Abstract
The integrated molecular interactions of proteins can create active biological networks whose material properties and actions can impact a variety of physiological processes. Chief among these is the ability to generate and respond to physical forces. The cytoskeleton plays a key role in this behavior, characterized by active self-reorganization to control a cell’s shape and mediate its physical interactions. This review discusses our current understanding of how the material properties of the cytoskeleton and its physical interactions with the extracellular environment impact cell migration.
Keywords: Cytoskeleton, Contractility, Adhesion, Migration, Mechanosensing
Introduction
Cells depend on biochemical signaling [1] and mechanical signaling [2,3] to regulate their interactions with the extracellular environment. The cytoskeleton, comprised of collections of filamentous proteins and their associated regulatory and binding proteins, is the foundation of these two signaling networks [4]. In addition to acting as a material that responds to externally applied forces [5], the cytoskeleton generates its own forces which are applied to the cell’s extracellular environment, whether that be the extracellular matrix (ECM)[6], or other cells [7,8].
While the individual molecular interactions underlying many of these physiological processes are well understood [9], their aggregated effects can precipitate starkly different collective behavior and interactions [10,11]. Simply mixing two types of filaments can create new architectures, such as the curved shapes that are produced by combining actin with septins [12]. The addition of crosslinkers, meanwhile, can shift the contraction of a network from isotropic to uniaxial through modulation of the stiffness of actin bundles [13]. Just the application of a force at one end of an actin filament can impact the activity of a formin at the other end of the filament [14]. Similarly, networks grown under an applied load self-organize to be globally stiffer, without changing the local material properties of the constituent filaments [15]. All of these structures and behaviors resemble those seen in vivo, where the cytoskeleton takes on specific architectures and organizations related to function [16,17].
With recent advances in imaging, it is possible to visualize the dynamics of the cytoskeleton in higher resolution [18], and more precisely measure mechanical interactions [19] and material properties [20,21] than ever before. These technological improvements provide important insights into local interactions between proteins and their spatial positioning within networks. The next challenge, however, is to understand how the macroscopic properties of cytoskeletal network behavior emerge from these integrated local molecular interactions across appropriate length and time scales. Here we summarize the current findings from the perspective of physics to understand force transmission as a network behavior as it relates to migration and invasion at the cellular scale.
Cell contractility is regulated by cell size
The dominant component of cell contractility is the product of non-muscle myosin II filaments pulling on the actin cytoskeleton [22]. These forces are then transmitted to the extracellular environment through integrin-based adhesions for cell-ECM interactions, or cadherin-based adhesions for cell-cell interactions. A number of different techniques have been developed to measure these types of forces [6], with recent advancements increasing the detection limit of the measurements [23] and adding the ability to resolve the spatial orientation of the applied forces [19].
A number of different metrics have been used to describe cellular force generation (See Box 1 for definitions and relations of terms related to force generation). In adherent cells the distribution of traction forces is highly heterogeneous and dependent on the spatial distribution of ligands [24,25] and the material properties of the extracellular environment [26]. Using micropatterning to constrain cell shape on substrates of different stiffness, we showed that both stress (force per unit area) and strain (relative displacement) are functions of the material properties of the substrate [26]. Cells generate larger traction stresses on stiffer substrates, but they result in smaller displacements (Figure 1). On soft substrates, the converse is true. The contractile energy (i.e. the total mechanical work done – see Box 1), however, is independent of the substrate stiffness [26]. Thus cells of the same size use the same amount of energy to deform the substrate (Figure 1). This suggests that when gauging the response of cells to changes in substrate stiffness, measurements of traction stress alone reveal more about the material properties of the substrate than they do about the contractile state of the cell.
Box 1. Lexicon of Force Generation.
Stress – A measure of force applied per unit area. Typically measured in pascals (Pa), where 1 Pa = 1 N/m2.
Strain – A measure of deformation, typically caused by a force, relative to the equilibrium length of an object. Strain is unitless and typically measured as a percent ΔL/L.
Displacement – A measure of distance between an initial and final position. Displacements have units of length (e.g. m) and are used to calculate the strain.
Stiffness – A measure of how resistant a material is to deformation. For objects (i.e. 2D and 3D materials) stiffness is often referred to as a modulus and measured in units of Pa.
Work (or Strain Energy) – A measure of the energy used to apply a force over a distance. For a constant force, work is defined in 1D as
where F is the applied force and d is the distance it is applied over. For a 2D system, such as used in traction force microscopy, the work is defined as the integral over the area of the traction stress multiplied by the displacement
where T(r) is the traction stress and u(r) is the displacement at position (W3 < W2).
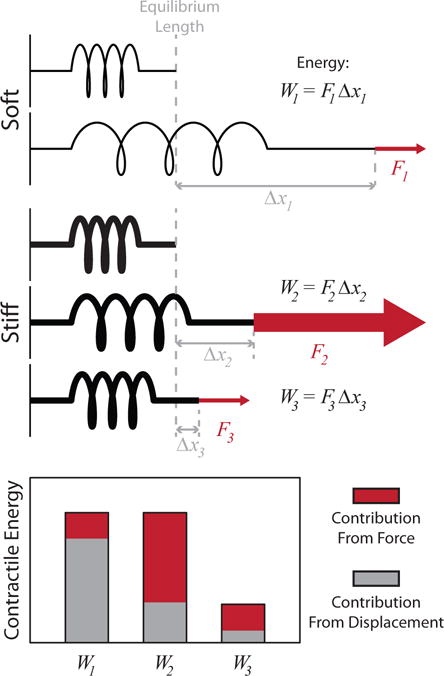
A cartoon illustrating the relationship between displacement, force, and contractile energy. The same amount of energy is used to deform the springs in cases 1 and 2. For the soft spring, a small force is applied over a long distance. In the stiff spring, a large force is applied over a short distance. The work done in each case is equivalent (W1 = W2 = F1Δx1 = F2Δx2). Conversely, in case 3, a small force results in only a small displacement, and therefore requires less energy (W2 < W2).
Figure 1.
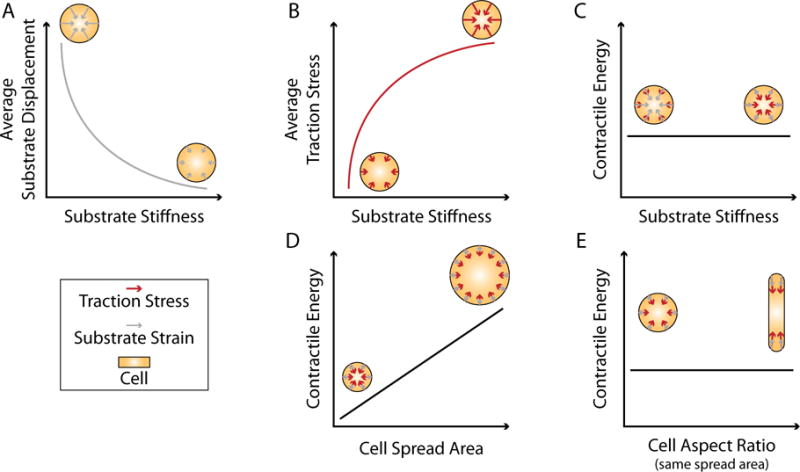
Relationships between displacements, stress, contractile energy and cell geometry. (A-C) For cells of the same size, (A) the average substrate displacement decreases and (B) the average traction stress increases as functions of substrate stiffness. (C) The contractile energy, meanwhile, is independent of the material properties of the substrate. Cells of the same size use the same amount of energy to deform the substrate regardless of stiffness. (D) Larger cells do more work than smaller cells, and thus the contractile energy increases as a function of cell spread area. (E) For a given cell area, however, the contractile energy is not sensitive to the shape of a cell. The shape will affect the distribution of stresses on the substrate but not the magnitude of the energy expended.
Measuring the contractile energy, on the other hand, reflects the entire output of the cell, accounting for both stress and strain. Unsurprisingly, the total contractile energy is sensitive to the overall size of the cell, with larger cells having larger cytoskeletons, and therefore a larger number of active motors doing work [26,27] (Figure 1). For a given spread area, however, the total contractile energy is independent of cell geometry [26,28] (Figure 1). This is in contrast to measurements like the average stress which are dependent upon cellular morphology and adhesion distribution. The scaling of contractile energy with cell area also suggests that cells actively maintain a contractility set point. Recently, two reports used optogenetic approaches to modulate RhoA, the GTPase that controls the contractile signaling pathway [29,30]. When RhoA is activated cells become more contractile, but then relax back to their initial contractile states when the stimulation is removed. This behavior is consistent with previous results using incubation and washout of myosin inhibitor drugs, which causes the contractility to initially decrease before recovering to their initial state [31,32]. In each case, perturbations to the contractile state of the cell result in the cell trying to re-establish its initial contractile state when the perturbation is removed. The contractile energy per unit area can therefore serve as a metric to compare contractile behavior across perturbations to cells and even different cell types [9].
Cytoskeletal architecture and ECM geometry regulate force transmission
While the contractile energy tells us about the mechanical state of the cell, to understand migration we must understand how cells spatially and temporally regulate force generation. The cytoskeleton consists of a number of different filamentous proteins (e.g. actin, microtubules, intermediate filaments, septins) and motor proteins (e.g. myosins, kinesins, dyenins). Because the actomyosin cytoskeleton is the only one directly coupled to the extracellular environment, the primary sources for forces that are transmitted to the substrate are myosin motors pulling on actin filaments connected to adhesions [33], and actin polymerization dynamics [34,35]. In each case, the forces are generated in the cytoskeleton and transmitted via transmembrane proteins to the ECM (via integrins) or to other cells (via cadherins). Cell morphology and the distribution of adhesions therefore play important roles in spatially regulating force transmission. Both integrins [36] and cadherins [37] act as catch bonds (bonds that increase their lifetime as a function of applied force), and thus their behavior can change as a function of applied load. In integrins this feature is speculated to play a role in stiffness sensing, by changing the force applied to the bonds [38,39]. On soft substrates the applied forces deform the substrate more than the integrin, leading to shorter bond lifetimes. On stiff substrates, the strain in the substrate is reduced, which puts more tension on the integrin and leads to longer bond lifetimes. The forces in either scenario can be generated through myosin activity [38,39], or actin polymerization [40]. These forces also drive actin retrograde flow which plays an important role in orienting integrins [41] and proteins within the focal adhesion [42].
While actin-polymerization forces can play a significant role in adhesion formation [43], the majority of forces generated by the cell that are capable of deforming the extracellular environment are the product of myosin activity transmitted through adhesions [22]. Since cells are unable to exert large stresses (e.g. > 100 Pa) in the absence of adhesions [35], the geometry of the ECM plays a significant role in the distribution of traction stresses [26,44,45]. The positioning of adhesions can in turn influence cytoskeletal organization [25] and overall cell morphology [45]. The cytoskeletal organization, therefore, plays an important role in directing force transmission across the network [28,30,46]. Simply changing the organization of the ECM influences both cytoskeletal organization and the distribution of traction stresses [25,26,44]. This interplay of cell shape, ECM organization, and cytoskeletal organization is thought to play an important role during development, such as during ventral furrow formation in Drosophila [47], and gastrulation in zebrafish [48].
It is important to note that while an adhesion connects the cytoskeleton to the extracellular environment, an ECM connection is not sufficient to guarantee that an adhesion is under tension. For example, many adhesions far from the cell edge are coupled mechanically (i.e. able to support tension) between the cytoskeleton and the substrate, but are not actively under tension [30,49,50]. Using an optogenetic approach, we showed that when contraction was induced, these adhesions could still transmit force to the substrate [30]. Interestingly, it was the coupling between the adhesions and the stress fibers, and not the stress fibers themselves, that exhibited the largest strains during these locally-induced contractions [30]. This phenomenon could potentially be related to the behavior of the proteins within the adhesions. For instance, vinculin, an adhesion protein that is present in both cell-ECM and cell-cell adhesions, couples the cytoskeleton to the adhesion plaque and also displays catch-bond behavior, but only in a single direction [51]. This intriguing finding suggests that organization and geometry of proteins in the adhesion plaques could play important roles in mediating force transmission.
Migration modes depend on ECM geometry and coupling efficiency
By regulating adhesion distribution, the geometry of the ECM significantly impacts modes of migration. Cells on fibrillar structures tend to take on more elongated morphologies, independent of the stiffness of the matrix [52]. In contrast, cells on planar surfaces tend to spread out more [53]. When cells are confined to migrate along linear strips of ECM, they migrate significantly faster than cells on planar substrates of the same material properties [27,45,54,55]. This holds true in 3D as well, as cells migrating on fibers display significantly faster migration rates than those on planar substrates [56]. While this may be related to the geometry of the ECM, it may also simply be a function of having fewer adhesions to turn over during the migration process [56].
In addition to ECM geometry, migration behavior also depends on adhesion stability, cell contractility, and ECM material properties. Decoupling these interconnected parameters (Figure 2) has proven challenging and there are unlikely to be universal relationships. This difficulty is particularly evident in the context of invasion, which typically sees perturbations to a combination of these parameters and additionally requires changes in the integrity of the basement membrane. Generally, however, increasing the substrate stiffness promotes adhesion formation, thus promoting increases in spread area and contractility [38,53]. Increases in myosin activity can increase cell stiffness and reduce adhesion turnover, making cells less invasive [57]. Conversely, reduction of myosin activity promotes spreading [35] and increases invasion [58], potentially by decreasing adhesion lifetime [59]. Similarly, destabilizing adhesions through knockdown of the formin Dia1 reduces invasion [60], while stabilizing them through upregulation of paxillin enhances it [61]. Finally, cells are also able to remodel the ECM as they migrate, changing its material properties [62] and organization [63], which can in turn promote invasion [64]. Each of these parameters feeds into the others (Figure 2). It is the overall balance of these interactions that regulates the behavior of cells, with deviations leading to aberrant behaviors such as invasion.
Figure 2.
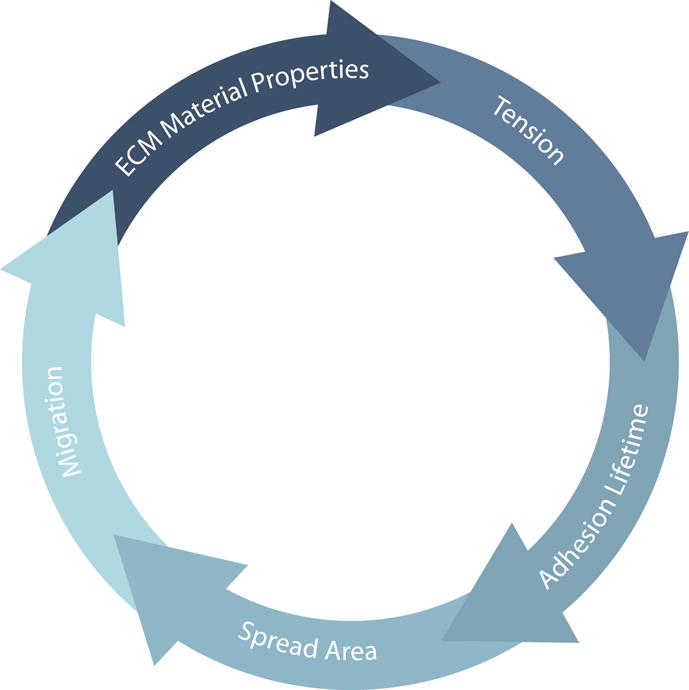
Migration and invasion are a balance of a number of different physical interactions. The material properties of the ECM regulate how much tension is transmitted to adhesions. The tension across adhesion bonds directly regulates their lifetime. Cells that are able to adhere to the substrate can increase their spread area. Cells that are able to spread can change their shape and migrate. Cells that migrate can remodel the ECM and change its material properties. While these processes are depicted as a simple loop, they are in reality interrelated, feeding into each other. It is their balance that regulates physiological function. Changes in cell behavior, whether related to misregulated biochemical signaling or changes in physical interactions, break this delicate balance and can promote aberrant behavior such as invasion.
It is important to note, however, that not all migration mechanisms require specific adhesion. It has been recently shown that cells can utilize alternative physical interactions to migrate in the absence of specific adhesion with the substrate. Typically these scenarios involve the cell deforming themselves more than the substrate [65]. When confined in channels, cells can use friction generated by actomyosin-driven flows in the cytoskeleton to generate propulsion [35]. This mechanism relies on the cell creating pressure against its confinement, akin to shimmying up a chimney. A similar mechanism that captures pressure differentials mediated by the nucleus between the front and rear of the cell has also recently been proposed [66]. While these mechanisms can allow cells to migrate through constricted environments, they may also lead to side effects such as DNA damage [67]. In less adherent environments, changes in cortical contractility can shift cells from bleb-driven to actomyosin-flow-driven migration [55,68,69]. Surprisingly, these modes of migration appear to be available to a wide range of cell types [68]. They are thought to play especially important roles in leukocyte migration in vivo, where ECM matrix composition and distribution can take on a number of different forms as cells leave the blood stream and migrate to sites of inflammation [65]. These studies show that cells adapt their migration mechanisms to their different environments.
Potential Roles of Mechanics in Signaling
While physical interactions are clearly part of the processes that result in migration and invasion, there is strong evidence that these interactions can regulate biochemical signaling as well. Changes in the physical environment have been shown to effect YAP/TAZ nuclear translocation [38,70] and SRF/Mkl1 [71] activity, potentially through LINC complexes [72]. While the direct mechanisms behind these interactions are unclear, an intriguing possibility is that the changes in the mechanical interactions between the cell and its extracellular environment could alter a cell’s metabolism. Mechanical interactions, just like biochemical interactions, require energy. Recently it was shown that metabolism was upregulated as a function of collagen density in the matrix [73] and that force applied to E-cadherin could activate energy production [74]. It was also recently shown that cancer cells, unlike wild-type cells, were unable to respond to compression of the ECM [75]. Together these results suggest that the cell’s ability to regulate the energy consumption by mechanical interactions may play a vital role in regulating its behavior. Without a doubt, the connections between mechanical and biochemical signaling networks will continue to provide the foundation for a number of interesting avenues of research in the years to come.
Acknowledgments
This work was supported by an National Science Foundation CAREER Award (#1749302).
This work was supported in part by the University of Rochester Program for Advanced Immune Bioimaging Pilot — P01 AI102851 (NIH/NIAID) and the University of Rochester School of Medicine and Dentistry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lawson CD, Ridley AJ. Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol. 2018;217:447–457. doi: 10.1083/jcb.201612069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iskratsch T, Wolfenson H, Sheetz MP. Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat Rev Mol Cell Biol. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Helvert S, Storm C, Friedl P. Mechanoreciprocity in cell migration. Nat Cell Biol. 2018;20:8–20. doi: 10.1038/s41556-017-0012-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 5.Pegoraro AF, Janmey P, Weitz DA. Mechanical Properties of the Cytoskeleton and Cells. Cold Spring Harb Perspect Biol. 2017;9:a022038. doi: 10.1101/cshperspect.a022038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roca-Cusachs P, Conte V, Trepat X. Quantifying forces in cell biology. Nat Cell Biol. 2017;19:742–751. doi: 10.1038/ncb3564. [DOI] [PubMed] [Google Scholar]
- 7.Petridou NI, Spiró Z, Heisenberg C-P. Multiscale force sensing in development. Nat Cell Biol. 2017;19:581–588. doi: 10.1038/ncb3524. [DOI] [PubMed] [Google Scholar]
- 8.Yap AS, Duszyc K, Viasnoff V. Mechanosensing and Mechanotransduction at Cell-Cell Junctions. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a028761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murrell M, Oakes PW, Lenz M, Gardel ML. Forcing cells into shape: the mechanics of actomyosin contractility. Nat Rev Mol Cell Biol. 2015;16:486–498. doi: 10.1038/nrm4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronceray P, Broedersz CP, Lenz M. Fiber networks amplify active stress. Proc Natl Acad Sci USA. 2016;113:2827–2832. doi: 10.1073/pnas.1514208113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broedersz CP, Mackintosh FC. Modeling semiflexible polymer networks. Rev Mod Phys. 2014;86:995. [Google Scholar]
- 12.Mavrakis M, Azou-Gros Y, Tsai F-C, Alvarado J, Bertin A, Iv F, Kress A, Brasselet S, Koenderink GH, Lecuit T. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat Cell Biol. 2014;16:322–334. doi: 10.1038/ncb2921. [DOI] [PubMed] [Google Scholar]
- 13.Stam S, Freedman SL, Banerjee S, Weirich KL, Dinner AR, Gardel ML. Filament rigidity and connectivity tune the deformation modes of active biopolymer networks. Proc Natl Acad Sci USA. 2017;114:E10037–E10045. doi: 10.1073/pnas.1708625114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Zimmermann D, Homa KE, Hocky GM, Pollard LW, La Cruz De EM, Voth GA, Trybus KM, Kovar DR. Mechanoregulated inhibition of formin facilitates contractile actomyosin ring assembly. Nat Commun. 2017;8:703. doi: 10.1038/s41467-017-00445-3. Illustrated that mechanical information could propagate from one end of an actin filament to the other, thereby affecting protein behavior at a distance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Bieling P, Li T-D, Weichsel J, McGorty R, Jreij P, Huang B, Fletcher DA, Mullins RD. Force Feedback Controls Motor Activity and Mechanical Properties of Self-Assembling Branched Actin Networks. Cell. 2016;164:115–127. doi: 10.1016/j.cell.2015.11.057. Showed that actin filaments organize into networks with different material properties when grown under an applied load, without changing the mechanical properties of the constituent filaments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svitkina T. The Actin Cytoskeleton and Actin-Based Motility. Cold Spring Harb Perspect Biol. 2018;10:a018267. doi: 10.1101/cshperspect.a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suarez C, Kovar DR. Internetwork competition for monomers governs actin cytoskeleton organization. Nat Rev Mol Cell Biol. 2016;17:799–810. doi: 10.1038/nrm.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritz-Laylin LK, Riel-Mehan M, Chen B-C, Lord SJ, Goddard TD, Ferrin TE, Nicholson-Dykstra SM, Higgs H, Johnson GT, Betzig E, et al. Actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. Elife. 2017;6:e26990. doi: 10.7554/eLife.26990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brockman JM, Blanchard AT, Pui-Yan V, Derricotte WD, Zhang Y, Fay ME, Lam WA, Evangelista FA, Mattheyses AL, Salaita K. Mapping the 3D orientation of piconewton integrin traction forces. Nat Methods. 2018;15:115–118. doi: 10.1038/nmeth.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scarcelli G, Polacheck WJ, Nia HT, Patel K, Grodzinsky AJ, Kamm RD, Yun SH. Noncontact three-dimensional mapping of intracellular hydromechanical properties by Brillouin microscopy. Nat Methods. 2015;12:1132–1134. doi: 10.1038/nmeth.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Fiore A, Yun SH, Kim H, Scarcelli G. Line-scanning Brillouin microscopy for rapid non-invasive mechanical imaging. Sci Rep. 2016;6:35398. doi: 10.1038/srep35398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Y, Rossier O, Gauthier NC, Biais N, Fardin M-A, Zhang X, Miller LW, Ladoux B, Cornish VW, Sheetz MP. Cytoskeletal coherence requires myosin-IIA contractility. J Cell Sci. 2010;123:413–423. doi: 10.1242/jcs.058297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kronenberg NM, Liehm P, Steude A, Knipper JA, Borger JG, Scarcelli G, Franze K, Powis SJ, Gather MC. Long-term imaging of cellular forces with high precision by elastic resonator interference stress microscopy. Nat Cell Biol. 2017;19:864–872. doi: 10.1038/ncb3561. [DOI] [PubMed] [Google Scholar]
- 24.Tseng Q, Wang I, Duchemin-Pelletier E, Azioune A, Carpi N, Gao J, Filhol O, Piel M, Théry M, Balland M. A new micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. Lab Chip. 2011;11:2231–2240. doi: 10.1039/c0lc00641f. [DOI] [PubMed] [Google Scholar]
- 25.Théry M, Racine V, Piel M, Pépin A, Dimitrov A, Chen Y, Sibarita J-B, Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc Natl Acad Sci USA. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oakes PW, Banerjee S, Marchetti MC, Gardel ML. Geometry regulates traction stresses in adherent cells. Biophys J. 2014;107:825–833. doi: 10.1016/j.bpj.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leal-Egaña A, Letort G, Martiel J-L, Christ A, Vignaud T, Roelants C, Filhol O, Théry M. The size-speed-force relationship governs migratory cell response to tumorigenic factors. Mol Biol Cell. 2017;28:1612–1621. doi: 10.1091/mbc.E16-10-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kassianidou E, Brand CA, Schwarz US, Kumar S. Geometry and network connectivity govern the mechanics of stress fibers. Proc Natl Acad Sci USA. 2017;114:2622–2627. doi: 10.1073/pnas.1606649114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valon L, Marín-Llauradó A, Wyatt T, Charras G, Trepat X. Optogenetic control of cellular forces and mechanotransduction. Nat Commun. 2017;8:14396. doi: 10.1038/ncomms14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oakes PW, Wagner E, Brand CA, Probst D, Linke M, Schwarz US, Glotzer M, Gardel ML. Optogenetic control of RhoA reveals zyxin-mediated elasticity of stress fibres. Nat Commun. 2017;8:15817. doi: 10.1038/ncomms15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labouesse C, Verkhovsky AB, Meister J-J, Gabella C, Vianay B. Cell Shape Dynamics Reveal Balance of Elasticity and Contractility in Peripheral Arcs. Biophys J. 2015;108:2437–2447. doi: 10.1016/j.bpj.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aratyn-Schaus Y, Oakes PW, Gardel ML. Dynamic and structural signatures of lamellar actomyosin force generation. Mol Biol Cell. 2011;22:1330–1339. doi: 10.1091/mbc.E10-11-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz US, Gardel ML. United we stand - integrating the actin cytoskeleton and cell-matrix adhesions in cellular mechanotransduction. J Cell Sci. 2012;125:3051–3060. doi: 10.1242/jcs.093716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Bergert M, Erzberger A, Desai RA, Aspalter IM, Oates AC, Charras G, Salbreux G, Paluch EK. Force transmission during adhesion-independent migration. Nat Cell Biol. 2015;17:524–529. doi: 10.1038/ncb3134. Demonstrated that cells could migrate in the absence of adhesions using friction generated by actomyosin flow. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong F, García AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manibog K, Li H, Rakshit S, Sivasankar S. Resolving the molecular mechanism of cadherin catch bond formation. Nat Commun. 2014;5:3941. doi: 10.1038/ncomms4941. [DOI] [PubMed] [Google Scholar]
- 38.Oria R, Wiegand T, Escribano J, Elosegui-Artola A, Uriarte JJ, Moreno-Pulido C, Platzman I, Delcanale P, Albertazzi L, Navajas D, et al. Force loading explains spatial sensing of ligands by cells. Nature. 2017;196:395. doi: 10.1038/nature24662. [DOI] [PubMed] [Google Scholar]
- 39*.Elosegui-Artola A, Oria R, Chen Y, Kosmalska A, Pérez-González C, Castro N, Zhu C, Trepat X, Roca-Cusachs P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol. 2016;18:540–548. doi: 10.1038/ncb3336. Demonstrates how a mechanical clutch can be used to sense substrate stiffness. [DOI] [PubMed] [Google Scholar]
- 40.Oakes PW, Bidone TC, Beckham Y, Skeeters AV, Ramirez-San Juan GR, Winter SP, Voth GA, Gardel ML. Lamellipodium is a myosin-independent mechanosensor. Proc Natl Acad Sci USA. 2018;115:2646–2651. doi: 10.1073/pnas.1715869115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordenfelt P, Moore TI, Mehta SB, Kalappurakkal JM, Swaminathan V, Koga N, Lambert TJ, Baker D, Waters JC, Oldenbourg R, et al. Direction of actin flow dictates integrin LFA-1 orientation during leukocyte migration. Nat Commun. 2017;8:2047. doi: 10.1038/s41467-017-01848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swaminathan V, Kalappurakkal JM, Mehta SB, Nordenfelt P, Moore TI, Koga N, Baker DA, Oldenbourg R, Tani T, Mayor S, et al. Actin retrograde flow actively aligns and orients ligand-engaged integrins in focal adhesions. Proc Natl Acad Sci USA. 2017;114:10648–10653. doi: 10.1073/pnas.1701136114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pontes B, Monzo P, Gole L, Le Roux A-L, Kosmalska AJ, Tam ZY, Luo W, Kan S, Viasnoff V, Roca-Cusachs P, et al. Membrane tension controls adhesion positioning at the leading edge of cells. J Cell Biol. 2017;216:2959–2977. doi: 10.1083/jcb.201611117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tseng Q, Duchemin-Pelletier E, Deshiere A, Balland M, Guillou H, Filhol O, Théry M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci USA. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez-San Juan GR, Oakes PW, Gardel ML. Contact guidance requires spatial control of leading-edge protrusion. Mol Biol Cell. 2017;28:1043–1053. doi: 10.1091/mbc.E16-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mak M, Zaman MH, Kamm RD, Kim T. Interplay of active processes modulates tension and drives phase transition in self-renewing, motor-driven cytoskeletal networks. Nat Commun. 2016;7:10323. doi: 10.1038/ncomms10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Chanet S, Miller CJ, Vaishnav ED, Ermentrout B, Davidson LA, Martin AC. Actomyosin meshwork mechanosensing enables tissue shape to orient cell force. Nat Commun. 2017;8:15014. doi: 10.1038/ncomms15014. Demonstrated that mechanical and geometrical restrictions play important roles in orienting the cytoskeleton during development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morita H, Grigolon S, Bock M, Krens SFG, Salbreux G, Heisenberg C-P. The Physical Basis of Coordinated Tissue Spreading in Zebrafish Gastrulation. Dev Cell. 2017;40:354–366.e4. doi: 10.1016/j.devcel.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothenberg KE, Neibart SS, LaCroix AS, Hoffman BD. Controlling Cell Geometry Affects the Spatial Distribution of Load Across Vinculin. Cell Mol Bioeng. 2015;8:1–19. [Google Scholar]
- 50.Morimatsu M, Mekhdjian AH, Chang AC, Tan SJ, Dunn AR. Visualizing the interior architecture of focal adhesions with high-resolution traction maps. Nano Lett. 2015;15:2220–2228. doi: 10.1021/nl5047335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51**.Huang DL, Bax NA, Buckley CD, Weis WI, Dunn AR. Vinculin forms a directionally asymmetric catch bond with F-actin. Science. 2017;357:703–706. doi: 10.1126/science.aan2556. Demonstrated that vinculin only behaves as a catch-bond when forces are loaded in one direction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker BM, Trappmann B, Wang WY, Sakar MS, Kim IL, Shenoy VB, Burdick JA, Chen CS. Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments. Nat Mater. 2015;14:1262–1268. doi: 10.1038/nmat4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charrier EE, Pogoda K, Wells RG, Janmey PA. Control of cell morphology and differentiation by substrates with independently tunable elasticity and viscous dissipation. Nat Commun. 2018;9:449. doi: 10.1038/s41467-018-02906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doyle AD, Kutys ML, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Micro-environmental control of cell migration – myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. J Cell Sci. 2012;125:2244–2256. doi: 10.1242/jcs.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maiuri P, Rupprecht J-F, Wieser S, Ruprecht V, Bénichou O, Carpi N, Coppey M, De Beco S, Gov N, Heisenberg C-P, et al. Actin Flows Mediate a Universal Coupling between Cell Speed and Cell Persistence. Cell. 2015 doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 56.Doyle AD, Carvajal N, Jin A, Matsumoto K, Yamada KM. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat Commun. 2015;6:8720. doi: 10.1038/ncomms9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Surcel A, Ng WP, West-Foyle H, Zhu Q, Ren Y, Avery LB, Krenc AK, Meyers DJ, Rock RS, Anders RA, et al. Pharmacological activation of myosin II paralogs to correct cell mechanics defects. Proc Natl Acad Sci USA. 2015;112:1428–1433. doi: 10.1073/pnas.1412592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen-Ngoc K-V, Silvestri VL, Georgess D, Fairchild AN, Ewald AJ. Mosaic loss of non-muscle myosin IIA and IIB is sufficient to induce mammary epithelial proliferation. J Cell Sci. 2017;130:3213–3221. doi: 10.1242/jcs.208546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou DW, Lee TT, Weng S, Fu J, García AJ. Effects of substrate stiffness and actomyosin contractility on coupling between force transmission and vinculinpaxillin recruitment at single focal adhesions. Mol Biol Cell. 2017;28:1901–1911. doi: 10.1091/mbc.E17-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fessenden TB, Beckham Y, Perez-Neut M, Ramírez-San Juan G, Chourasia AH, Macleod KF, Oakes PW, Gardel ML. Dia1-dependent adhesions are required by epithelial tissues to initiate invasion. J Cell Biol. 2018;217:1485–1502. doi: 10.1083/jcb.201703145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mekhdjian AH, Kai F, Rubashkin MG, Prahl LS, Przybyla LM, McGregor AL, Bell ES, Barnes JM, Dufort CC, Ou G, et al. Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix. Mol Biol Cell. 2017;28:1467–1488. doi: 10.1091/mbc.E16-09-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nam S, Lee J, Brownfield DG, Chaudhuri O. Viscoplasticity Enables Mechanical Remodeling of Matrix by Cells. Biophys J. 2016;111:2296–2308. doi: 10.1016/j.bpj.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oudin MJ, Jonas O, Kosciuk T, Broye LC, Guido BC, Wyckoff J, Riquelme D, Lamar JM, Asokan SB, Whittaker C, et al. Tumor Cell-Driven Extracellular Matrix Remodeling Drives Haptotaxis during Metastatic Progression. Cancer discovery. 2016;6:516–531. doi: 10.1158/2159-8290.CD-15-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bordeleau F, Mason BN, Lollis EM, Mazzola M, Zanotelli MR, Somasegar S, Califano JP, Montague C, LaValley DJ, Huynh J, et al. Matrix stiffening promotes a tumor vasculature phenotype. Proc Natl Acad Sci USA. 2017;114:492–497. doi: 10.1073/pnas.1613855114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paluch EK, Aspalter IM, Sixt M. Focal Adhesion-Independent Cell Migration. Annu Rev Cell Dev Biol. 2016;32:469–490. doi: 10.1146/annurev-cellbio-111315-125341. [DOI] [PubMed] [Google Scholar]
- 66.Petrie RJ, Harlin HM, Korsak LIT, Yamada KM. Activating the nuclear piston mechanism of 3D migration in tumor cells. J Cell Biol. 2017;216:93–100. doi: 10.1083/jcb.201605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, Tewari M, Bennett RR, Harding SM, Liu AJ, et al. DNA Damage Follows Repair Factor Depletion and Portends Genome Variation in Cancer Cells after Pore Migration. Curr Biol. 2017;27:210–223. doi: 10.1016/j.cub.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68**.Liu Y-J, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuzé M, Takaki T, Voituriez R, Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. Demonstrated that a wide variety of cells are able to use low adhesive modes of migration. [DOI] [PubMed] [Google Scholar]
- 69.Ruprecht V, Wieser S, Callan-Jones A, Smutny M, Morita H, Sako K, Barone V, Ritsch-Marte M, Sixt M, Voituriez R, et al. Cortical Contractility Triggers a Stochastic Switch to Fast Amoeboid Cell Motility. Cell. 2015;160:673–685. doi: 10.1016/j.cell.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 71.Willer MK, Carroll CW. Substrate stiffness-dependent regulation of SRF/Mkl1 requires the inner nuclear membrane protein Emerin. J Cell Sci. 2017 doi: 10.1242/jcs.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thakar K, May CK, Rogers A, Carroll CW. Opposing roles for distinct LINC complexes in regulation of the small GTPase RhoA. Mol Biol Cell. 2017;28:182–191. doi: 10.1091/mbc.E16-06-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morris BA, Burkel B, Ponik SM, Fan J, Condeelis JS, Aguirre-Ghiso JA, Castracane J, Denu JM, Keely PJ. Collagen Matrix Density Drives the Metabolic Shift in Breast Cancer Cells. EBioMedicine. 2016;13:146–156. doi: 10.1016/j.ebiom.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74*.Bays JL, Campbell HK, Heidema C, Sebbagh M, Demali KA. Linking E-cadherin mechanotransduction to cell metabolism through force-mediated activation of AMPK. Nat Cell Biol. 2017;19:724–731. doi: 10.1038/ncb3537. Showed that tension across E-cadherin could lead directly to changes in cell metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang L, Carrington LJ, Erdogan B, Ao M, Brewer BM, Webb DJ, Li D. Biomechanics of cell reorientation in a three-dimensional matrix under compression. Exp Cell Res. 2017;350:253–266. doi: 10.1016/j.yexcr.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


