 in control animals.
Notes: Administration of a single dose of AuNR-CLPFFD/siRNA complex (ip) to control animals. Treatment: 1) saline solution (Saline); 2) naked siRNA (siRNA), or 3) AuNR-CLPFFD/siRNA complex (Complex), 1 hour after birth. Total protein extracts were obtained 24 hours after the treatment. PARP-1 protein levels were quantified by Western blots, in mesencephalon, hippocampus, and telencephalon. The quantification showed that in mesencephalon and telencephalon, the AuNR-CLPFFD/siRNA complex treatment decreased PARP-1 levels. We also showed the decreasing levels of PARP-1 in rats treated with naked siRNA administration. Further study will be needed to explain the nature of this effect (aP<0.05 compared to the saline control; Student’s t-test; n=6 for each condition).
Abbreviations: AuNR, gold nanorod; ip, intraperitoneal.