Abstract
Pathogenicity islands (PAIs) are large genomic regions that contain virulence genes, which aid pathogens in establishing infections. While PAIs in clinical strains (strains isolated from a human infection) are well-studied, less is known about the occurrence of PAIs in strains isolated from the environment. In this study we describe three PAIs found in environmental Vibrio vulnificus and Vibrio parahaemolyticus strains, as well as a genomic fitness island found in a Vibrio diabolicus strain. All four islands had markedly different GC profiles than the rest of the genome, indicating that all of these islands were acquired via lateral gene transfer. Genes on the PAIs and fitness island were characterized. The PAI found in V. parahaemolyticus contained the tdh gene, a collagenase gene, and genes involved in the type 3 secretion system II (T3SS2). A V. vulnificus environmental strain contained two PAIs, a small 25 kbp PAI and a larger 143 kbp PAI. Both PAIs contained virulence genes. Toxin–antitoxin (TA) genes were found in all three species: on the V. diabolicus fitness island, and on the V. parahaemolyticus and V. vulnificus PAIs.
Keywords: Vibrio parahaemolyticus, Vibrio vulnificus, Pathogenicity islands
Introduction
Vibrio parahaemolyticus and Vibrio vulnificus can cause illnesses in humans, with an estimated 80,000 cases occurring annually in the United States (Scallan et al. 2011; CDC 2017). The hospitalization and mortality rates of V. parahaemolyticus gastroenteritis are 22% and 1%, respectively (Scallan et al. 2011). Although cases are usually mild and tend to resolve themselves after 1–3 days, V. parahaemolyticus is responsible for the majority of vibriosis cases (Scallan et al. 2011). V. vulnificus cases are less common; only about 100 occur each year in the United States. However, the hospitalization and mortality rates of this bacterium are much higher, at 92% and 35%, respectively (Scallan et al. 2011). V. vulnificus also causes sepsis and necrotizing fasciitis if it enters the body through an open wound. The majority of reported V. vulnificus cases are from wound infections (45%) and septicemia (43%); only 5% are gastroenteritis (Scallan et al. 2011). The mortality rate of V. vulnificus when it invades the bloodstream (sepsis) increases to 60%. Pathogenesis of both species is complex, and while some virulence factor genes have been implicated, the mechanisms underlying V. vulnificus and V. parahaemolyticus virulence are not well understood (Broberg et al. 2011; Lovell 2017; Klein and Lovell 2016).
Pathogenicity islands (PAIs), a subgroup of genomic islands that aid in and contribute to pathogenesis, have been found in clinical strains of both V. vulnificus and V. parahaemolyticus. PAIs are large chromosomal regions that are flanked by tRNA genes, and are usually associated with mobile genetic elements, such as phage, plasmid, integron, and transposon genes. A genomic island must contain at least one virulence gene, or gene that contributes to pathogenesis, to be considered a PAI. The size of PAIs ranges from 10 to 200 kbp (Schmidt and Hensel 2004; Hacker and Kaper 2000; Hacker and Carniel 2000) and the average Vibrio genome is 4.5 mbp (Pipes et al. in preparation), meaning that a single PAI could make up as much as 4% of a Vibrio genome. PAIs are flanked by highly conserved tRNA genes that act as both integration and excision sites. The majority (~ 75%) of PAIs discovered have tRNA flanking sequences (Schmidt and Hensel 2004; Hacker and Kaper 2000). Additionally, tRNA loci are often found on extrachromosomal elements, such as plasmids and bacteriophages. This indicates that the most likely mechanism for extrachromosomal element insertion is homologous recombination between the extrachromosomal element tRNA and PAI flanking tRNA loci (Hacker and Kaper 2000).
There is considerable evidence that PAIs are acquired horizontally via one or more lateral transfer events. Within some PAIs there is evidence of one large transfer event, while other PAIs are more “mosaic-like.” The “mosaic-like” composition of certain PAIs is caused by multiple, independent lateral transfer events (Hacker and Kaper 2000; Schmidt and Hensel 2004). PAIs usually differ in codon usage biases and have a markedly lower or higher GC content than the rest of the genome (Schmidt and Hensel 2004; Hacker and Kaper 2000; Hacker et al. 1997; Hacker and Carniel 2000). This supports the idea that recognizable PAIs are incorporated into a genome via lateral gene transfer from a dissimilar or unrelated organism (donor) having differing GC content and codon usage than the recipient (Schmidt and Hensel 2004). However, PAI GC content may not differ from that of the core genome if the donor and recipient microorganisms are closely related (Hacker and Kaper 2000). Dissimilarities in base composition confirm that detectable lateral transfer of PAIs must have been of recent origin, as insufficient time for genetic drift has passed (Schmidt and Hensel 2004).
PAIs have been found in clinical strains of V. vulnificus and V. parahaemolyticus (e.g. Makino et al. 2003; Wang et al. 2006; Sugiyama et al. 2008; Quirke et al. 2006; Cohen et al. 2007). Nine PAIs have been identified in V. parahaemolyticus, with VPAI-1 and VPAI-7 (V. parahaemolyticus pathogenicity island one and V. parahaemolyticus pathogenicity island seven) being the most studied (Ceccarelli et al. 2013). VPAI-1 is a 22 kbp island that is found on chromosome 1 in some strains, and chromosome 2 in others (Wang et al. 2006; Chen et al. 2011). This observation provides evidence for the mobility of this genomic island. VPAI-7 is the largest Vibrio genomic island found to date. This island contains the virulence factors TDH (thermostable direct hemolysin) and type III secretion system 2 (T3SS2) (Makino et al. 2003; Sugiyama et al. 2008). Other names for VPAI-7 include VPaIα or tdhVPA (Xu et al. 2017) and parts of VPAI-7 have been found in other Vibrio species, such as Vibrio mimicus (Gennari et al. 2011).
Genomic islands have been found in V. vulnificus clinical strains YJ016 and CMCP6, with 14 regions ranging in size from 14 to 117 kpb. A superintergon (SI) and nine V. vulnificus genomic islands (VVI–I to VVI–IX) have been found in these clinical strains. PAIs have not been detected in environmentally derived V. vulnificus strains (Quirke et al. 2006). V. vulnificus VVI–I has been found in the Vibrio cholerae biotype El Tor and O139 serogroup. The functional role of this island has not been determined but its presence in V. cholerae supports the idea that these regions can be transferred to other closely related species (O’Shea et al. 2004).
Work on Vibrio PAIs is heavily skewed toward clinical strains, with the pathogenic potential of naturally-occurring (environmental) strains rarely considered. In this study, we characterized four genomic islands found in environmental Vibrio strains: a PAI within a V. parahaemolyticus strain, two novel PAIs within a V. vulnificus strain, and a novel fitness island found in a Vibrio diabolicus strain. Environmental Vibrio strains, and the PAIs within them, could serve as reservoirs for virulence genes.
Materials and methods
Strain isolation
Environmental V. parahaemolyticus and V. diabolicus strains were isolated previously (Gutierrez West et al. 2013; Klein et al. 2014) from the pristine North Inlet salt marsh estuary near Georgetown, SC, USA (33°20′N, 79°12′W). Environmental V. vulnificus strains were also isolated near Georgetown, SC; however, they were isolated from lower salinity waters in Winyah Bay and the Waccamaw River. Water samples were plated on CHROMagar Vibrio (DRG International, NJ, USA) for isolation of V. vulnificus strains following the US Food and Drug Administration protocol (DePaola and Kaysner 2004). Vibrio strains were routinely cultivated on saline Luria Agar (SLA; per L; 10 g tryptone, 5 g yeast extract, 27 g NaCl, 15 g Bacto Agar). V. parahaemolyticus TS-8-11-4 and V. diabolicus JBS-8-11-1 were deposited into the DSMZ Public Culture Collection and were assigned their respective accession numbers: DSM 107522 and DSM 107521.
Whole genome sequencing
Genomic DNA was isolated through the Wizard Genomic DNA Purification kit following the protocol for Gram negative organisms (Promega, Madison, WI, USA). After DNA was extracted, DNA quantity was measured via Quibit fluorimetry. Libraries were prepared and then sequenced using an Illumina MiSeq (V3 26300 base) at the Indiana University Center for Genomic Studies as a part of the Genome Consortium for Active Teaching NextGenSequencing Group (GCAT-SEEK) shared run (Buonaccorsi et al. 2011, 2014). Sequencing reads were filtered (median phred score 0.20), trimmed (phred score 0.16), and assembled using the paired-end de novo assembly option in NextGENe V2.3.4.2 (SoftGenetics, State College, PA, USA). The assembled genomes were uploaded to the Rapid Annotation with Subsystem Technology (RAST) web service (Aziz et al. 2008; Overbeek et al. 2005, 2014) for analysis, guided contig reordering and assembly improvement. Genomes were aligned based on completed sequences using dotplot comparisons. Whole genome sequence data obtained from this work were submitted to the NCBI GenBank and assigned the accession numbers: PKQA00000000, PKPY00000000, and PKPZ00000000.
PAI detection and characterization
The fully sequenced genomes were uploaded to TUBIC (Tiajin University Bioinformatics Center) to determine their GC profiles (http://tubic.tju.edu.cn/). This tool displays GC content variation across a genome and can be useful for identifying genomic regions that differ from the rest of the genome in GC content (Gao and Zhang 2006). Genomic islands that were detected via TUBIC were isolated and the island nucleotide sequence was uploaded to RAST to identify and characterize the specific genes found on the genomic islands (http://rast.nmpdr.org/). NCBI GenBank was also used to characterize genomic island genes (http://www.ncbi.nlm.nih.gov/genbank/). Gene sequences of interest were edited and maximum-likelihood trees were constructed using the Kimura 2-parameter model with Mega version 7. DNAPlotter was used to visualize the circular chromosomes of the Vibrio strains (Carver et al. 2009).
Results
V. parahaemolyticus island
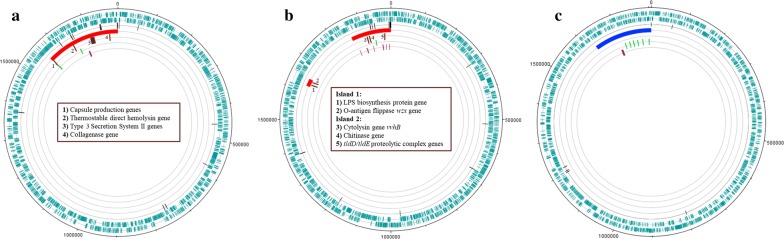
Vibrio parahaemolyticus strain TS-8-11-4 was isolated from salt marsh sediments (Gutierrez West et al. 2013; Klein et al. 2014) at the pristine North Inlet estuary in South Carolina, USA. This strain had a genome of 4.98 mbp; chromosome 1 was 3.19 mbp in length and chromosome 2 was 1.78 mbp in length. The majority of the genome contained a GC content of 45.57%, which is typical for V. parahaemolyticus (Farmer and Janda 2005). However, this strain contained a 223 kbp island that had a markedly lower GC content (41.5%) not typical of V. parahaemolyticus. The majority (69%) of genes on the TS-8-11-4 PAI could not be assigned specific identities and were thus designated hypothetical. The genomic island of V. parahaemolyticus TS 8-11-4 was on the second chromosome of and it harbored virulence genes (Fig. 1a). The virulence factor genes that were found on this island included the thermostable direct hemolysin gene, genes involved in the type three secretion system II (T3SS2), a collagenase gene, as well as capsule production genes.
Fig. 1.
a–c Circular presentation of the second chromosome of a Vibrio parahaemolyticus environmental strain TS-8-11-4, b Vibrio vulnificus environmental strain WR-2-BW, and c Vibrio diabolicus environmental strain JBS-8-11-1. Track 1, forward coding sequences; track 2, reverse coding sequences; track 3, tRNA genes; track 4, red, pathogenicity islands, blue, genomic fitness islands; track 5, virulence and virulence-associated genes; track 6, genes involved in toxin–antitoxin systems; track 7, mobile genetic elements. Virulence and virulence-associated genes are numbered and are defined via the center text boxes
V. vulnificus islands
Vibrio vulnificus strain WR-2-BW was isolated near Georgetown, SC from Waccamaw River waters. Its genome (4.96 mpb) contained two chromosomes, the first chromosome larger (2.96 mbp) than the second (1.99 mbp). The average GC content of V. vulnificus ranges from 46 to 48% (Farmer and Janda 2005), and the average GC content of strain WR-2-BW was 46.83%. Two regions within the genome had GC contents that were markedly lower from the rest of the genome. The first region had a GC content of 38.2% and the second region had a GC content of 42.5%; both of which are lower than the typical GC content of V. vulnificus strains. These regions were found on the second chromosome (Fig. 1b). The first region was a 25 kbp island and the second region was a 143 kbp island. The 25 kbp island was 30 genes in length, and had two genes that had virulence-related functions, which include a putative LPS biosynthesis protein gene and an O-antigen flippase wzx gene. The 143 kbp island contained the cytolysin gene vvhB, a chitinase gene, tldD/tldE proteolytic complex genes, and Type IV secretory pathway components. The 143 kbp genomic island was comprised of 160 genes in total, 63% of which were characterized as hypothetical or had unknown function.
V. diabolicus island
Vibrio diabolicus strain JBS-8-11-1 was isolated previously from North Inlet salt marsh sediments (Gutierrez West et al. 2013; Klein et al. 2014). Its genome (5.04 mbp) was comprised of two chromosomes, the first (3.23 mbp) being larger than the second (1.81 mbp). Its GC content was typical of other V. diabolicus genomes (44.91%) (Goudenege et al. 2014), except for a 182 kbp island, located on chromosome 2, which had a GC content of 40.8%. Eighty-two percentage of the island consisted of hypothetical genes. This island harbored no known virulence genes; it is hereafter referred to as a fitness island (Fig. 1c). Three genes, a phage DNA synthesis gene, a phage DNA replication gene, and a gene encoding a phage capsid protein, were located very close to each other on the fitness island. Thirteen genes involved in toxin–antitoxin (TA) systems were located on the fitness island.
Discussion
The genomic island of V. parahaemolyticus TS 8-11-4 was deemed a PAI due to the presence of virulence genes on this island, despite its environmental origin (Schmidt and Hensel 2004; Hasan et al. 2010; Dobrindt et al. 2004). The thermostable direct hemolysin gene (tdh) was found on this island, as well as genes involved in the type three secretion system II (T3SS2). Both the tdh gene and T3SS2 complex are the two major virulence factors implicated in V. parahaemolyticus pathogenesis (Makino et al. 2003; Park et al. 2004; Yanagihara et al. 2010). A collagenase gene was found on the island; collagenase is thought to be involved in V. parahaemolyticus virulence (Gode-Portratz et al. 2011). The genomic island of V. parahaemolyticus strain TS-8-11-4 is a PAI, and more specifically, because it contains tdh and T3SS2 genes, we designate this island as a VPAI-7 (VPaIα or tdhVPA) (Makino et al. 2003; Sugiyama et al. 2008; Xu et al. 2017).
Four genes involved in capsule production, as well as one integrase gene, and a Na+/H+ antiporter (nhaA) were also found on this PAI. Capsules aid pathogens in evasion of host immune defenses, establishing infections, and survival in harsh environments, such as the stomach. V. parahaemolyticus virulence is correlated with capsule production (Broberg et al. 2011; Letchumanan et al. 2014). One capsule gene had high homology with Gram positive capsule production genes. This is interesting because vibrios are Gram negative organisms, so this gene may have been acquired laterally. An integrase gene was found near the center of the island. Integrase genes are associated with PAIs and function to integrate foreign DNA into the genome (Hacker and Kaper 2000). Usually VPAI-7 does not contain an integrase gene, but a few transposon genes instead (Ceccarelli et al. 2013). Finally, we determined that a nhaA gene is located on this genomic island. nhaA genes encode Na+/H+ antiporters, which transport ions to balance pH. Na+/H+ antiporters aid V. cholerae in environmental persistence (Vimont and Berche 2000) and are essential for Yersinia pestis virulence (Minato et al. 2013).
Similar to V. parahaemolyticus, the two islands found for the V. vulnificus WR-2-BW strain are characterized as PAIs due to the presence of virulence genes and virulence-related genes. Two of these genes had virulence-related functions, a putative LPS biosynthesis protein gene and an O-antigen flippase wzx gene. These genes are virulence-associated factors, as they do not directly cause host cell damage, but they do contribute to pathogenesis, aiding in the establishment of infections. Lipopolysaccharide (LPS) is a main component of the outer membrane of Gram negative bacteria, and is a known pyrogen (fever-producing agent) (McPherson et al. 1991; Jones and Oliver 2009). Phylogenies show that the LPS biosynthesis protein gene from V. vulnificus WR-2-BW was closely related to an LPS biosynthesis protein gene from a Vibrio coralliilyticus species. The O-antigen flippase wzx gene is part of the major class of O-antigen gene clusters, and it encodes a hydrophobic protein with 12 potential transmembrane segments (Liu et al. 1996).
A cytolysin secretion gene, vvhB, was found also found on the 143 kbp V. vulnificus island. Cytolysins lyse erythrocytes by forming small pores in the cytoplasmic membrane or binding to cholesterol to interrupt potassium and sodium ion channels (Choi et al. 2002). In V. vulnificus, the expression and mechanism of cytolysins vvhA and vvhB are not fully understood, however, they are both believed to play a role in pathogenicity (Choi et al. 2002). They are homologous to a known V. cholerae El Tor hemolysin (Choi et al. 2002; Yamamoto et al. 1990). Phylogenies show that the vvhB gene in the V. vulnificus WR-2-BW strain was 99% identical to other V. vulnificus vvhB genes from other strains.
Other genes of interest on the 143 kbp PAI include a chitinase gene, tldD/tldE proteolytic complex genes, and type IV secretory pathway components. In Escherichia coli, it was shown that the TldD and TldE proteins could be involved in regulating gyrase function as well as aiding in proteolytic activity (Allali et al. 2002). The chitinase gene had a 99% blast identity score to the chitinase gene found in the V. vulnificus YJ016 strain; however, the chitinase gene in YJ016 is located on the first chromosome and WR-2-BW’s chitinase gene is located in the second chromosome. Chitinous exoskeletal materials of invertebrates can be a source of carbon and nitrogen for bacteria; vibrios in particular have a well-known association with marine copepods (Kaneko and Colwell 1975; Lovell 2017). V. cholerae has a well-studied association with copepods, which commonly serve as a vector of cholera infections in Bangladesh water systems (Tamplin et al. 1990). Chitinase has been identified as part of the mechanism for adsorption and attachment to copepods, which relates to its ability to colonize its host and degrade the host exoskeleton, increasing the overall ecological fitness of the vibrios (Huq et al. 1983; Nalin et al. 1979; Bhowmick et al. 2006).
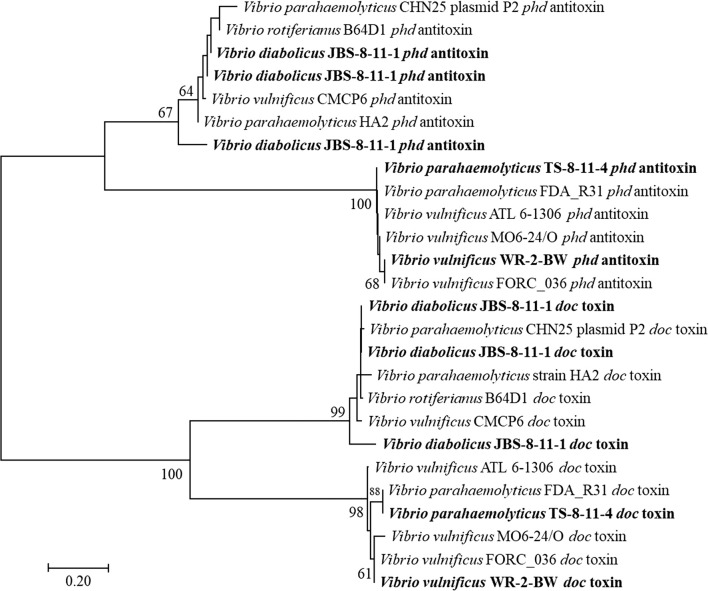
Vibrio diabolicus had a large genomic island that did not contain any virulence factors or virulence associated genes, which we defined as a fitness island, as it contained genes that would aid the organism in persistence in the environment. Toxin–antitoxin (TA) systems are found either on plasmids, genomic islands, or within the chromosome and are made up of closely linked toxin and antitoxin genes. The encoded labile antitoxin protects the host from the stable toxin, while competitor cells that do not have the TA system (and respective antitoxin) are eliminated (Hayes 2003; Van Melderen and Saavedra 2009). Sometimes TA systems are referred to as “addiction modules” because the host cell is dependent on the antitoxin (Van Melderen and Saavedra 2009). The toxin and respective antitoxin loci are usually found neighboring each other, often overlapping (Hayes 2003). Seven type II TA toxins were found on JBS-8-11-1’s fitness island, along with their neighboring respective antitoxins. Type I TAs include RNA antitoxins, while type II TAs have protein antitoxins (Hayes 2003). The relE, yafQ, and yoeB toxin genes encode mRNA interferase endoribonucleases; all three of these toxin genes were detected on this fitness island. The doc toxin gene (death on curing) inhibits translation by blocking translation elongation at the 30S ribosomal subunit (Lui et al. 2008); three copies of the doc toxin gene and three copies of its antitoxin partner gene, phd (prevent host death) were found on JBS-8-11-1’s fitness island. doc toxin genes and phd antitoxin genes are widespread in vibrios and were also found on V. parahaemolyticus strain TS-8-11-4’s PAI as well as V. vulnificus strain WR-2-BW’s PAI (Fig. 2).
Fig. 2.
Maximum-likelihood phylogeny (Kimura 2-parameter model) of doc toxin genes and phd antitoxin genes. Bold indicates sequences obtained from this study. The bootstrap values represent 1000 replications, and values of less than 50 are not shown. The reference sequences were acquired from NCBI GenBank
Lateral gene transfer in environmental strains
PAIs are present in environmental Vibrio strains and are most likely acquired via lateral gene transfer. All four of the islands described here have significant lower GC content than the rest of the genome, providing evidence that these islands originated from a foreign source and were transferred into these genomes relatively recently. Additional evidence includes mobile genetic elements, such as phage and plasmid genes, integrases, and transposons. Virulence loci on VPAI-7 have been detected in environmental species that do not cause human infections: Vibrio mimicus, Vibrio harveyi, and Vibrio natriegens (Gennari et al. 2011; Klein et al. 2014). Clearly, lateral transfer of individual virulence loci and/or entire PAIs is occurring between and among environmental vibrios. It is well documented that V. cholerae enters a natural competency state in the presence of chitin or under low-nutrient conditions (Hazen et al. 2010; Metzger and Blokesch 2016); however, less is known about uptake of exogenous DNA by other Vibrio species. Further studies examining the rates of lateral transfer among vibrios in the environment are needed. Vibrios survive, persist and can undergo rapid population expansions (bloom) in coastal ecosystems. Consequently, the pathogenicity loci (and potential of said loci to be transferred laterally) of naturally occurring environmental strains are clearly important.
Authors’ contributions
All authors (SK, SP, and CL) conceived and designed the experiments. SK was responsible for sample collection and strain isolation. SP performed the whole genome sequence annotation and assembly. Both authors (SK, SP) used bioinformatic techniques and tools to identify and characterize PAIs in environmental Vibrio genomes. CL directed the project and obtained funding; all authors (SK, SP, and CL) wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank the Belle W. Baruch Institute for access to the pristine North Inlet Estuary near Georgetown, SC, USA. We would also like to thank Dr. Daniel Tufford for access to environmental V. vulnificus strains and Dr. Bob Friedman for helpful bioinformatic discussions.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Data will be made available through publication and GenBank.
Compliance with ethical standards
This article does not contain any studies with human participants or animals performed by the author.
Consent for publication
All authors listed on this manuscript have read and agreed to the publication of this research.
Funding
Genomic sequencing was supported by the office of the Vice President for Research, University of South Carolina, 2015 South Carolina Floods Research Initiative.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- PAI
pathogenicity island
- TA
toxin–antitoxin
- LPS
lipopolysaccharide
- T3SS2
type three secretion system II
- tdh
thermostable direct hemolysin gene
Contributor Information
Savannah Klein, kleinsl@email.sc.edu.
Shannon Pipes, spipes@email.sc.edu.
Charles R. Lovell, Phone: 803-777-5153, Email: lovell@biol.sc.edu
References
- Allali N, Afif H, Couturier M, Van Melderen L. The highly conserved TldD and TldE proteins of Escherichia coli are involved in microcin B17 processing and in CcdA degradation. J Bacteriol. 2002;184(12):3224–3231. doi: 10.1128/JB.184.12.3224-3231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick R, Ghosal A, Chatterjee NS. Effect of environmental factors on expression and activity of chitinase genes of vibrios with special reference to Vibrio cholerae. J Appl Micobiol. 2006;103:97–108. doi: 10.1111/j.1365-2672.2006.03253.x. [DOI] [PubMed] [Google Scholar]
- Broberg CA, Calder TJ, Orth K. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 2011;13:992–1001. doi: 10.1016/j.micinf.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaccorsi VP, Boyle MD, Grove D, Praul C. GCATSEEKquence: genome consortium for active teaching of undergraduates through increased faculty access to next-generation sequencing data. CBE Life Sci Educ. 2011;10:342–345. doi: 10.1187/cbe.11-08-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaccorsi VP, Peterson M, Lamendella G, Newman JD, Trun N. Vision and change through the genome consortium for active teaching using next-generation sequencing (GCAT-SEEK) CBE Life Sci Educ. 2014;13:1–2. doi: 10.1187/cbe.13-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25(1):119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D, Hasan NA, Huq A, Colwell RR. Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front Cell Infect Microbiol. 2013 doi: 10.3389/fcimb.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2017) Vibrio species causing vibriosis. Atlanta, GA. https://www.cdc.gov/vibrio/index.html
- Chen Y, Stine OC, Badger JH. Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence. BMC Genomics. 2011;12:294. doi: 10.1186/1471-2164-12-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Park NY, Kim D, Chung HJ, Ryu S, Choi SH. Promoter analysis and regulatory characteristics of vvhBA encoding cytolytic hemolysin of Vibrio vulnificus. J Biol Chem. 2002;277:47292–47299. doi: 10.1074/jbc.M206893200. [DOI] [PubMed] [Google Scholar]
- Cohen AL, Oliver JD, DePaola A, Fiel EJ, Boyd EF. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl Environ Microbiol. 2007;17(73):5553–5565. doi: 10.1128/AEM.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaola A, Kaysner CA (2004) Vibrio. Bacteriological analytical manual online. U.S. Food and Drug Administration, Washington, DC. http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070830.htm
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Revi Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- Farmer JJ, Janda JM. Family I. Vibrionaceae Véron, 1965, 5245AL. In: Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s manual of systematic bacteriology. 2. NY: New York; 2005. pp. 491–555. [Google Scholar]
- Gao F, Zhang C. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006;34:W686–W691. doi: 10.1093/nar/gkl040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennari M, Ghinidi V, Carbulutto G, Lleo MM. Virulence genes and pathogenicity islands in environmental Vibrio strains nonpathogenic to humans. FEMS Microbiol Ecol. 2011;82:563–573. doi: 10.1111/j.1574-6941.2012.01427.x. [DOI] [PubMed] [Google Scholar]
- Gode-Portratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, McCarter L. Surfing sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol. 2011;79(1):240–263. doi: 10.1111/j.1365-2958.2010.07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudenege D, Boursicot V, Versigny T, Bonnetot S, Ratiskol J, Sinquin C, LaPointe G, Roux F, Delbarre-Ladrat C. Genome sequence of Vibrio diabolicus and identification of the exopolysaccharide HE800 biosynthesis locus. Appl Microbiol Biotechnol. 2014;98:10165–10176. doi: 10.1007/s00253-014-6086-8. [DOI] [PubMed] [Google Scholar]
- Gutierrez West CK, Klein SL, Lovell CR. The virulence factor genes tdh, trh and tlh occur at high frequency in Vibrio parahaemolyticus isolated from a pristine estuary. Appl Environ Microbiol. 2013;79:2247–2252. doi: 10.1128/AEM.03792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Carniel E. Review: ecological fitness, genomic islands and bacterial pathogenicity: a darwinian view of the evolution of microbes. EMBO Rep. 2000;2(5):376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annual Rev Microbiol. 2000;23(6):1089–1097. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- Hacker J, Blum-Oehler G, Muldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- Hasan NA, Grim CJ, Haley BJ, Chun J, Alam M, Taviani E, Hoq M, Munk AC, Saunders E, Brettin TS, Bruce DC, Challocombe JF, Detter CJ, Han CS, Xie G, Nair B, Huq A, Colwell RR. Comparative genomics of clinical and environmental Vibrio mimicus. PNAS. 2010;107(49):21134–21139. doi: 10.1073/pnas.1013825107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F. Tonxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- Hazen TH, Pan L, Gu J, Sobecky PA. The contribution of mobile genetic elements to the evolution and ecology of vibrios. FEMS Microbiol Ecol. 2010;74:485–499. doi: 10.1111/j.1574-6941.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK, Oliver JD. Vibrio vulnificus: disease and pathogenesis. Infect Immun. 2009;77(5):1723–1733. doi: 10.1128/IAI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Colwell RR. Adsorption of Vibrio parahaemolyticus onto chitin and copepods. Appl Environ Microbiol. 1975;29(2):269–274. doi: 10.1128/am.29.2.269-274.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Lovell CR. The hot oyster: levels of virulent Vibrio parahaemolyticus strains in individual oysters. FEMS Microbiol Ecol. 2016;93(2):fiw232. doi: 10.1093/femsec/fiw232. [DOI] [PubMed] [Google Scholar]
- Klein SL, West CK, Mejia DM, Lovell CR. Genes similar to the Vibrio parahaemolyticus virulence related genes tlh, tdh, and vscC2 occur in other Vibrionaceae species isolated from a pristine estuary. Appl Environ Microbiol. 2014;80:595–602. doi: 10.1128/AEM.02895-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchumanan V, Kok-Gan C, Lee LH. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and molecular identification techniques. Front Microbiol. 2014;5:705–718. doi: 10.3389/fmicb.2014.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Cole RA, Reeves PR. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J Bacteriol. 1996;178(7):2102–2107. doi: 10.1128/jb.178.7.2102-2107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell CR. Ecological fitness and virulence features of Vibrio parahaemolyticus in estuarine environments. Appl Microbiol Biotechnol. 2017;101:1781–1794. doi: 10.1007/s00253-017-8096-9. [DOI] [PubMed] [Google Scholar]
- Lui M, Zhang Y, Inouye M, Woychik NA. Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. PNAS. 2008;105:5885–5890. doi: 10.1073/pnas.0711949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K, Oshima K, Kurokawa K, Yokoyama K, Takayuki U, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet. 2003;361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- McPherson VL, Watts JA, Simpson LM, Oliver JD. Physiological effects of the lipopolysaccharide of Vibrio vulnificus on mice and rats. Microbios. 1991;7(272–273):141–149. [PubMed] [Google Scholar]
- Metzger LC, Blokesch M. Regulation of competence-mediated horizontal gene transfer in the natural habitat of Vibrio cholerae. Curr Opin Microbiol. 2016;30:1–7. doi: 10.1016/j.mib.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Minato Y, Ghosh A, Faulkner WJ, Lind EJ. Na+/H+ antiporter is essential for Yersinia pestis virulence. Infect Immun. 2013;81(9):3163–3172. doi: 10.1128/IAI.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalin DR, Daya V, Reid A, Levine MM, Cisneros L. Adsorption and growth of Vibrio cholerae on chitin. Infec and Immun. 1979;25(2):768–770. doi: 10.1128/iai.25.2.768-770.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea YA, Finnan S, Reen FJ, Morrissey JP, O’Gara F, Boyd EF. The Vibrio seventh pandemic island-II is a 269 kb genomic island present in Vibrio cholerae El Tor and O139 serogroup isolates that shows homology to a 434 kb genomic island in V. vulnificus. Microbiol. 2004;150:4053–4063. doi: 10.1099/mic.0.27172-0. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. The SEED and the Rapid Annotation of microbial genomesusing Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, Honda T. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun. 2004;72:6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirke AM, Reen FJ, Boyd EF. Genomic island identification in Vibrio vulnificus reveals significant genome plasticity in this human pathogen. Bioinformatics. 2006;22(8):905–910. doi: 10.1093/bioinformatics/btl015. [DOI] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17(1):14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Iida T, Izutsu K, Park KS, Honda T. Precise region and the character of the pathogenicity island in clinical Vibrio parahaemolyticus strains. J Bacteriol. 2008;190(5):1835–1837. doi: 10.1128/JB.01293-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol. 1990;56:1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L, Saavedra DM. Bacterial toxin–antitoxin systems: more than just selfish entities? PLoS Genet. 2009;9(3):e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimont S, Berche P. NhaA, a Na+/H+ antiporter involved in environmental survival of Vibrio cholerae. J Bacteriol. 2000;182(10):2937–2944. doi: 10.1128/JB.182.10.2937-2944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wong MML, O’Toole D, Mak MMH, Wu RSS, Kong RYC. Identification of a DNA methyltransferase gene carried on a pathogenicity island-like element (VPAI) in Vibrio parahaemolyticus and its prevalence among clinical and environmental isolates. Appl Environ Microbiol. 2006;72(6):4455–4460. doi: 10.1128/AEM.02095-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Gonzalez-Escalona N, Drees KP, Sebra RP, Cooper VS, Jones SH, Whistler CA. Parallel evolution of two clades of an Altantic-endemic pathogenic lineage of Vibrio parahaemolyticus by independent acquisition of related pathogenicity islands. Appl Environ Microbiol. 2017;83(18):e01168-17. doi: 10.1128/AEM.01168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Wright AC, Kaper JB, Morris JG. The cytolysin gene of Vibrio vulnificus: sequence and relationship to the Vibrio cholerae El Tor hemolysin gene. Infect Immun. 1990;58(8):2706–2709. doi: 10.1128/iai.58.8.2706-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara I, Nakahira K, Yamane T, Kaieda S, Mayanagi K, Hamada D, Fukui T, Ohnishi K, Kajiyama S, Toshiyuki S, Sato M, Ikegami T, Ikeguchi M, Honda T, Hashimoto H. Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin. J Biol Chem. 2010;285:16267–16274. doi: 10.1074/jbc.M109.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available through publication and GenBank.