Abstract
Psoriasis is a chronic inflammatory skin disease with a strong genetic background and is triggered by environmental factors. Available evidence supports CD6, a lymphocyte surface receptor mostly expressed by T cells, as a putative target in autoimmunity. Accordingly, a humanized anti-CD6 antibody has been assayed for the treatment of certain autoimmune disorders, including psoriasis. Here, we present novel evidence in mice and humans for a direct involvement of CD6 in psoriasis pathophysiology. First, an attenuated form of imiquimod-induced psoriasis-like skin inflammation was demonstrated in CD6-deficient mice, as deduced from lower epidermal thickness and local reduced production of pro-inflammatory cytokines, namely, interleukin-17A. Thus, isolated CD4+CD62L+ T cells from CD6-deficient mice displayed decreased in vitro T-helper type 17 polarization. Second, a statistically significant association between CD6 single-nucleotide polymorphisms (rs17824933, rs11230563 and rs12360861) and more severe forms of psoriasis was demonstrated in a cohort of 304 patients at three public hospitals from the metropolitan area of Barcelona. Taken together, these results provide new supportive evidence of the contribution of the CD6 lymphocyte receptor in psoriasis at both experimental and clinical levels.
Keywords: ALCAM, CD5, CD6, lymphocyte, psoriasis
Introduction
Psoriasis (Ps) is a chronic immune-mediated systemic disease mainly characterized by cutaneous manifestations that affects more than 125 million people worldwide.1 Clinically, Ps may adopt multiple phenotypes, with plaque-type Ps being the most extended form of the disease.1 The pathophysiology of Ps involves abnormal keratinocyte proliferation and dermal and epidermal infiltration by innate (e.g., neutrophils) and adaptive (e.g., T cells) immune cells. Ps is considered a multifactorial disease with a strong genetic, albeit polygenic, component, which interplays with different environmental factors (namely, trauma, infection and drugs).1 The most important heritability factor is found within the PSORS1 region mapping the human leukocyte antigen, with smaller contributions from a multitude of other genetic loci mainly identified by genome-wide association studies (GWAS).2 The latter includes gene candidates mostly from immune response effectors/regulators and keratinocyte differentiation-related genes.3 Despite new discoveries regarding the genetic basis of Ps, the interpretation of gene-variation associations with biologic functional effects remains unsolved.The CD6 lymphocyte receptor has recently been implicated in the pathogenesis of some autoimmune diseases4 and is currently under clinical evaluation as a putative therapeutic target for some of these diseases,5 including Ps.6 CD6 is a type I glycoprotein of the scavenger receptor cysteine-rich (SRCR) superfamily, expressed by all T cells, a subset of natural killer cells and the B1a-cell subset.7 The main reported CD6 ligand is CD166/ALCAM (activated leukocyte cell adhesion molecule), which is a broadly distributed member of the immunoglobulin cell adhesion superfamily.8 However, additional interactions with well-conserved bacterial components,9,10 such as galectin 1 and 3,11 and CD318 (also named CDCP1 for CUB domain-containing protein 1 or TRASK for transmembrane and associated with Src kinases),12 have also been reported. Even though the function of CD6 remains incompletely understood, recent in vitro and in vivo evidence suggests roles in T-cell development13 and in the modulation of peripheral T- and B1a-cell-mediated immune responses.14,15,16 CD6 has a cytoplasmic tail devoid of intrinsic catalytic activity, but contains consensus motifs for Tyr and Thr/Ser17,18 phosphorylation. CD6 also interacts with intracellular signaling effectors, such as mitogen-activated protein kinases,19 SH2 domain-containing leukocyte protein of 76 kDa,20 T-cell-specific adaptor protein21 and syntenin.22 These characteristics, together with its physical association with the TCR/CD3 (T-cell receptor/cluster of differentiation 3) complex,23 position CD6 well to positively or negatively modulate signaling by a key receptor.24 Although CD6 was initially considered a T-cell co-stimulatory molecule based on the co-mitogenic properties of some anti-CD6 monoclonal antibodies (mAbs), later evidence from CD6 transfectants has supported a putative role for CD6 as an attenuator of TCR/CD3-mediated signaling,24 a fact that has been recently confirmed by data from CD6-deficient (CD6−/−) mice.13,14 Specifically, studies of such deficient mice have shown that (i) CD6 behaves as a negative modulator of TCR/CD3 signaling in the thymus, setting the threshold for the final steps of T-cell selection and maturation,13 and (ii) CD6 contributes to the proper generation and function of the inducible peripheral T-regulatory (Treg) cell population.13,15 Despite the current availability of a humanized anti-CD6 mAb for therapeutic purposes6 and the availability of genetically modified CD6 mice,13,14,15,16 in vivo evidence for the direct involvement of CD6 in mouse and/or human Ps has yet to be reported. To this end, we investigated the phenotype of CD6−/− mice in an experimental Ps-like model induced by imiquimod (IMQ).25 Because (i) no human CD6 deficiency has been reported so far and (ii) data from experimental models of the disease should be cautiously translated into the clinic, we investigated the putative influence of previously reported CD6 single-nucleotide polymorphisms (SNPs) on the clinical characteristics of a cohort of 304 psoriatic patients. The three CD6 SNPs included in the analysis have been linked to enhanced susceptibility to multiple sclerosis (MS).4 Additional SNPs involving the functionally related CD5 and CD166/ALCAM genes were also investigated. CD5 encodes a lymphocyte receptor closely related to CD6. Variants of CD5 have been associated with severe forms of systemic lupus erythematosus26 and a better prognosis in certain types of cancers.27,28 Likewise, CD166/ALCAM variants have been associated with increased risk and progression of MS29 and breast cancer.30,31 The results obtained from these studies provide novel human genetic and mouse experimental evidence supporting the involvement of CD6 in the pathogenesis of Ps.
Results
Human CD6 polymorphism impact Ps severity
Psoriatic patients (N=304) and controls (N=305) were genotyped for CD6 variants and investigated for association with Ps susceptibility and/or prognosis. The genetic analysis included CD6 SNPs (rs17824933, rs11230563 and rs12360861) previously associated with increased risk of MS,32,33 as well as SNPs from CD5 (rs2229177 and rs2241002) and ALCAM (rs6437585 and rs579565), two functionally related genes associated with increased severity and risk of SLE and MS,26,29 respectively (Figure 1).
Figure 1.
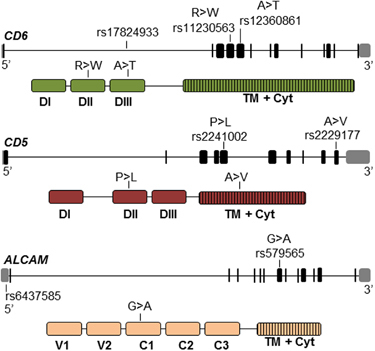
Schematic location at genomic and protein levels of CD6, CD5 and ALCAM polymorphisms analyzed. Upper pictures represent the genomic organization of the genes with exons represented as black boxes, introns as solid lines and 5′- and 3′-untranslated regions as gray boxes. Lower pictures represent the extracellular and intracellular domain organization of protein products. The relative position of SNPs and non-synonymous amino-acid changes are marked. ALCAM, activated leukocyte cell adhesion molecule; Cyt, cytoplasmic domain; SNP, single-nucleotide polymorphism; TM, transmembrane domain.
In a first analysis, the genotypic frequencies of the CD6, CD5 and ALCAM SNPs from psoriatic patients and controls were shown to be in Hardy–Weinberg equilibrium (P>0.02). Moreover, no statistically significant differences between patients and controls were observed for any of the SNPs, thus discarding the possibility of any being a susceptibility marker for Ps (Supplementary Table S1).
In a second analysis, we evaluated whether any of the genetic variants had an effect on Ps severity. To this end, the allelic and genotypic frequencies of the CD6, CD5 and ALCAM SNPs were compared among patients receiving or not receiving systemic treatment (categorized as moderate–severe or mild disease subgroups, respectively)34,35,36,37,38 and among patients displaying high (>7) or low (⩽7) Psoriasis Area and Severity Index (PASI) values 39 according to previous well-established criteria.34,35,36,37,38,39 Shown in Supplementary Table S2 are the median and the interquartile ranges for the clinical and demographic characteristics of patients involved in the study. Kruskal–Wallis (continuous variables) or χ 2 (frequencies) analyses showed statistically significant associations of moderate–severe Ps with earlier disease onset (P=0.0007), arthritis occurrence (P=0.0023), higher PASI (P<0.0001) and longer disease duration (P=0.0056) (Supplementary Table S2). Subsequently, multivariate genetic analyses were adjusted for age of onset, duration of the disease and presence of psoriatic arthritis. Sex was considered a confounding variable and was also adjusted. Because PASI is also considered a disease severity outcome, the same clinical variables were adjusted for PASI analyses.
Univariate analyses revealed statistically significant associations of the rs12360861A CD6 allele with moderate–severe Ps (Table 1, top) and the rs17824933G, rs11230563C and rs12360861A CD6 alleles with high (>7) PASI (Table 1, bottom). The latter association was maintained in subsequent multivariate analyses. By contrast, no significant relationships were observed for CD5 and ALCAM SNPs regarding any indicator of Ps severity (Supplementary Table S3).
Table 1.
CD6 SNPs allele distribution in patients displaying ‘mild’ and ‘moderate-severe’ (top) or PASI scores ⩽7 and >7 (bottom)
| SNP | Allele | Mild Ps No. (%) | Moderate-severe Ps No. (%) | ORa (95% CI)b | P value | Adj.c ORa (95% CI)b | Adj. c P value |
|---|---|---|---|---|---|---|---|
| rs17824933 | C | 182 (80.5) | 271 (72.8) | ||||
| G | 44 (19.5) | 101 (27.2) | 1.54 (1.03–2.30) | 0.0343 | 1.44 (0.93–2.21) | 0.1009 | |
| rs11230563 | C | 110 (51.9) | 213 (60.5) | ||||
| T | 102 (48.1) | 139 (39.5) | 0.70 (0.50–0.99) | 0.0453 | 0.68 (0.47–0.99) | 0.0463 | |
| rs12360861 | G | 60 (28.6) | 70 (20.0) | ||||
| A | 150 (71.4) | 280 (80.0) | 0.63 (0.42–0.93) | 0.0206 | 0.68 (0.44–1.05) | 0.0811 |
| SNP | Allele | PASI⩽7 No. (%) | PASI>7 No. (%) | ORa (95% CI)b | P-value | Adj.c ORa (95% CI)b | Adj.cPvalue |
|---|---|---|---|---|---|---|---|
| rs17824933 | C | 226 (80.7) | 227 (71.4) | ||||
| G | 54 (19.3) | 91 (28.6) | 1.68 (1.14–2.46) | 0.0082 | 1.59 (1.05–2.42) | 0.0284 | |
| rs11230563 | C | 127 (49.2) | 196 (64.1) | ||||
| T | 131 (50.8) | 110 (35.9) | 0.54 (0.39–0.76) | 0.0004 | 0.53 (0.37–0.78) | 0.0010 | |
| rs12360861 | G | 75 (29.3) | 55 (18.1) | ||||
| A | 181 (70.7) | 249 (81.9) | 0.53 (0.36–0.79) | 0.0019 | 0.56 (0.36–0.85) | 0.0074 |
aOR=Odds Ratio
bCI=Confidence Interval
cAdj.=Adjusted; multivariate analysis adjusted by onset of disease, arthropathy occurrence, gender and disease length.
Univariate and multivariate analyses also showed statistically significant associations for individual and/or pooled genotype frequencies of CD6 SNPs with moderate–severe Ps (Table 2, top) and/or high (>7) PASI (Table 2, bottom). Again, no statistically significant association was found for any of the CD5 and ALCAM SNPs.
Table 2.
CD6 SNP genotypic frequency distribution among patients displaying ‘mild’ or ‘moderate—severe’ Ps (top) as well as low (⩽7) or high (>7) PASI scores (bottom)
| SNP | Genotype | Mild Ps, no. (%) | Moderate–severe Ps no. (%) | OR (95% CI) | P-value | Adj. a OR (95% CI) | Adj.a P-value |
|---|---|---|---|---|---|---|---|
| Rs17824933 | CC | 76 (67.3) | 97 (52.2) | ||||
| CG | 30 (26.5) | 76 (40.9) | 1.99 (1.18–3.33) | 0.009 | 1.78 (1.00–3.18) | 0.049 | |
| GG | 7 (6.2) | 13 (6.9) | 1.45 (0.55–3.83) | 0.447 | 1.25 (0.43–3.68) | 0.681 | |
| CG+GG | 37 (32.7) | 89 (47.8) | 1.88 (1.16–3.07) | 0.0103 | 1.68 (0.98–2.90) | 0.061 | |
| Rs11230563 | CC | 26 (24.5) | 72 (40.9) | ||||
| CT | 58 (54.7) | 69 (39.2) | 0.43 (0.24–0.76) | 0.0036 | 0.42 (0.22–0.80) | 0.008 | |
| TT | 22 (20.8) | 35 (19.9) | 0.57 (0.29–1.15) | 0.119 | 0.60 (0.27–1.32) | 0.203 | |
| CT+TT | 80 (75.5) | 104 (59.1) | 0.47 (0.28–0.80) | 0.0056 | 0.47 (0.26–085) | 0.013 | |
| Rs12360861 | GG | 51 (48.6) | 108 (61.7) | ||||
| AG | 48 (45.7) | 63 (36) | 0.32 (0.09–1.17) | 0.0834 | 0.42 (0.11–1.64) | 0.213 | |
| AA | 6 (5.7) | 4 (2.3) | 0.62 (0.38–1.02) | 0.0618 | 0.75 (0.43–1.30) | 0.304 | |
| AG+AA | 54 (51.4) | 67 (38.3) | 0.59 (0.36–0.96) | 0.0322 | 0.71 (0.41–1.22) | 0.211 |
| SNP | Genotype | PASI⩽7, no. (%) | PASI>7, no. (%) | OR (95% CI) | P-value | Adj. a OR (95% CI) | Adj. a P- value |
|---|---|---|---|---|---|---|---|
| Rs17824933 | CC | 93 (66.4) | 80 (50.3) | ||||
| CG | 40 (28.6) | 66 (41.5) | 1.92 (1.17–3.14) | 0.0097 | 1.88 (1.07–3.29) | 0.0288 | |
| GG | 7 (5.0) | 13 (8.2) | 2.16 (0.82–5.67) | 0.1185 | 1.85 (0.63–5.42) | 0.2622 | |
| CG+GG | 47 (33.6) | 79 (49.7) | 1.95 (1.22–3.12) | 0.0051 | 1.87 (1.10–3.20) | 0.0219 | |
| Rs11230563 | CC | 30 (23.3) | 68 (44.5) | ||||
| CT | 67 (51.9) | 60 (39.2) | 0.40 (0.23–0.69) | 0.0010 | 0.42 (0.23–0.78) | 0.0063 | |
| TT | 32 (24.8) | 25 (16.3) | 0.35 (0.18–0.68) | 0.0020 | 0.34 (0.16–0.74) | 0.0069 | |
| CT+TT | 99 (76.7) | 85 (55.6) | 0.38 (0.23–0.64) | 0.0002 | 0.40 (0.22–0.71) | 0.0020 | |
| Rs12360861 | GG | 60 (46.9) | 99 (65.1) | ||||
| AG | 60 (46.9) | 51 (33.6) | 0.15 (0.03–0.74) | 0.0194 | 0.22 (0.04–1.13) | 0.0694 | |
| AA | 8 (6.2) | 2 (1.3) | 0.52 (0.32–0.84) | 0.0082 | 0.56 (0.32–0.97) | 0.0377 | |
| AG+AA | 68 (53.1) | 53 (34.9) | 0.47 (0.29–0.77) | 0.00023 | 0.52 (0.30–0.89) | 0.0017 |
Abbreviations: Adj., adjusted; CI, confidence interval; OR, odds ratio; PASI, Psoriasis Area and Severity Index; SNP, single-nucleotide polymorphism.
aMultivariate analysis adjusted by onset of disease, arthropathy occurrence, gender and disease length.
Further haplotype analyses showed that the CD6 SNPs were linked to each other with D′ values >95% and r values of −0.47 (rs17824933 vs rs11230563), −0.31 (rs17824933 vs rs12360861) and 0.64 (rs11230563 vs rs12360861), thus confirming that they were in high linkage disequilibrium (LD) and constituted an LD block.
Four inferred CD6 haplotypes accounted for 99.6% of estimated haplotypes in psoriatic patients (Table 3). Further haplotype analyses revealed that the allele combination rs17824933C, rs11230563T and rs12360861A together with the allele combination rs17824933C, rs11230563T and rs12360861G (Supplementary Table S2, bottom) was significantly linked to lower PASI (P=0.0086 and P=0.013, respectively). The significance of the latter association was maintained in multivariate analyses. Additionally, the frequency of the haplotype carrying the three risk CD6 SNPs in the control population was compared with those in patients with ‘mild’ and ‘severe’ Ps (Supplementary Table S4) and patients displaying low (⩽7) and high (>7) PASI scores (Supplementary Table S5). Univariate analyses in each case revealed no statistically significant associations between any of the studied patient subgroups and the controls.
Table 3.
CD6 total patient haplotype frequency distribution and further analysis between patients displaying ‘mild’ and ‘severe’ Ps (top), as well as low (⩽7) vs high PASI scores (bottom)
| Rs17824933 | Rs11230563 | Rs12360861 | % | OR (95% CI) | P-value | Adj.a ORa (95% CI) | Adj.a P-value |
|---|---|---|---|---|---|---|---|
| C | C | G | 33.0 | ||||
| G | C | G | 24.2 | 1.31 (0.82–2.08) | 0.26 | 1.12 (0.68–1.84) | 0.660 |
| C | T | A | 23.6 | 0.61 (0.38–0.99) | 0.046 | 0.62 (0.36–1.05) | 0.074 |
| C | T | G | 19.2 | 0.99 (0.61–1.60) | 0.95 | 0.77 (0.45–1.31) | 0.330 |
| C | C | G | 32.5 | ||||
| G | C | G | 24.6 | 0.04 (−0.12 to 0.21) | 0.59 | 0.01 (−0.16 to 0.17) | 0.930 |
| C | T | A | 23.9 | −0.23 (−0.41 to 0.06) | 0.008 | −0.2 (−0.37 to 0.02) | 0.033 |
| C | T | G | 19.0 | 0.64 (−0.36 to 1.64) | 0.013 | −0.27 (−0.45 to 0.09) | 0.003 |
Abbreviations: Adj., adjusted; CI, confidence interval; OR, odds ratio; PASI, Psoriasis Area and Severity Index.
aMultivariate analysis adjusted by onset of disease, arthropathy occurrence, gender and disease length.
Finally, the statistical analysis was further extended to include potential CD6 SNP genotype associations with earlier/later onset of Ps, which is often linked with irregular courses and having strong tendencies to become generalized.40,41,42 The results showed a significant relationship between the age of Ps onset and the rs17824933 SNP (P=0.0075 and P=0.0095, the latter for the pooled genotype), with the earliest onset linked to the heterozygous combination (Supplementary Figure S1).
CD6-deficient mice develop attenuated IMQ-induced Ps
A potential in vivo role of CD6 in Ps development was investigated by subjecting wild-type (CD6+/+) and CD6-deficient (CD6−/−) mice13 to the Ps-like skin inflammation model induced by the topical application of the synthetic TLR7/8 agonist IMQ.25 The C57BL/6 strain was chosen for this purpose because it provides a better genetic background for modeling Ps disease mechanisms.43 Skin examination at the end of the experimental period (day 7) showed that CD6−/− mice had skin inflammation to a significantly lesser extent than CD6+ / + controls, as demonstrated by epidermal thickness assessments (Figure 2a, left). In line with these findings, lower splenomegaly (Figure 2a, right) and decreased relative local mRNA expression of different pro-inflammatory cytokines (interleukin-17A (IL-17A), IL-17F, IL-22, IL-6 and interferon-γ (IFN-γ)) in skin lesions, which was significant for IL-17A and IL-22 (Figure 2b), were found in CD6−/− mice compared with CD6+ / + controls. By contrast, no differences were observed for the immunosuppressive effectors IL-10 and FoxP3 (Figure 2b).
Figure 2.
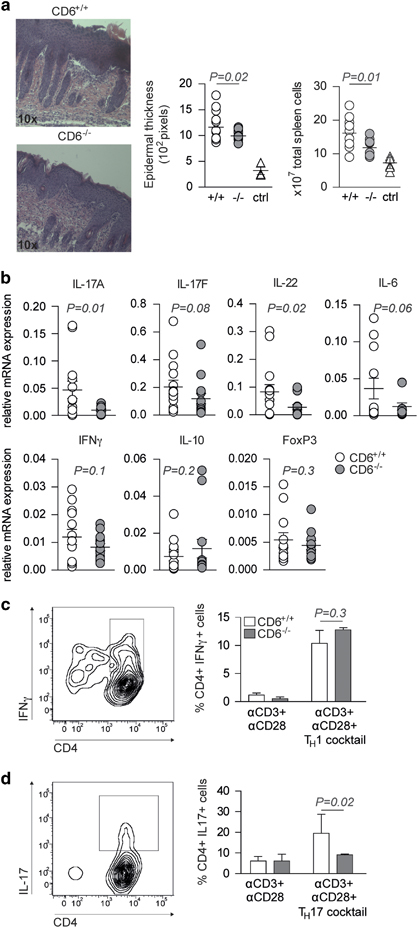
CD6−/− mice show attenuated IMQ-induced Ps-like inflammation and in vitro TH17 polarization. (a, left) Representative hematoxylin and eosin stain (x10) of day 7 skin lesions from IMQ-treated CD6−/− and CD6+/+ mice. (Right) Dot-plot showing the means of epidermal thickness and total spleen cells from IMQ- or Vaseline (ctrl)-treated mice (N=8). A representative experiment of three performed is shown. (b) Relative cytokine mRNA expression levels in skin lesions from IMQ-treated CD6−/− and CD6+/+ mice. (c, left) Representative contour plot showing the gating strategy used to detect in vitro polarized TH1 cells. (Right) percentages (mean±s.e.m.) of CD4+IFNγ+ cells following stimulation of CD6−/− and CD6+/+ naïve T cells under TH1-polarizing and -non-polarizing conditions. (d) Same analyses as in (c) for TH17 polarization. Statistical analyses were performed by nonparametric t-tests for unpaired data (level of significance set at P<0.05). ctrl, Control; IFN, interferon; IMQ, imiquimod; Ps, psoriasis; TH17, T-helper type 17.
In light of the attenuated local TH17 inflammatory response observed in CD6−/− mice following IMQ-induced Ps, in vitro TH1 and TH17 polarization assays were performed. To this end, purified CD4+CD62Lhi T cells from CD6−/− and CD6+/+ mice were cultured under TH1 and TH17 polarization conditions and further analyzed by flow cytometry for intracellular IFN-γ and IL-17 expression. As illustrated in Figures 2c and d, non-significant differences were detected between the two groups of mice concerning the percentage of CD4+IFN-γ+ cells. By contrast, a significantly lower percentage of CD4+IL-17+ cells was observed for CD6−/− T cells than for CD6+/+ controls. This result was consistent with the in vivo data from the IMQ-induced Ps-like model and suggested that CD6 deficiency implied defective TH17 but TH1 polarization in C57BL/6 mice.
Discussion
The present work provides novel genetic and experimental insight supporting the putative involvement of the CD6 lymphocyte receptor in the pathophysiology of Ps. As far as we know, available GWAS have not provided evidence for the involvement of CD6 polymorphisms in Ps. This was not unexpected because GWAS analyses of complex/polygenic human diseases (such as Ps) have relied on the proper density coverage of the trait loci with preselected common SNPs, most of which have no known functional implications.44 This limitation can be partly overcome by gene-specific candidate-driven analyses. We focused on genetic variants of CD5 and CD6, two contiguous genes on human chromosome 11q.13 coding for highly related lymphocyte surface receptors at structural and functional levels.45 In fact, these genes are believed to be derived from the duplication of a common ancestral gene. The CD5 and CD6 receptors are physically associated with the antigen-specific TCR and contribute to the fine tuning of activation and differentiation signals mediated by this relevant complex.45 Our analysis also included genetic variants of CD166/ALCAM, thus providing a more comprehensive analysis on the contribution of functionally related genes to Ps.
The genetic analyses showed that all three CD6 SNPs were positively associated with Ps severity, as inferred from the need for systemic therapy and higher PASI (>7) scores. This conclusion was derived not only from the genotypic and allelic analyses but also from haplotype analyses, which showed significant associations with the two severity outcomes. Taking into account the link between the age of onset and the disease severity, also confirmed in our patient cohort, it was not unexpected that one of the CD6 SNPs (rs17824933) was found to be associated with an earlier form of disease. Altogether, the results argued in favor of a contribution of the CD6 receptor in Ps pathophysiology and the potential usefulness of CD6 genotyping in Ps prognosis. The fact that CD5 and CD166/ALCAM SNPs were found to have neutral effects also supported a specific effect of CD6 as a disease-modifier gene in Ps. However, the putative influence of genetic variations in CD318/CDCP1/TRASK, a very recently reported CD6 ligand expressed on many epithelial cells, and some hematopoietic and mesenchymal stem cells should be further explored.12
Only a few reports have linked CD6 gene variation to certain autoimmune conditions in different human populations. Thus, a 19-base pair insertion within the CD6 gene was associated with good response to tumor necrosis factor-α inhibitors in a cohort of Danish rheumatoid arthritis patients.46 Recently, the CD6 rs11230563 has been associated with susceptibility to Behçet’s disease in a Chinese Han population.47 However, the most important and well-studied association of CD6 polymorphism has been with MS in which TH17 cells play an important pathogenic role. The first report on this regard found that the intronic rs177824933G allele was associated with an increased risk of developing MS.4 Other risk alleles within the CD6 gene (rs11230563 and rs12360861) were soon found and validated as independent risk factors for MS.4 Thus, the short list of autoimmune disorders linked to CD6 polymorphism were subsequently extended to Ps, wherein TH17 cells have been found to play a relevant pathogenic role.48,49,50 Interestingly, the Danish Nationwide Cohort Study linked these two inflammatory diseases and revealed Ps as a severity-dependent risk factor for MS.51
Further evidence involving CD6 in Ps physiopathology comes from the observation that CD6−/− mice exhibit attenuated IMQ-induced Ps-like skin inflammation, likely resulting from defective peripheral TH17 polarization, which has been reported in experimental autoimmune encephalomyelitis (EAE).14 These data support CD6, a component of the TCR-complex signaling machinery, as a modifier of the homeostasis of TH17 cells, a critical pathogenic subset in Ps (and EAE) physiopathology. The molecular basis linking CD6 deficiency and altered TH17 differentiation remain unknown; however, similar alterations have been reported for the TH114,52 and Treg15 cell subsets. Intriguingly, CD6 is involved in T-cell development. There is evidence that natural TH17 cells (nTH17) may arise from the thymus, analogous to natural Treg cells (nTreg).53 Concomitant to the already detected defects in inducible TH17 and Treg polarization of naïve peripheral T cells,14,15 defects in nTH17 development should also be explored in CD6-deficient mice, a fact that could be ignored for nTreg cells.13
Although no mouse model can accurately reproduce all aspects of human Ps,54 the data from the IMQ model in CD6−/− mice correlate with our human genetic studies and highlight emerging evidence on the potential role played by CD6 in the regulation of T-cell-driven autoimmune responses.13,14,15,16
None of the CD6 SNPs studied have been reported to induce deficient overall CD6 surface expression. The rs17824933G allele decreases the expression of full-length CD6 and increases that of the CD6ΔD3 isoform,55 which lacks the CD166/ALCAM-binding domain and is unable to co-localize with the TCR at the center of the immunological synapse.56 In contrast, the rs11230563C allele is associated with higher CD6 expression.33 It would be interesting to analyze whether any of the CD6 SNPs may affect their interactions with the recently reported CD6 ligand, CD318/CDCP1/TRASK, which could impair overall CD6 functionality.12
In conclusion, the results presented in our genetic association studies of Ps patients and CD6−/− mice in an experimental model of Ps support a potential role for CD6 in Ps pathogenesis. Future studies are warranted to validate the role of CD6 in Ps physiopathology and further assess whether CD6 genotyping would help predict patient response to distinct therapies, including anti-CD6 mAb treatment currently in clinical development.
Materials and methods
Subjects
Psoriatic patients of European descent (N=304) attending three Dermatology Departments from three University Hospitals (Barcelona) were included in the study. Inclusion criteria were age above 18 years and a minimum follow-up of 6 months. All patients were managed according to the clinical guidelines of the Spanish Psoriasis Group from the Spanish Academy of Dermatology and Venereology.57
Additionally, 305 unrelated volunteers from the Blood and Tissue Bank of Barcelona were included as controls (42±13.6 years, 18–68 age range; 47% female). The study was approved by the three local Hospital Ethics Committees, and written informed consent was obtained from all participants before inclusion and blood extraction.
Clinical assessment
Epidemiologic, demographic and clinical data, including age, gender, age of disease onset (early onset <40 or late onset ⩾40), disease duration, family history of Ps, presence of psoriatic arthritis, progressive (persistent disease flare of at least 6 months) or intermittent clinical course, highest Ps area and severity index (PASI, ⩽7 or >7)39 and therapies received, were retrospectively collected from medical records. Patients who received at least one systemic treatment (methotrexate, cyclosporine or acitretin), biological agent (etanercept, adalimumab, infliximab, efalizumab and/or ustekinumab) or phototherapy (oral psoralen with UVA—PUVA− or narrow-band UVB—nbUVB−) were included in the moderate–severe Ps group, whereas the rest of the patients were classified as mild.34,35,36,37,38
Genotyping
Genomic DNA was purified from EDTA-anticoagulated peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen, Venio, The Netherlands) and subjected to real-time (RT)-PCR using TaqMan probes following the manufacturer's recommendations (ThermoFisher, Barcelona, Spain): CD6 rs17824933 (assay number: C_33967506_10), CD6 rs11230563 (assay number: C_31727142_10), CD6 rs12360861 (assay number: C_25922320_10), CD5 rs2229177 (assay number: C_3237272_10), CD5 rs2241002 (assay number: C_25472293_20) and ALCAM rs6437585 (assay number: C_29281365_20), all from ThermoFisher (Barcelona, Spain). Primers for PCR amplification and further sequence-base typing (PCR-SBT) ALCAM rs579565 (Hs00666884_CE and Hs00666884_CE) were from ThermoFisher.
IMQ-induced Ps mouse model
Eight- to 12-week-old CD6−/− and CD6+/+ C57BL/6 females13 received a daily topical dose of 62.5 mg of commercially available IMQ cream (5%, Aldara; 3M Pharmaceuticals, MEDA, Barcelona, Spain) on shaved backs for 6 consecutive days.25 Control mice were likewise treated with a control vehicle (Vaseline; Braun, Barcelona, Spain). At day 7, mice were killed, and skins were harvested for further analyses. This protocol was reviewed and approved by the University of Barcelona Animal Experimentation Ethical Committee (Barcelona, Spain).
Quantitative RT-PCR
Total mRNA was extracted from whole back skin biopsies using the PureLinkTM RNA Mini Kit (Ambion, Life Technologies, Barcelona, Spain). Synthesis of cDNA was performed by reverse transcription from 1 μg of total RNA (High Capacity cDNA Kit; Life Technologies). Cytokine mRNA levels were assessed by quantitative RT-PCR analysis (Taqman Fast Universal PCR Master Mix; Life Technologies) using a 7900HT Fast Real-time PCR System (Applied Biosystems, Bleiswijk, The Netherlands) and the following Taqman probes (Life Technologies): IL-17A (Mm00439618_m1), IL-17F (Mm00521423_m1), IL-22 (Mm01226722_g1), IFN-γ (Mm01168134_m1), IL-10 (Mm01288386_m1), FoxP3 (Mm00475162_m1), IL-6 (Mm01210733_m1), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Mm99999915_g1). Cytokine mRNA expression was adjusted by GAPDH expression as 2ΔCt, where ΔCt= Ct (GAPDH)−Ct (gene of interest).
TH1 and TH17 in vitro polarization
Electronically sorted Tconv cells (CD4+CD62Lhi, 105 cells per well) from CD6−/− and CD6+/+ spleens were cultured for 3 days in ‘U’-bottomed plate-bound anti-CD3 mAb (145-2C11; 2 μg/ml; Tonbo Bioscience, San Diego, CA, USA) with soluble anti-CD28 mAb (37.51; 5 μg/ml; Tonbo Bioscience) in the presence of TGF-β (5 ng/ml; ImmunoTools, Friesoythe, Germany) and IL-6 (30 ng/ml; ImmunoTools), IL-1β (10 ng/ml; ImmunoTools), IL-23 (10 ng/ml; ImmunoTools), anti-IL-4 mAb (5 μg/ml; BioLegend, San Diego, CA, USA), anti-IFNγ mAb (5 μg/ml; Tonbo Bioscience) and anti-IL-2 mAb (5 μg/ml; eBioscience, Barcelona, Spain) for TH17 induction. IL-12 (10 ng/ml; R&D Systems, Minneapolis, MN, USA) and anti-IL-4 mAb (10 ng/ml; ImmunoTools) were used for TH1 induction. TH1 and TH17 cell detection was performed by intracellular cytokine staining according to the manufacturer’s indications (Fixation/Permeabilization Kit with BD GolgiStop, BD Bioscience, Madrid, Spain) and further analysis by flow cytometry on a FACS Canto II equipped with CellQuest (BD Bioscience) and FlowJo 8.7 Software (Ashland, OR, USA).
Statistical analyses
Genotypic and haplotype statistical associations among the SNPs and the disease outcomes (‘mild’ vs ‘moderate–severe’ Ps and PASI) were tested by χ 2-test (frequencies). Univariate and multivariate logistic regression models were estimated to report unadjusted and adjusted odds ratio and their 95% confidence intervals, respectively, using ‘epiR’ and ‘SNPassoc’ packages of R software, version 3.3.3 (Available at http://www.r-project.org/). Disease onset (dichotomized as ⩽40 and >40 years), arthropathy occurrence, gender and disease duration were included in the multivariate logistic regression models. The distribution of alleles in controls and patients was tested to fit to the Hardy–Weinberg equilibrium using the bioinformatics tool SNPStats (http://bioinfo.iconcologia.net/SNPstats).58 The same tool was used in haplotype association and LD parameters R and D' analyses. Because the analysis was performed for three genes, Bonferroni’s correction was applied to the asymptotic P-values, with P⩽0.02 considered significant.
Electronic supplementary material
Acknowledgements
We thank Francesc Calafell and Elena Bosch (Institute of Evolutionary Biology, University Pompeu Fabra, Barcelona) for statistical analysis support and critically reviews of the manuscript. The work is supported by grants from the Spanish Ministerio de Economía y Competitividad (Plan Nacional I+D+i, SAF2013-46151-R and SAF2016-80535-R to FL), co-financed by the European Development Regional Fund ‘A way to achieve Europe’ ERDF. FA is supported by the Sara Borrell fellowship CD15/00016 from Instituto de Salud Carlos III.
Conflict of interest
The authors declare no conflict of interest
Electronic supplementary material
Supplementary Information for this article can be found on the Cellular & Molecular Immunology website 10.1038/cmi.2017.119
References
- 1.Greb JE, Goldminz AM, Elder JT, Lebwohl MG, Gladman DD, Wu JJ, et al. Psoriasis. Nat Rev Dis Prim. 2016;2:16082. doi: 10.1038/nrdp.2016.82. [DOI] [PubMed] [Google Scholar]
- 2.Hwang ST, Nijsten T, Elder JT. Recent highlights in psoriasis research. J Invest Dermatol. 2017;137:550–556. doi: 10.1016/j.jid.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Ray-Jones H, Eyre S, Barton A, Warren RB. One SNP at a time: moving beyond GWAS in psoriasis. J Invest Dermatol. 2016;136:567–573. doi: 10.1016/j.jid.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 4.Kofler DM, Farkas A, von Bergwelt-Baildon M, Hafler DA. The link between CD6 and autoimmunity: genetic and cellular associations. Curr Drug Targets. 2016;17:651–665. doi: 10.2174/1389450117666160201105934. [DOI] [PubMed] [Google Scholar]
- 5.Hernández P, Moreno E, Aira LE, Rodríguez PC. Therapeutic targeting of CD6 in autoimmune diseases: a review of cuban clinical studies with the antibodies IOR-T1 and itolizumab. Curr Drug Targets. 2016;17:666–677. doi: 10.2174/1389450117666160201114308. [DOI] [PubMed] [Google Scholar]
- 6.Krupashankar DS, Dogra S, Kura M, Saraswat A, Budamakuntla L, Sumathy TK, et al. Efficacy and safety of itolizumab, a novel anti-CD6 monoclonal antibody, in patients with moderate to severe chronic plaque psoriasis: results of a double-blind, randomized, placebo-controlled, phase-III study. J Am Acad Dermatol. 2014;71:484–492. doi: 10.1016/j.jaad.2014.01.897. [DOI] [PubMed] [Google Scholar]
- 7.Sarukhan A, Martinez-Florensa M, Escoda-Ferran C, Carrasco E, Carreras E, Lozano F. Pattern recognition by CD6: a scavenger-like lymphocyte receptor. Curr Drug Targets. 2015;17:640–650. doi: 10.2174/1389450116666150316224308. [DOI] [PubMed] [Google Scholar]
- 8.Brown MH. CD6 as a cell surface receptor and as a target for regulating immune responses. Curr Drug Targets. 2016;17:619–629. doi: 10.2174/1389450116666150825120536. [DOI] [PubMed] [Google Scholar]
- 9.Sarrias M-R, Farnós M, Mota R, Sánchez-Barbero F, Ibáñez A, Gimferrer I, et al. CD6 binds to pathogen-associated molecular patterns and protects from LPS-induced septic shock. Proc Natl Acad Sci USA. 2007;104:11724–11729. doi: 10.1073/pnas.0702815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Florensa M, Consuegra-Fernández M, Martínez VG, Cañadas O, Armiger-Borràs N, Bonet-Roselló L, et al. Targeting of key pathogenic factors from gram-positive bacteria by the soluble ectodomain of the scavenger-like lymphocyte receptor CD6. J Infect Dis. 2014;209:1077–1086. doi: 10.1093/infdis/jit624. [DOI] [PubMed] [Google Scholar]
- 11.Escoda-Ferran C, Carrasco E, Caballero-Baños M, Miró-Julià C, Martínez-Florensa M, Consuegra-Fernández M, et al. Modulation of CD6 function through interaction with Galectin-1 and -3. FEBS Lett. 2014;588:2805–2813. doi: 10.1016/j.febslet.2014.05.064. [DOI] [PubMed] [Google Scholar]
- 12.Enyindah-Asonye G, Li Y, Ruth JH, Spassov DS, Hebron KE, Zijlstra A, et al. CD318 is a ligand for CD6. Proc Natl Acad Sci USA. 2017;114:E6912–E6921. doi: 10.1073/pnas.1704008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orta-Mascaró M, Consuegra-Fernández M, Carreras E, Roncagalli R, Carreras-Sureda A, Alvarez P, et al. CD6 modulates thymocyte selection and peripheral T cell homeostasis. J Exp Med. 2016;213:1387–1397. doi: 10.1084/jem.20151785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Singer NG, Whitbred J, Bowen MA, Fox DA, Lin F. CD6 as a potential target for treating multiple sclerosis. Proc Natl Acad Sci USA. 2017;114:2687–2692. doi: 10.1073/pnas.1615253114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consuegra-Fernández M, Martínez-Florensa M, Aranda F, de Salort J, Armiger-Borràs N, Lozano T, et al. Relevance of CD6-mediated interactions in the regulation of peripheral T-cell responses and tolerance. Front Immunol. 2017;8:594. doi: 10.3389/fimmu.2017.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enyindah-Asonye G, Li Y, Xin W, Singer NG, Gupta N, Fung J, et al. CD6 receptor regulates intestinal ischemia/reperfusion-induced injury by modulating natural IgM-producing B1a cell self-renewal. J Biol Chem. 2017;292:661–671. doi: 10.1074/jbc.M116.749804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobarg J, Whitney GS, Palmer D, Aruffo A, Bowen MA. Analysis of the tyrosine phosphorylation and calcium fluxing of human CD6 isoforms with different cytoplasmatic domains. Eur J Immunol. 1997;27:2971–2980. doi: 10.1002/eji.1830271133. [DOI] [PubMed] [Google Scholar]
- 18.Bonet L, Farnós M, Martínez-Florensa M, Martínez VG, Lozano F. Identification of functionally relevant phoshorylatable serine clusters in the cytoplasmic region of the human CD6 lymphocyte surface receptor. FEBS Lett. 2013;587:2205–2213. doi: 10.1016/j.febslet.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 19.Ibáñez A, Sarrias M-R, Farnós M, Gimferrer I, Serra-Pagès C, Vives J, et al. Mitogen-activated protein kinase pathway activation by the CD6 lymphocyte surface receptor. J Immunol. 2006;177:1152–1159. doi: 10.4049/jimmunol.177.2.1152. [DOI] [PubMed] [Google Scholar]
- 20.Breuning J, Brown MH. T cell costimulation by CD6 is dependent on bivalent binding of a GADS/SLP-76 complex. Mol Cell Biol 2017; 37; doi: 10.1128/MCB.00071-17. [DOI] [PMC free article] [PubMed]
- 21.Hem CD, Ekornhol M, Granum S, Sundvold-Gjerstad V, Spurkland A. CD6 and linker of activated T cells are potential interaction partners for T cell-specific adaptor protein. Scand J Immunol. 2017;85:104–112. doi: 10.1111/sji.12513. [DOI] [PubMed] [Google Scholar]
- 22.Gimferrer I, Ibáñez A, Farnós M, Sarrias M-R, Fenutría R, Roselló S, et al. The lymphocyte receptor CD6 interacts with syntenin-1, a scaffolding protein containing PDZ domains. J Immunol. 2005;175:1406–1414. doi: 10.4049/jimmunol.175.3.1406. [DOI] [PubMed] [Google Scholar]
- 23.Gimferrer I, Calvo M, Mittelbrunn M, Farnós M, Sarrias MR, Enrich C, et al. Relevance of CD6-mediated interactions in T cell activation and proliferation. J Immunol. 2004;173:2262–2270. doi: 10.4049/jimmunol.173.4.2262. [DOI] [PubMed] [Google Scholar]
- 24.Santos RF, Oliveira L, Carmo AM. Tuning T cell activation: the function of CD6 at the immunological synapse and in T cell responses. Curr Drug Targets. 2016;17:630–639. doi: 10.2174/1389450116666150531152439. [DOI] [PubMed] [Google Scholar]
- 25.van der Fits L, Mourits S, Voerman JSA, Kant M, Boon L, Laman JD, et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol. 2009;182:5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 26.Cenit MC, Martínez-Florensa M, Consuegra M, Bonet L, Carnero-Montoro E, Armiger N, et al. Analysis of ancestral and functionally relevant CD5 variants in systemic lupus erythematosus patients. PLoS One. 2014;9:e113090. doi: 10.1371/journal.pone.0113090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potrony M, Carreras E, Aranda F, Zimmer L, Puig-Butille J-A, Tell-Martí G, et al. Inherited functional variants of the lymphocyte receptor CD5 influence melanoma survival. Int J Cancer. 2016;139:1297–1302. doi: 10.1002/ijc.30184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delgado J, Bielig T, Bonet L, Carnero-Montoro E, Puente XS, Colomer D, et al. Impact of the functional CD5 polymorphism A471V on the response of chronic lymphocytic leukaemia to conventional chemotherapy regimens. Br J Haematol. 2016;177:147–150. doi: 10.1111/bjh.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner M, Bilinska M, Pokryszko-Dragan A, Sobczynski M, Cyrul M, Kusnierczyk P, et al. ALCAM and CD6—multiple sclerosis risk factors. J Neuroimmunol. 2014;276:98–103. doi: 10.1016/j.jneuroim.2014.08.621. [DOI] [PubMed] [Google Scholar]
- 30.Zhou P, Du L-F, Lv G-Q, Yu X-M, Gu Y-L, Li J-P, et al. Functional polymorphisms in CD166/ALCAM gene associated with increased risk for breast cancer in a Chinese population. Breast Cancer Res Treat. 2011;128:527–534. doi: 10.1007/s10549-011-1365-x. [DOI] [PubMed] [Google Scholar]
- 31.Varadi V, Bevier M, Grzybowska E, Johansson R, Enquist-Olsson K, Henriksson R, et al. Genetic variation in ALCAM and other chromosomal instability genes in breast cancer survival. Breast Cancer Res Treat. 2012;131:311–319. doi: 10.1007/s10549-011-1765-y. [DOI] [PubMed] [Google Scholar]
- 32.De Jager PL, Jia X, Wang J, de Bakker PIW, Ottoboni L, Aggarwal NT, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swaminathan B, Cuapio A, Alloza I, Matesanz F, Alcina A, García-Barcina M, et al. Fine mapping and functional analysis of the multiple sclerosis risk gene CD6. PLoS One. 2013;8:e62376. doi: 10.1371/journal.pone.0062376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahlehoff O, Skov L, Gislason G, Lindhardsen J, Kristensen SL, Iversen L, et al. Cardiovascular disease event rates in patients with severe psoriasis treated with systemic anti-inflammatory drugs: a Danish real-world cohort study. J Intern Med. 2013;273:197–204. doi: 10.1111/j.1365-2796.2012.02593.x. [DOI] [PubMed] [Google Scholar]
- 35.Gelfand JM, Shin DB, Neimann AL, Wang X, Margolis DJ, Troxel AB. The risk of lymphoma in patients with psoriasis. J Invest Dermatol. 2006;126:2194–2201. doi: 10.1038/sj.jid.5700410. [DOI] [PubMed] [Google Scholar]
- 36.Gelfand JM, Neimann AL, Shin DB, Wang X, Margolis DJ, Troxel AB. Risk of myocardial infarction in patients with psoriasis. JAMA. 2006;296:1735. doi: 10.1001/jama.296.14.1735. [DOI] [PubMed] [Google Scholar]
- 37.Gelfand JM, Troxel AB, Lewis JD, Kurd SK, Shin DB, Wang X, et al. The risk of mortality in patients with psoriasis: results from a population-based study. Arch Dermatol. 2007;143:1493–1499. doi: 10.1001/archderm.143.12.1493. [DOI] [PubMed] [Google Scholar]
- 38.Kurd SK, Troxel AB, Crits-Christoph P, Gelfand JM, Catalin MP, Hywel W, et al. The risk of depression, anxiety, and suicidality in patients with psoriasis: a population-based cohort study. Arch Dermatol. 2010;146:891–895. doi: 10.1001/archdermatol.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt J, Wozel G. The psoriasis area and severity index is the adequate criterion to define severity in chronic plaque-type psoriasis. Dermatology. 2005;210:194–199. doi: 10.1159/000083509. [DOI] [PubMed] [Google Scholar]
- 40.Henseler T, Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol. 1985;13:450–456. doi: 10.1016/S0190-9622(85)70188-0. [DOI] [PubMed] [Google Scholar]
- 41.Ferrándiz C, Pujol RM, García-Patos V, Bordas X, Smandía JA. Psoriasis of early and late onset: a clinical and epidemiologic study from Spain. J Am Acad Dermatol. 2002;46:867–873. doi: 10.1067/mjd.2002.120470. [DOI] [PubMed] [Google Scholar]
- 42.Langley RGB, Krueger GG, Griffiths CEM. Psoriasis: epidemiology, clinical features, and quality of life. Ann Rheum Dis 2005; 64 (Suppl 2): ii18–ii23; discussion ii24–ii25. [DOI] [PMC free article] [PubMed]
- 43.Swindell WR, Michaels KA, Sutter AJ, Diaconu D, Fritz Y, Xing X, et al. Imiquimod has strain-dependent effects in mice and does not uniquely model human psoriasis. Genome Med. 2017;9:24. doi: 10.1186/s13073-017-0415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu L, Schaid DJ, Sicotte H, Wieben ED, Li H, Petersen GM. Case-only exome sequencing and complex disease susceptibility gene discovery: study design considerations. J Med Genet. 2015;52:10–16. doi: 10.1136/jmedgenet-2014-102697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarrias MR, Gronlund J, Padilla O, Madsen J, Holmskov U, Lozano F. The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit Rev Immunol. 2004;24:1–38. doi: 10.1615/CritRevImmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- 46.Krintel SB, Essioux L, Wool A, Johansen JS, Schreiber E, Zekharya T, et al. CD6 and syntaxin binding protein 6 variants and response to tumor necrosis factor alpha inhibitors in Danish patients with rheumatoid arthritis. PLoS One. 2012;7:e38539. doi: 10.1371/journal.pone.0038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng M, Zhang L, Yu H, Hu J, Cao Q, Huang G, et al. Genetic polymorphisms of cell adhesion molecules in Behcet’s disease in a Chinese Han population. Sci Rep. 2016;6:24974. doi: 10.1038/srep24974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamzaoui K. Th17 cells in Behçet’s disease: a new immunoregulatory axis. Clin Exp Rheumatol. 2011;29:S71–S76. [PubMed] [Google Scholar]
- 49.Dos Passos GR, Sato DK, Becker J, Fujihara K. Th17 cells pathways in multiple sclerosis and neuromyelitis optica spectrum disorders: pathophysiological and therapeutic implications. Mediat Inflamm. 2016;2016:5314541. doi: 10.1155/2016/5314541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marinoni B, Ceribelli A, Massarotti MS, Selmi C. The Th17 axis in psoriatic disease: pathogenetic and therapeutic implications. Auto Immun Highlights. 2014;5:9–19. doi: 10.1007/s13317-013-0057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Egeberg A, Mallbris L, Gislason GH, Skov L, Hansen PR. Risk of multiple sclerosis in patients with psoriasis: a Danish Nationwide Cohort Study. J Invest Dermatol. 2016;136:93–98. doi: 10.1038/JID.2015.350. [DOI] [PubMed] [Google Scholar]
- 52.Bughani U, Saha A, Kuriakose A, Nair R, Sadashivarao RB, Venkataraman R, et al. T cell activation and differentiation is modulated by a CD6 domain 1 antibody Itolizumab. PLoS One. 2017;12:e0180088. doi: 10.1371/journal.pone.0180088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zúñiga LA, Jain R, Haines C, Cua DJ. Th17 cell development: from the cradle to the grave. Immunol Rev. 2013;252:78–88. doi: 10.1111/imr.12036. [DOI] [PubMed] [Google Scholar]
- 54.Hawkes JE, Gudjonsson JE, Ward NL. The snowballing literature on imiquimod-induced skin inflammation in mice: a critical appraisal. J Invest Dermatol. 2017;137:546–549. doi: 10.1016/j.jid.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kofler DM, Severson CA, Mousissian N, De Jager PL, Hafler DA. The CD6 multiple sclerosis susceptibility allele is associated with alterations in CD4+ T cell proliferation. J Immunol. 2011;187:3286–3291. doi: 10.4049/jimmunol.1100626. [DOI] [PubMed] [Google Scholar]
- 56.Castro MAA, Oliveira MI, Nunes RJ, Fabre S, Barbosa R, Peixoto A, et al. Extracellular isoforms of CD6 generated by alternative splicing regulate targeting of CD6 to the immunological synapse. J Immunol. 2007;178:4351–4361. doi: 10.4049/jimmunol.178.7.4351. [DOI] [PubMed] [Google Scholar]
- 57.Puig L, Bordas X, Carrascosa JM, Daudén E, Ferrándiz C, Hernanz JM, et al. Consensus document on the evaluation and treatment of moderate-to-severe psoriasis. Spanish Psoriasis Group of the Spanish Academy of Dermatology and Venereology. Actas Dermosifiliogr. 2009;100:277–286. doi: 10.1016/S0001-7310(09)70821-X. [DOI] [PubMed] [Google Scholar]
- 58.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


