Abstract
As a component of p53-dependent lncRNA (long non-coding RNA), PANDAR (the promoter of CDKN1A antisense DNA damage activated RNA) participates in the epigenetic regulation in human cancer. However, the involvement of PANDAR in cancer chemoresistance is unknown. In this study, we report that PANDAR serves as a negative regulator of cisplatin sensitivity in human ovarian cancer via PANDAR-SRFS2-p53 feedback regulation in nuclear. Our data showed that among the drugs commonly used in ovarian cancer therapy, cisplatin induces higher levels of PANDAR compared with doxorubicin and paclitaxel. We also proved that PANDAR exhibited higher expression in cisplatin-resistant ovarian cancer tissues and cells, compared with cisplatin-sensitive ones, and this expression pattern depends on wild-type p53 (wt-p53), not mutant-p53 (mt-p53). In vitro and in vivo, PANDAR overexpression improved cell survival rate and tumor growth in response to cisplatin, while depletion of PANDAR leads to a reduced tumor growth. Further investigation revealed that PANDAR-reduced cisplatin sensitivity was likely or partly due to the PANDAR-binding protein SFRS2 (arginine/serine-rich 2), a splicing factor with the ability to negative regulate p53 and its phosphorylation at Serine 15 (Ser15). This feedback regulation of PANDAR–SFRS2–p53 leads to a reduced transactivation of p53-related pro-apoptotic genes, such as PUMA (p53-upregulated modulator of apoptosis). In addition, in platinum-treated patients with relapsed ovarian cancer, resistant period was positively correlated with the expression of PANDAR and SFRS2, and inversely associated with expression of p53-Ser15 and PUMA in these clinical tissues. Last but not least, the role of PANDAR in chemoresistance was confirmed in patients with ovarian cancer. These findings reveal a novel regulatory maneuver of cancer cells in response to chemostress, and might shed light on overcoming cisplatin resistance in ovarian cancer.
Introduction
Ovarian cancer (OC) continues to kill more than 150,000 women every year worldwide1. It is usually advanced when diagnosed. Staging is surgical. Treatment requires cytoreduction and chemotherapy. Chemotherapy is essential for the management of cancer progression1. However, drug resistance can lead to treatment failure2. Hence, a better understanding of chemoresistance in ovarian cancer therapeutics is urgently needed.
Cisplatin, the basic anticancer drug of chemotherapy, often develop drug resistance in ovarian cancer treatment2. To date, the mechanism of cisplatin resistance has been elusive3. Although the tumor suppressor p53 phosphorylation at Serine 15 (Ser15) and Serine 20 (Ser20) were identified as the key to cisplatin resistance in OC3,4, it still lacks a clear regulatory mechanism during this process. Serine-rich and arginine-rich proteins (SR proteins), a family of RNA-binding proteins, were initially discovered as regulators of alternative splicing5. Recent studies have revealed that SR proteins are involved in p53 and its phosphorylation and acetylation6,7. For instance, in response to ribosomal disturbances, SFRS1 (arginine/serine-rich 1) interacts with MDM2 (murine double minute 2) to inhibit p53 degradation6. p53 post-translational turnover is regulated by another member of SR family, SFRS2 (arginine/serine-rich 2), also called SC35 or SRFS2. SFRS2 depletion from mouse embryonic fibroblasts could result in p53 hyperphosphorylation6. However, whether SFRS2 regulates p53 phosphorylation in human OC remains unclear.
Long non-coding RNAs (lncRNAs), with 200–100,000 nt in size, has been found to regulate various cellular mechanisms, including cisplatin resistance8, through interacting with proteins and co-factors9. PANDAR, the Promoter of CDKN1A Antisense DNA damage Activated RNA, was first reported as the most upregulated p53-dependent lncRNA responding to drug-induced cell apoptosis10. The roles of PANDAR are diverse according to the cellular location and interaction partners. For instance, when bound to the SAFA (the scaffold attachment factor A) protein in cardiomyocytes, PANDAR regulates cellular senescence11. In this study, we found a matching sequence of PANDAR (167bp–176bp) containing 5′-CCAG-3′, which is reported as the high-affinity binding sequence recognized by SFRS2 and could now be found in all SELEX (Systematic Evolution of Ligands by Exponential Enrichment) consensus sequences and in all identified SFRS2-specific ESEs (exon-splicing enhancers)12. In line with these observations, we reason that whether PANDAR could interact with SFRS2 in OC cells.
To fill the above gaps, we studied the role of PANDAR in cisplatin sensitivity and discovered that cisplatin-induced PANDAR expression counter-regulates nuclear p53 and its phosphorylation at Ser15 via interacting with SFRS2, which in turn, attenuates cisplatin sensitivity in ovarian cancer chemotherapy.
Results
Inverse association between PANDAR expression and cisplatin sensitivity in OC
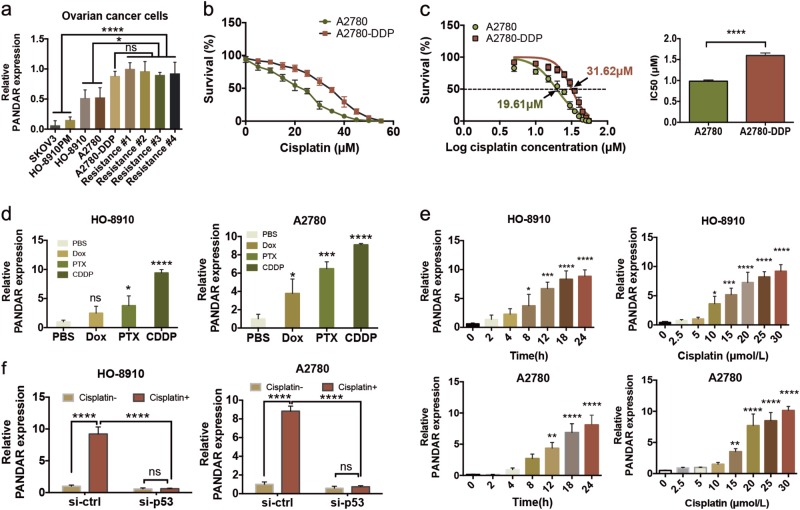
To investigate whether lncRNA PANDAR was associated with ovarian cancer chemosensitivity, we examined PANDAR expression profile in cisplatin-sensitive and cisplatin-resistant cells of OC (Fig. 1). First, we detected the expression profiles of wt-p53 and mt-p53 in OC cell lines, in which PANDAR expression was largely determined. Data showed that wt-p53 was positive in OC cell lines except SKOV3, and wt-p53 was only seen in the cytoplasm of A2780-DDP and HO-8910PM cells (Supplementary Fig. S1a, b), indicating that PANDAR roles in ovarian cancer chemoresistance could be sought among A2780, HO-8910, HO-8910PM, and A2780-DDP cell lines. We also isolated primary cells from the recurrent OC samples without p53 mutation (Supplementary Fig. S1c, Table 1), namely Resistance #1, #2, #3, #4, and then measured PANDAR expression level in these recurrent cells, cisplatin-resistant cell line (A2780-DDP), and cisplatin-sensitive cell lines (A2780, HO-8910, HO-8910PM, and SKOV3). Data showed PANDAR level was higher in resistant OC cells compare with cisplatin-sensitive cells, but there was no significance among those chemoresistant cells (Fig. 1a). Cell survival rate (Fig. 1b) and IC50 (Fig. 1c) in A2780 and A2780-DDP cell lines were measured with an increasing cisplatin treatment, validating A2780-DDP cells are more prone to survive compared with A2780 cells in response to cisplatin. These observations suggest that PANDAR may play a role in platinum-based resistance in OC. To confirm this, we measured PANDAR levels in A2780 and HO-8910 cells following treatments by chemo-drugs doxorubicin (Dox), paclitaxel (PTX), and cisplatin (CDDP), as they were commonly used in clinical ovarian cancer chemotherapeutics. We found that cisplatin induced the highest expression of PANDAR among other drugs (Fig. 1d) in a dosage-dependent and time-dependent manner (Fig. 1e). The induction of PANDAR by cisplatin was also p53 dependent (Fig. 1f). These results suggest that cisplatin-induced PANDAR may dictate cisplatin resistance of OC without p53 mutation.
Fig. 1. Inverse association between PANDAR-expression and cisplatin sensitivity in ovarian cancer cells.
a QRT-PCR analysis of PANDAR expression in the chemosensitive ovarian cancer cell lines, A2780 cisplatin-resistant cell lines (A2780-DDP) and primary recurrent ovarian cancer cells of the 3rd generation. b CCK-8 assay of cell survival rate in A2780 and A2780-DDP cells after treated with an increasing dose (0–55 μM) of cisplatin for 24 h. c The half maximal inhibitory concentration (IC50) was calculated from Fig. 1b via three independent experiments using GraphPad 6.2 software. d QRT-PCR of PANDAR expression in A2780 and HO-8910 cells after treated with phosphate buffer saline (PBS, 5 µM), doxorubicin (DOX, 5 µM), paclitaxel (PTX, 25 nM), or cisplatin (CDDP, 20 µM) for 24 h. PBS serves as control. e PANDAR expression in A2780 and HO-8910 cells after treated with 20 µM cisplatin for indicated time (left) or treated with indicated concentrations (right) of cisplatin for 24 h. f PANDAR expression in A2780 and HO-8910 cells with TP53 silenced after treated with or without 20 µM dose of cisplatin for 24 h. Vehicle groups were treated with PBS. PBS groups are normalized to 1. Data presents the mean ± S.D. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns non-significant. Two-tailed Student’s t-test. n = 3 independent experiments
Table 1.
Relative expression of PANDAR, p53-Ser15, and PUMA in patients with newly diagnosed and recurrent ovarian cancer
| No | Age (years) |
Classification | FIGO stage |
Differentiation | Platinum sensitive (N/R) | p53 mutation | PANDAR Exp. (N/R) | p53-Ser15 Exp. (N/R) | PUMA Exp. (N/R) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 50 | OCCC | IIB | N/A | Yes/ No | No | Low/ high | High/ low | High/ low |
| 2 | 51 | ENOC | IIIC | Poor | Yes/ No | No | Low/ high | High/ low | High/ low |
| 3 | 49 | ENOC | IIIA | Poor | Yes/ No | No | Low/ high | High/ low | High/ low |
| 4 | 53 | HGSOC | IIC | Poor | Yes/ No | No | Low/ high | High/ low | High/ low |
| 5 | 80 | HGSOC | IIIB | Poor | No/ No | Yes | Low/ low | Low/ low | Low/ low |
| 6 | 62 | HGSOC | IIIC | Moderate | No/ No | Yes | Low/ low | Low/ low | Low/ low |
| 7 | 56 | HGSOC | IIIC | Poor | No/ No | Yes | Low/ low | Low/ low | Low/ low |
Low, below the median of the intergrated optical density. High, above the median of the intergrated optical density
HGSOC high-grade serous ovarian cancer, OCCC ovarian ovarian clear cell carcinomas, ENOC endometrioid ovarian cancer, Exp expression, N newly diagnosed, R recurrence
PANDAR attenuates cisplatin sensitivity in chemosensitive OC cells
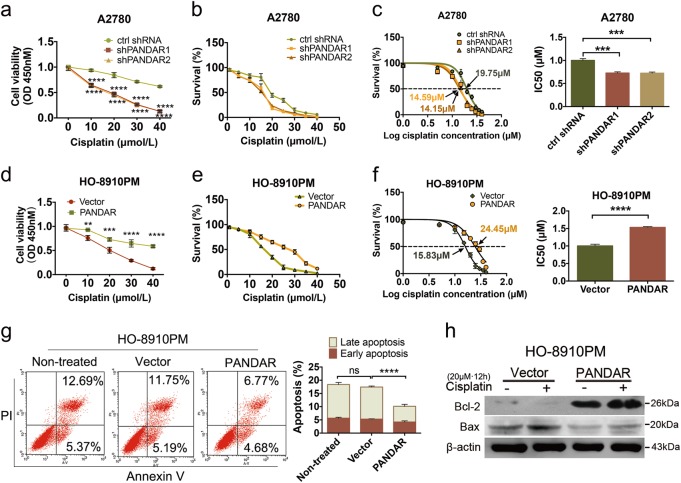
To further investigate the biological functions of PANDAR in ovarian cancer, we created isogenic PANDAR Venus knock-in HO-8910PM cell line and PANDAR Venus knockdown (shPANDAR) A2780 cell line. We observed a 320-fold increase of PANDAR expression in PANDAR knock-in HO-8910PM cell line (Supplementary Fig. S2a), while a 33-fold and 3-fold decrease of PANDAR expression in A2780-PANDAR-knockdown and A2780-DDP-PANDAR-knockdown cell lines was detected (Supplementary Fig. S2a, b). Next, we measured cell viability, as well as survival rate and the IC50 of PANDAR knock-in and knock-down cells in response to cisplatin (Fig. 2a). We found that A2780-PANDAR-knockdown cells displayed a decreased survival ability compared with control shRNA cells (Fig. 2a, b). The IC50 was also downregulated after PANDAR knockdown (Fig. 2c). Conversely, HO-8910PM cells with PANDAR overexpression exhibited a stronger survival ability in response to high doses of cisplatin, compared with cells infected with empty vectors (Fig. 2d, e). Moreover, IC50 in PANDAR-overexpressing cells was almost 1.5-fold of that in Vector cells (Fig. 2f). On the other hand, apoptosis population was decreased after PANDAR overexpression in HO-8910PM cells (Fig. 2g), characterized with a slight downregulation of early apoptosis (Annexin V+, PI−) and a significant reduction of late apoptosis (Annexin V+, PI+), compared with non-treated cells and vector-infected cells, suggesting that PANDAR overexpression reduces the sensitivity of OC cells in response to cisplatin. To further reconfirm this suggestion, we detected and quantified Bax and Bcl-2 expression levels in that the increased ratio of Bax/Bcl-2 is an indicator of apoptosis initiation13. As expected, Bax was downregulated in PANDAR-overexpressed cells whereas Bcl-2 level was upregulated, especially after a 12-h cisplatin treatment (Fig. 2h). The Bax/Bcl-2 ratio upon cisplatin treatment was significantly downregulated in HO-8910PM-PANDAR overexpressing cells (Supplementary Fig. S2c), compared with the empty vector groups. More importantly, cisplatin-induced Bax/Bcl-2 ratio was limited in PANDAR-overexpressing cells, indicating that PANDAR overexpression reduces cisplatin sensitivity. These data suggest that PANDAR functions as an oncogene in OC cells and reduces cisplatin sensitivity.
Fig. 2. PANDAR reduces cisplatin sensitivity in chemosensitive ovarian cancer cells.
a CCK-8 assay of cell viability in A2780-PANDAR-knockdown cells (shPANDAR) after treated with indicated doses of cisplatin for 24 h, ctrl shRNA cells serve as controls. b Cell survival rate was measured from a through three independent experiments. c The half maximal inhibitory concentration (IC50) was calculated from b using GraphPad 6.2 software. d Cell viability in HO-8910PM-PANDAR cells after treated with indicated doses of cisplatin for 24 h via CCK-8 analysis, HO-8910PM-Vector cells serve as controls. e Cell survival rate was calculated from d through three independent experiments. f The half maximal inhibitory concentration (IC50) was calculated from e using GraphPad 6.2 software. g Flow cytometry assay (FCM) of cell apoptosis in chemosensitive HO8910PM-PANDAR-overexpressing cells with or without a 24-h treatment of 20 μmol/L cisplatin. Early apoptotic population in the lower right gate is characterized with Annexin V (+) and PI (−), and late apoptotic population in the upper right gate is characterized with Annexin V (+) and PI (+). h Protein expression of Bax and Bcl-2 in HO-8910PM-PANDAR overexpressing cells with a 12-h treatment of 20 μM cisplatin, Non-treated Vector cells serve as controls. Vector and ctrl shRNA groups are normalized to 1. Data presents the mean ± S.D. *p < 0.05, **p < 0.01, ****p < 0.0001, ns non-significant, two-tailed Student’s t-test. n = 3 independent experiments. A-V Annexin V
PANDAR reduces cisplatin sensitivity in a mouse model
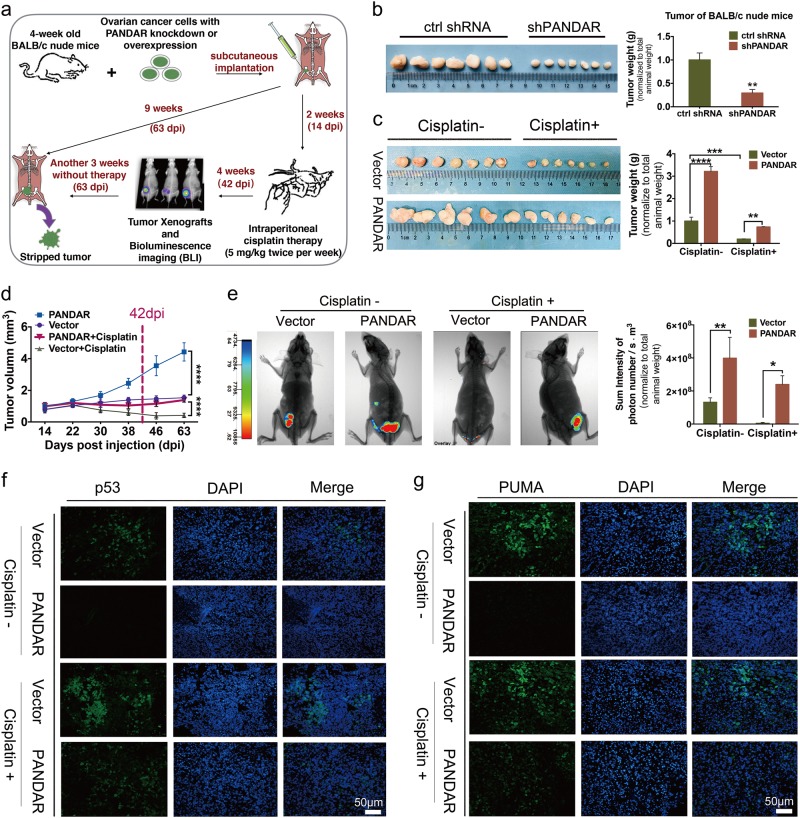
To substantiate the role of PANDAR in tumor growth and cisplatin resistance in addition to in vitro, we created mouse xenograft models via subcutaneous injections of HO-8910PM-Vector-GFP and HO-8910PM-PANDAR-GFP cells, A2780-ctrl shRNA and -shPANDAR cells. The schematic diagram was shown in Fig. 3a. Mice implanted with shPANDAR cells developed smaller tumors compared with control mice (Fig. 3b). At the first 2 weeks without cisplatin treatment, mice implanted with PANDAR-overexpressing cells started to develop tumors as early as 5 days post injection (dpi), compared with tumor onset at 8 dpi in control mice (data not shown). After 4 consecutive weeks of cisplatin therapy starting at 14 dpi, the average volume of tumor-like nodules from mice implanted with PANDAR-overexpressing cells was more than twofold larger than that the control mice (Fig. 3c), regardless of cisplatin treatment. Furthermore, a strike-back growth tendency occurred after cisplatin withdrawal (63 dpi) (Fig. 3d). Besides, by utilizing bioluminescence imaging (BLI) and its analysis system at 42 dpi, alive mice with PANDAR-overexpressing tumor with a larger subcutaneous tumor volume was observed, compared to accordingly control mice (Fig. 3e). Concomitantly, the downregulation of p53 and its upregulated modulator PUMA were observed in PANDAR-overexpressing nodules by immunofluorescence staining (Fig. 3f, g) and accordingly quantification (Supplementary Fig. S2d). Overall tumor growth profile during the cisplatin therapy revealed that PANDAR overexpression effectively attenuates cisplatin sensitivity.
Fig. 3. PANDAR promotes tumor growth and reduces cisplatin sensitivity in mice with tumor implantation.
a Schematic diagram showing the process of subcutaneous tumor transplantation in mice with PANDAR over-expression or knock-down (shPANDAR) cells. b Subcutaneous tumor derived from mice with A2780-shPANDAR cells and -ctrl shRNA cells transplanted after 9 weeks. No treatment applied during this time. Tumor weight was normalized to total animal weight. c Subcutaneous tumor derived from mice with HO8910PM-PANDAR cells and -Vector cells at the 63-day post injection (63 dpi). Cisplatin treatment was applied during this time. Tumor weight is normalized to total animal weight. d Tumor growth curves of mice injected with PANDAR-overexpressing cells after treated with cisplatin since 14 dpi. Non-treated mice with vector cells serve as controls. e Bioluminescence imaging (BLI) of tumor and accordingly quantification (intensity of photon number) in BALB/c nude mice implanted with HO-8910PM-PANDAR-GFP or HO-8910PM-Vector-GFP cells at 42 dpi after cisplatin therapy. f Immunofluorescent staining of p53 on subcutaneous tumor sections from mice implanted with HO-8910PM-PANDAR or -Vector cells at 63 dpi after a 4-week cisplatin therapy. Nuclei are stained in blue. Scale bar: 50 µm. g Immunofluorescent staining of PUMA on subcutaneous tumor sections from mice implanted with HO-8910PM-PANDAR or HO-8910PM-Vector cells at 63 dpi after a 4-week cisplatin therapy. Nuclei are stained in blue. Scale bar: 50 µm. Vector group is normalized to 1. Data presents the mean ± S.D. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns non-significant, two-tailed Student’s t-test. n = 7 mice per group
SFRS2 is a target protein of PANDAR in ovarian-cancer cell nucleus
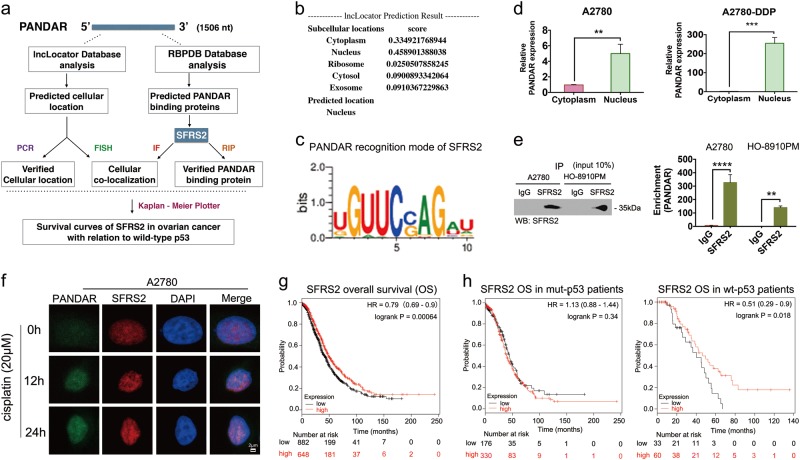
To investigate how PANDAR attenuate cisplatin sensitivity, we sought for PANDAR-target proteins at the RNA-binding Proteins Database (RBPDB) and identified SFRS2 as the best candidate of PANDAR-binding proteins (Fig. 4a). First of all, SFRS2 is the top among the predicted binding proteins for PANDAR, and the nuclear location of SFRS2 accords with predicted location of PANDAR (Fig. 4b). Moreover, SFRS2 has been reported to be an inhibitor of p53 and its phosphorylation6,7, which might involve in PANDAR generation. The most effective evidence is that the matching sequence of 5′-CCAG-3′ within 167 bp–176 bp of PANDAR (Fig. 4c, Supplementary Materials), which has been reported as the high-affinity binding sequence for SFRS2 in all identified SFRS2-specific ESEs (exon-splicing enhancers)12. Therefore, we detected SFRS2 expression and PANDAR location in OC cell. As predicted in the lncLocator Database, PANDAR expression in nuclear is far more than that in cytoplasm either in cisplatin-sensitive (A2780) or in cisplatin-resistant (A2780-DDP) OC cell (Fig. 4d), the internal control of nuclear and cytoplasm was U6 and GAPDH, respectively (Supplementary Fig. S2e). Then we detected endogenous PANDAR RNA co-immunoprecipitated with SFRS2 in cellular lysates from OC cells. In this part, the interaction was not detected when an isogenic IgG antibody was used, whereas a robust and specific interaction between SFRS2 and PANDAR was read in the isogenic SFRS2 antibody group (Fig. 4e). To explore the cellular location of this interaction, we detected PANDAR and SFRS2 expression in A2780 cells treated with cisplatin. Data showed an increasing co-localization of PANDAR and SFRS2 in nucleus over time (Fig. 4f, Supplementary Fig. S2f). To further investigate the role of this candidate protein (SFRS2) in OC, we drew Kaplan–Meier curves of SFRS2 focusing on overall survival (OS) probability. Data showed that high expression of SFRS2 might develop a better progression compared with low-expression group (Fig. 4g). Interestingly, the SFRS2-OS in OC depends on wild-type p53, but not mutant-p53 (Fig. 4h). These results indicate that SFRS2 is a PANDAR-target gene in OC, and the following function in cisplatin sensitivity might be associated with SFRS2-regulated p53.
Fig. 4. Identification of SFRS2 as a PANDAR-targeting protein in ovarian cancer cellular nucleus.
a Schematic of prediction and consensus experiments of PANDAR-binding protein. b Prediction of PANDAR cellular location using lncLocator Database (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/). c Logo of matching sequence or motif between PANDAR and SFRS2 downloaded from RBPDB database (http://rbpdb.ccbr.utoronto.ca/experiments.php?exp_id=791). d QRT-PCR assay of PANDAR expression in the nucleus and cytoplasm from A2780 and A2780-DDP cells after a 24 h treatment of 20 μM cisplatin. e Cellular extracts from A2780 and HO-8910PM cells treated with 20 μM cisplatin for 24 h, and RNA immunoprecipitation (RIP) assay showing immunoprecipitation (IP) with control IgG or SFRS2 antibody by western blotting (left) and qRT-PCR (right) performed with isolated RNA and the following reverse transcription (RT) using primers for lncRNA PANDAR. f Fluorescence in situ hybridization (FISH) assay of PANDAR RNA (green) in A2780 cells distributes in discrete foci through the nucleus and cytoplasm at the beginning of treatment with 20 μM cisplatin (0 h). The foci increased and focused in the nucleus after cisplatin treatment for 12 and 24 h. Co-localization of SFRS2 protein (red) in A2780 cells via immunocytochemistry performed with SFRS2 antibodies and showed an increasing discreted foci through the nucleus after cisplatin treatment (12 h, 24 h), and is not detected in cytoplasm. Scale bar: 2 μm. g Kaplan–Meier Plotter of overall survival (OS) regarding SFRS2 (200753_x_at) in ovarian cancer patients (http://kmplot.com/analysis/index.php?p=service&start=1). Kaplan–Meier Plotter of SFRS2 (200753_x_at) overall survival (OS) in ovarian cancer patients with (right) or without (left) p53 mutation. Control groups are normalized to 1. Data presents the mean ± S.D. **p < 0.01, ***p < 0.001, ****p < 0.0001, two-tailed Student’s t-test. n = 3 independent experiments
SFRS2-mediated p53 and its phosphorylation at Serine 15 is required for PANDAR-regulated cisplatin sensitivity
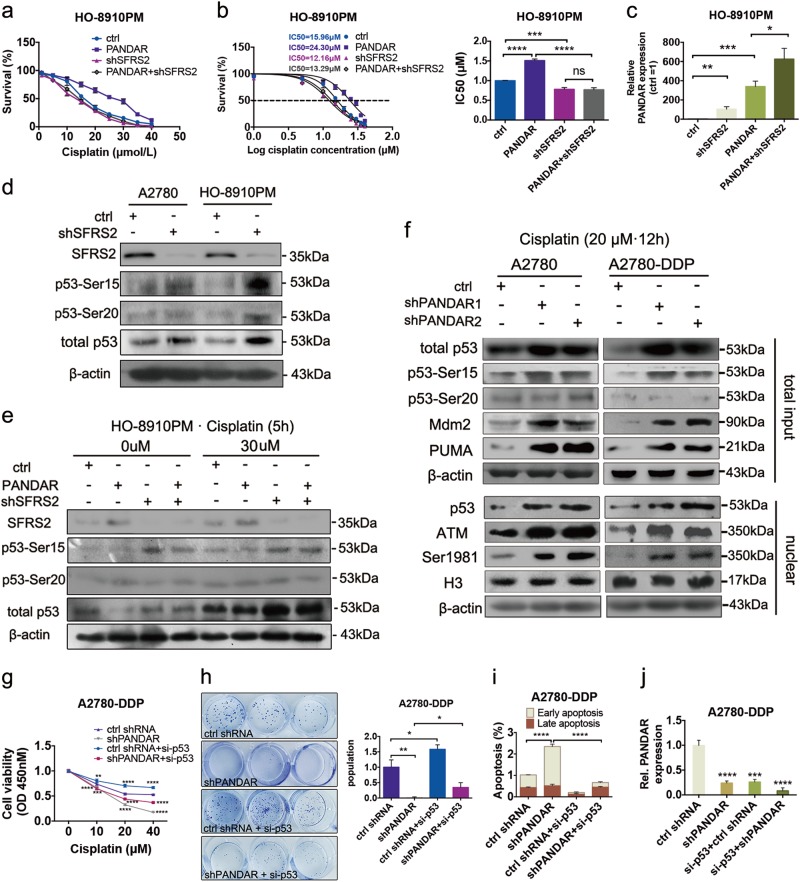
To further explore the role of SFRS2 in PANDAR-regulated cisplatin sensitivity in OC, we knocked down SFRS2 gene (shSFRS2) in PANDAR-overexpressing cells, and then detected cell survival rate and the IC50. Data showed that depletion of SFRS2 could downregulate cell survival rate (Fig. 5a) and accordingly IC50 (Fig. 5b), regardless of PANDAR overexpression. This indicating that PANDAR-downregulated cisplatin sensitivity may be SFRS2-dependent. Interestingly, PANDAR expression was upregulated after SFRS2 knockdown (Fig. 5c), suggesting that SFRS2 reduced PANDAR expression in direct or indirect way. Recent studies have reported that SFRS2 downregulated p53 and its phosphorylation in mouse fibroblasts6,7. We wonder if this could be detected in OC. Next, we measured p53 and its phosphorylation at Serine 15 and Serine 20 in SFRS2-knockdown cells, the two important N-terminal Serine in cisplatin resistance of OC4. Indeed, after SFRS2 knocked down, p53 expression and p53 phosphorylation at Serine 15 (p53-Ser15) were significantly induced compared with control cells, whereas p53-Ser20 was slightly increased (Fig. 5d). We next focused on the p53 and p53-Ser15 and -Ser20 in PANDAR-overexpressing cells with or without SFRS2 knockdown. Data showed that the levels of SFRS2 and p53 reduced in PANDAR-overexpressing cells compared with control group (Fig. 5e), although the alteration of p53-Ser15 level was not in a very significant way, the downregulated level of p53 and p53-Ser15 by PANDAR was rescued when SFRS2 knocked down (Fig. 5e). Whereas p53-Ser20 level changed not so much among all these groups (Fig. 5e). These results above indicate that PANDAR, as a p53-dependent lncRNA, may counter-regulate SFRS2-related p53 expression and p53-Ser15 to mediate cisplatin sensitivity. To confirm this suggestion, we detected p53 and p53-related pro-apoptotic genes expression in PANDAR-knockdown cells. Indeed, p53 and p53-Ser15 was significantly upregulated after PANDAR knockdown, either in cisplatin-sensitive cells (A2780) or cisplatin-resistant cells (A2780-DDP) (Fig. 5f, accordingly quantification in Supplementary Fig. S2g). Meanwhile, the p53-mediated apoptotic genes were upregulated, such as PUMA and Mdm2 (murine double minute 2) (Fig. 5f), BAX (BCL2 associated X) and NOXA (NADPH oxidase), but this induction drew back after p53 was silenced (Supplementary Fig. S3a). Data showed a further activation of these p53-activated genes after PANDAR knock-down. Given that these apoptosis genes transcription was partly due to p53-Ser15 in nuclear14, we detected nuclear p53 and the specific phosphatase ATM (ataxia-telangiectasia mutated protein kinase), in which the monomerization and its autophosphorylation at Serine 1981 are critical steps to p53 phosphorylation at Ser1515. As is shown in Fig. 5f, nuclear p53, ATM and its autophosphorylation at Ser1981 were all upregulated in PANDAR-knockdown cells, compared with control cells. These data suggest that PANDAR reduced cisplatin sensitivity may be partly due to SFRS2-downregulated p53 and its associated apoptotic genes transcription. To verify this hypothesis, we silenced p53 in PANDAR knockdown cells and then measured cell viability with an increasing cisplatin treatment. As shown in Fig. 5g, cisplatin sensitivity in cisplatin-resistant cells (A2780-DDP) was upregulated when PANDAR knocked down, but this was rescued after p53 was silenced. Similarly, colonies of A2780-DDP cells after a 10-day cisplatin treatment were dramatically reduced after PANDAR knockdown, while this ability of resistance to cisplatin regained when p53 silenced simultaneously (Fig. 5h). Apoptosis (Supplementary Fig. S3b) and accordingly analysis (Fig. 5i) also exhibited a recovered ability of resistance to cisplatin in A2780-DPP cells after p53 silenced in PANDAR-knockdown cells. Besides, PANDAR level was also downregulated in p53-silenced cells (Fig. 5j). p53 expression was upregulated in PANDAR-overexpressing cells and was downregulated in PANDAR-knockdown OC cells (Supplementary Fig. S3c). The above data indicate there may be a feedback loop between PANDAR and p53, that PANDAR counter-regulated the level of SFRS2-associated p53 and p53-Ser15, leading to a reduced transactivation of apoptosis genes expression in OC.
Fig. 5. SFRS2-mediated p53 and its phosphorylation at Serine 15 is required for PANDAR-regulated cisplatin sensitivity.
a CCK-8 assay of cell viability in HO-8910PM-PANDAR cells after treated with indicated doses (1–40 μM) of cisplatin for 24 h with or without SFRS2 knockdown (shSFRS2). Cell survival rate was calculated through three independent experiments. b Cell survival rate and the subsequent half maximal inhibitory concentration (IC50) were calculated from a using GraphPad 6.2 software. c QRT-PCR assay of PANDAR expression in HO-8910PM-PANDAR cells with or without SFRS2 knockdown (shSFRS2). d Protein expression performed with western blotting assay showing an increased level in p53, phosphor-p53 at Ser15 and -Ser20 in A2780 and HO-8910PM cells with SFRS2 knockdown (shSFRS2). e Protein expression levels of SFRS2, p53, phosphor-p53 at Ser15 and Ser20 in HO-8910PM-PANDAR cells with or without SFRS2 knockdown (shSFRS2) after 0 or 30 μM cisplatin treatment for 5 h. Non-treated cells serve as controls. f Proteins expression in total and nuclear fractions from A2780 and A2780-DDP cells with PANDAR knockdown (shPANDAR1, shPANDAR2) after treated with cisplatin for 12 h. H3 protein serves as nuclear control. g Cell viability via CCK-8 assay in A2780-DDP-PANDAR knockdown (shPANDAR) cells with or without p53 silenced after treated with indicated doses (0–40 μM) of cisplatin for 24 h. h Colony formation and accordingly quantification in A2780-DDP-PANDAR knockdown (shPANDAR) cells with or without p53 silenced after incubated with cisplatin for 10 days. i Apoptosis measurement in A2780-DDP-PANDAR knockdown (shPANDAR) cells with or without p53 silenced after incubated with 20 μM cisplatin for 24 h. j QRT-PCR assay of PANDAR expression in A2780-DDP-PANDAR knockdown (shPANDAR) cells with or without p53 silenced. Control groups are normalized to 1. Data presents the mean ± S.D. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, two-tailed Student’s t-test. n = 3 independent experiments
The feedback loop between PANDAR and p53 contributes to the clinicopathology of cisplatin resistance in OC patients
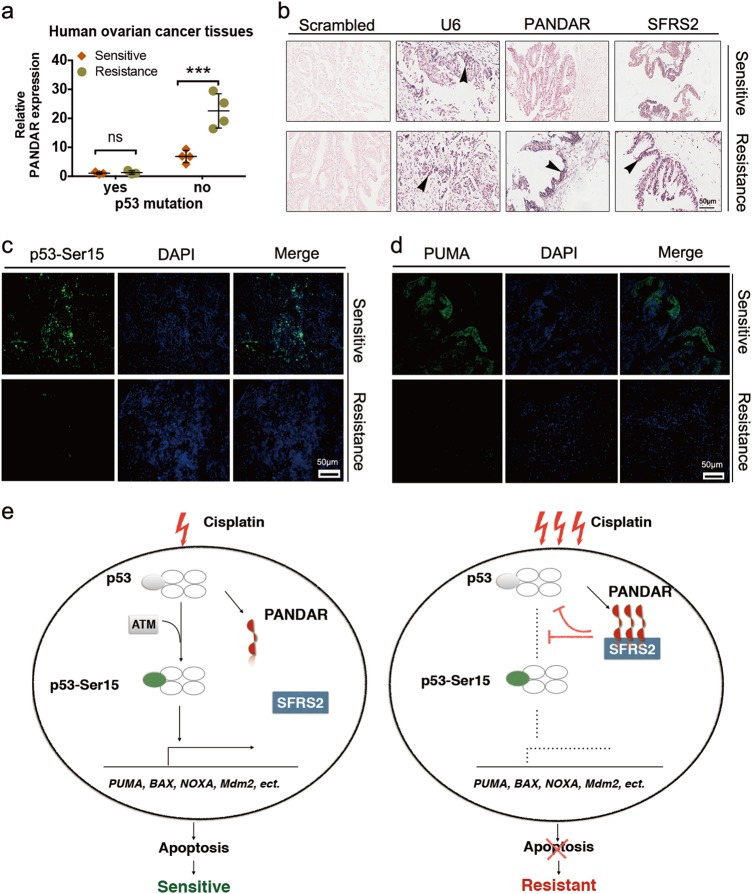
We further examined PANDAR-mediated chemoresistance in patients by tissue biopsy from 25 OC patients under platinum-based neoadjuvant and adjuvant therapy followed by complete surgical resection. Unfortunately, seven patients relapsed after the cisplatin-based or its analog carboplatin-based chemotheraputics within 6 months (Table 1), in which chemoresistance was identified as the leading cause16. The PANDAR expression regarding p53 mutation was measured in specimens from these seven patients before and after recurrence (Supplementary Fig. S4a). Data showed that PANDAR expression in OC tissues depends on wt-p53, but not mutant-p53, regardless of platinum-sensitivity. As is quantified by qRT-PCR, 4 of these 7 recurrent patients without p53 mutation exhibited higher levels of PANDAR in platinum-resistant cancer tissues compared with their newly diagnosed periods (platinum-sensitive) (Fig. 6a). Next, we examined the interplay among PANDAR, SFRS2, p53-Ser15, and PUMA in OC tissues collected from their sensitive and resistant periods, separately. Data showed that four patients with wt-p53 exhibited higher levels of PANDAR and SFRS2 (Fig. 6b) and lower levels of p53-Ser15 (Fig. 6c) and PUMA (Fig. 6d) in their resistant tissues, compared with their sensitive tissues from newly diagnosed periods. TEM images on cancer tissue biopsies also showed a better condition of cancer tissue in platinum-resistant period, compared with platinum-sensitive tissues (Supplementary Fig. S4b). These results are translational and indicated the significance of PANDAR and SFRS2 co-regulated p53-mediated apoptosis in clinical chemoresistance. Accordingly, we propose a schematic model of underlying mechanism regarding cisplatin resistance (Fig. 6e).
Fig. 6. The feedback profile of PANDAR–SFRS2–p53 in ovarian cancer patients with chemoresistance and recurrence.
a QRT-PCR quantification of PANDAR expression in tissues from ovarian cancer patients collected at newly diagnosed stage (Sensitive) and after disease progression during platinum-based therapy (Resistance). P53 missense mutation is in accord with post-surgical pathology reports. Data presents the mean ± S.D. n = 7 patients determined by Student’s t-test. ***p < 0.001, ns, non-significant, p53 mutation, positive mutant-p53 more than 80%. b LNA ISH analysis of lncRNA PANDAR with LNA probes and IHC assay of SFRS2 protein with SFRS2 antibody in matched ovarian cancer tissues before platinum-based therapy (Sensitive) and after disease progression during platinum-based treatment (Resistance). Representative LNA ISH and IHC images are shown. Scrambled groups serve as negative controls. U6 groups serve as positive controls. Black arrow head indicates the positive expression of RNA or protein. Scale bar: 50 μm. c, d Representative images of p53-Ser15 (c) and PUMA (d) immunofluorescent staining on tissues from patients at platinum-sensitive stage and after platinum-resistant ovarian cancer diagnosis. Scale bar: 50 µm. Data presents the mean ± S.D. n = 10 independent fields per group determined by Student’s t-test. **p < 0.01, ***p < 0.001. e Schematic model of underlying mechanisms regarding cisplatin resistance. With the sequential stimulation of cisplatin, a certain amount of p53-dependent PANDAR is upregulated to interact with SFRS2, leading to a reduction of p53 and its phosphorylation at Ser15, which in turn inactivated p53-mediated genes expression, such as PUMA, BAX, NOXA, Mdm2, and downregulated subsequent apoptosis
Discussion
In this study, we identified cisplatin upregulates p53-dependent lncRNA PANDAR in OC cells, which is responsible for the transition from chemorsensitivity to chemoresistance upon cisplatin treatment. The mechanism involved in this transition is based on a regulatory feedback loop of PANDAR–SFRS2–p53 in nuclear. This feedback regulation may cast a promising target for future advances in ovarian cancer chemotherapy.
In this feedback regulation, we demonstrated that SFRS2 deregulates p53 and its phosphorylation. Given that ATM (ataxia-telangiectasia mutated protein kinase), ATR (ATM-related and Rad3-related protein kinase), and DNA-PKs (DNA-dependent protein kinase catalytic subunit) are the specific phosphatases in DNA damage-induced p53 phosphorylation at Serine 1515, it becomes a compelling question for the further to investigate whether SFRS2 has a role in the activation of those phosphatases.
Based on the wild-type TP53 status, lncRNA PANDAR acts as an oncogene to promote cisplatin resistance in OC. Hence, choosing the appropriate cell lines with wild-type p53 protein is the first step in this study. Wild-type p53 was detected in OC chemosensitive cell lines (A2780, HO-8910, HO-8910PM), and the chemresistant cell line (A2780-DDP) in cellular nucleus, partly in cytoplasm, except SKOV3, which was p53 null as reported17 and showed in this study (Supplementary Fig. S1a, b). While two of those OC cell lines, namely HO-8910PM and A2780-DDP, also expressed with mutant-p53 in cytoplasm, indicating that p53 mutation occurs in these two cell lines. However, PANDAR functions involved in the feedback regulation of p53 in HO-8910PM and A2780-DDP were in accord with the other cell lines without p53 mutation (Figs. 2 and 5). This accordance may due to wild-type p53 expression in nucleus of these cell lines. It needs further investigation that if the co-expression of mutant-type and wild-type p53 in ovarian cancer cells was due to the alternative splicing by the splicing factor SFRS2.
The involvement of p53 in DNA damage resistance in OC is largely depended on TP53 status and tumor histologic type18–20. In high-grade serous ovarian cancer (HGSOC), for example, TP53 mutation occurs in approximately 95% of this kind of epithelial ovarian tumor and thus leads to a poor prognosis and a high rate of chemoresistance21. While ovarian clear cell carcinoma (OCCC) has relatively normal genomes with rare TP53 mutations22, and are less sensitive to chemotherapy23. Therefore, a better understanding of wild-type p53 in regulating ovarian cancer may shed light on more effective chemotherapeutics. Recently, new treatments targeting DNA damage repair has recently opened the door in OC16, especially for modulators of p53-related mechanisms24,25. The previous studies have identified p53 phosphorylation at Ser15 and/or Ser20 increased the ability of p53 to facilitate cisplatin-induced apoptosis in OC4. Until now, only two proteins have been reported in human OC resistance to cisplatin via regulating p53 and its post-transcriptional modification, namely AKT24 and in this paper, SFRS2. The cellular location and the consensus mode for these two proteins has common features in regulating cisplatin resistance. In OC cells, wild-type p53 and its phosphorylation at Ser15 were both downregulated by AKT24 and SFRS2, but only SFRS2 involves in p53 feedback regulation (Fig. 6e). Moreover, the dependent on wild-type p53 status of SFRS2 role in OC progression (Fig. 4h) was first determined in this study. In 1999, Elizabeth and colleagues suggested that SFRS2 might function as a tumor suppressor gene in OC26. This suggestion was confirmed in this study that the high expression level of SFRS2 developed a better overall survival (OS) probability in patients with OC, compared with its low expression level group (Fig. 4g). However, in the DNA damage response to cisplatin, SFRS2 was identified as an oncogene interacting with lncRNA PANDAR to promote cisplatin resistance in ovarian cancer cells (Fig. 6e). This discrepancy may due to the feedback regulation of PANDAR and p53 induced by cisplatin.
In the past years, the interaction of SFRS2 with lncRNAs in cancer cellular nucleus has been reported27, but it still lacks a statement that how this protein could identify lncRNAs in human cancer cell. Our study filled this gap and revealed the matching sequence of 5′-CCAG-3′ in the lncRNA PANDAR (Fig. 4c, Supplementary Materials) on the basis of the high-affinity binding consensus sequence 5′-SSNG-3′ (S = C/G) for SFRS2 discovered by Daubner et al.12.
The function profile of lncRNA PANDAR can vary within different cell types and diseases. In breast cancer, for instance, PANDAR has been shown to negatively regulate cell apoptosis28, while in non-small cell lung cancer (NSCLC), PANDAR positively regulates cell apoptosis29. In this study, PANDAR attenuates cisplatin-induced apoptosis and IC50 in OC cells (Fig. 2), this pro-survival role of PANDAR in OC is also shown in other types of epithelial cancers, such as in clear cell renal cell carcinoma and bladder cancer, where PANDAR upregulation is associated with poor prognosis and tumorigenesis30–34. Inspired by these observations, our study also provided a rationale that tumor heterogeneity serves as an inherent property of lncRNA35. Besides, PANDAR location in ovarian cancer cell also varies with the extended response time during cisplatin treatment (Fig. 4f, Supplementary Fig. S2f), further investigation might focus on how this dynamic cellular location move forward.
To our knowledge, this is the first time a generic lncRNA feedback profile has been investigated in order to identify the features of wild-type p53 post-translational modifications in drug-induced chemoresistance in cancer therapy. This study also highlights the importance of seeking for the high-affinity matching sequence of lncRNA for RNA-binding proteins in molecular mechanism exploration. Last but not least, the insights of this study provide a rationale to develop PANDAR as a chemoresistant biomarker and a target for molecular therapy in OC despite the mutation of p53.
Materials and Methods
Reagents
Cisplatin used for this study was provided by the 2nd Affiliated Hospital of Harbin Medical University.
Patients
All biospecimens used in this study were provided by the 2nd Affiliated Hospital of Harbin Medical University. All specimens were obtained with informed consent under the approval of the Harbin Medical University of Medicine Institutional Ethics Review Board protocol.
Mouse colonies
Four-week-old female BALB/c nude mice used in this study were purchased from SLAC Laboratory Animal CO. LTD (Shanghai, China). Mice were housed under pathogen-free at the animal facility of the 2nd Affiliated Hospital of Harbin Medical University. All study protocols were approved by the Institutional Animal Care and Use Committee at the 2nd Affiliated Hospital of Harbin Medical University.
Tumor xenografts and bioluminescence imaging
The previously described HO-8910PM-Vector and HO-8910PM-PANDAR cells expressing green fluorescent protein (GFP)36,37 were injected subcutaneously into the lower abdominal flank of BALB/c nude mice. Two weeks after the HO-8910PM-PANDAR cell injection, mice were treated with PBS (cisplatin-) or cisplatin (5 mg/kg) by intraperitoneal injection twice a week38. Four weeks after cisplatin treatments, animals were housed with no disturbance for another 3 weeks. The subcutaneous tumor size was measured by a caliper every 4 day. At the 42-day post injection (42 dpi), subcutaneous tumor in alive mice were observed using a BLI system. Briefly, in vivo imaging based on GFP fluorescence detection (excitation: 488 nm, emission: 510 nm) was captured by a Kodak Image Station 4000 Multi-Modal Imaging System (IS4000MM) equipped with an X-ray unit or a Kodak Image Station 2000 (TAMAR-Laboratory Supplies Ltd. ISRAEL)39. After killing, subcutaneous tumors were dissected out at 63 dpi. The stripped tumor weight was measured using an electronic balance and the volume was determined by caliper measurements of tumor dimensions using the prolate ellipsoid geometric model: (length × width2)/229,40.
Cell culture studies
Human ovarian cancer cell lines SKOV3, HO-8910, HO-8910PM, A2780, and cisplatin-resistant cell line A2780/DDP were obtained from the Chinese Academy of Sciences Committee on Type Culture Collection Cell Bank (Shanghai, China). Recurrent ovarian cancer cells were obtained from patients with postoperative pathological confirmation (Table 1 No.4–7, Supplementary Fig. S1c). All cells were cultured in RPMI (Roswell Park Memorial Institute) 1640 medium supplemented with 10% fetal bovine serum (Coring Cellgro) and 1% penicillin/streptomycin (Beyotime, Shanghai, China) in a humid incubator containing 5% CO2 at 37 °C.
Lentivirus infection and stable cell line generation
Lentivirus containing overexpressed lncRNA PANDAR or shRNA that knocks down human PANDAR were constructed by subcloning the synthesized PANDAR open reading frame (ORF) into the pLVX or pLVX-GFP vector. Constructs were confirmed by sequencing. Lentivirus was produced using 293T cells, and viral packaging was conducted using ViraPower Lentiviral Packaging mix (Invitrogen, K497500). The packed virus was concentrated by an ultracentrifugation/a Lenti-X concentrator (Clontech, 631231). PANDAR overexpression or knockdown efficacy was confirmed by quantitative real-time PCR (Supplementary Fig. S2). 1 × 108 TU/mL of Lentivirus, 10 μg/mL Polybrene, and 1 mL Enhanced Infection Solution (Eni.S) were infected 1 × 105 cells per well on 6-well plates. To establish stable cell lines containing overexpressing or knockdown PANDAR, after lentivirus infection, cells were selected by adding 1 μg/mL puromycin for 48 h. PANDAR shRNA sequences are provided in Supplementary Table 2. Lentivirus concentrations were chosen based on preliminary studies.
Small interfering RNA and DNA plasmids transduction
Small interfering RNA (siRNA) and p53 (p. Arg273His) DNA plasmids were transduced into ovarian cancer cells using siRNA Transfection Reagent (Cat. no. sc-29528; Santa Cruz, Biotechology, Inc.) and Lipofectamine 2000 CD Transfection Reagent (Cat. no.12566014; Invitrogen Thermo Fisher Scientific Inc.) according to manufacturer′s protocol. The Stealth RNAiTM negative control duplex (cat. no. 12935–200) and siRNA duplex oligoribonucleotides targeting human p53 (Cat. no. 13750047) were obtained from Ambion (Thermo Fisher Scientific, Inc.).
Subcellular fractionation location
Separation of the nuclear and cytosolic RNA fractions was performed using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek Corp.; Thorold, Canada) according to the manufacturer’s instructions.
Quantitative real-time PCR
Total RNA from fresh clinical samples or ovarian cancer cells was dissolved in TRIzol (Cat. No. 15596018, Invitrogen Thermo Fisher Scientific Inc.). Separated nucleoplasmic RNA was extracted from cancer cells by a Cytoplasmic & Nuclear RNA Purification Kit (Cat. No. 21000, Norgen Biotek Corp, Thorold, Canada). RNA was synthesized to cDNA using a Transcriptor First Strand cDNA Synthesis Kit (Cat. No. 04 879 030 001, Roche Molecular Systems, Inc.). Real-time PCR was performed using SYBR green reagants (Cat. No. 11 418 033 001, Roche Molecular Systems, Inc.) in a real-time PCR system (Bio-Rad). GAPDH and U6 were used as internal/ cytoplasmic and nuclear RNA control, respectively. Data were analyzed using a LightCycler 96 Instrument software (Roche Diagnostics Corporation, Indianapolis, IN, USA). Relative gene expression was calculated using the 2−ΔΔCt method41. Primer sequences are shown in Supplementary Table 2.
Immunofluorescent staining
Cultured cells/neurons were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and processed for immunostaining as described previously42. Fluorescent microscopic images were captured on an Axiovert 200 inverted microscope (Zeiss, Thornwood, NY) equipped with a cooled CCD camera (SensiCam; Cooke, Auburn Hills, MI). Images to be directly compared were processed in an identical manner with Slidebook imaging software (Intelligent Imaging Innovations, Denver, CO) and Adobe Photoshop (version 7.0.1. or CS2 9.0.2; Adobe Systems, San Jose, CA).
Immunohistochemistry, LNA-based in situ hybridization, fluorescence in situ hybridization, and immunocytochemistry
For immunohistochemistry (IHC), paraffin embedded sections were deparaffinized, rehydrated, followed by antigen retrieval. After primary and secondary antibody (listed in Supplementary Table 1) incubation, the slide was finally incubated with diaminobenzidine (DAB) (Dako, USA), and counterstained with hematoxylin (Sigma Chemical Co, USA).
LNA-based in situ hybridization (LNA ISH) was performed by using miRCURY LNATM miRNA ISH Optimization Kit (Exiqon, Denmark) as previously described43 with minor modification. Briefly, the sections were deparaffinized and deproteinated using proteinase K (15 μg/mL, Roche) at 37 °C for 10 min. The endogenous peroxidases were blocked in 1% H2O2 for 30 min, and sections were pre-hybridized at 62 °C for 30 min in formamide-free Exiqon ISH buffer (Exiqon, Denmark). Sections were then hybridized with DIG-labeled LNA probes (listed in Supplementary Table 2) for lncRNA PANDAR (50 nM) at 62 °C overnight. Slides are then stringently washed, incubated with alkaline phosphatase-conjugated anti-DIG Fab fragments (Roche, USA) for 60 min and then detected by NBT/BCIP reagent (Invitrogen, USA). Sections were finally counterstained with nuclear fast red staining solution (Sigma Chemical Co, USA).
High resolution images were captured with an Aperio ScanScope AT Turbo (Aperio, USA) equipped with Aperio ImageScope software (Aperio, USA). Assessment of the staining was based on the staining intensity and the percentage of positively stained cells using Image-Pro Plus 6.0 software (Media Cybernetics, Inc., USA).
Fluorescence in situ hybridization (FISH) and Immunocytochemistry was performed as previously described44. To prepare the antisense probe of lncRNA PANDAR, the cDNA fragment was amplified by using primers (5′-ctgcccagaagcaaacaggactc-3′ and 5′-tttgggagaccgaggcagacaga-3′) and was subcloned to pEASY®-T1 Cloning Vector (TransGen Biotech Co., Ltd, Beijing, China). Then A digoxigenin (DIG)-UTP labeled RNA antisense probe was synthesized using a DIG RNA labeling kit (Roche, Germany) according to the manufacturer’s instructions. The A2780 and HO-8910 cells were cultured on 24-well chamber slides and fixed with 4% PFA for 20 min at RT. was performed in hybridization buffer. Hybridization was performed at 60 °C overnight in hybridization buffer with probes added at the final concentration 400 ng/mL after prehybridization at 60 °C for 1 h. The slides were washed in PBS three times and incubated with the anti-digoxigenin fluorescein Fab fragments (Roche) diluted 1:200 for 4–5 h at RT. After washing with PBS for five times, the slides were blocked with 5% BSA in PBS for 1 h, and Anti-SC-35(SFRS2) antibody (Abcam) was diluted 1:200 in blocking buffer and incubated at 4 °C overnight. Next that, the slides were incubated with Alexa Flour568 (Invitrogen) at 37 °C for 1 h, washed with PBS and counterstained with DAPI for 10 min at RT. The signals were imaged with using a fluorescence microscope (Confocal).
RIP assays
RIP experiments were performed using a Magna RIPTM RNA-binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. The SFRS2 antibody for RIP assays was obtained from Abcam Biotechnology.
Nucleoplasmic protein separation
The Nucleoplasmic protein was obtained and separated from ovarian cancer cells using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Cat. No. 78833; Thermo Fisher Scientific Inc.) according to the manufacturer’s protocol.
Western blotting
Total protein lysates from cells were lysed in RIPA lysis buffer (Cat. No. P0013B, Beyotime, Shanghai, China) supplemented with complete Protease Inhibitor Cocktail (Cat. No. B14000, Biotool, Switzerland). Protein concentration was determined using a bicinchoninic acid (BCA) kit (Cat. No. 23227, Thermo Fisher Scientific Inc.). Samples were heated and reduced and separated on polyacrylamide gels. Separated proteins were transferred to PVDF membranes. After immunoblotting with primary and secondary antibodies with HRP conjugations, blots were reacted with enhanced chemiluminescence (ECL) reagent (Cat. No. RPN2232, GE Healthcare, Jiangsu, China). Protein semiquantification was determined using densitometry by Image J. Information of antibodies is shown in Supplementary Table 1.
Apoptosis assay
Apoptosis was examined using FITC/ PI Annexin V Apoptosis Detection Kit I (Cat. No. 556547, BD Pharmingen) or PE/ 7-AAD Annexin V Apoptosis Detection Kit I (Cat. No. 559763, BD Pharmingen) according to the manufacturer’s protocol.
CCK-8 assay
Cell viability was measured using the Cell Counting Kit (CCK-8/WST-8) (Cat. No. CK04, DOJINDO, Japan). Briefly, cells were plated at a density of 5 × 103 cells per well on 96-well plates and subjected to indicated different treatments. Following a 24-h incubation at 37 °C, cells were incubated for an additional 3 h with CCK-8 reagent. Cell viability was read at 450 nm on a multi-label plate reader (Bio-Rad) based on color changes due to the formation of formazan product.
Clonogenic assay
After indicated treatments, 100 number of tumor cells were plated in 12-well-plates to generate single colonies. After incubated 10 d at 37 °C, cells were fixed in 4% paraformaldehyde for 30 min, followed by 1% crystal violet staining for 15 min. After washed by water for three times, samples were photographed, and the number of visible colonies was counted by ImageJ software.
Transmission electron microscopy
Cells were fixed with 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) and then post-fixed with 1% OsO4 for 2 h. Cells were dehydrated using a gradient series of ethanol (30, 50, 70, 90, and 100%). Cell were then incubated with LR White resin (Sigma, 62661) twice for 1 h, and subsequently embedded in LR White resin45. The solidified blocks were cut into 60-nm sections and stained with uranyl acetate and lead citrate. Samples were observed and imaged under a transmission electron microscope (TEM, Hitachi H-7600; Hitachi High-Technologies Corporation, Tokyo, Japan). Ten fields were selected by the presence of cytoplasm shrinkage, nuclear membrane shrinkage and/ or nuclear chromatin in the outer nuclear layer gathered towards the center with uneven distribution, and the results were averaged.
Statistical analyses
All data were exported to GraphPad Prism v6.2 (GraphPad Software) for statistical analyses. Values represent the mean ± standard deviation (S.D.). Statistical significance was determined based on p-values obtained from an unpaired two-tailed Student’s t-test or two-way ANOVA.
Electronic supplementary material
Acknowledgements
This work was supported by Applied Technology Research and Development Projects of Heilongjiang Province (GA14C101-06, 2011-068, 12521306), the Harbin Medical University Scientific Research Innovation Fund (2017LCZX37, YJSCX2015-63HYD), and the China Postdoctoral Science Foundation (20110491107). Special thanks to Professor Daling Zhu, Miss Zhou and her friend Judie for their hard work of manuscript revision. Many thanks to Hailian Ma, Liting Huo, Xin Yue, Luyao Wang, and Hao Lu for their work of tissue collection. Thanks to Dr. Yue Li, Professor Fu Chen and colleagues for their hard work in FISH and IHC experiments. Also, thanks to Xue Guan for her technical guidance of Bioluminescence imaging.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by R. Aqeilan
Contributor Information
Meimei Liu, Email: mm7723@163.com.
Zongfeng Zhang, Email: viaac1973@163.com.
Peiling Li, Phone: +86 (451) 86605343, Email: peilley@126.com.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-1148-y).
References
- 1.Di Lorenzo G, et al. Imaging and therapy of ovarian cancer: clinical application of nanoparticles and future perspectives. Theranostics. 2018;8:4279–4294. doi: 10.7150/thno.26345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan J. Targeting resistance. Cell. 2016;166:523. doi: 10.1016/j.cell.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Hu HM, et al. A quantitative chemotherapy genetic interaction map reveals factors associated with PARP inhibitor resistance. Cell Rep. 2017;23:918–929. doi: 10.1016/j.celrep.2018.03.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser M, Bai T, Tsang BK. Akt promotes cisplatin resistance in human ovarian cancer cells through inhibition of p53 phosphorylation and nuclear function. Int. J. Cancer. 2008;122:534–546. doi: 10.1002/ijc.23086. [DOI] [PubMed] [Google Scholar]
- 5.Kim E, et al. SRSF2, mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kędzierska H, Piekiełko-Witkowska A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 2017;53:396. doi: 10.1016/j.canlet.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Xiao R, et al. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol. Cell Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y, et al. Emerging role of long non-coding RNAs in cisplatin resistance. Onco Targets Ther. 2018;11:3185–3194. doi: 10.2147/OTT.S158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, et al. LncRNA-dependent mechanisms of androgen receptor-regulated gene activation programs. Nature. 2013;191:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavan KP, et al. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat. Commun. 2014;5:5323. doi: 10.1038/ncomms6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daubner GM, et al. A syn–anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. Opt. Fib Commun. Confer. 2012;4:3. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed JC. Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell Death Differ. 2006;13:1378–1386. doi: 10.1038/sj.cdd.4401975. [DOI] [PubMed] [Google Scholar]
- 14.Morachis JM, Murawsky CM, Emerson BM. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev. 2010;24:135–147. doi: 10.1101/gad.1856710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paull TT. Mechanisms of ATM activation. Annu. Rev. Biochem. 2015;84:711. doi: 10.1146/annurev-biochem-060614-034335. [DOI] [PubMed] [Google Scholar]
- 16.Pujade LE. New treatments in ovarian cancer. Ann. Oncol. 2017;28:57–60. doi: 10.1093/annonc/mdx442. [DOI] [PubMed] [Google Scholar]
- 17.Ince TA, et al. Characterization of twenty-five ovarian tumour cell lines that phenocopy primary tumours. Nat. Commun. 2015;6:7419. doi: 10.1038/ncomms8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S, et al. p53 amyloid formation leading to its loss of function: implications in cancer pathogenesis. Cell Death Differ. 2017;24:10. doi: 10.1038/cdd.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu F, et al. Suppression of p53 potentiates chemosensitivity in nutrient-deprived cholangiocarcinoma cells via inhibition of autophagy. Oncol. Lett. 2017;14:1959–1966. doi: 10.3892/ol.2017.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duffy MJ, Synnott NC, Crown J. Mutant p53 as a target for cancer treatment. Eur. J. Cancer. 2017;83:258–265. doi: 10.1016/j.ejca.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 21.Bowtell DD, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer. 2015;15:668–679. doi: 10.1038/nrc4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SI, et al. Genomic landscape of ovarian clear cell carcinoma via whole exome sequencing. Gynecol. Oncol. 2017;148:2. doi: 10.1016/j.ygyno.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Ji JX, Wang YK, Cochrane DR, Huntsman DG. Clear cell carcinomas of the ovary and kidney: clarity through genomics. J. Pathol. 2018;244:550–564. doi: 10.1002/path.5037. [DOI] [PubMed] [Google Scholar]
- 24.Vidal A, et al. Lurbinectedin (PM01183), a new DNA minor groove binder, inhibits growth of orthotopic primary graft of cisplatin-resistant epithelial ovarian cancer. Clin. Cancer Res. 2012;18:5399–5411. doi: 10.1158/1078-0432.CCR-12-1513. [DOI] [PubMed] [Google Scholar]
- 25.Bellone S, et al. Exceptional response to pembrolizumab in a metastatic, chemotherapy/radiation resistant ovarian cancer patient harboring a CD274/PD-L1-genetic rearrangement. Clin. Cancer Res. 2018;1085:2017. doi: 10.1158/1078-0432.CCR-17-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalikin LM, et al. An integrated physical and gene map of human distal chromosome 17q24–proximal 17q25 encompassing multiple disease loci. Genomics. 1999;57:36–42. doi: 10.1006/geno.1998.5740. [DOI] [PubMed] [Google Scholar]
- 27.John NH, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sang Y, et al. LncRNA PANDAR regulates the G1/S transition of breast cancer cells by suppressing p16(INK4A) expression. Sci. Rep. 2016;6:22366. doi: 10.1038/srep22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han L, et al. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng W, Fan H. Long non-coding RNA PANDAR correlates with poor prognosis and promotes tumorigenesis in hepatocellular carcinoma. Biomed. Pharmacother. 2015;72:113–118. doi: 10.1016/j.biopha.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Yi X, et al. An increase in long non-coding RNA PANDAR is associated with poor prognosis in clear cell renal cell carcinoma. BMC Cancer. 2017;17:373. doi: 10.1186/s12885-017-3339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y, et al. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis. 2016;5:e192. doi: 10.1038/oncsis.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T, Yang P, Wang H, He ZY. Silence of long noncoding RNA PANDAR switches low-dose curcumin-induced senescence to apoptosis in colorectal cancer cells. Onco. Targets Ther. 2017;10:483–491. doi: 10.2147/OTT.S127547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhan Y, et al. Up-regulation of long non-coding RNA PANDAR is associated with poor prognosis and promotes tumorigenesis in bladder cancer. J. Exp. Clin. Canc Res. 2016;35:83. doi: 10.1186/s13046-016-0354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 36.Azevedo A, Prado AF, Issa JP, Gerlach RF. Matrix metalloproteinase 2 fused to GFP, expressed in E. coli, successfully tracked MMP-2 distribution in vivo. Int. J. Biol. Macromol. 2016;89:737–745. doi: 10.1016/j.ijbiomac.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, et al. Delphinidin-3-glucoside suppresses breast carcinogenesis by inactivating the Akt/HOTAIR signaling pathway. BMC Cancer. 2016;16:423. doi: 10.1186/s12885-016-2465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang Y, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2014;33:378–386. doi: 10.1038/onc.2012.575. [DOI] [PubMed] [Google Scholar]
- 39.Backer MV, et al. Molecular imaging of VEGF receptors in angiogenic vasculature with single-chain VEGF-based probes. Nat. Med. 2007;13:504–509. doi: 10.1038/nm1522. [DOI] [PubMed] [Google Scholar]
- 40.Kim M, et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. 2016;13:149–168. doi: 10.1080/15548627.2016.1239676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmittgen TD. Real-time quantitative PCR. Methods. 2001;25:383–385. doi: 10.1006/meth.2001.1260. [DOI] [PubMed] [Google Scholar]
- 42.Osborn M, Weber K. Immunofluorescence and immunocytochemical procedures with affinity purified antibodies: tubulin-containing structures. Method Cell Bio. 1982;24:97. doi: 10.1016/S0091-679X(08)60650-0. [DOI] [PubMed] [Google Scholar]
- 43.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat. Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 44.Silahtaroglu AN, et al. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat. Protoc. 2007;2:2520–2528. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- 45.Wang ZL. Transmission electron microscopy of shape-controlled nanocrystals and their assemblies. J. Phys. Chem. B. 2000;104:1153–1175. doi: 10.1021/jp993593c. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.