Summary
Biological systems require precise copper homeostasis enabling metallation of cuproproteins while preventing metal toxicity. In bacteria, sensing, transport and storage molecules act in coordination to fulfill these roles. However, there is not yet a kinetic schema explaining the system integration. Here, we report a model emerging from experimental and computational approaches that describes the dynamics of copper distribution in Pseudomonas aeruginosa. Based on copper uptake experiments, a minimal kinetic model describes well the copper distribution in the wild type bacteria but is unable to explain the behavior of the mutant strain lacking CopA1, a key Cu+ efflux ATPase. The model was expanded through an iterative hypothesis-driven approach, arriving to a mechanism that considers the induction of compartmental pools and the parallel function of CopA and Cus efflux systems. Model simulations support the presence of a periplasmic copper storage with a crucial role under dyshomeostasis conditions in P. aeruginosa. Importantly, the model predicts not only the interplay of periplasmic and cytoplasmic pools but also the existence of a threshold in the concentration of external copper beyond which cells lose their ability to control copper levels.
Keywords: Pseudomonas aeruginosa, copper, homeostasis, computational modeling, system identification
Abbreviated Summary
Copper distribution across periplasm and cytoplasm of Pseudomonas aeruginosa was experimentally determined in wild type and ΔcopA1 strains and the data was fit to computational models of increasing mechanistic complexity. The best fit was by a model where the periplasmic cuproproteins are induced by cytoplasmic copper levels, suggesting that the periplasm has an important role in maintaining copper homeostasis. The model also predicts a critical copper concentration beyond which homeostasis is no longer supported.
Introduction
Copper is an essential trace element used as a cofactor in redox enzymes (Fraústo da Silva and Williams, 2001; Cobine et al., 2006; Tavares et al., 2006). However, copper in its “free” form is highly toxic and leads to mismetallation of proteins, disassembly of Fe-S centers, and generation of free radicals (Macomber and Imlay, 2009; Dupont et al., 2011). Therefore, organisms have evolved molecular systems targeting the metal to cuproproteins while limiting high intracellular copper levels. Understanding of bacterial mechanisms of copper homeostasis is relevant in light of their importance for pathogenic virulence. The antibacterial properties of copper and role in innate immunity are well established (Hodgkinson and Petris, 2012; Ladomersky and Petris, 2015; Djoko et al., 2015). Studies on bacterial response to copper stress have revealed various essential components such as sensing transcriptional regulators, membrane transporters and chaperones that control Cu+ levels and localization (Rensing and Grass, 2003; Osman and Cavet, 2008; Argüello et al., 2013). Deletion of some of these proteins leads to a consequent copper dyshomeostasis. In addition proteins that bind copper with high capacity and might act as storage pools have been proposed recently (Vita et al., 2016). Nevertheless, there is still a lack of a general model to describe and predict the accumulation and distribution of copper in Gram-negative bacteria in kinetics terms.
Several proteins that play a role in copper sensing, transfer, and distribution between compartments have been identified in the periplasm, cytoplasm, inner, and outer membranes of Pseudomonas aeruginosa (Yoneyama and Nakae, 1996; Teitzel et al., 2006; Caille et al., 2007; Frangipani and Haas, 2009; Thaden et al., 2010; González-Guerrero et al., 2010; Elsen et al., 2011; Quintana et al., 2017). Extracellular Cu2+ likely enters the periplasm through OprC, an outer membrane porin (Yoneyama and Nakae, 1996). While OprC binds Cu2+, the specificity of transport as well as the putative presence of external reductases generating Cu+ has not been established. The inner membrane CcoA transports into the cytoplasm the periplasmic copper destined to metalate cytochrome c oxidase (Ekici et al., 2012), while novel members of the Major Facilitator Superfamily might also enable copper influx through this membrane (Quintana et al., 2017). Copper is likely supplied to cytoplasmic cuproproteins and other putative copper binging pools via cytoplasmic chaperones, as has been proposed for the role of chaperones in delivering the metal to inner membrane Cu+ exporters (P1B-ATPases) and transcriptional sensors (Cobine et al., 1999; González-Guerrero et al., 2009). Two cytoplasmic Cu+ chaperones, CopZ1 and CopZ2, have been identified in P. aeruginosa although their singular roles have not been established (Quintana et al., 2017). Two Cu+-ATPases, CopA1 and CopA2, mediate cytoplasmic copper export in these bacteria. CopA2 provides copper for cytochrome c oxidase, while CopA1 functions as a primary exporter to the periplasm, guarding against excess buildup of cytoplasmic Cu+ (González-Guerrero et al., 2010). Cellular copper might also be exported by the P. aeruginosa Cus system, CusA, CusB and CusC, spanning from the inner to the outer membrane (Thaden et al., 2010; Kim et al., 2011; Quintana et al., 2017). This efflux system has been proposed to transport copper from the cytoplasmic and periplasmic copper pools to the extracellular space in E. coli (Kim et al., 2010; Long et al., 2010; Kim et al., 2011). However, P. aeruginosa does not have a periplasmic chaperone CusF that might deliver copper to CusB (Kim et al., 2010; Quintana et al., 2017), suggesting that cytoplasmic Cu+ might be the substrate of this Cus system. Regarding periplasmic copper, this is likely exported through the outer membrane protein PcoB (Cha and Cooksey, 1991; Lee et al., 2002). Expression of these transporters and chaperones, as well as other proteins of unknown functions, is controlled by a two component periplasmic copper sensor (CopS/CopR) and the cytoplasmic sensor CueR (Thaden et al., 2010; Quintana et al., 2017).
Using genome-wide transcriptomic analysis, we have proposed an integrated copper homeostatic network (Quintana et al., 2017). This considers the requirement of specific influx and efflux systems and subcellular metal chaperones; as well as transcriptional regulation of the various functional elements in each compartment. However, these observations do not explain how the interactions of the metal distribution components function in a coordinated manner to timely respond to environmental changes. Mathematical models are increasingly used to understand complex biological systems and test different mechanisms that can explain the behavior at system level. Previously developed mathematical models of copper homeostasis in archaea and Gram-positive bacteria (Pécou et al., 2006; Pang et al., 2013) have not addressed the dynamics of Cu+ movements via bona fide transporters and chaperones nor the distribution across periplasmic and cytoplasmic pools. We hypothesized that mathematical analysis of copper uptake kinetics might provide evidence on the necessary elements (influx and efflux transporters and compartmental pools) that explain the organization of the reported homeostatic network. Here we present a predictive mathematical model, consistent with experimental observations, that advances our understanding of the mechanisms of copper homeostasis in P. aeruginosa.
Results
Modeling copper uptake kinetics
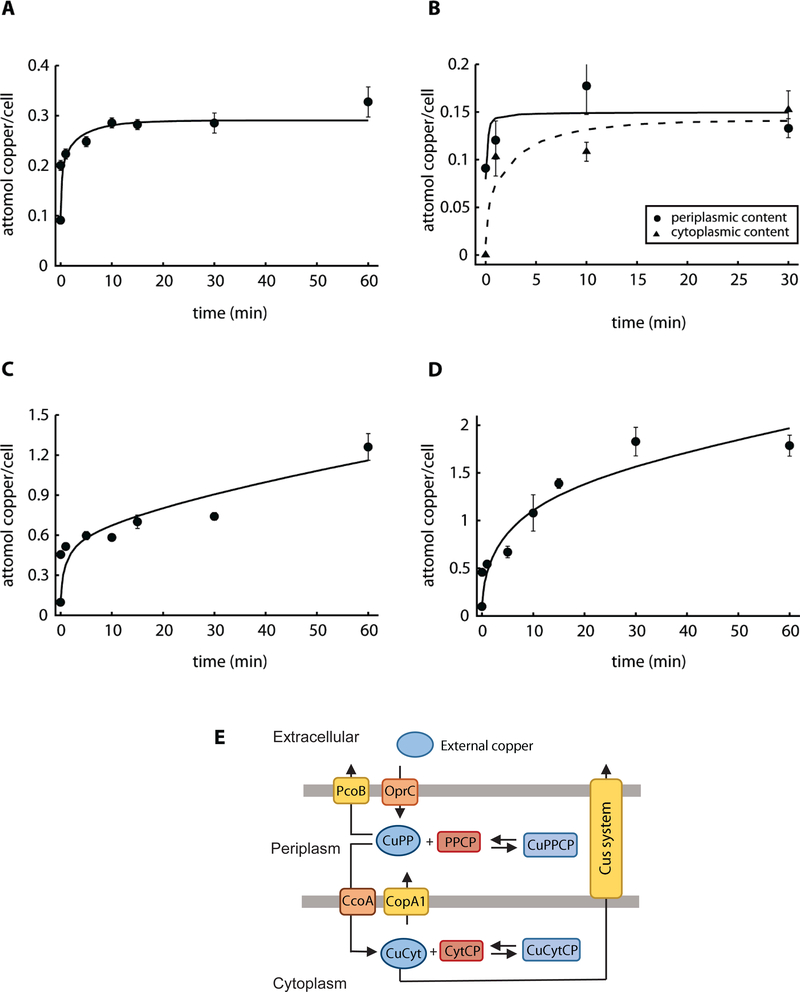
Our goal was to develop a mathematical model that can describe the copper uptake kinetics at various concentrations of extracellular metal, as well as the distribution in the two cellular compartments, periplasm and cytoplasm. We expected that simulations can predict the copper homeostasis not only for the wild type strain but also under conditions of dyshomeostasis; i.e. when an element of the system is knocked out via mutagenesis. We have previously shown that P. aeruginosa viability and growth rate are not compromised by external copper concentrations up to 0.5 mM Cu2+, while incubation at higher levels leads to eventual cell death (Quintana et al., 2017). In the same work, copper uptake kinetics at this “threshold” concentration was also studied. These data served for an initial assessment of the proposed model (Figure 1A). Nonetheless, towards enhancing the scope of our approach, the compartmental distribution was estimated at pre-steady (1 min, 10 min) and steady state (30 min) (Fig. 1B). We also analyzed Cu2+ uptake kinetics of wild type P. aeruginosa in the presence of 2 mM Cu2+ (Fig. 1C) and 4 mM Cu2+ (Fig. 1D) in the culture medium. Cell viability started to decrease in cells exposed to 2 mM CuSO4 after 1 h treatment, whereas toxicity is evident as soon as 10 min after the addition of 4 mM CuSO4 (data not shown). Cells exposed to 2 mM Cu2+ showed a modest but noticeable increase in Cu levels beyond the initial fast metal uptake. Alternatively, exposure to 4 mM Cu2+ lead to a marked steady increase of intracellular metal levels over the course of the experiment. Then, it is apparent the presence of a “switch” from tolerance to toxicity at around 1 attomol of copper per cell (Fig. 1C), as higher steady state levels lead to cell death (Quintana et al., 2017).
Figure 1. Experimental data and best fit by Model I.
A) Copper uptake kinetics in the presence of 0.5 mM CuSO4 (Quintana et al. 2017). B) Compartmental distribution of copper levels 1, 10 and 30 min after the addition of 0.5 mM CuSO4, solid line is for periplasmic and dashed line is for cytoplasmic fraction. C, D) Copper uptake kinetics in the presence of 2 mM and 4 mM CuSO4. Solid lines represent the best fit to the data using Model I (parameter values in Table 1). Experimental data are the mean ± standard error of 3 independent replicates. E) Parsimonious model of copper uptake kinetics in P. aeruginosa PAO1 (Model I). CuPP, periplasmic copper bound to chaperones, and CuCyt, cytoplasmic copper bound to chaperones. PPCP, periplasmic and outer membrane proteins; CytCP cytoplasmic and inner membrane proteins. Equilibrium copper binding is represented by arrows pointing both ways. Single arrows represent the direction of transport.
We initiated the modeling of the uptake kinetics using a parsimonious Model I (Fig. 1E). A fixed total cell volume of 10−15 L is adopted in the model, composed of a periplasmic compartment (20% of the total volume) and a cytoplasmic compartment (80% of the total volume) (Kumar and Imlay, 2013). For modeling purposes, based on the virtual absence of free (unbound) copper in the cell (Changela et al., 2003; Smirnova et al., 2012), we assumed all copper to be bound to proteins (copper chaperones and cuproproteins). We use the term copper chaperones to refer to those cuproproteins that bind copper as a means to hand it off/on transporters. We term cuproproteins those proteins that have copper bound for a long time scale and do not make it available to transport until it dissociates and binds to chaperone. Molecular elements of the model could be predicted based in our earlier transcriptional analysis; i.e. chaperones CopZ1 and CopZ2, transporters CcoA and CopA1, etc. However, given the uncertainties on the identity of periplasmic chaperones, as well as on the roles of both cytoplasmic chaperones (CopZ1 and CopZ2), the species CuPP and CuCyt represent Cu+ bound to periplasmic and cytoplasmic chaperone molecules respectively (Fig 1E). To reduce the model complexity all cuproproteins were grouped into one pool per compartment; PPCP represents all periplasmic and outer membrane cuproproteins, and CytCP represents all cytoplasmic and inner membrane cuproproteins. It was considered that Cu+ binding to these proteins follows mass action kinetics. For simplicity, the model does not explicitly consider the apo forms of the chaperones; this is, their concentration is not limiting. The Cu+ binding and dissociation with cuproproteins (PPCP and CytCP) is also represented by mass action, however in this case we represent explicitly the apo version of the proteins. Our experimental results suggested that before cells are exposed to Cu2+ (t = 0), most of intracellular copper exists in the periplasm (Fig. 1B). However, in order to undertake experimental constraints, we allowed the initial concentration of cytoplasmic copper to vary between 0 to 30% of the total during parameter estimation. Then, the initial concentrations of copper bound to cuproproteins (CuPPCP and CuCytCP) were obtained through parameter estimation while the concentrations of copper bound to chaperones (CuCyt and CuPP) were obtained by subtracting those estimated values from the total compartmental copper. The initial concentrations of PPCP and CytCP were also parameters estimated to fit the data. We assumed that extracellular Cu2+ (CuExt) enters the periplasm (CuPP) through the influx protein (OprC) and exported via an efflux protein (PcoB). Periplasmic and cytoplasmic Cu+ exchange is through dedicated influx and efflux transporters (CcoA and CopA). In addition, we included in the model the export of cytoplasmic Cu+ into the medium via the Cus-system present in P. aeruginosa. Considering the different transmembrane systems, their transport rates were defined by Michaelis-Menten equations. The resulting differential equations for transport and equilibrium copper exchange were integrated in a full model (see Supplemental Information) that yielded parameter values (Table 1) that fit well the experimental Cu+ uptake curves in a range of external Cu2+ concentrations (Fig. 1A, 1C, 1D), as well as the compartmental distribution (Fig. 1B).
Table 1 -.
Best fit parameter values for Models I, II, and III.
| Parameter | Role | Best fit value | Units | ||
|---|---|---|---|---|---|
| Model I | Model II | Model III | |||
| vOprC | Limiting rate of copper import from medium to periplasm | 7.83 | 2.44 | 6.84 | mmol/min |
| KmOprC | Michaelis constant for copper import from medium to periplasm | 0.546 | 0.328 | 1.66 | mM |
| vPcoB | Limiting rate of copper export from periplasm to medium | 7.30 | 2.20 | 3.68 | mmol/min |
| KmPcoB | Michaelis constant for copper export from periplasm to medium | 0.33 | 0.232 | 0.390 | mM |
| vCcoA | Limiting rate for copper import from periplasm to cytoplasm | 2.72 | 1.19×106 | 4.43 | mmol/min |
| KmCcoA | Michaelis constant for copper import from periplasm to cytoplasm | 22.1 | 36.7 | 9.50 | μM |
| vCopA1 | Limiting rate for copper export from cytoplasm to periplasm | 0.665 | 0.127 | 0.713 | mmol/min |
| KmCopA1 | Michaelis constant for copper export from cytoplasm to periplasm | 6.00 | 5.42 | 0.476 | μM |
| kasCuPPCP | On constant for copper to periplasmic cuproproteins | 595.3 | 283 | 6.98×104 | 1/M min |
| kdsCuPPCP | Off constant for copper from periplasmic cuproproteins | 2.24×10−6 | 1.31×10−6 | 1.26×10−6 | 1/min |
| kasCuCytCP | On constant for copper to cytoplasmic cuproproteins | 4.19×108 | 8.33×108 | 4.70×108 | 1/M min |
| kdsCuCytCP | Off constant for copper from cytoplasmic cuproproteins | 97.9 | 98.4 | 98.6 | 1/min |
| vCusSystem | Limiting rate for copper export from cytoplasm to medium | 1.02 | 1.23 | 0.390 | mmol/min |
| KmCusSystem | Michaelis constant for copper export from cytoplasm to medium | 33.4 | 0.337 | 38.9 | μM |
| kdPPCP | First order rate constant for periplasmic cuproproteins degradation (turnover) | NA | NA | 0.005 | 1/min |
| ksPPCP | Limiting rate for synthesis of periplasmic cuproproteins | NA | NA | 14.4 | μmol/min |
| KaCuCytPPCP | Activation constant by copper towards synthesis of periplasmic cuproproteins | NA | NA | 0.0380 | μM |
| vCopA2 | Limiting rate for copper export from cytoplasm to periplasm through CopA2 | NA | 5.46×105 | NA | mmol/min |
| kmCopA2 | Michaelis constant for copper export from cytoplasm to periplasm through CopA2 | NA | 0.139 | NA | μM |
The resulting kinetic parameter values (Table 1) are within expected transport rates and equilibrium affinity constants (González-Guerrero et al., 2008; González-Guerrero et al., 2010; Xiao et al., 2011; Argüello et al., 2012; Quintana et al., 2017). Importantly, the model can describe the copper distribution between the periplasmic and cytoplasmic compartments (Fig. 1E) and marks the parallel transport by the CopA and Cus-system. In this direction, exclusion of the Cus-system prevented fitting of the uptake curves.
Effects of dyshomeostasis on copper uptake kinetics
Previous results in our laboratory showed that a P. aeruginosa mutant strain lacking CopA1 (ΔcopA1) has increased intracellular Cu+ levels that result in an increase sensitivity to the metal (González-Guerrero et al., 2010). To aid the mathematical modeling of copper distribution, we studied the kinetics of copper accumulation in ΔcopA1 after exposure to 0.5 mM CuSO4 as well as the metal compartmental distribution (Fig. 2). We previously observed that P. aeruginosa, as well as mutant strains lacking periplasmic (ΔcopR) and cytoplasmic (ΔcueR) Cu+ sensors, growth at normal rates at the sub-lethal concentrations of 0.5 mM external copper (Quintana et al., 2017). A similar behavior was observed in the ΔcopA1 strain and this allowed the study of copper levels per cell. Determination of the copper uptake kinetics showed metal levels rose faster in the mutant strain than in the wild type bacteria reaching steady state after 60 min (Fig. 2A; compare to Fig. 1A). As previously shown, mutant complementation with the copA1 gene reestablished the wild type phenotype (González-Guerrero et al., 2010). Interestingly, the compartmental response of the mutant strain was distinct from that of the wild type and relatively faster overload of the periplasm compared to cytoplasm was observed (Fig. 2B). Our minimal Model I was incompatible with these experimental data as it predicted an increase of the cytoplasmic pool. This is, it was not possible to obtain good global fits against the WT and ΔcopA1 data with the minimal model. We then used the parameter values obtained just using WT data to simulate the ΔcopA1 mutant by setting the VCopA1 parameter to zero; the results predicted that the cytoplasm would accumulate very high copper concentration while the periplasmic copper would decrease below its initial concentration. This contradicts the experimental results suggesting the presence of additional homeostatic components.
Figure 2. Effect of dyshomeostasis on copper uptake kinetics in P. aeruginosa PAO1.
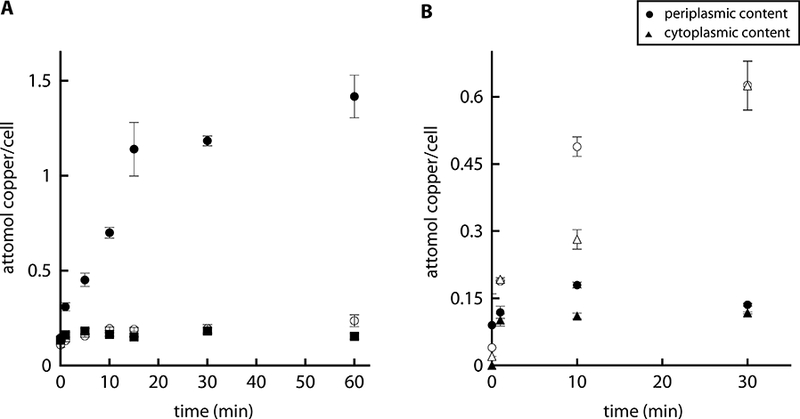
Copper uptake kinetics in the insertional mutant ΔcopA1 (black circles) and the corresponding complemented strain (black squares) after addition of 0.5 mM CuSO4. Empty circles correspond to WT. B) Compartmental distribution of copper levels in the insertional mutant ΔcopA1 (white shape) and WT (black shape) 1, 10 and 30 min after the addition of 0.5 mM CuSO4. Experimental data are the mean ± standard deviation of 3 independent replicates.
Looking for an alternative model, we considered that P. aeruginosa has two Cu+-ATPases (CopA1 and CopA2) responsible for cytoplasmic Cu+ efflux (González-Guerrero et al., 2010). The rationale behind this hypothesis was that if CopA2 could transport copper at significant higher rate from cytoplasm to periplasm in the absence of CopA1 then the periplasm would accumulate more copper compared to the cytoplasm. A new Model II was therefore created to include two cytoplasm to periplasm efflux systems (Fig. 3A). Model II was able to fit the behavior of the wild type strain (Fig. 3B, 3C, 3C, 3D). However, Model II also failed to describe the behavior of the ΔcopA1 mutant strain. Although only copA1 is apparently regulated by copper (González-Guerrero et al., 2010; Quintana et al., 2017), a variation of Model II was tested where the VCopA2 parameter was allowed to increase with the cytoplasmic copper concentration, as if transcriptionally upregulated. However, this modification still did not lead to the satisfactory fitting of the data.
Figure 3. Experimental data and best fit by Model II.
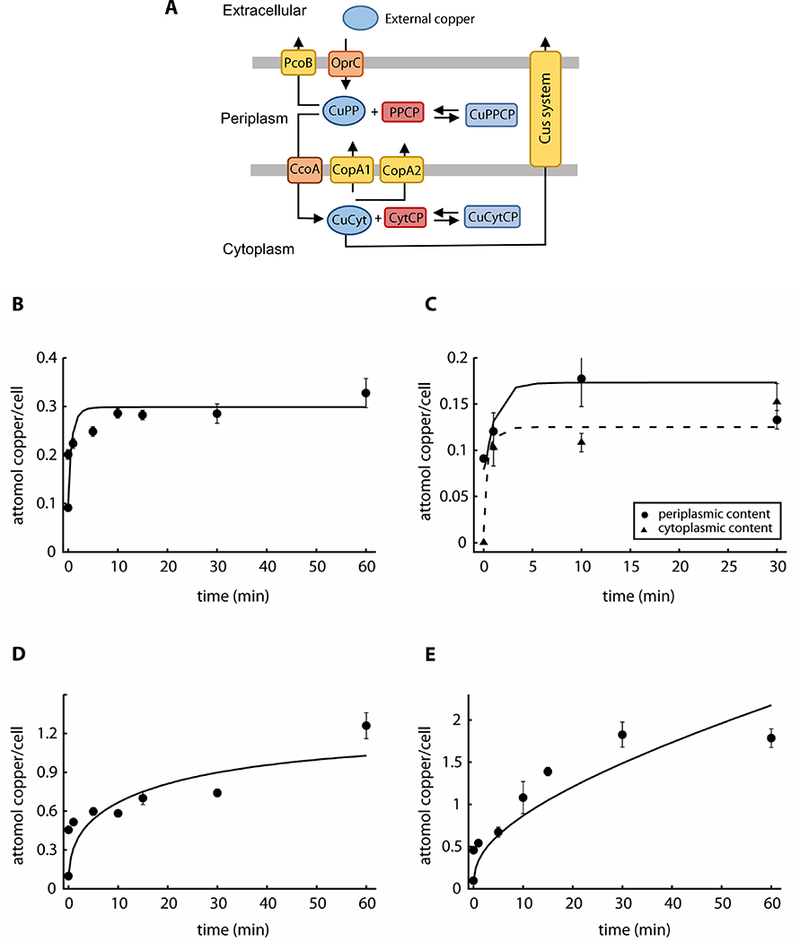
A) Modified model, including CopA2, of copper uptake kinetics in P. aeruginosa WT PAO1 (Model II). CuPP, periplasmic copper bound to chaperones, and CuCyt, cytoplasmic copper bound to chaperones. PPCP, periplasmic and outer membrane proteins; CytCP cytoplasmic and inner membrane proteins. Equilibrium copper binding is represented by arrows pointing both ways. Single arrows represent the direction of transport. B) Copper uptake kinetics in the presence of 0.5 mM copper (Quintana et al. 2017). C) Compartmental distribution of copper levels 1, 10 and 30 min after the addition of 0.5 mM CuSO4, solid line is for periplasmic and dashed line is for cytoplasmic fraction. D, E) Copper uptake kinetics in the presence of 2 mM and 4 mM CuSO4. Solid lines represent the best fit to the data using Model II (parameter values in Table 1). Experimental data are the mean ± standard deviation of 3 independent replicates.
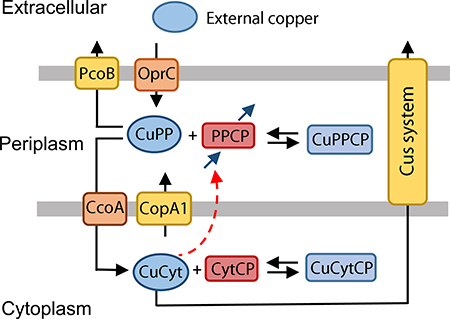
An alternative mechanism that could explain the higher periplasmic copper levels in the ΔcopA1 strain could be an increased expression of the periplasmic cuproproteins. Supporting this hypothesis, we have observed high levels of periplasmic cuproproteins in the ΔcopA1 albeit determinations were performed in the absence of Cu2+ stress (Raimunda et al., 2012). Following this line of thinking, gene expression would be increased in the cytoplasm after sensing of a high copper level in the periplasm by CopS/R, to ultimately induce higher synthesis of periplasmic cuproproteins. Testing this hypothesis, we created Model III where the periplasmic pool of cuproproteins (PPCP) is induced by the cytoplasmic copper (CytCP) (Fig. 4). Here, we also represent the synthesis and degradation of PPCP; the degradation rate of PPCP is assumed to be a first order process, while the synthesis rate is assumed to be a sigmoidal function of [CuCyt]:
| 3 |
where V is a basal rate of synthesis, h is a Hill coefficient, KCuCyt is an affinity constant for the activation of the synthesis rate, and VolPP is the volume of the periplasm. We then proceeded to fit this model against both the wild type and the ΔcopA1 strains data. Interestingly, Model III was able to simultaneously reproduce the response from both ΔcopA1 and wild type strains (Fig. 5A, 5C, 5D, 5E), including the compartmental distribution (Fig. 5B, 5F). Moreover, the model accurately predicted the steady-state conditions for WT after 5–10 min (Fig. 5A, 5C, 5D) and the increase in copper levels in ΔcopA1 even 60 min after exposure (Fig. 5E).
Figure 4. Model explaining the role of periplasmic copper pool in P. aeruginosa PAO1 (Model III).
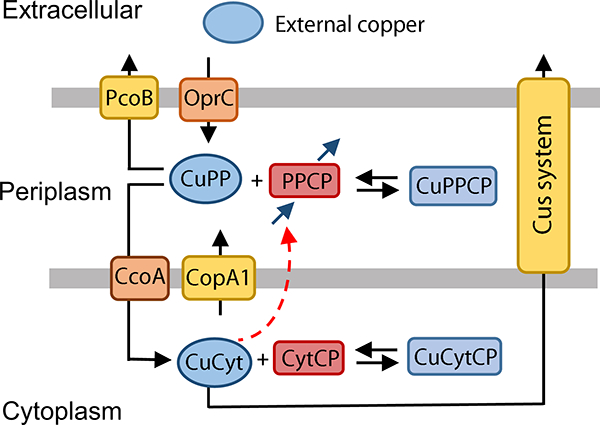
CuPP, periplasmic copper bound to chaperones, and CuCyt, cytoplasmic copper bound to chaperones. PPCP, periplasmic and outer membrane proteins; CytCP cytoplasmic and inner membrane proteins. Black arrows represent transport and binding, blue arrows represent synthesis and degradation of PPCP and red dashed arrow represents induction of PPCP synthesis by CuCyt. Note that synthesis of PPCP happens in the cytoplasm and the protein is appropriately transported to the periplasm, but for clarity the synthesis arrow is included in the periplasm in this diagram.
Figure 5. Experimental data and best fit by Model III.
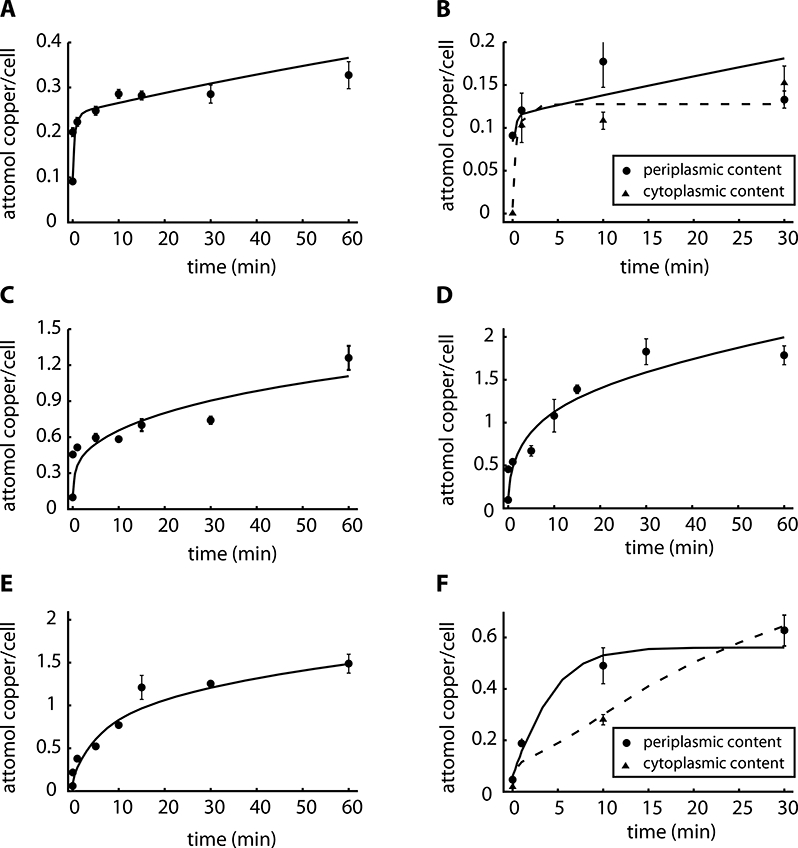
A) Copper uptake kinetics in the presence of 0.5 mM CuSO4 (Quintana et al. 2017). B) Compartmental distribution of copper levels 1, 10 and 30 min after the addition of 0.5 mM CuSO4, solid line is for periplasmic and dashed line is for cytoplasmic fraction. C, D) Copper uptake kinetics in the presence of 2 mM and 4 mM CuSO4. E) Copper uptake kinetics of the insertional mutant ΔcopA1 in the presence of 0.5 mM copper. F) Compartmental distribution of copper levels in the insertional mutant ΔcopA1 1, 10 and 30 min after the addition of 0.5 mM CuSO4, solid line is for periplasmic and dashed line is for cytoplasmic fraction. Solid lines represent the best fit to the data using Model III (parameter values in Table 1). Experimental data are the mean ± standard deviation of 3 independent replicates.
For each of the models we characterized parameter identifiability using likelihood profiles (Raue et al., 2009). As depicted in Figures S1-S2, most parameters of the Models I and II are not identifiable (those that have flat curves), the only exception being KmOprC and some of the initial concentrations. However, in the case of the Model III (Figure S3), most parameters are identifiable within 90% confidence intervals. The dissociation rate constants kdsCuPPCP and kdsCuCytCP are not identifiable for all three models because they can be counterbalanced by the respective association constants (to determine these parameters well would require binding studies with purified proteins). The other non-identifiable parameter, common to all three models, is KmPcoB which suggests that the export of copper from periplasm through the outer membrane may not be essential to explain the experimental observations. Overall, the analysis indicated that the inclusion of ΔcopA1 mutant data in Model III is what constrained the model parameters to become identifiable. This provides further confirmation that Model III is the best of the three in describing copper transport and homeostasis, and suggests a prominent role of the periplasm in protecting from higher environmental copper.
Model Predictions
One of the advantages of computational modeling is the capability to simulate experiments where predictions can be tested. Toward this goal, we used Model III to simulate the P. aeruginosa response of external copper concentrations higher than 4 mM (Fig. 6). Model III suggests that, in these conditions, intracellular copper accumulation continuously increases without attaining a steady state (Fig. 6A). This is P. aeruginosa loses its homeostatic control above a threshold of 2–4 mM Cu2+ leading to intracellular metal levels above 1 attomol/cell. In the case of the ΔcopA1 strain, this extracellular Cu2+ threshold is even lower since the internal copper increases at faster rate (Fig. 6B). This is in agreement with the experimental observation that the ΔcopA1 strain is much more sensitive than wild type P. aeruginosa to high levels of extracellular Cu2+ (González-Guerrero et al., 2010). Moreover, Model III predicts that the periplasmic copper pool becomes larger than the cytoplasmic at 1 mM external Cu2+. However, at highly toxic external copper conditions, i.e. 4 mM, cytoplasmic copper increases dramatically compared to the periplasmic concentration (Fig. 6B). On the other hand, the ΔcopA1 strain accumulates high copper in the cytoplasm at both 1 mM and 4 mM external copper (Fig. 6C). Thus, the model predicts that P. aeruginosa respond to increased copper initially inducing periplasmic storage. However, beyond a certain concentration of external copper, tolerance mechanisms are saturated and copper is re-distributed, i.e. cytoplasmic storage mechanisms are induced. This threshold is lower when copper homeostasis is impaired, as it occurs in ΔcopA1.
Figure 6. Predicting the effects of copper toxicity using Model III.
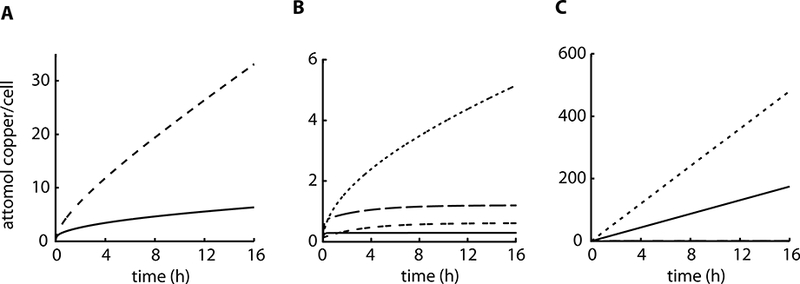
A) Computational prediction of total cellular copper accumulation in P. aeruginosa WT strain for 4 mM (solid line) and 6 mM (dashed line) CuSO4. B) Computational prediction of copper levels in the P. aeruginosa WT strain in the presence of 1 mM and 4 mM CuSO4 in the periplasm (dashed line for 1 mM and dashed-dotted line for 4 mM) and for the cytoplasm (solid line for 1 mM and dotted line for 4 mM). C) Computational prediction of copper levels in the P. aeruginosa ΔcopA1 strain in the presence of 1 mM and 4 mM CuSO4 in the periplasm (dashed line for 1 mM and dashed-dotted line for 4 mM) and for the cytoplasm (solid line for 1 mM and dotted line for 4 mM; note that the periplasm concentrations are very small).
Discussion
To shed light on the mechanisms of copper homeostasis in P. aeruginosa, we have previously established protein components (transporters, chaperones, transcriptional regulators) that enable metal uptake, distribution and efflux (Quintana et al., 2017). Along this line, we reported the regulons of periplasmic and cytoplasmic metal sensors that control the copper levels in each cellular compartment. However, this system description did not provide a quantitative explanation of the dynamic interplay among these homeostatic elements. Here, we present a mathematical model that using a single set of parameters is able to replicate the observed copper membrane fluxes and compartmental distribution in P. aeruginosa. Moreover, this model is able to describe the system under conditions of dyshomeostasis by exposure to high copper or deletion of a key protein.
Our strategy employed a bottom-up modeling approach starting with a minimal Model I. This considered all the transmembrane transporters identified in P. aeruginosa and pooled copper chaperones and cuproproteins into single compartmental pools. Departing from a parsimonious Model I, mechanistic complexity was expanded and tested against experimental observations. The proposal of mechanistic hypotheses at each step led to a minimal model that is consistent with the observations. Moreover, this iterative approach reduced the possibility of over-parameterization and data overfitting.
As the experimental basis for system simulations, we analyzed the accumulation and intracellular distribution of copper in P. aeruginosa exposed to increasing external metal concentrations, both in wild type and ΔcopA1 strains. Three kinetic models representing different mechanisms were tested in their capability to reproduce copper uptake data. Two of these models were unable to simultaneously explain the experimental observations from the two strains, indicating that the hypothetical systems were not consistent with the cellular response. Thus, we rejected the minimal Model I (Fig 1E) unable to simulate the copper compartmental distribution in the ΔcopA1 strain; as well as the Model II where CopA2 would not undertake the function of CopA1 when the latter is deleted (Fig. 3A). Model III, which includes an induction of periplasmic cuproproteins by cytoplasmic copper is able to fit, with a single set of parameter values, all of our experimental data. Indeed we found out that not only is Model III better at fitting the experimental observations, but it also has better parameter identifiability. This may seem surprising since Model III is more complex than the two other models. In this case the extra complexity added in Model III was important in constraining the parameter values. This also gives validity to the approach adopted here: starting with simple models and gradually adding complexity until a good fit and identifiability is obtained.
The final Model III supported hypothetical and experimental observations. The model was consistent with the need of specific metal influx transporters in the outer and inner membranes. The outer membrane porin OprC, down regulated in the presence of high copper, appears as the logical candidate for influx transport across this membrane (Quintana et al., 2017). We assumed that CcoA mediates transport across the inner membrane (Ekici et al., 2012). However, required for metallation of cytochrome c oxidase, CcoA is not down regulated in conditions of metal stress and it is likely that a member of the Major Facilitator Superfamily encoded by PA5030 is a major contributor to copper influx (Quintana et al., 2017). Regarding copper efflux, Model III supports two significant characteristics of the system. First, it is congruent with the previously reported roles of CopA1 maintaining cytoplasmic copper levels and CopA2 supplying the metal for metallation of cytochrome c oxidase (González-Guerrero et al., 2010). Further studies are required to understand the integration of CcoA, CopA2 and perhaps specific chaperones in the metallation of respiration complexes. Second, the model points to the parallel function of a CopA1-PcoB tandem and the Cus-system to control the cell copper levels. While it appears to be a larger contribution of CopA1 relative to Cus (compare estimated Vmax), the transport via Cus appears essential to explain the experimental observations.
As expected, the model describes well the cellular metal accumulation which could be simulated assuming the absence of free copper. Importantly, it revealed an unexpected important early role of the periplasmic compartment in controlling copper levels. This is mediated by the upregulation of the periplasmic pool of cuproproteins (PPCP). The proposed model formally assumes that a cytoplasmic cuproprotein is responsible for the induction of the periplasmic cuproprotein pool. If the present hypothesis is correct, deletion of this protein (likely CopR) should result in phenotypes severely affected in copper homeostasis.
We have previously reported that azurin (PA4922) is the major cupropotein in the periplasm of P. aeruginosa (Raimunda et al., 2013). Moreover, the expression of azu is increased in the ΔcopA1 and ΔcopA2 stains compared to wild type strain. However, rather than having a metallochaperone role azurin has been reported to serve as an electron transfer carrier redox protein (Gray and Winkler, 1996), then other periplasmic proteins regulated by the CopS/R system might serve as copper sinks. In this direction, we observed the upregulation of a cupredoxin-like protein (PA2807), also with potential redox activity and multicopper oxidase PcoA (Djoko et al., 2008). The upregulation of several periplasmic redox enzymes not only supports the proposed role of oxidases producing a “less-toxic” Cu(II) (Grass and Rensing, 2001), but provide a molecular framework for the early accumulation of periplasmic Cu upon copper stress.
It is also notable that Model III does not include changes in the level of the cytoplasmic pool of cuproproteins. However this is because of the induction of periplasmic cuproproteins forms a negative feedback which stabilizes the cytoplasmic copper pool. Indeed the cytoplasm is an important compartment for copper sensing, distribution, and storage in bacteria (Padilla-Benavides et al., 2013; Philips et al., 2015; Vita et al., 2015). We have been unable to identify a cytoplasmic storage protein in P. aeruginosa, since no change in expression of likely candidates (metallothionein, Csp3, or glutathione biosynthesis genes) is observed during high-copper stress (Quintana et al., 2017).
In summary, we present a mathematical model that explains the dynamic function of influx and efflux systems in coordination with periplasmic and cytoplasmic metalloprotein copper pools toward achieving copper homeostasis. Importantly, the established parameters can simulate conditions of dyshomeostasis by exposure to high metal levels and the removal of system components.
Experimental procedures
Bacterial strains and growth conditions
P. aeruginosa PAO1 wild type and PW7626 (PA3920/copA1 insertional mutant, ΔcopA1) strains were obtained from the Comprehensive P. aeruginosa Transposon Mutant Library at the University of Washington Genome Center (Jacobs et al., 2003). The ΔcopA1 strain was complemented with the copA1 gene under control of its own promoter region as described (González-Guerrero et al., 2010). Cells were grown at 37ºC in Luria-Bertani (LB) medium, supplemented with irgasan (25 μg/ml), tetracycline (60 μg/ml), or kanamycin (35–50 μg/ml) as required.
Copper uptake assays
Copper uptake was measured using cells in mid-log phase in antibiotic-free LB medium. Upon addition of 0.5, 2, or 4 mM CuSO4, aliquots were taken at indicated times for cell counting and copper levels determinations. Metal was measured in cells treated with 1 mM DTT and 1 mM bathocuproinedisulfonic acid (BCS) and harvested by centrifugaton. Pellets were washed twice with 150 mM NaCl. Cells were mineralized with 15.6 M HNO3 (trace metal grade) for 1 h at 80ºC and overnight at 20ºC. Digestions were completed by adding 2 M H2O2. Copper levels were measured by atomic absorption spectroscopy (AAS). When required, samples were diluted with 300 mM HNO3 before analysis by AAS.
Copper content in cell fractions
Subcellular fractions, namely periplasm, cytoplasm, and total membrane fraction, were obtained using cells grown to mid-log phase in antibiotic-free LB medium. Aliquots (1.8 ml) were collected at indicated time points after addition of 0.5 mM CuSO4, treated with 1 mM DTT and 1 mM BCS and harvested by centrifugation. Pellets were washed twice with 1 ml 50 mM Tris-HCl, pH 7.6 and were kept on ice for no longer than 1 h prior to cell fractionation. Fractionation was performed following Ize et al. (2014) with modifications. The periplasmic fractions were obtained by resuspending cells in 0.5 ml of 200 mM MgCl2, 50 mM Tris-HCl, pH 7.6 and incubation for 10 min at room temperature with gentle shaking. After 5 min incubation on ice, periplasmic fraction was collected by centrifugation (8000 g for 10 min at 4ºC). The resulting osmotically shocked cells and spheroplasts were washed with 1 ml 50 mM Tris-HCl, pH 7.6 and disrupted by sonication. Cells debris and unlysed cells were separated from cytoplasm and membrane fraction by centrifugation (2000 g for 15 min at 4ºC). The procedure yielded >93±2 % cell lysis (n=5). Controls using anti-His-tag antibodies and cells expressing cytoplasmic His-tagged proteins could not detect the presence of cytoplasmic proteins in the periplasmic fraction (data not shown). Protein content was quantified in different fractions by Bradford (Bradford, 1976). Copper levels were measured by AAS as described above.
Model development, simulations, and parameter estimation
All three models consist of six differential equations, the full models are provided in the Supplementary Text, and Supplementary Files in COPASI and SBML (Hucka et al., 2003) formats. The models were also deposited in the BioModels database (Chelliah et al., 2015) and assigned identifiers MODEL1804120001, MODEL1804120002, and MODEL1804120003. Computational analyses were carried out using the open source software COPASI (Hoops et al., 2006), version 4.21. The kinetic models are based on differential equations and solved using the LSODA integration algorithm (Petzold, 1983). Parameter values were estimated with COPASI’s parameter estimation task which minimizes the sum of squares of residuals (SSR) between model predictions and experimental data; minimization of the SSR was carried out with the simulated annealing algorithm (Corana et al., 1987) followed by the Hooke-Jeeves algorithm (Hooke and Jeeves, 1961). Parameter identifiability was carried out by profile likelihood analysis (Raue et al., 2009) using a previously described procedure (Schaber, 2012). All computations ran on desktop computers under Windows and Linux operating systems.
Supplementary Material
Acknowledgements
This work was supported by a grant from the National Institutes of Health (R01GM114949 to J.M.A.). P.M. thanks the National Institutes of Health for funding work on COPASI (RO1GM080219). The Authors declare no conflicts of interest.
References
- Argüello JM, Raimunda D, and González-Guerrero M (2012) Metal transport across biomembranes: Emerging models for a distinct chemistry. J Biol Chem 287: 13510–13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüello JM, Raimunda D, and Padilla-Benavides T (2013) Mechanisms of Copper Homeostasis in Bacteria. Front Cell Infect Microbiol 3: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Caille O, Rossier C, and Perron K (2007) A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J Bacteriol 189: 4561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JS, and Cooksey DA (1991) Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci 88: 8915–8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changela A, Chen K, Xue Y, Holschen J, Outten CE, O’Halloran TV, and Mondragon A (2003) Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301: 1383–7. [DOI] [PubMed] [Google Scholar]
- Chelliah V, Juty N, Ajmera I, Ali R, Dumousseau M, Glont M, et al. (2015) BioModels: ten-year anniversary. Nucleic Acids Res 43: D542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine P, Wickramasinghe WA, Harrison MD, Weber T, Solioz M, and Dameron CT (1999) The Enterococcus hirae copper chaperone CopZ delivers copper(I) to the CopY repressor. FEBS Lett 445: 27–30. [DOI] [PubMed] [Google Scholar]
- Cobine PA, Pierrel F, and Winge DR (2006) Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim Biophys Acta 1763: 759–772. [DOI] [PubMed] [Google Scholar]
- Corana A, Marchesi M, Martini C, and Ridella S (1987) Minimizing multimodal functions of continuous variables with the “simulated annealing” algorithm. ACM Trans Math Softw 13: 262–280. [Google Scholar]
- Djoko KY, Ong CL, Walker MJ, and McEwan AG (2015) The Role of Copper and Zinc Toxicity in Innate Immune Defense against Bacterial Pathogens. J Biol Chem 290: 18954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoko KY, Xiao Z, and Wedd AG (2008) Copper resistance in E. coli: the multicopper oxidase PcoA catalyzes oxidation of copper(I) in Cu(I)Cu(II)-PcoC. Chembiochem Eur J Chem Biol 9: 1579–1582. [DOI] [PubMed] [Google Scholar]
- Dupont CL, Grass G, and Rensing C (2011) Copper toxicity and the origin of bacterial resistance-new insights and applications. Metallomics 3: 1109–1118. [DOI] [PubMed] [Google Scholar]
- Ekici S, Yang H, Koch HG, and Daldal F (2012) Novel transporter required for biogenesis of cbb3-type cytochrome C oxidase in Rhodobacter capsulatus. MBio 3: e00293–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsen S, Ragno M, and Attree I (2011) PtrA is a periplasmic protein involved in Cu tolerance in Pseudomonas aeruginosa. J Bacteriol 193: 3376–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangipani E, and Haas D (2009) Copper acquisition by the SenC protein regulates aerobic respiration in Pseudomonas aeruginosa PAO1. FEMS Microbiol Lett 298: 234–40. [DOI] [PubMed] [Google Scholar]
- Fraústo da Silva JJR, and Williams RJP (2001) The Biological Chemistry of the Elements : the Inorganic Chemistry of Life. 2nd ed., Oxford University Press, Oxford. [Google Scholar]
- González-Guerrero M, Eren E, Rawal S, Stemmler TL, and Argüello J (2008) Cu+ transporting ATPases: Structure of the Two Transmembrane Cu+ Transport Sites. J Biol Chem 283: 29753–29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guerrero M, Hong D, and Argüello JM (2009) Chaperone-mediated Cu+ delivery to Cu+ transport ATPases. Requirement of nucleotide binding. J Biol Chem 284: 20804–20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guerrero M, Raimunda D, Cheng X, and Argüello JM (2010) Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol Microbiol 78: 1246–1258. [DOI] [PubMed] [Google Scholar]
- Grass G, and Rensing C (2001) CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys Res Commun 286: 902–908. [DOI] [PubMed] [Google Scholar]
- Gray HB, and Winkler JR (1996) Electron transfer in proteins. Annu Rev Biochem 65: 537–61. [DOI] [PubMed] [Google Scholar]
- Hodgkinson V, and Petris MJ (2012) Copper homeostasis at the host-pathogen interface. J Biol Chem 287: 13549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooke R, and Jeeves TA (1961) “Direct search” solution of numerical and statistical problems. J Assoc Comput Mach 8: 212–229. [Google Scholar]
- Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, et al. (2006) COPASI–a COmplex PAthway SImulator. Bioinformatics 22: 3067–3074. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19: 524–531. [DOI] [PubMed] [Google Scholar]
- Ize B, Viarre V, and Voulhoux R (2014) Cell fractionation. Methods Mol Biol 1149: 185–91. [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Nat Acad Sci USA 100: 14339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EH, Nies DH, McEvoy MM, and Rensing C (2011) Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol 193: 2381–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EH, Rensing C, and McEvoy MM (2010) Chaperone-mediated copper handling in the periplasm. Nat Prod Rep 27: 711–9. [DOI] [PubMed] [Google Scholar]
- Kumar SR, and Imlay JA (2013) How Escherichia coli Tolerates Profuse Hydrogen Peroxide Formation by a Catabolic Pathway. J Bacteriol 195: 4569–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomersky E, and Petris MJ (2015) Copper tolerance and virulence in bacteria. Metallomics 7: 957–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Grass G, Rensing C, Barrett SR, Yates CJ, Stoyanov JV, and Brown NL (2002) The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem Biophys Res Commun 295: 616–620. [DOI] [PubMed] [Google Scholar]
- Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, and Yu EW (2010) Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467: 484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, and Imlay JA (2009) The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106: 8344–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D, and Cavet JS (2008) Copper homeostasis in bacteria. Adv Appl Microbiol 65: 217–47. [DOI] [PubMed] [Google Scholar]
- Padilla-Benavides T, McCann CJ, and Argüello JM (2013) The mechanism of Cu+ transport ATPases: interaction with CU+ chaperones and the role of transient metal-binding sites. J Biol Chem 288: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang WL, Kaur A, Ratushny AV, Cvetkovic A, Kumar S, Pan M, et al. (2013) Metallochaperones Regulate Intracellular Copper Levels. PLoS Comput Biol 9: e1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pécou E, Maass A, Remenik D, Briche J, and Gonzalez M (2006) A mathematical model for copper homeostasis in Enterococcus hirae. Math Biosci 203: 222–239. [DOI] [PubMed] [Google Scholar]
- Petzold L (1983) Automatic selection of methods for solving stiff and nonstiff systems of ordinary differential equations. SIAM J Sci Stat Comput 4: 136–148. [Google Scholar]
- Philips SJ, Canalizo-Hernandez M, Yildirim I, Schatz GC, Mondragón A, and O’Halloran TV (2015) TRANSCRIPTION. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science 349: 877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana J, Novoa-Aponte L, and Argüello JM (2017) Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. J Biol Chem 292: 15691–15704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimunda D, Khare T, Giometti C, Vogt S, Argüello JM, and Finney L (2012) Identifying metalloproteins through X-ray fluorescence mapping and mass spectrometry. Metallomics 4: 921–927. [DOI] [PubMed] [Google Scholar]
- Raimunda D, Padilla-Benavides T, Vogt S, Boutigny S, Tomkinson KN, Finney LA, and Argüello JM (2013) Periplasmic response upon disruption of transmembrane Cu transport in Pseudomonas aeruginosa. Metallomics 5: 144–151. [DOI] [PubMed] [Google Scholar]
- Raue A, Kreutz C, Maiwald T, Bachmann J, Schilling M, Klingmüller U, and Timmer J (2009) Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 25: 1923–1929. [DOI] [PubMed] [Google Scholar]
- Rensing C, and Grass G (2003) Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev 27: 197–213. [DOI] [PubMed] [Google Scholar]
- Schaber J (2012) Easy parameter identifiability analysis with COPASI. Biosystems 110: 183–185. [DOI] [PubMed] [Google Scholar]
- Smirnova G, Muzyka N, and Oktyabrsky O (2012) Transmembrane glutathione cycling in growing Escherichia coli cells. Microbiol Res 167: 166–172. [DOI] [PubMed] [Google Scholar]
- Tavares P, Pereira AS, Moura JJ, and Moura I (2006) Metalloenzymes of the denitrification pathway. J Inorg Biochem 100: 2087–2100. [DOI] [PubMed] [Google Scholar]
- Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, and Parsek MR (2006) Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J Bacteriol 188: 7242–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaden JT, Lory S, and Gardner TS (2010) Quorum-sensing regulation of a copper toxicity system in Pseudomonas aeruginosa. J Bacteriol 192: 2557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita N, Landolfi G, Basle A, Platsaki S, Lee J, Waldron KJ, and Dennison C (2016) Bacterial cytosolic proteins with a high capacity for Cu(I) that protect against copper toxicity. Sci Rep 6: 39065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita N, Platsaki S, Baslé A, Allen SJ, Paterson NG, Crombie AT, et al. (2015) A four-helix bundle stores copper for methane oxidation. Nature 525: 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Brose J, Schimo S, Ackland SM, La Fontaine S, and Wedd AG (2011) Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: detection probes and affinity standards. J Biol Chem 286: 11047–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama H, and Nakae T (1996) Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 142: 2137–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.