Abstract
Biological control using bacteriophages is a promising approach for mitigating the devastating effects of coral diseases. Several phages that infect Vibrio coralliilyticus, a widespread coral pathogen, have been isolated, suggesting that this bacterium is permissive to viral infection and is, therefore, a suitable candidate for treatment by phage therapy. In this study, we combined functional and genomic approaches to evaluate the therapeutic potential of BONAISHI, a novel V. coralliilyticus phage, which was isolated from the coral reef in Van Phong Bay (Vietnam). BONAISHI appears to be strictly lytic for several pathogenic strains of V. coralliilyticus and remains infectious over a broad range of environmental conditions. This candidate has an unusually large dsDNA genome (303 kb), with no genes that encode known toxins or implicated in lysogeny control. We identified several proteins involved in host lysis, which may offer an interesting alternative to the use of whole bacteriophages for controlling V. coralliilyticus. A preliminary therapy test showed that adding BONAISHI to an infected culture of Symbiodinium sp. cells reduced the impact of V. coralliilyticus on Symbiodinium sp. photosynthetic activity. This study showed that BONAISHI is able to mitigate V. coralliilyticus infections, making it a good candidate for phage therapy for coral disease.
Keywords: phage therapy, coral disease, Vibrio coralliilyticus, viral genomics, phage–host interactions
Introduction
Coral reefs are one of the most productive and diversified ecosystems on the planet (Connell, 1978) and they provide a wealth of ecological services as well as being economically important, supporting fisheries, tourism, and medical applications (Moberg and Folke, 1999; Hughes et al., 2003; Cooper et al., 2014). The health of these ecosystems is severely threatened by the combined effect of local anthropogenic pressures and global changes (Jackson et al., 2001; Hughes et al., 2003; Pandolfi et al., 2003; Bellwood et al., 2004). Over-exploitation of marine species, pollution, and increased sea surface temperature are associated with the emergence of coral diseases, which are contributing to the decline of coral reefs worldwide.
Several studies have identified Vibrio spp. (⋎-Proteobacteria) as causative agents of coral bleaching for multiple coral species and in multiple locations (Ushijima et al., 2014; reviewed in Mera and Bourne, 2018). Vibrio coralliilyticus (V. coralliilyticus) has emeregd as an important bacterial pathogen model for understanding the establishement and propagation of coral disease (Sussman et al., 2008; O'Santos et al., 2011; Garren et al., 2014; Pollock et al., 2015). Studies have shown that V. coralliilyticus infection is temperature dependent and infects the coral endosymbiont Symbiodinum through the production of proteases that inhibit photosynthesis. This results in the loss of the endosymbiont from the coral tissues and ultimately leads to coral bleaching (Ben-Haim et al., 2003; Sussman et al., 2008, 2009; Cohen et al., 2013). With the increasing devastation of coral reefs, the development of new tools and strategies to control pathogens and treat diseased corals is becoming a major issue. Currently, biocontrol strategies, such as phage therapy, are being seriously evaluated for mitigating coral diseases (Efrony et al., 2009; Teplitski and Ritchie, 2009; Atad et al., 2012; Cohen et al., 2013).
The potential of viruses (more specifically bacteria viruses also referred to as bacteriophages or phages) as therapeutic agents to treat infectious diseases has been known for a long time (d'Herelle, 1926; Duckworth, 1976; Duckworth and Gulig, 2002). The idea of phage therapy arose from the early discovery that a given virus usually infects a single host species, leaving the rest of the microbial community untouched. Moreover, viruses are obligate intracellular organisms and, therefore, their production is self-regulated and limited by the availability of suitable hosts. Over the past decade, there have been promising in vitro and in situ trials of phage therapy for corals. One used BA3, a virulent bacteriophage that infect the causative agent of white plague disease (Efrony et al., 2007, 2009), to inhibit the progression of the disease and its transmission to healthy corals (Atad et al., 2012). Although this research is still at an early stage, this promising result in the open sea suggest that in situ phage therapy for coral diseases is achievable (Atad et al., 2012). The isolation and characterization of new bacteriophages is, therefore, essential to increase the collection of potential candidates for therapeutic assays.
The therapeutic value of a candidate bacteriophage relies on the characterization of viral properties such as the virion stability, growth kinetics, viral yield, and host range. Understanding the lifestyle of the candidate phage is probably the key to their use in therapy. Only virulent bacteriophages, which replicate through a lytic cycle and kill their host after infection, will be suitable candidates. Temperate bacteriophages, which replicate using a lysogenic cycle, may improve host fitness through gene transfer. It is, however, difficult to distinguish between virulent and temperate phage because temperate viruses can switch to a lytic cycle in response to environmental changes, such as temperature, pH salinity, UV, pollution, or nutrient availability (Jiang and Paul, 1996; Williamson and Paul, 2006, reviewed in Howard-Varona et al., 2017). Furthermore, infection dynamics can be highly variable, even between two closely related hosts (Holmfeld et al., 2014; Dang et al., 2015). Over the past decade, genomics has greatly improved our understanding of host—virus interactions, and is the key to establishing whether a candidate is an obligate lytic bacteriophage (Howard-Varona et al., 2017). Bacteriophage genome sequencing is also essential for evaluating the safety of a candidate (absence of toxins and temperate phage hallmarks), and to provide information on the candidate's evolution history and ecology (Lobocka et al., 2014).
In this study, we report a novel bacteriophage, hereafter referred to as Vibrio phage BONAISHI that infects the model coral pathogen V. coralliilyticus. We studied it using a combination of functional, genomic, and metagenomic approaches to evaluate the potential of this phage for mitigating disease caused by V. coralliilyticus.
Material and methods
Virus isolation
Seawater samples were collected from coral surrounding water off Whale Island (Van Phong Bay, Vietnam). A 50 mL aliquot was supplemented with 10% (v/v) Marine Broth (MB, Difco) and the mixture was enriched with 1 mL Vibrio coralliilyticus LMG20984 (YB1) culture and incubated for 48 h at 25°C (Brussaard et al., 2016). The sample was clarified (7,000 g, 15 min) and the supernatant was filtered through 0.2 μm PES filters to separate the viral community. A 100 μL aliquot of the filtrate was added to 900 μL YB1 culture and incubated for 30 min at 25°C. The mixture was included in molten agar (Marine Broth supplemented with 0.6% noble agar) and spread on a Marine Agar plate. After 48 h incubation, a translucent plaque indicating host lysis was removed from the bacterial lawn, eluted in 0.22 μm filtered Salt Marine (SM) buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl pH 8.0) and combined with a host culture in a plaque assay (Brussaard et al., 2016). This procedure was repeated twice to ensure the clonality of the bacteriophage. Finally, the clonal phage suspension and host culture were used in a plaque assay giving confluent lysis. The plaques were eluted in SM buffer, the eluent was clarified by centrifugation (7,000 g, 30 min, 4°C), and the supernatant was filtered through 0.22 μm and stored at 4°C until use.
Transmission electronic microscopy
A 10 μL aliquot of the phage suspension was loaded onto a Formvar/carbon film coated 400 mesh copper grid (Euromedex). After 5 min incubation, the grid was blotted with filter paper and stained with 2% uranyl acetate for 30 s, blotted again to remove excess dye and air dried for 30 min (Ackermann and Heldal, 2010). The specimen was imaged using JEOL 1400 transmission electron microscope operating at 120 keV at a magnification of 80,000X.
Environmental range of infectivity
The tolerance of BONAISHI to temperature and pH ranges was evaluated by monitoring the loss of infectivity of a freshly produced suspension by spot test. For evaluating the temperature range, 100 μL of viral suspension (5 x 108 PFU/mL) was incubated at temperatures from 4 to 70°C for 30 min in a dry bath. The samples were then cooled for 5 min at 4°C and virus infectivity was assessed by spot test. Briefly, 5 μL of the treated viral suspension was spotted on a host lawn obtained by plating a 1:4 mixture of host culture in molten agar onto a marine agar plate. For evaluating the pH range, 100 μL of viral suspension were added to 900 μL SM buffer adjusted at pH ranging from 2 to 10. The samples were incubated for 24 h at 4°C and then spot-tested as described above.
Host range
A selection of 43 bacterial strains (Table S1) related to the original host V. coralliilyticus YB1 were used to determine the host specificities of BONAISHI. The ability of BONAISHI phage to infect these strains was determined by pairwise infection. The bacterial strains were grown in Marine Broth media (DIFCO) overnight. Each culture was included in molten agar (Marine Broth supplemented with 0.6% noble agar) and spread on a Marine Agar plate. A freshly produced suspension of BONAISHI was serially diluted in SM buffer and a 5 μL drop of each dilution was spotted on a bacterial lawn. After 24–48h incubation at 25°C, the formation of translucent spots, indicative of host lysis, was recorded.
Life strategy
The growth cycle of BONAISHI was tested on two V. coralliilyticus strains including the original host LMG 20984 (YB1) and the alternate host LMG 23696 (P1) (Middelboe et al., 2010). Both host cultures were grown in MB and divided into four 25 mL sub-cultures. One sub-culture served as control and the remaining 3 were inoculated with a freshly produced BONAISHI suspension at multiplicity of infection (MOI) of 0.1 as determined by flow cytometry (see below). All cultures were incubated at 25°C for 48 h. Samples for viral and bacterial counts were taken every hour for 10 h, and then every 4 h for 48 h. Samples were immediately fixed with glutaraldehyde (0.5% final concentration) for 10 min at 4°C, flash frozen in liquid nitrogen, and stored at −80°C until flow cytometry analysis (see below). Bacterial host and virus counts were used to calculate the phage latent period and burst size. The latent period corresponds to the time elapsed between the viral inoculation and the release of virions. The burst-size, which corresponds to the number of virions produced per infected host cell, was determined by the ratio of the net increase in virus concentration over the net decrease in host concentration during the first burst.
Flow cytometry
For determining the bacterial abundance, samples were diluted in autoclaved 0.2 μm filtered TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and stained with a SYBR Green (10,000-fold dilution of commercial solution) for 15 min in the dark at ambient temperature. For determining the viral abundance, 100–1,000-fold diluted samples were stained with SYBR Green (20,000-fold dilution of commercial solution) for 10 min in the dark at 80°C. Analyses were performed using a FACS Canto II equipped with an argon-laser (455 nm). The trigger was set on the green fluorescence and the sample was delivered at a rate of 0.06 mL min−1 and analyzed for 1 min (Brussaard, 2004). Viral and bacterial counts were corrected for a blank consisting of TE-buffer with autoclaved 0.2 μm filtered seawater at the corresponding dilution.
Genome extraction and sequencing
A 1 L viral suspension was concentrated by ultrafiltration using a 30 kDa PES membrane (Vivaflow 50, Vivascience) and centrifugal concentrator (Vivaspin 20, 30 kDa, PES) to a final volume of 2 mL. The concentrate was subsequently purified on linear sucrose gradient (10–40 % in 0.2 μm SM) by ultracentrifugation (SW41.Ti rotor, 96,808 g, 30 min at 4°C). The BONAISHI particles formed a well resolved band that was extracted, dialyzed against SM buffer using a centrifugal concentrator (Vivaspin 20, 30 kDa, PES) and stored at 4°C until use. The genome of the purified phage suspension was extracted using a DNAeasy Blood and Tissue kit (QUIAGEN, Valencia, CA) according to the manufacturer's protocol. Samples were sent to GATC Biotech, and sequenced using PACBIO RS II (17,131 mean read length). Raw read sequences assembled as a single contig using HGAP software (Chin et al., 2013). The final draft assembly was 303, 340 bp with an average coverage of 1,720x and an average base quality score of 86%.
Bioinformatic analysis
Phage genome annotation
Putative coding DNA sequences (CDS) in the BONAISHI genome were predicted using Glimmer (Delcher et al., 1999) and Genemark (Besemer et al., 2001). The coordinates of each translated open reading frames (ORF) were also inspected manually. ORF smaller than 200 base pairs (bp) were removed from the analysis. The predicted amino acid sequences were assigned manually by BLASTP and PSI-BLAST (cutoff e-value < 10−5) against the NCBI non-redundant database (January 2018) and InterProScan 5 (Jones et al., 2014), as well as the fully automated RAST server annotation service (Aziz et al., 2008). BLASTP was used to search for putative toxins in the databases MvirDB (Zhou et al., 2006), VirulenceFinder (Chen et al., 2005), Vibrio-base (Choo et al., 2014), and t3DB (Lim et al., 2009; Wishart et al., 2015) toxin databases. The IntegrallDB (Moura et al., 2009) and ACLAME (Leplae et al., 2006) databases were used to check for prophage-like sequences. tRNAScanSE (Lowe and Eddy, 1997) and Aragorn (Laslett and Canback, 2004) were used to check for tRNA. The genome map was produced using Artemis (Rutherford et al., 2000) and DNA Plotter (Carver et al., 2009). The BONAISHI genome sequence has been submitted to the GenBank database under accession number MH595538.
Terminase large subunit (TerL) protein phylogeny
The amino acids sequence of the terminase large subunit from 63 jumbo phages including BONAISHI were used for phylogenetic analysis. Sequences were trimmed to 406 bp, the minimum sequence length of Aeromonas phage px29, using BioEdit (Hall, 1999). Sequences were aligned by Muscle and the tree was constructed by Maximum Likelihood with 1,000 bootstrap iterations using Mega 6.06 (Tamura et al., 2013).
Host genome analysis
The clustered regularly interspaced short palindromic repeats (CRISPR), in the pathogenic V. coralliilyticus strains P1 and YB1 genome, were searched for genetic signatures of viral resistance mechanisms using CRISPRfinder (Grissa et al., 2007). Genetic exchange between the phage BONAISHI and its bacterial hosts was checked by homology between the phage ORFs and the ORFs of V. coralliilyticus P1 (AEQS00000000) and YB1 (ACZN00000000) using BLASTP.
Metagenomic analysis
To determine the distribution of BONAISHI, the genome was used to recruit homologous reads from 56 coral-associated virome in Metavir (Roux et al., 2011), and 137 CAMERA Broad Phage metagenomes (Seshadri et al., 2007) and viral contigs in IMG/VR (Paez-Espino et al., 2017, Table S2). These samples comprised a wide range of marine environments including tropical and temperate pelagic ecosystems, healthy and diseased coral reefs including slurry from individual coral colonies, coral mucus, and the water from coral reefs. We also carried out recruitments in prokaryote metagenome from 4 coral atolls (Dinsdale et al., 2008) to check whether BONAISHI genome sequences were inserted into prokaryote genomes. Each reads served as a query and was assigned to a (single) best-matching hit by BLASTN and TBLASTX if the alignments met the following criteria: e-value < 10−3, alignment length > 50, bitscore > 40. BLASTN parameters were set to: open gap cost = 8, extend gap cost = 6, match reward = 5, mismatch penalty = −4, word size = 8. Reads were recruited from each metagenome in order to determine the fraction of recruited reads that can be assigned to BONAISHI.
Preliminary treatment of diseased symbiodinium
Culture of Symbiodinium sp. cells (Clade A1) originally extracted from the scleractinian coral Galaxea fascicularis (Goiran et al., 1996) were maintained in the laboratory in F/2 medium (Guillard and Ryther, 1962) prepared from Guillard's Marine Water Enrichment Solution (Sigma-Aldrich G9903). Cultures were maintained at 25°C under 100 μmol photons m−2 s−1 of white light provided by fluorescent tubes (Mazda 18WJr/865) using a 12:12 light:dark cycle. One day prior to the therapy assay, exponentially growing Symbiodinium sp. cultures were transferred to 30°C under the same light conditions. A 20 mL aliquot was concentrated at 5,000 g for 10 min at 30°C (VIVASPIN 20, PES, 30 kDa). The retentate was gently resuspended in 20 ml EDTA free F/2 medium and the procedure was repeated twice to wash the culture. The Symbiodinium sp. abundance was determined by flow cytometry and adjusted to 104 cells mL−1. V. coralliilyticus YB1 was cultured overnight in MB and then purified in the same way. Bacterial abundance was determined by flow cytometry and adjusted to 107 cells mL−1. A freshly produced suspension of BONAISHI was purified by sucrose gradient and diluted in SM buffer. The viral abundance was determined by flow cytometry.
For the therapy assay, the algal culture was split into 3 equal sub-cultures. One sub-culture served as control, while two of the subcultures were inoculated with an equal volume of V. coralliilyticus YB1. Of these, one was also inoculated with 108 phages mL−1. All three treatments were sampled at 0, 5, 20, and 60 min to determine the photosystem II quantum yield of Symbiodinium sp. cells using a pulse amplitude modulated fluorimeter (Phyto-PAM, Walz) connected to a chart recorder (Labpro, Vernier). After 5 min relaxation in darkness, the non-actinic modulated light (450 nm) was turned on in order to measure the fluorescence basal level, F0. A saturating red light pulse (655 nm, 4 000 μmol quanta m−2 s−1, 400 ms) was applied to determine the maximum fluorescence level in the dark adapted sample, FM. The maximal photosystem II fluorescence quantum yield of photochemical energy conversion, FV/FM, was calculated using the following formula:
| (1) |
Results
Morphology
The Vibrio phage BONAISHI formed relatively large, round plaques on V. coralliilyticus YB1 and produced high titer suspension. TEM microscopy showed that BONAISHI has an isometric capsid of 120 nm in diameter connected to a 190 nm long, contractile tail (Figure 1). This indicates that BONAISHI belongs to the order of the Caudovirales and the family of the Myoviridae.
Figure 1.
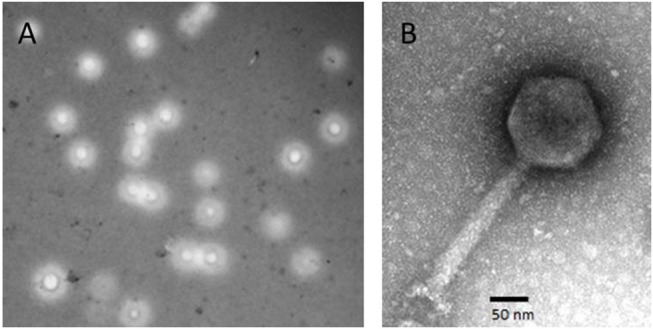
Morphology of Vibrio phage BONAISHI. (A) BONAISHI forms large, round plaque on a lawn of V. coralliilyticus YB1 on 0.6% soft agar. (B) Transmission electron micrographs of a negatively stained particle of bacteriophage BONAISHI. The icosahedral head (120 nm in diameter) and the long, contractile tail (190 nm in length) suggest that BONAISHI belongs to the Myoviridae family.
Tolerance to environmental factors
The incubations showed that BONAISHI can tolerate a pH ranging from 3 to 10 and temperatures ranging from 4 to 50°C without loss of infectivity (Table 1). This high tolerance suggests that the phage is very stable in the environment.
Table 1.
Tolerance of Vibrio phage BONAISHI to temperature and pH.
| Treatments | Infectivity |
|---|---|
| TEMPERATURE (°C) | |
| 4 | + |
| 15 | + |
| 20 | + |
| 25 | + |
| 30 | + |
| 35 | + |
| 40 | + |
| 45 | + |
| 50 | + |
| 60 | - |
| 70 | - |
| pH | |
| 2 | - |
| 3 | + |
| 4 | + |
| 5 | + |
| 6 | + |
| 7 | + |
| 8 | + |
| 9 | + |
| 10 | + |
Host specificities
Spot tests using a broad range of potential hosts indicated that BONAISHI was able to infect and lyse several strains of V. coralliilyticus of interest (Table S1). Besides V. coralliilyticus YB1, BONAISHI can infect another known coral pathogen, V. coralliilyticus P1, as well as V. coralliilyticus LMG21348, isolated from a bleached coral colony (Pocillopora damicornis), and V. coralliilyticus 1H13, isolated from the mucus of a Fungia specimen. This phage did not lyse of any of the closely related species in the test, suggesting that it is species-specific.
Life cycle
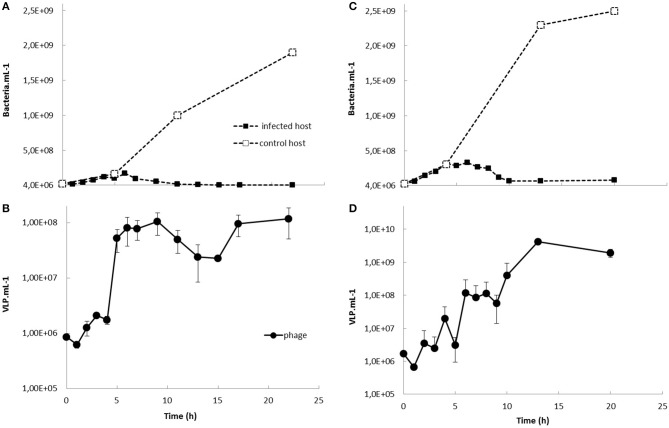
One-step growth experiments showed that the phage readily propagated on each of its hosts with a latent period of 2–3 h (strains P1 and YB1) and burst size of 8 (P1) and 19 (YB1) (Figure 2). Nearly all the virions produced (96%) were infectious virions per infected cell. The infected host culture collapsed rapidly and there was total lysis 10 h after inoculation.
Figure 2.
One-step growth experiments of BONAISHI on Vibrio coralliilyticus strain P1 (A,B) and YB1 (C,D). Bacteria and bacteriophage counts were assessed by flow cytometry upon staining with the nucleic acid dye SYBR Green (Life technology) according to Brussaard (2004).
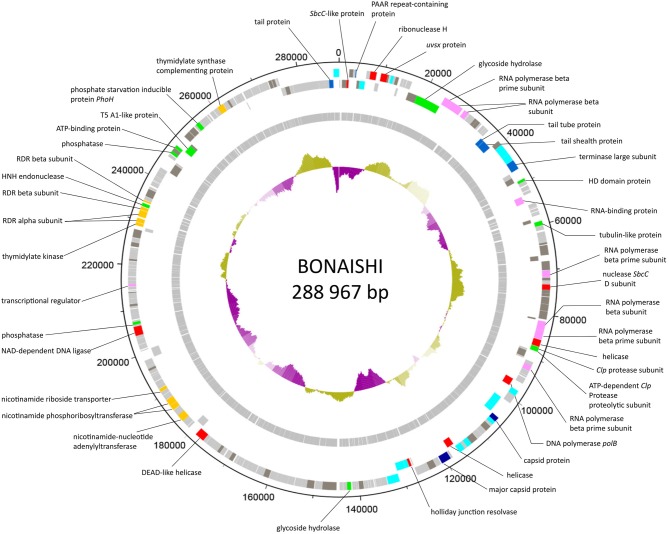
General features of BONAISHI genome
The genome of BONAISHI consists of a large double-stranded circularly permuted DNA sequence of 303,340 base-pair (bp) with a % G+C content of 42.5% (Figure 3). Terminal duplications at the extremities of the assembled sequence are 14,373 bp, giving a non-redundant genome of 288,967 bp. Glimmer and Genemark predicted 301 putative ORFs, which comprised 93.8% of the total sequence and were mostly oriented in a single direction. Most ORFs initiate translation at an ATG start codon except for 6 ORFs, 4 of which have a GTG start codon and the other 2 a TTG start codon. We did not find any tRNA in the genome. Of the 301 predicted ORFs, 110 ORFs (36.4%) had significant homologs in public databases and a biochemical function could be assigned for 62 of these (Table 2, Figure 3). Blast searches revealed that most of the ORFs (66/110) with significant homologs were closely related to other Myoviridae (Table 2). Most best-hits corresponded to members of the genus Phikzvirus, which comprises myoviruses with a large genome (> 200 kb), including Pseudomonas phages Phikz and PhiPA3, Erwinia phage Ea35-70, Ralstonia phage RSF1 and RSL2 and Vibrio phage pVa21, VP4B, pTD1, Phabio, and Noxifer. Although many gene functions could be assigned, most of the predicted genes are ORFans that are unique to BONAISHI.
Figure 3.
Genome map of BONAISHI. The 110 protein-coding genes are shown as colored blocks. Protein-coding genes that are transcribed in the forward and reverse direction are located on the external and internal circle, respectively. The functional assignments of the protein-coding genes are derived from blastp matches in NCBI nr and expert databases. We confidently discriminated proteins involved in general virion structure (light blue), head, and tail structure and assembly (dark blue), DNA replication,recombination and repair (red), nucleotide metabolism (orange), DNA transcription (pink), and host phage interactions (green). Genes marked “h.p.” made strong matches to hypothetical protein sequences in the database with no identified functions (dark gray). The GC content of the genome sequence is indicated by the internal purple or green histograms.
Table 2.
Summary table of Vibrio phage BONAISHI predicted proteins that contained relevant annotation information as determined from significant BLASTP hits (e-value < e-3) against the GenBank non-redundant and CAZY databases.
| Predicted function | START | STOP | Size (n) | Best-hit | Classification | E-value | Score (bits) | N°accession | |
|---|---|---|---|---|---|---|---|---|---|
| ORF01c | hypothetical protein | 653 | 1753 | 1100 | Pseudocercospora musae | Fungi | 2.00E-07 | 66.00 | KNG49823.1 |
| ORF02c | SbcC-like protein | 1750 | 2184 | 435 | Pseudomonas phage 201phi2-1 | Myoviridae | 1.00E-17 | 88.00 | YP_001956973 |
| ORF03 | conserved hypothetical protein | 2241 | 3119 | 879 | Pseudomonas phage Phikz | Myoviridae | 2.00E-32 | 130.00 | NP_803730 |
| ORF05 | PaaR repeat-containing protein | 3722 | 4018 | 297 | Vibrio cholerae | ⋎-proteobacteria | 3.00E-21 | 89.00 | WP_057643503 |
| ORF06c | conserved hypothetical protein | 4055 | 4657 | 603 | Pseudomonas phage Phikz | Myoviridae | 1.00E-25 | 203.00 | AAL83062 |
| ORF07c | virion structural protein | 4667 | 5962 | 1296 | Pseudomonas phage KTN4 | Myoviridae | 7.00E-32 | 147.00 | ANM44952.1 |
| ORF10 | ribonuclease H | 6889 | 8271 | 1383 | Pseudomonas phage 201phi2-1 | Myoviridae | 3.00E-25 | 117.00 | YP_001956963 |
| ORF12 | uvsx protein | 9269 | 10744 | 1476 | Pseudomonas phage 201phi2-1 | Myoviridae | 1.00E-89 | 290.00 | YP_001956960 |
| ORF13 | conserved hypothetical protein | 10746 | 11108 | 363 | Pseudomonas phage phiPA3 | Myoviridae | 3.00E-28 | 108.00 | AEH03597 |
| ORF15 | virion structural protein | 11460 | 12125 | 666 | Pseudomonas phage phiPA3 | Myoviridae | 2.00E-27 | 113.00 | AEH03595 |
| ORF17 | hypothetical protein | 12558 | 13256 | 699 | Erwinia phage Ea35-70 | Myoviridae | 8.00E-09 | 64.00 | YP_009004948 |
| ORF22c | conserved hypothetical protein | 16270 | 18384 | 2115 | Ralstonia phage RSL2 | Myoviridae | 3.00E-74 | 261.00 | BAQ02568 |
| ORF23c | glycoside hydrolase | 18393 | 24110 | 5718 | Ralstonia phage RP31 | Myoviridae | 2.00E-70 | 277.00 | BAW19303.1 |
| ORF24 | RNA polymerase beta prime subunit | 24183 | 25850 | 1668 | Pseudomonas phage OBP | Myoviridae | 1.00E-110 | 346.00 | YP_004958184 |
| ORF25 | RNA polymerase beta subunit | 25847 | 28741 | 2895 | Ralstonia phage RSF1 | Myoviridae | 1.00E-102 | 358.00 | BAS04832 |
| ORF26 | RNA polymerase beta subunit | 29242 | 30369 | 1128 | Pseudomonas phage 201phi2-1 | Myoviridae | 2.00E-61 | 223.00 | YP_001956996 |
| ORF29 | conserved hypothetical protein | 32135 | 32860 | 726 | Pseudomonas phage Noxifer | Myoviridae | 3.00E-21 | 110.00 | ARV77361.1 |
| ORF30 | virion structural protein | 32871 | 33641 | 771 | Ralstonia phage RSF1 | Myoviridae | 3.00E-31 | 124.00 | BAS04828 |
| ORF36c | tail tube protein | 36168 | 37040 | 873 | Pseudomonas phage 201phi2-1 | Myoviridae | 2.00E-24 | 108.00 | YP_001956757 |
| ORF37c | tail shealth protein | 37089 | 39137 | 2049 | Erwinia phage Ea35-70 | Myoviridae | 2.00E-83 | 286.00 | YP_009004971 |
| ORF38 | hypothetical protein | 39208 | 40278 | 1071 | Erwinia phage Ea35-70 | Myoviridae | 3.00E-09 | 67.00 | YP_009004972 |
| ORF39 | virion structural protein | 40288 | 42819 | 2532 | Pseudomonas phage 201 phi2-1 | Myoviridae | 2.00E-55 | 226.00 | YP_001956754.1 |
| ORF40 | virion structural protein | 42831 | 44483 | 1653 | Pseudomonas phage 201phi2-1 | Myoviridae | 3.00E-27 | 125.00 | YP_001956753 |
| ORF41 | terminase large subunit | 44535 | 46691 | 2157 | Pseudomonas phage 201phi2-1 | Myoviridae | 1.00E-142 | 442.00 | YP_001956731 |
| ORF43c | conserved hypothetical protein | 47798 | 49027 | 1230 | Pseudomonas phage Noxifer | Myoviridae | 4.00E-37 | 164.00 | ARV77197.1 |
| ORF44 | HD domain protein | 49121 | 49720 | 600 | Salmonella phage SPN3US | Myoviridae | 3.00E-23 | 101.00 | AEP84084 |
| ORF49c | RNA-binding protein | 52606 | 54225 | 1620 | Actinomyces massiliensis | Actinobacteria | 2.00E-73 | 251.00 | WP_017194325 |
| ORF58c | conserved hypothetical protein | 58317 | 59075 | 759 | Erwinia phage Ea35-70 | Myoviridae | 1.00E-22 | 102.00 | YP_009005001 |
| ORF59 | tubulin-like protein | 59178 | 60167 | 990 | Erwinia phage Ea35-70 | Myoviridae | 3.00E-21 | 101.00 | YP_009005002 |
| ORF68 | hypothetical protein | 64017 | 64424 | 408 | Ralstonia phage RSL2 | Myoviridae | 7.00E-06 | 52.00 | BAQ02532 |
| ORF69 | conserved hypothetical protein | 64596 | 66782 | 2187 | Erwinia phage Ea35-70 | Myoviridae | 1.00E-105 | 345.00 | YP_009005012 |
| ORF70c | hypothetical protein | 66814 | 67875 | 1062 | Ralstonia phage RSF1 | Myoviridae | 9.00E-08 | 63.00 | BAS05022 |
| ORF71 | hypothetical protein | 68148 | 70115 | 1967 | ⋎-protebacteria bacterium | ⋎-proteobacteria | 1.00E-15 | 94.00 | OUV32520.1 |
| ORF72 | RNA polymerase beta prime subunit | 70204 | 71697 | 1494 | Erwinia phage Ea35-70 | Myoviridae | 3.00E-44 | 171.00 | YP_009005015 |
| ORF73 | conserved hypothetical protein | 71795 | 72433 | 639 | Erwinia phage Ea35-70 | Myoviridae | 9.00E-15 | 79.00 | YP_009005021 |
| ORF76 | nuclease SbcC subunit | 73326 | 74498 | 1173 | Pseudomonas phage PhipA3 | Myoviridae | 1.00E-46 | 172.00 | AEH03486 |
| ORF77 | conserved hypothetical protein | 74495 | 75289 | 795 | Erwinia phage PhiEaH1 | Siphoviridae | 9.00E-23 | 103.00 | YP_009010069 |
| ORF78 | hypothetical protein | 75301 | 75972 | 672 | Uncultured bacterium | Bacteria | 2.00E-06 | 56.00 | EKD22589 |
| ORF79 | hypothetical protein | 76106 | 77656 | 1551 | Pseudomonas phage phiPA3 | Myoviridae | 9.00E-06 | 59.00 | AEH03489 |
| ORF80 | conserved hypothetical protein | 77691 | 79211 | 1521 | Pseudomonas phage phiPA3 | Myoviridae | 1.00E-16 | 93.00 | AEH03490 |
| ORF81 | hypothetical protein | 79251 | 81056 | 1806 | Gossypium arboreum | Magnoliopsida | 1.00E-06 | 63.00 | KHG21929 |
| ORF82c | hypothetical protein | 81109 | 81456 | 348 | Pseudomonas phage PA7 | Myoviridae | 5.00E-09 | 59.00 | AFO71119 |
| ORF83 | RNA polymerase beta subunit | 81518 | 83584 | 2067 | Pseudomonas phage Phabio | Myoviridae | 8.00E-66 | 260.00 | ARV76743.1 |
| ORF84 | RNA polymerase beta prime subunit | 83584 | 85557 | 1974 | Ralstonia phage RSF1 | Myoviridae | 1.00E-50 | 196.00 | BAS05006 |
| ORF85 | helicase | 85657 | 87198 | 1542 | Pseudomonas phage OBP | Myoviridae | 1.00E-40 | 176.00 | AEV89521.1 |
| ORF86 | Clp protease subunit | 87276 | 87848 | 573 | Bacillus cereus | Bacilli | 5.00E-05 | 52.00 | WP_048520069 |
| ORF87 | ATP-dependent Clp protease proteolytic subunit | 87848 | 88339 | 492 | Dactylosporangium aurantiacum | Actinobacteria | 1.00E-27 | 110.00 | WP_033356707 |
| ORF89c | conserved hypothetical protein | 88833 | 90389 | 1557 | Pseudomonas phage PA7 | Myoviridae | 2.00E-15 | 89.00 | AFO71110 |
| ORF92 | RNA polymerase beta prime subunit | 91656 | 92915 | 1260 | Ralstonia phage RSF1 | Myoviridae | 8.00E-49 | 180.00 | BAS04991 |
| ORF96c | DNA polymerase polB | 96090 | 97820 | 1731 | Ralstonia phape RP12 | Unclassified virus | 6.00E-107 | 343.00 | BAW19225.1 |
| ORF97 | virion structural protein | 97895 | 99190 | 1296 | Erwinia phage PhiEaH1 | Siphoviridae | 2.00E-23 | 111.00 | YP_009010288 |
| ORF99c | virion structural protein | 101147 | 104005 | 2859 | Pseudomonas phage 201phi2-1 | Myoviridae | 1.00E-93 | 327.00 | YP_001956873 |
| ORF100c | virion structural protein | 104007 | 105122 | 1116 | Ralstonia phage RSL2 | Myoviridae | 2.00E-49 | 180.00 | BAQ02702 |
| ORF101 | capsid protein* | 105166 | 106242 | 1077 | Ralstonia phage RSF1 | Myoviridae | 3.00E-22 | 105.00 | BAS04975 |
| ORF102 | virion structural protein | 106257 | 107141 | 885 | Ralstonia phage RSF1 | Myoviridae | 4.00E-07 | 60.00 | BAS04974 |
| ORF104 | hypothetical protein | 107713 | 109107 | 1394 | ⋎-protebacteria bacterium | ⋎-proteobacteria | 1.00E-12 | 83.00 | OUV32343.1 |
| ORF107 | conserved hypothetical protein | 111644 | 113257 | 1614 | Ralstonia phage RSF1 | Myoviridae | 8.00E-22 | 108.00 | BAS04969 |
| ORF108 | virion structural protein | 113257 | 114459 | 1203 | Pseudomonas phage PhiPA3 | Myoviridae | 8.00E-12 | 77.00 | AEH03528 |
| ORF110 | virion structural protein | 115149 | 116531 | 1383 | Pseudomonas phage PhiPA3 | Myoviridae | 2.00E-28 | 126.00 | AEH03530 |
| ORF111c | helicase | 116571 | 118181 | 1611 | Pseudomonas phage 201phi2-1 | Myoviridae | 2.00E-48 | 1884.00 | YP_001956921 |
| ORF113 | major capsid protein | 118843 | 121035 | 2193 | Erwinia phage Ea35-70 | Myoviridae | 2.00E-23 | 116.00 | YP_009005109 |
| ORF115 | conserved hypothetical protein | 122460 | 124031 | 1572 | Ralstonia phage RSL2 | Myoviridae | 1.00E-46 | 179.00 | BAQ02643 |
| ORF118c | holliday-junction resolvase | 127427 | 127987 | 561 | Pseudomonas phage Phabio | Myoviridae | 4.00E-18 | 99.00 | ARV76843.1 |
| ORF119c | virion structural protein | 128029 | 128865 | 837 | Pseudomonas phage 201phi2-1 | Myoviridae | 8.00E-44 | 159.00 | YP_001956947 |
| ORF120c | virion structural protein | 128878 | 130947 | 2070 | Pseudomonas phage phiPA3 | Myoviridae | 7.00E-72 | 256.00 | AEH03570 |
| ORF121 | virion structural protein | 131049 | 133649 | 2601 | Pseudomonas phage phabio | Myoviridae | 4.00E-61 | 245.00 | ARV76832.1 |
| ORF133 | hypothetical protein | 139060 | 140100 | 1041 | Vibrio tasmaniensis | ⋎-proteobacteria | 2.00E-11 | 73.00 | WP_017112059 |
| ORF138 | glycoside hydrolase | 141805 | 142716 | 912 | Aureimonas altamirensis | α-proteobacteria | 2.00E-49 | 174.00 | BAT26087 |
| ORF141 | hypothetical protein | 145070 | 148201 | 3132 | Psychromonas ingrahamii | ⋎-proteobacteria | 1.00E-08 | 72.00 | WP_011768462.1 |
| ORF143 | hypothetical protein | 150013 | 151644 | 1631 | Vibrio phage s4-7 | Unclassified virus | 2.00E-06 | 63.00 | AOQ26845.1 |
| ORF151 | hypothetical protein | 160192 | 161433 | 1242 | Colwellia phage 9A | Siphoviridae | 4.00E-08 | 66.00 | YP_006489231 |
| ORF159 | hypothetical protein | 168857 | 169801 | 945 | Escherichia phage phAPEC8 | Myoviridae | 4.00E-10 | 68.00 | YP_007348452 |
| ORF163 | hypothetical protein | 172164 | 172847 | 684 | Ruegeria halocynthiae | α-proteobactérie | 1.00E-19 | 91.00 | WP_037312174 |
| ORF167 | dead-like helicase | 176707 | 178806 | 2100 | Erwinia phage Ea35-70 | Myoviridae | 1.00E-114 | 367.00 | YP_009004923 |
| ORF172 | nicotinamide-nucleotide adenylyltransferase | 182827 | 184476 | 1650 | Vibrio phage 11895-B1 | Myoviridae | 1.00E-106 | 331.00 | YP_007673553 |
| ORF176 | nicotinamide-nucleotide adenylyltransferase | 185996 | 187114 | 1119 | Thiorhodococcus drewsii | ⋎-proteobacteria | 3.00E-46 | 170.00 | WP_007039048 |
| ORF177 | nicotinamide phosphoribosyltransferase | 187169 | 188668 | 1500 | Vibrio nigripulchritudo | ⋎-proteobacteria | 3.00E-59 | 210.00 | WP_022562194 |
| ORF181 | nicotinamide riboside transporter | 190786 | 191502 | 717 | Vibrio phage 11895-B1 | Myoviridae | 3.00E-76 | 238.00 | YP_007673552 |
| ORF189 | hypothetical protein | 196049 | 196543 | 717 | Kaistia granuli | α-proteobacteria | 1.00E-08 | 57.00 | WP_018183972 |
| ORF198 | NAD-dependent DNA ligase | 204242 | 206257 | 2016 | Vibrio maritimus | ⋎-proteobacteria | 0.00E+00 | 608.00 | WP_042496716 |
| ORF200 | phosphatase | 206698 | 207330 | 633 | Vibrio phage VH7D | Myoviridae | 6.00E-19 | 89.00 | YP_009006310 |
| ORF206 | hypothetical protein | 210803 | 211318 | 516 | Psychromonas aquimarina | ⋎-proteobacteria | 7.00E-34 | 126.00 | WP_028862581 |
| ORF209 | hypothetical protein | 213895 | 214209 | 315 | Enterovibrio calviensis | ⋎-proteobacteria | 6.00E-07 | 52.00 | WP_017007757 |
| ORF212 | transcriptional regulator | 215240 | 215764 | 525 | Leptolyngbya sp. PCC 7375 | cyanobacteria | 1.00E-05 | 53.00 | EKV01169 |
| ORF220 | hypothetical protein | 220875 | 221303 | 429 | Shewanella sp. phage 1/4 | Myoviridae | 6.00E-11 | 64.00 | YP_009100318 |
| ORF227 | hypothetical protein | 224066 | 224998 | 933 | Pseudomonas phage PhiPA3 | Myoviridae | 9.00E-06 | 57.00 | AEH03433 |
| ORF229 | hypothetical protein | 226086 | 227921 | 1835 | Vibrio phage RYC | Unclassified virus | 7.00E-70 | 243.00 | BAV81012.1 |
| ORF231 | thymidylate kinase | 228796 | 229458 | 663 | Desulfosporosinus acidiphilus | Clostridia | 8.00E-42 | 150.00 | WP_014828008 |
| ORF232 | ribonucleotide-diphosphate reductase subunit alpha | 229535 | 230446 | 912 | Aeromonas molluscorum 848 | ⋎-proteobacteria | 1.00E-132 | 385.00 | EOD53957 |
| ORF233 | ribonucleotide-diphosphate reductase subunit alpha | 230844 | 232277 | 1434 | Neisseria meningitidis | β-proteobacteria | 0.00E+00 | 635.00 | WP_049227356 |
| ORF234 | ribonucleotide-diphosphate reductase subunit beta | 232355 | 233062 | 708 | Thiomicrospira sp. Kp2 | ⋎-proteobacteria | 3.00E-96 | 294.00 | WP_040727751 |
| ORF235 | HNH endonuclease | 233205 | 233927 | 723 | Bacillus pumilus | Bacilli | 7.00E-32 | 123.00 | WP_051149989 |
| ORF236 | ribonucleotide-diphosphate reductase subunit beta | 234231 | 234617 | 387 | Shigella sonnei | ⋎-proteobacteria | 2.00E-36 | 129.00 | CSE34793 |
| ORF238 | hypothetical protein | 235508 | 237232 | 1725 | Polyangium brachysporum | β-proteobacteria | 2.00E-23 | 114.00 | WP_047195109 |
| ORF249c | hypothetical protein | 242652 | 244886 | 2235 | Vibrio mimicus | ⋎-proteobacteria | 4.00E-08 | 67.00 | WP_001015571 |
| ORF253 | phosphatase | 246359 | 247015 | 657 | Verrucomicrobium spinosum | verucomicrobia | 2.00E-22 | 97.00 | WP_009962909 |
| ORF254 | hypothetical protein | 247012 | 247842 | 831 | Shewanella sp. | ⋎-proteobacteria | 6.00E-06 | 57.00 | WP101034114.1 |
| ORF255 | ATP-binding protein | 247870 | 248553 | 684 | Vibrio phage RYC | Unclassified virus | 1.00E-36 | 137.00 | BAV81012.1 |
| ORF256c | T5 A1-like protein | 248608 | 250479 | 1872 | Caulobacter phage phiCbK | Siphoviridae | 3.00E-83 | 281.00 | YP_006988022 |
| ORF257c | hypothetical protein | 250484 | 250927 | 444 | Pseudoalteromonas (multispecies) | ⋎-proteobacteria | 9.00E-19 | 86.00 | WP_024591352 |
| ORF258 | conserved hypothetical protein | 251087 | 252592 | 1506 | Campylobacter phage CP30A | Myoviridae | 3.00E-20 | 102.00 | YP_006908082 |
| ORF259 | conserved hypothetical protein | 252655 | 253968 | 1314 | Campylobacter phage CP30A | Myoviridae | 2.00E-18 | 96.00 | YP_006908082 |
| ORF261 | phosphate starvation protein PhoH | 254632 | 255627 | 996 | Corynebacterium glucuronolyticum | Actinobacteria | 6.00E-49 | 175.00 | WP_005389286 |
| ORF269 | thymidylate synthase-complementing protein | 260653 | 262095 | 1443 | Parcubacteria | Parcubacteria | 5.00E-62 | 217.00 | KKR42866 |
| ORF272 | conserved hypothetical protein | 263205 | 263723 | 519 | Cronobacter phage vB_CsaM_GAP32 | Myoviridae | 9.00E-15 | 77.00 | YP_006987447 |
| ORF289 | conserved hypothetical protein | 273177 | 273917 | 740 | Vibrio phage vB_VhaS-a | Unclassified virus | 4.00E-18 | 100.00 | ANO57550.1 |
| ORF290 | conserved hypothetical protein | 273997 | 274665 | 668 | Vibrio phage vB_VhaS-a | Unclassified virus | 1.00E-14 | 88.00 | ANO57549.1 |
| ORF295c | hypothetical protein | 276639 | 277370 | 731 | Ralstonia phage RP12 | Myoviridae | 9.00E-10 | 73.00 | BAW19047.1 |
| ORF300 | hypothetical tail protein | 286648 | 287631 | 983 | Pseudomonas phage Phabio | Myoviridae | 7.00E-15 | 90.00 | ARV76834.1 |
| ORF301 | virion structural protein | 287726 | 288967 | 1241 | Pseudomonas phage Noxifer | Myoviridae | 3.00E-07 | 65.00 | ARV77324.1 |
Gene annotation
Proteins involved in virion structure and assembly
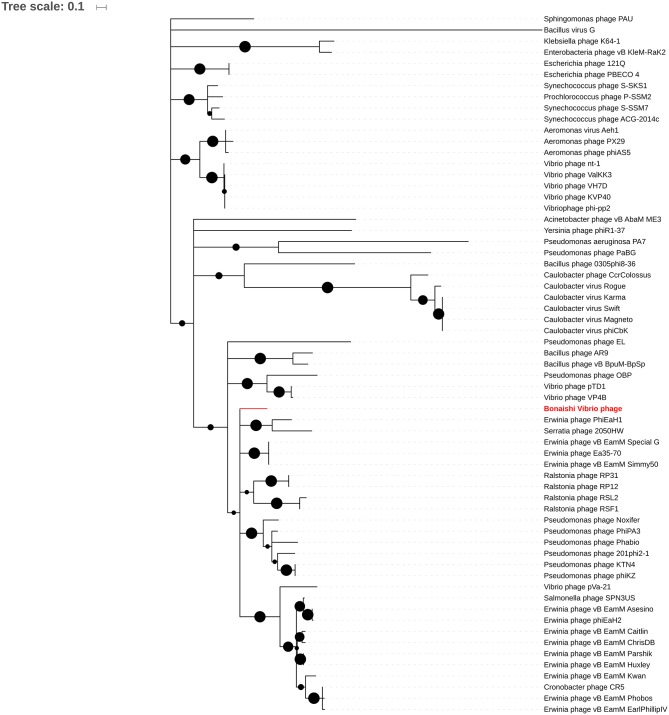
The predicted proteins involved in the virion structure and assembly included a major and an accessory capsid protein (ORF 101 and 113), several tail components including a tail tube (ORF 36), a tail shealth (ORF 37), and an accessory tail protein (ORF 300), as well as several conserved structural proteins (ORFs 7, 15, 30, 39, 40, 97, 99, 100, 102, 108, 110, 119, 120, 121, and 301). We also identified a terminase large subunit (ORF 41) involved in DNA packaging. All these structural components shared strong homologies with proteins encoded by other members of Phikzvirus genus (Table 1, Figure S1). Phylogenetic analyses based on the amino acid sequence of the terminase large subunit classified BONAISHI as a singleton (Figure 4). The closest sequences belong to Vibrio phage pVa-21 and a cluster with Cronobacter phage CR5, Erwinia phage vB EamM, phiEaH2 and Salmonella phage SPN3US. The second closest cluster comprised Phikzvirus Pseudomonas phage KTN4, Phikz, Noxifer, PA3, Phabio, and 201 phi2-1.
Figure 4.
Phylogenetic analysis of 63 Jumbo phages based on the amino acids sequence of the terminase large subunit. Sequences were aligned by Muscle and the tree was constructed by Maximum Likelihood method with a bootstrap of 1,000 using Mega 6.0 (Tamura et al., 2013) as in Yuan and Gao (2017). Bootstrap values higher than 0.5 are represented by black circles, ranging from 0.5 (smaller circle) to 1 (bigger circle).
Proteins involved in dna replication, recombination and repair
The BONAISHI genome encoded at least 10 proteins involved in DNA replication, recombination, and repair. These included a DNA polymerase B (ORF 96), putative DnaB, and DEAD-like helicases (ORF 85 and 111), NAD-dependent DNA ligase (ORF 198), SbcC-like nucleases (ORF2 and 76), a ribonuclease H (ORF 10), Holliday junction resolvase (ORF 118), uvsx recombinase (ORF12). ORF 235 corresponded to the HNH family of homing endonuclease located between genes encoding β subunits of ribonucleotide diphosphate reductase.
Proteins involved in nucleotide metabolism and dna modification
We were able to assign a putative function to 10 enzymes involved into nucleotide metabolism. Predicted proteins included two ribonucleotide diphosphate reductase (RDR) α subunits (ORFs 232 and 233) and two RDR β subunits (ORFs 234 and 236), 4 proteins of the pyridine nucleotide salvage pathway corresponding to a nicotinamide riboside transporter (ORF 181), a nicotinamide phosphoribosyltransferase (ORF 177) as well as nicotinamide mononucleotide adenylyltransferases (ORF 172 and 176). BONAISHI also encodes 2 proteins involved in thymidine biosynthesis one of which is a thymidylate synthase complementing protein (ORF 269) and the other a thymidylate kinase (ORF 231). We did not identify any gene for protein involved in DNA modification.
Proteins involved in dna transcription
A transcription regulator related to the PadR family (ORF 212) was found as well as two sets of multisubunit RNA polymerase genes (β- and β'- RNAP subunits). ORFs 24, 25, 26, 92 corresponded to the Phikz virion-associated RNAP and ORFs 72, 83, 84 shared significant homologies with the Phikz genes encoding the early expressed RNAP (Table 2, Figure S1). No homologs of the early expressed Phikz β- RNAP subunits were found in BONAISHI genome. ORF 49 was assigned to an RNA binding protein.
Lysis, host-phage interaction, and lysogeny
We were able to annotate two proteins (ORFs 23 and 138) involved into host-virus interactions. There were good blastp hits on proteins of unknown function in the NCBI nr database that could be expanded to known glycoside hydrolases (GH) in the expert database CAZY (Carbohydrate Active enZYmes db, http://www.cazy.org). These included a glycoside hydrolase with a conserved endopeptidase domain (ORF 23) affiliated to the GH23 family, which mostly include lysozymes. The protein encoded by ORF 138 belongs to the glycoside hydrolase family GH19, which comprises chitinases and lysozymes. Both enzyme groups catalyze the hydrolysis of polysaccharides containing N-Acetylglucosamine, but the gene sequence does not discriminate between the two. No genes implicated in lysogeny establishment or control (integrase/excisionase, transposases, ParA/ParB genes, attachment sites, transcription repressor, etc.) were detected in the BONAISHI genome, even using prophage expert databases, which confirms that it is exclusively lytic. In addition, blastp searches using BONAISHI hosts genome did not find evidence of the presence of BONAISHI as prophage. Only three genes involved in DNA replication and recombination (ORF 225), nucleotide metabolism (ORF 192) and gene encoding for a phosphate starvation protein showed high score but the similarity was relatively low (<77%).
Miscellaneous proteins
We identified a phosphate starvation protein PhoH (ORF 261). This protein has been reported in many other Myoviridae. For therapeutic applications of this phage, we also looked for potential toxin encoding genes using BLASTP on 4 toxin expert databases. No homologs to currently known toxins were detected.
Metagenomic analysis
The global distribution of BONAISHI was investigated using marine viromes from various waters including coral mucus and the water from coral reefs (Table S2). The highest number of recruited reads was from the virome collected at ALOHA station in the North Pacific subtropical gyre (CAM_SMPL_00823): BONAISHI recruited 1.04% of the reads with a mean identity of 58.23%. None of the recruited reads exceeded 85% identity. We also used prokaryotic metagenomes to test whether the BONAISHI genome is found as a prophage in bacterial hosts. No bacterial reads were recruited.
Preliminary therapy assays of diseased symbiodinium
Symbiodinium sp. control culture showed optimal photosynthetic activity with the quantum yield Fv/Fm between 0.57 and 0.59 during the course of the experiment. As expected, inoculation with V. coralliilyticus caused a rapid photoinhibition with a 50% decrease in the quantum yield 60 min after inoculation (Figure 5). BONAISHI was able to significantly counteract the bacterial algicidal activity (t-test, p < 0.001) as, with both V. coralliilyticus and BONAISHI, the quantum yield was only 14% lower than the control 60 min after inoculation.
Figure 5.
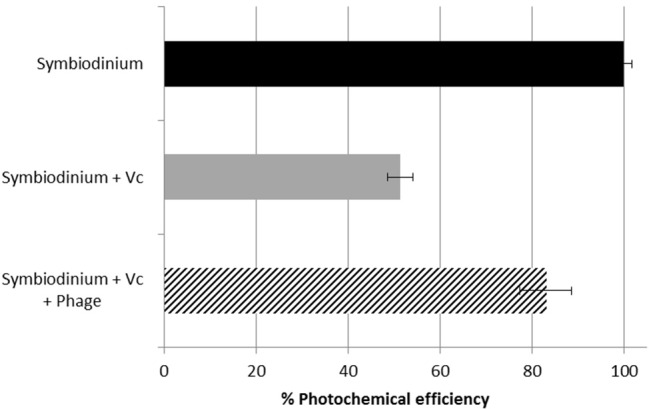
Quantum yield (Fv/Fm) of the photosystem II of control Symbiodinium sp. culture (black), Symbiodinium sp. inoculated with V. coralliilyticus YB1 pathogen (gray), and Symbiodinium sp. co-inoculated with V. coralliilyticus YB1 pathogen and Vibrio phage BONAISHI (hashed) after 60 min incubation. Results are expressed as the % of Fv/Fm in the control culture. As reported in previous study, the inoculation of V. coralliilyticus YB1 pathogen induced a rapid decline in Symbiodinium sp. photochemical efficiency. The addition of BONAISHI rapidly counteracted the impact of V. coralliilyticus YB1 on the efficiency of Symbiodinium photochemical activity.
Discussion
In recent years, Vibrio coralliilyticus has been used as a model pathogen to gain insights into the establishement and propagation of coral diseases (Sussman et al., 2008; O'Santos et al., 2011; Garren et al., 2014; Pollock et al., 2015). The use of phages to control V. coralliilyticus has been reported recently (Efrony et al., 2007; Cohen et al., 2013) but if phage therapy is to become a practical approach, fundamental knowledge on pathogen-virus interactions must be investigated in detail to evaluate, on the one hand, the therapeutic potential of the candidate phage and, on the other hand, the suitability of the host for phage therapy.
The detection of virus-derived genes in the Vibrio coralliilyticus P1 and YB1 genomes (Weynberg et al., 2015) shows that these pathogenic strains have interacted with phages during the course of their evolutionary history. Past interactions events with viruses can lead to the development of resistance mechanisms to escape phage infection, which may, in turn, limit the application of phage therapy. We, however, did not detect any of the distinctive genetic signatures of viral resistance mechanisms, such as the insertion of short palindromic sequences (CRISPR, data not shown) in V. coralliilyticus P1 and YB1. Although other resistance mechanisms exist, the recurrent isolation of phages that infect V. coralliilyticus YB1 and/or P1 (Efrony et al., 2007; Cohen et al., 2013; Ramphul et al., 2017) supports the idea that V. coralliilyticus pathogens are permissive to viral infection and are suitable candidates for treatment by phage therapy.
The Vibrio phage BONAISHI isolated from coastal waters in the South China Sea is distinct from the known V. coralliiltyicus phages YB2, YC, CKB-S1, CKB-S2, RYC (Efrony et al., 2007; Cohen et al., 2013; Ramphul et al., 2017). Although all these phages belong to the order of Caudovirales (tailed bacteriophages), BONAISHI has an unusually large genome, 303 kbp, rather than 11 kbp to 158 kbp. With such a large genome, BONAISHI is a novel jumbo phage (or giant phage). These are tailed phages with a dsDNA genome larger than 200 kb (Hendrix, 2009; Yuan and Gao, 2017). Jumbo phages have often been isolated in recent years and they mostly infect Gram-negative bacteria, including the genera Synechoccocus, Vibrio, Pseudomonas, Caulobacter, Erwinia, and Aeromonas (Yuan and Gao, 2017). As observed for most jumbo phages, the large genome of BONAISHI is packaged in a large head connected to a long, contractile tail. The genome of jumbo phages typically includes core genes involved in virion structure and assembly, DNA replication, and nucleotide metabolism, including several genes encoding multisubunit RNAP. In BONAISHI, the core genes are scattered throughout the genome sequence and most of them share significant homology with other jumbo phages affiliated to the Phikzvirus genus. BONAISHI is the first marine representative of this divergent group within the Myoviridae. Interestingly, many Phikzviruses are considered to be promising biocontrol agents for plant-pathogenic bacteria (Ralstonia solanacearum, Erwinia amylovora) and some of them are already found in commercial products for phage therapy (Fujiwara et al., 2011; Bhunchoth et al., 2016).
The life history traits and genomic analysis showed that Vibrio phage BONAISHI is a good candidate for biological control of V. coralliilyticus. Firstly, BONAISHI appears to be structurally stable as it can withstand a wider range of pH (3–10) and temperature (4–45°C) than in the environment where it would be used. Second, it readily infects and lyses several pathogenic strains of V. coralliilyticus but no related species. Thirdly incubation experiments showed that the replication cycle is fast (latent period < 3 h). The presence of genes encoding virion-associated RNAP (ORFs 24, 25, 26, and 92) and early-expressed RNAP (ORFs 72, 83, and 84) in the BONAISHI genome may, at least partly, explain the rapid cycle (Ceyssens et al., 2014). In Phikzviruses, these two sets of RNAP may operate in concert during the replication cycle (Ceyssens et al., 2014; Yuan and Gao, 2017). The virion-associated RNAP may be injected into the host cell to start immediate gene expression whereas the early expressed RNAP may function during the middle and late phases of phage gene expression. The consecutive action of these enzymes, unique to Phikzviruses, confers the ability to produce viral progeny independently of the host transcription apparatus (Ceyssens et al., 2014; Yuan and Gao, 2017). In addition, the absence of detectable tRNA in BONAISHI genome suggests that it is well adapted to the translation machinery of its hosts, which is a critical process for efficient phage propagation. Finally, the genomic analysis did not identify any temperate phage hallmarks such as integration mediating enzymes, or genome architecture or sequence similarity with known temperate phages. Furthermore, the absence of homology between BONAISHI gene sequences and bacteria reads from coral metagenomes supports the idea that this phage does not integrate into the host genome. These results suggest very strongly that the Vibrio phage BONAISHI is a strictly lytic phage that is species specific and stable, although it appears to be relatively rare in the environment.
Another important issue for therapeutic applications of phages is to ensure that the candidate does not perform specialized or generalized transduction (Duckworth and Gulig, 2002). Given that BONAISHI appears to be strictly lytic based on the growth experiments and the genome analysis, it is unlikely that this candidate will perform specialized transduction of host DNA. Specialized transduction is restricted to temperate phages and occurs when the prophage is not cleanly excised during induction and includes the flanking bacterial genes which are then packaged in the viral progeny. Our candidate, however, may be able to perform generalized transduction. In this type of transduction, random segments of degraded host chromosome are mistakenly packaged instead of the phage DNA and may be transmitted by horizontal gene transfer. Phages that use a headful DNA packaging mechanism, such as many jumbo phages including BONAISHI, may be able to perform generalized transduction. However, it is, to the best of our knowledge, impossible to predict the frequency of generalized transduction based purely on the genome analysis. For example, giant bacteriophages with similar headful DNA packaging mechanisms can have very different transduction rates as, for example, the T4 and T4G bacteriophages (Young et al., 1982; Young and Edlin, 1983). The ability of BONAISHI to perform generalized transduction would, therefore, require proper laboratory investigation.
An alternative to avoid potential issue with phage-mediated gene transfer is, rather than using whole bacetriophages, to use bacteriolytic proteins encoded by phages, among which the most notable are phage-encoded peptidoglycan hydrolases (PGH, see review by Roach and Debarbieux, 2017). PGHs, also called endolysins, degrade the cell peptidoglycan from within and contribute to the release of progeny and cell burst. A second type of PGH can be associated with the virion and initiate cell wall penetration through localized peptidoglycan or lipopolysaccharide degradation during the infection process. Both types of PGH are already used as bacteriocins in animal models of human infection and disease (see Roach and Debarbieux, 2017 and references therein). Jumbo phages typically encode more proteins for the lysis of the host cell wall including endolysin, glycoside hydrolase and chitinase, which are often bound to the virion than small genome phages (Yuan and Gao, 2017). In BONAISHI genome, we identified two glycoside hydrolases distantly related to known enzymes that belong to the families GH19 and GH23 using the expert database CAZY. Although the catalytic activities of these molecules cannot be determined based solely on the genome analysis, their overexpression and characterization might provide interesting tools for controlling V. coralliilyticus infection.
A preliminary assay suggests that BONAISHI is a promising candidate for treating V. coralliilyticus infection. Studies investigating the action of V. coralliilyticus on coral symbionts showed that photosynthesis was inactivated by the expression of a Zn-metalloprotease (Sussman et al., 2009). Our experiments on Symbiodinum cultures infected by V. coralliilyticus showed that BONAISHI phage treatment was effective: adding the phage to the infected cultures rapidly reduced Symbiodinium PSII inactivation. As reported in previous studies, phage addition probably lysed the bacterial pathogen, stopping Zn-metalloprotease production and further damage to Symbiodinium sp. cells (Cohen et al., 2013). Future studies should now focus on the effectiveness of the treatment either under realistic field conditions or in mesocosms to start including bacteriophages (and/or derived compounds) in an integrated management program to mitigate the damage caused by the infectious agents responsible for coral diseases. We recommend genome sequencing and analysis of any future phage candidate as a prerequisite to any field test to ensure safe environmental applications as this provides essential information on the phage replication cycle and host-virus interactions.
Author contributions
A-CB, YB, and TB, designed the study. LJ, JM, and A-CB performed the experiments and analyzed the results. LJ, SH, CD, and EC performed the bioinformatics analyses. CF-P provided and helped with the diseased Symbiodium cultures. LJ and A-CB wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported jointly by the EC2CO PATRICIA Project, the TOTAL Foundation, and the ANR CALYPSO (ANR-15-CE01-0009). We would like to thank Andrew Millard for discussion on phage transduction, Simon Roux for his help with metagenomic analysis, Gurvan Michel for his assistance with CAZYmes annotation, Pei Ge for her technical assistance, and Tony Tebby for the manuscript editing. We would also like thank the three reviewers for their constructive comments on a previous version of this manuscript. We are also very grateful to Michel Galey, Alexandre Portier, and all the staff from Whale Island Resort for their hospitality and help during our stay.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02501/full#supplementary-material
References
- Ackermann H.-W., Heldal M. (2010). Basic electron microscopy of aquatic viruses, in Manual of Aquatic Viral Ecology, eds Wilhelm S. W., Weinbauer M. G., Suttle C. A. (Waco, TX: ASLO; ), 182–192. [Google Scholar]
- Atad I., Zvuloni A., Loya Y., Rosenberg E. (2012). Phage therapy of the white plague-like disease of Favia favus in the Red Sea. Coral Reefs 31, 665–670. 10.1007/s00338-012-0900-5 [DOI] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., Dejongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwood D. R., Hughes T. P., Folke C., Nyström M. (2004). Confronting the coral reef crisis. Nature 429, 827–833. 10.1038/nature02691 [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y., Thompson F. L., Thompson C. C., Cnockaert M. C., Hoste B., Swings J., et al. (2003). Vibrio coralliilyticus sp. nov. a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 53, 309–315. 10.1099/ijs.0.02402-0 [DOI] [PubMed] [Google Scholar]
- Besemer J., Lomsadze A., Borodovsky M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory region. Nucleic Acids Res. 29, 2607–2618. 10.1093/nar/29.12.2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhunchoth A., Blanc-Mathieu R., Mihara T., Nishimura Y., Askora A., Phironrit N., et al. (2016). Two asian jumbo phages, ϕRSL2 and ϕRSF1, infect Ralstonia solanacearum and show common features of ϕKZ-related phages. Virology 494, 56–66. 10.1016/j.virol.2016.03.028 [DOI] [PubMed] [Google Scholar]
- Brussaard C. P. D. (2004). Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol 70, 1506–1513. 10.1128/AEM.70.3.1506-1513.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard C. P. D., Baudoux A.-C., Rodriguez-Varela F. (2016). Marine viruses, in The Marine Microbiome–an Untold Resource of Biodiversity and Biotechnological Potential. eds. Stal L. J., Cretoiu S. M. C. (Springer International Publishing; ), 305–32. [Google Scholar]
- Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. (2009). DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25, 119–120. 10.1093/bioinformatics/btn578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceyssens P. J., Minakhin L., Van den Bossche A., Yakunina M., Klimuk E., Blasdel B., et al. (2014). Development of giant bacteriophage phi KZ is independent of the host transcription apparatus. J. Virol. 88:105010 10.1128/JVI.01347-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. H., Yang J., Yu J., Yao Z. J., Sun L. L., Shen Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33:D325–D328. 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., Heiner C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10, 563–569 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Choo S. W., Heydari H., Tan T. K., Siow C. C., Beh C. Y., Wee W. Y., et al. (2014) VibrioBase: a model for next-generation genome annotation database development. Sci. World J. 2014:569324. 10.1155/2014/569324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y., Joseph Pollock F., Rosenberg E., Bourne D. G. (2013). Phage therapy treatment of the coral pathogen Vibrio coralliilyticus. Microbiol. Open 2, 64–74. 10.1002/mbo3.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell J. H. (1978). Diversity in tropical rain forests and coral reefs. Science 199, 1302–1310. 10.1126/science.199.4335.1302 [DOI] [PubMed] [Google Scholar]
- Cooper E. L., Hirabayashi K., Strychar K. B., Sammarco P. W. (2014). Corals and their potential applications to integrative medicine. Evid. Based Complement. Alternat. Med. 2014:184959. 10.1155/2014/184959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang V. T., Howard-Varona C., Schwenck S., Sullivan M. B. (2015). Variably lytic infection dynamics of large B acteroidetes podovirus phi38: 1 against two Cellulophaga baltica host strains. Environ. Microbiol. 17, 4659–4671. 10.1111/1462-2920.13009 [DOI] [PubMed] [Google Scholar]
- Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641. 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Herelle F. (1926). The Bacteriophage and Its Behavior. Baltimore, MD: Williams & Wilkins, 490–497. [Google Scholar]
- Dinsdale E. A., Pantos O., Smriga S., Edwards R. A., Angly F., Wegley L., et al. (2008). Microbial ecology of four coral atolls in the Northern Line Islands. PLoS ONE 3:e1584. 10.1371/journal.pone.0001584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth D. H. (1976). Who discovered bacteriophage? Bacteriol. Rev. 40:793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth D. H., Gulig P. A. (2002). Bacteriophages. BioDrugs 16, 57–62. 10.2165/00063030-200216010-00006 [DOI] [PubMed] [Google Scholar]
- Efrony R., Atad I., Rosenberg E. (2009). Phage therapy of coral white plague disease: properties of phage BA3. Curr. Microbiol. 58, 139–145. 10.1007/s00284-008-9290-x [DOI] [PubMed] [Google Scholar]
- Efrony R., Loya Y., Bacharach E., Rosenberg E. (2007). Phage therapy of coral disease. Coral Reefs 26, 7–13. 10.1007/s00338-006-0170-1 [DOI] [Google Scholar]
- Fujiwara A., Fujisawa M., Hamasaki R., Kawasaki T., Fujie M., Yamada T. (2011). Biocontrol of Ralstonia solanacearum by treatment with lytic bacteriophages. Appl. Environ. Microbiol. 77, 4155–4162. 10.1128/AEM.02847-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren M., Son K., Raina J.-B., Rusconi R., Menolascina F., Shapiro O. H., et al. (2014). A bacterial pathogen uses dimethylsulfonioproprionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007. 10.1038/ismej.2013.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goiran C., Al-Moghrabi S., Allemand D., Jaubert J. (1996). Inorganic carbon uptake for photosynthesis by the symbiotic coral/dinoflagellate association I. Photosynthetic performances of symbionts and dependence on sea water bicarbonate. J. Exp. Mar. Biol. Ecol. 199, 207–225. 10.1016/0022-0981(95)00201-4 [DOI] [Google Scholar]
- Grissa I., Vergnaud G., Pourcel C. (2007). CRISPRfinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35(Suppl. 2), W52–7. 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard R., Ryther J. (1962). Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea. Can. J. Microbiol. 8, 229–239. 10.1139/m62-029 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Hendrix R. W. (2009). Jumbo Bacteriophages. Curr. Top. Microbiol. Immunol. 328, 229–240. 10.1007/978-3-540-68618-7_7 [DOI] [PubMed] [Google Scholar]
- Holmfeld K., Howard-Varona C., Solonenko N., Sullivan M. B. (2014). Contrasting genomic patterns and infection strategies of two co-existing Bacteroidetes podovirus genera. Environ. Microbiol. 16, 2501–13. 10.1111/1462-2920.12391 [DOI] [PubMed] [Google Scholar]
- Howard-Varona C., Hargreaves K. R., Abedon S. T., Sullivan M. B. (2017). Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11, 1511–1520. 10.1038/ismej.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. P., Baird A. H., Bellwood D. R., Card M., Connolly S. R., Folke C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–33. 10.1126/science.1085046 [DOI] [PubMed] [Google Scholar]
- Jackson J. B. C., Kirby M. X., Berger W. H., Bjorndal K. A., Botsford L. W., Bourque B. J., et al. (2001). Historical overfishing and the recent collapse of coastal ecosystems. Science 293, 629–637. 10.1126/science.1059199 [DOI] [PubMed] [Google Scholar]
- Jiang S. C., Paul J. H. (1996). Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 142, 27–38. 10.3354/meps142027 [DOI] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., et al. (2014). InterProScan 5: genome-sclae protein function classification. Bioinformatics 30, 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D., Canback B. (2004) ARAGORN, a program to detect tRNA genes tmRNA genes in nucleotide sequences. Nucleic Acids Res 32, 11–16. 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R., Lima-Mendez G., Toussaint A. (2006). A first global analysis of plasmid encoded proteins in the ACLAME database. FEMS Microbiol. Rev. 30, 980–994. 10.1111/j.1574-6976.2006.00044.x [DOI] [PubMed] [Google Scholar]
- Lim E., Pon A., Djoumbou Y., Knox C., Shrivastava S., Guo A. C., et al. (2009). T3DB: a comprehensively annotated database of common toxins and their targets. Nucleic Acids Res. 38(suppl. 1), D781–D786. 10.1093/nar/gkp934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobocka M., Hejnowicz M. S., Gagala U., Weber-Dabrowska B., Wegrzyn G., Dadlez M. (2014). The first step to bacteriophage therapy: how to choose the correct phage, in Phage Therapy: Current Research and Applications, eds Borysowski J., Miedzybrodsky R., Gorski A. (Norfolk, UK: Caister Academic Press; ), 23–67. [Google Scholar]
- Lowe T. M., Eddy S. R. (1997). rRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mera H., Bourne D. G. (2018). Disentangling causation: complex roles of coral associated microorganisms in disease. Environ. Microbiol. 20, 431–449. 10.1111/1462-2920.13958 [DOI] [PubMed] [Google Scholar]
- Middelboe M., Chan A. M., Bertelsen S. K. (2010) Isolation life cycle characterization of lytic viruses infecting heterotrophic bacteria cyanobacteria, in Manual of Aquatic Viral Ecology, edsWilhelm S. W., Weinbauer M. G., Suttle C. A. (Waco, TX: ASLO; ), 118–133. [Google Scholar]
- Moberg F., Folke C. (1999). Ecological goods and services of coral reef ecosystems. Ecol. Econ. 29, 215–233. 10.1016/S0921-8009(99)00009-9 [DOI] [Google Scholar]
- Moura A., Soares M., Pereira C., Leitão N., Henriques I., Correia A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. 10.1093/bioinformatics/btp105 [DOI] [PubMed] [Google Scholar]
- O'Santos E., Alves N., Dias G. M., Mazotto A. M., Vermelho A., Vora G. J., et al. (2011). Genomic and proteomic analyses of the coral pathogen Vibrio coralliilyticus reveal a diverse virulence repertoire. ISME J. 5, 1471–83. 10.1038/ismej.2011.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino P., Chen I.-M. A., Palaniappan K., Ratner A., Chu K., Szeto E., et al. (2017). IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses. Nucleic Acids Res. 45, D457–D465. 10.1093/nar/gkw1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi J. M., Bradbury R. H., Sala E., Hughes T. P., Bjorndal K. A., Cooke R. G., et al. (2003). Global trajectories of the long-term decline of coral reef ecosystems. Science 301, 955–958. 10.1126/science.1085706 [DOI] [PubMed] [Google Scholar]
- Pollock F. J., Krediet C. J., Garren M., Stocker R., Winn K., Wilson B., et al. (2015). Visualization of coral host–pathogen interactions using a stable GFP-labeled Vibrio coralliilyticus strain. Coral Reefs 34, 655–662. 10.1007/s00338-015-1273-3 [DOI] [Google Scholar]
- Ramphul C., Estela B., Dohra H., Suzuki T., Yoshimatsu K., Yoshinaga K., et al. (2017). Marine genomics genome analysis of three novel lytic Vibrio coralliilyticus phages isolated from seawater, Okinawa, Japan. Mar. Genomics 35, 69–75. 10.1016/j.margen.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Roach D. R., Debarbieux L. (2017). Phage therapy: awakening a sleeping giant. Emerg. Topics Life Sci. 1, 93–103. 10.1042/ETLS20170002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S., Faubladier M., Mahul A., Paulhe N., Bernard A., et al. (2011) Metavir: a web server dedicated to virome analysis. Bioinformatics 27, 3074–3075. 10.1093/bioinformatics/btr519. [DOI] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M. A., et al. (2000) Artemis: sequence visualization annotation. Bioinformatics 16, 944–945. 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Seshadri R., Kravitz S. A., Smarr L., Gilna P., Frazier M. (2007) CAMERA: a community resource for metagenomics. PLoS Biol 5:e75. 10.1371/journal.pbio.0050075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M., Mieog J. C., Doyle J., Victor S., Willis B. L., Bourne D. G. (2009). Vibrio zinc-metalloprotease causes photoinactivation of coral endosymbionts and coral tissue lesions. PLoS ONE 4:e4511. 10.1371/journal.pone.0004511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M., Willis B. L., Victor S., Bourne D. G. (2008). Coral pathogens identified for White Syndrome (WS) epizootics in the Indo-Pacific. PLoS ONE 3:e2393. 10.1371/journal.pone.0002393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M., Ritchie K. (2009). How feasible is the biological control of coral diseases? Trends Ecol. Evol. 24, 378–385. 10.1016/j.tree.2009.02.008 [DOI] [PubMed] [Google Scholar]
- Ushijima B., Videau P., Burger A. H., Shore-Maggio A., Runyon C. M., Sudek M., et al. (2014). Vibrio coralliilyticus strain OCN008 is an etiological agent of acute Montipora white syndrome. Appl. Environ. Microbiol. 80, 2102–2109. 10.1128/AEM.03463-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weynberg K. D., Voolstra C. R., Neave M. J., Buerger P., van Oppen M. J. H. (2015) From cholera to corals: viruses as drivers of virulence in a major coral bacterial pathogen. Sci. Rep. 5:17889. 10.1038/srep17889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S. J., Paul J. H. (2006). Environmental factors that influence the transition from lysogenic to lytic existence in the φHSIC/Listonella pelagia marine phage–host system. Microbial Ecol. 52, 217–225. 10.1007/s00248-006-9113-1 [DOI] [PubMed] [Google Scholar]
- Wishart D., Arndt D., Pon A., Sajed T., Guo A. C., Djoumbou Y., et al. (2015). T3DB: the toxic exposome database. Nucleic Acids Res. 43, D928–D934. 10.1093/nar/gku1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K. K., Edlin G. (1983). Physical and genetic analysis of bacteriophage T4: generalized transduction. Mol. Gen. Genet. 192, 241–246. 10.1007/BF00327673 [DOI] [PubMed] [Google Scholar]
- Young K. K., Edlin G., Wilson G. G. (1982). Genetic analysis of bacteriophage T4: transducing bacteriophages. J. Virol. 41, 345–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Gao M. (2017). Jumbo bacteriophages: an overview. Front. Microbiol. 8, 1–9. 10.3389/fmicb.2017.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C. E., Smith J., Lam M., Zemla A., Dyer M. D., Slezak T. (2006). MvirDB—a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res. 35(suppl. 1), D391–D394. 10.1093/nar/gkl791 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.