Abstract
Aberrant expression of G protein-coupled receptors (GPCRs) is frequently associated with tumorigenesis. G Protein-coupled receptor class C group 5 member A (GPRC5A) is a member of the GPCR superfamily, is expressed preferentially in lung tissues, and is regulated by various entities at multiple levels. GPRC5A exerts a tumor suppressive role in lung cancer and GPRC5A deletion promotes lung tumor initiation and progression. Recent advances have highlighted that GPRC5A dysregulation is found in various human cancers and is related to many tumor-associated signaling pathways, including the cyclic adenosine monophosphate (cAMP), nuclear factor (NF)-κB, signal transducer and activator of transcription (STAT) 3, and focal adhesion kinase (FAK)/Src signaling. This review aimed to summarize our updated view on the biology and regulation of GPRC5A, its expression in human cancers, and the linked signaling pathways. A better comprehension of the underlying cellular and molecular mechanisms of GPRC5A will provide novel insights into its potential diagnostic and therapeutic value.
1. Introduction
The early and accurate diagnosis of cancer is a long-standing problem which, if solved, can significantly improve the patient prognoses. For this purpose, modern molecular diagnosis is an advanced and essential detection technique. As cancer is the result of the accumulation of adverse disease-related molecular events, it is reasonable to stratify patients according to genetic alterations in one or more genes. This has become an important factor in clinical intervention [1, 2]. However, while a number of cancer biomarkers for molecular diagnosis have been described recently, the specificity and diagnostic capacity of currently available biomarkers are limited [3–5]. Therefore, there remains a requirement for novel biomarkers with high specificity and sensitivity.
G protein-coupled receptors (GPCRs) are one of the largest and most diverse superfamilies of receptors and play a key role in a broad variety of physiological processes [6]. GPCRs are characterized by a common structure of one bundle of seven transmembrane helices connected by three extracellular and three intracellular loops. The vast majority of ligands interact with the extracellular oriented part of the helices [7, 8]. Due to their broad physiological functions, aberrant GPCRs activation is frequently associated with disease initiation and progression. A number of studies have indicated the critical role of GPCRs in tumor proliferation, invasiveness, angiogenesis, metastasis, and drug resistance [9–16]. Notably, GPCRs are highly attractive targets in drug design, accounting for more than 30% of all commercially available pharmaceutical drugs [17, 18]. Recently, G protein-coupled receptor family C group 5 member A (GPRC5A), a member of class C orphan GPCRs, has been found to be dysregulated in several human cancers. It has been shown to have an important effect on tumor progression [19]. This review will focus on recent advances in GPRC5A research and on its role in human cancer.
2. The GPCR Family C Group 5
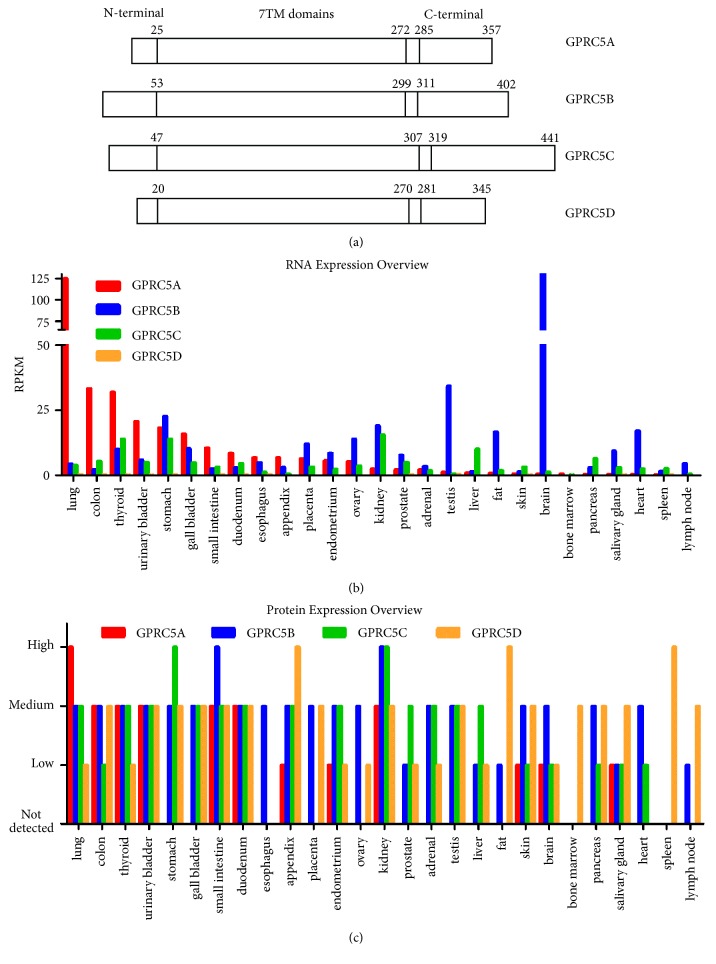
GPRC5A, also known as retinoic acid-induced protein 3 (RAI3) or retinoic acid-inducible gene (RAIG) 1, is a member of class C orphan GPCRs. GPRC5A was first described in the UMSCC-22B cell line as an all-transretinoic acid- (ATRA-) responsive gene [16], is located on 12p13.1, and encodes a 40 kDa protein. Three other members of this group, namely, GPRC5B (also known as RAIG2), GPRC5C (also known as RAIG3), and GPRC5D, were consequently identified [16, 20–22]. The four proteins share 31–42% amino acid sequence identity and have high sequence similarity within their transmembrane domains [22]. GPRC5A, GPRC5B, and GPRC5C can be induced by retinoic acid (RA) in a concentration- and time-dependent manner, whereas GPRC5D cannot. Unlike other members of the C family of GPCRs, whose ligand binding sites are located within the large N–terminal domain, the four members of group 5 possess very short N-terminals of 30–50 amino acids. Instead of binding to the N terminal domain, agonists can bind to the 7TM domains of the four proteins [22, 23]. The protein structure of GPRC5A–D is summarized in Figure 1(a). Interestingly, GPRC5A–D are expressed in a tissue-specific manner with GPRC5A being preferentially expressed in lung tissues; GPRC5B is predominately localized in tissues of the central nervous system, while GPRC5C and GPRC5D are observed in a variety of tissues (Figures 1(b) and 1(c)) [21, 22, 24].
Figure 1.
(a) The protein structure of GPRC5A–D. The amino acids of the four proteins are detailedly numbered. They have similar length of 7TM domain and have short N-ternimal of 20-53 amino acids as detailed in the text. (b) The mRNA expression profile of GPRC5A–D in different organs. Data was compiled from the RNA sequence conducted by Fagerberg L. et al. (c) The protein expression levels of GPRC5A–D (Data from the Human Protein Atlas http://www.proteinatlas.org/).
3. Regulation of GPRC5A Expression
3.1. Transcriptional Regulation
As summarized in a previously published review, the GPRC5A gene has many transcription factor binding sites; among these, RA is the most well studied. RA is a vitamin A-derived morphogen with many effects on cell growth and differentiation [25, 26]. GPRC5A has a RA response element (RARE) in its 5' upstream region, which binds the RA receptor (RAR)/retinoid X receptor (RXR) heterodimer. In the presence of RA, the inhibitory effect of the RAR/RXR heterodimer on the transcription of GPRC5A is relieved, resulting on the transcription of the gene [27, 28].
A custom-made cDNA microarray analysis showed that GPRC5A expression is induced when the levels of cyclic adenosine monophosphate (cAMP) increase. Specifically, a cAMP-responsive element (CRE) motif exists close to the transcription initiation site of GPRC5A. By upregulating cAMP levels, forskolin induces GPRC5A transcription and this effect can be strengthened by RA [29].
Furthermore, microarray and quantitative polymerase chain reaction (qPCR) assays conducted in four p53-mutant cell lines (MDA-MB-468, BT-20, BT-549, and SK-BR-3) and four p53 wild-type cell lines (MCF-7, T47D, ZR-75-1, and BT-474) have demonstrated that GPRC5A is a target of P53 and is suppressed by wild-type p53 [30]. In the same study, chromatin immunoprecipitation (ChIP) assays indicated that p53 binds to GPRC5A in a sequence-specific way in human ovarian tumor cell line 2774qw1. Additionally, overexpression of p53 in the p53-null human nonsmall cell lung cancer (NSCLC) H1299 cells led to increased GPRC5A expression, while p53 knockdown in the p53 wild-type human NSCLC A549 cells resulted in decreased expression of GPRC5A, indicating that GPRC5A is involved in the antitumor effect of p53 in NSCLC cells [31].
Additionally, GPRC5A expression is also related to BRCA1 status. In breast tumors with wild-type BRCA1, GPRC5A expression is higher than in BRCA1-mutated tumors. In vitro experiments show that knockdown of BRCA1 results in decreased expression of GPRC5A in MDA-MB-231 breast cancer cells, while the opposite results are obtained with BRCA1 overexpression [32].
3.2. Post-Transcriptional Regulation
Little is currently known about the post-transcriptional regulation of GPRC5A. MicroRNAs (miRNAs), small noncoding RNA molecules that regulate the expression of target genes in a sequence-dependent way, are important post-transcriptional regulators [33, 34]. A computational analysis conducted in a previous review using the RNA22 algorithm indicated that there are many putative miRNAs targeting GPRC5A, specifically 343 in the 5'untranslated region (UTR), 595 in the coding sequence (CDS), and 1170 in the 3' UTR [19]. Of these, miR-103a-3p has been extensively studied. miR-103a-3p has two target sites in the 5'UTR of GPRC5A, and in vitro studies have found that it suppresses the expression of GPRC5A mRNA and protein by binding to either of them [35]. Besides, miR-204 can inhibit GPRC5A expression via binding to its 3' UTR in gastric cancer (GC) [36].
RNA binding proteins (RBPs) also participate in the posttranscriptional regulation of GPRC5A. HuR, an RBP encoded by the ELAVL1 gene, was identified to upregulate GPRC5A expression via mRNA stabilization by binding to the 3' UTR of GPRC5A [37, 38]. Other crucial posttranscriptional regulators such as long noncoding RNAs (lncRNAs) are thought to significantly impact the regulation of GPRC5A, but evidence remains lacking [39–41].
3.3. Post-Translational Modification of GPRC5A
GPRC5A has several phosphorylation sites which have been found to be involved in many biological processes. Phosphorylation of serine (SER) 301 and 345 takes place during mitosis [42, 43]. The phosphorylation of GPRC5A in two conserved double-tyrosine (TYR) motifs, TYR-317/TYR-320, and TYR-347/TYR-350 is mediated by epidermal growth factor receptor (EGFR), leading to inactivation of the protein's tumor suppressive function [44, 45]. Furthermore, sequence analysis predicts that the arginine (ARG) 158 site of GPRC5A can be N-glycosylated. Additionally, several studies indicate that GPRC5A can be ubiquitinated at a number of sites, although the details remain to be clarified [46–53].
4. GPRC5A and Downstream Signaling Pathways
4.1. cAMP Signaling Pathway
GPRC5A is one of several genes whose expression increases when the cAMP level is elevated. As mentioned above, cAMP binds to the CRE motif in the GPRC5A gene inducing its transcription. In the same study, the authors found that, in the human thyroid epithelial cell line Nthy, GPRC5A expression is negatively correlated with intracellular cAMP and Gs alpha levels and that the suppression of GPRC5A results in inhibition of cell growth and induction of apoptosis [29]. These results suggest that there exists a negative feedback loop between cAMP and GPRC5A that also involves Gs alpha.
4.2. Nuclear Factor- (NF-) κB Signaling Pathway
NF-κB controls the expression of genes involved in many biological and pathological processes, and plays a critical role in inflammation and tumorigenesis. Dysregulation of NF-κB is related to pathological alterations in various cells including epithelial and stromal cells [54, 55]. In vivo studies have demonstrated that GPRC5A knockout mice are more sensitive to lipopolysaccharide- (LPS-) induced NF-κB signaling activation than are GPRC5A wild-type mice and that they have lower levels of proinflammatory cytokines and chemokines. Moreover, in vitro studies showed that GPRC5A knockout cells produce higher levels of chemokines and cytokines and promote broader macrophage migration through their conditioned medium compared to GPRC5A wild-type cells in a NF-κB dependent manner [56]. Additionally, they found that selective inhibition of NF-κB through the expression of the superrepressor IkBa in the GPRC5A knockout mice significantly alleviates the inflammation response and mice lung injury induced by LPS [57]. However, all of these results were based on the deletion of GPRC5A, and further in-depth studies are warranted to further explore the relationship.
4.3. Signal Transducer and Activator of Transcription (STAT) 3 Signaling Pathway
STATs are transcription factors that regulate cell growth, differentiation, survival and development by mediating the expression of target genes [58]. STAT3 is the best studied member of the STAT family. Aberrant activation of STAT3 has been identified in various human cancers, and correlates with poor prognosis in gastric, breast and lung cancer [59–65]. Recent studies suggest that GPRC5A is involved in the regulation of STAT3 signaling pathway. Knockdown of GPRC5A correlates with STAT3 activation in cancers such as lung cancer and head and neck squamous cell carcinoma (HNSCC), pointing to a tumor suppressive role for GPRC5A. Compared to GPRC5A wild-type cells, GPRC5A knockout cells have higher levels of activated-STAT3 and STAT3-regulated anti-apoptotic genes, independent of the presence of exogenous epidermal growth factor (EGF), resulting in enhancement of tumor progression [66, 67]. Contrarily, another study indicates that GPRC5A is positively correlated with STAT3, and that GPRC5A silencing is associated with suppression of STAT3 phosphorylation at TYR705 in human pancreatic cell lines [68]. These data suggest that in some cases GPRC5A may play an oncogenic role by activating STAT3 signaling and in others has a tumor suppressor role through STAT3 phosphorylation inhibition.
4.4. Focal Adhesion Kinase (FAK)/Src Signal Pathway
The regulation of cell-cell and cell-matrix adhesion plays a vital role in the integrity and homeostasis of epithelial tissue [69, 70], and interference with this process may contribute to tumor progression. The most important function of the FAK signal pathway is regulating cell adhesion [71–74]. GPRC5A silencing deregulates integrin β 1 (ITGB1) expression leading to restrained capacity of integrin-mediated cell adhesion. GPRC5A knockout interferes with the activation of the FAK/Src signaling pathway and the activity of downstream RhoA and Rac1 small GTPases [75].
5. GPRC5A and Its Role in Human Cancer
Although GPRC5A is predominately expressed in normal lung tissues, dysregulation of GPRC5A expression has been observed in a variety of human cancers (Table 1).
Table 1.
The dysregulation of GPRC5A in human cancers.
| Study | Year | Tumor type | Kind/num. of analyzed samples | Methods | Expression status | Potential clinical significant |
|---|---|---|---|---|---|---|
| Nagahata, T. | 2005 | BC | 25 primary BC tissues | qRT-PCR | up | None |
|
| ||||||
| Tao, Q. | 2007 | LC | 18 pairs of human LC tissues and adjacent normal tissues; microarray with 186 LC tissues and 17 normal lung samples | qRT-PCR; Microarray |
down | None |
|
| ||||||
| Dairkee, S. H. | 2009 | BC | 50 pairs of cDNAs from matched BC and normal breast tissues; 147 invasive BC and 44 normal breast tissues | CPA;IHC | up | No association with clinical parameters and prognosis |
|
| ||||||
| Cheng, L. | 2012 | GC | 25 paired GC tissues and matched adjacent non-tumor tissues | Microarray; qRT-PCR |
up | None |
|
| ||||||
| Fujimoto, J. | 2012 | NSCLC | 31 NBE,24 COPD,26 COPD with cancer and 474 NSCLC tissnes;6 NSCLC and matched normal lung tissues | Microarray; qRT-PCR |
down | Positively associated with adenocarcinoma histology; highly suppressed in NCSSLC |
|
| ||||||
| Liu, S. L. | 2013 | OSCC | 60 paired primary OSCC and adjacent normal specimens | IHC | down | Inversely correlated with the malignant grade |
|
| ||||||
| Subrungruanga, I. | 2013 | ICC | 18 paired ICC and matched normal tissues | Microarray; qRT-PCR |
up | None |
|
| ||||||
| Zougman, A. | 2013 | CC | 347 CC specimens | Microarray | up | Positively correlated with tumor recurrence |
|
| ||||||
| Kume, H. | 2014 | CRC | 33 primary CRC and 16 colon polyps | LC-MS/MS | up | None |
|
| ||||||
| Lin, X. | 2014 | NSCLC | 129 paired NSCLC and adjacent normal tissues | IHC | down | None |
|
| ||||||
| Sokolenko, A. P. | 2014 | BC | 17 BC with BRCA1 mutation and 94 BRCA1 non-mutation tissues | qRT-PCR | down in BRCA1 mutation samples | Inversely correlated with BRCA1 mutation |
|
| ||||||
| Zheng, J. | 2014 | HCC | 106 HCC | qRT-PCR;WB; IHC |
up | Positively correlated with advanced TNM stage, high serum AFP, vascular invasion, tumor recurrence, DFS and OS |
|
| ||||||
| Liu, H. | 2016 | GC | 30 paired GC and adjacent normal tissues;106 GC samples | qRT-PCR;WB; IHC |
up | Positively associated with tumor size, diffuse type, serosal invasion,lymph node metastasis and OS |
|
| ||||||
| Zhou, H. | 2016 | Pancreatic cancer | 46 normal pancreatic tissues,145 primary pancreatic tumors and 61 metastatic tumors;203 samples pancreatic tumors | Microarray; IHC |
up | None |
|
| ||||||
| Jahny, E. | 2017 | PDAC | 435 PDAC and 209 non-cancerous pancreatic tissues | Microarray | up | None |
|
| ||||||
| Liu, S. | 2017 | HNSCC | 86 paired HNSCC and adjacent normal tissues | IHC | down | Positively associated with tumor differentiation |
|
| ||||||
| Zhang, L. | 2017 | CRC | 57 paired CRC and adjacent normal tissues | qRT-PCR | up | Positively associated with tumor grade |
BC: Breast cancer; LC: lung cancer; GC: gastric cancer; NBE: normal bronchial epithelia; COPD: chronic obstructive pulmonary disease; NSCLC: nonsmall cell lung cancer; OSCC: oral squamous cell carcinoma; ICC: intrahepatic cholangiocarcinoma; CC: Colon cancer; CRC: colorectal cancer; HCC: hepatocellular carcinoma; PDAC: pancreatic ductal adenocarcinoma; HNSCC: head and neck squamous cell carcinoma; qRT-PCR: quantitative real-time PCR; CPA: cancer profiling array; IHC: immunohistochemistry; LC-MS/MS: liquid chromatography with tandem mass spectrometry; WB: Western blot; DFS: disease-free survival; OS: overall survival.
5.1. GPRC5A and Lung Cancer
GPRC5A exhibits a promising tumor suppressive role in lung cancer. Its expression, both at the mRNA and protein level, is much lower in lung cancer than in healthy lung tissue [66, 76–78]. According to recent reports, the expression of GPRC5A is the highest in disease-free normal bronchial epithelia (NBE), intermediate in cancer-free lungs from patients with chronic obstructive pulmonary disease (COPD) and the lowest in patients with COPD and lung cancer [77]. Moreover, homozygous GPRC5A knockout mice are more likely to spontaneously develop lung tumors than GPRC5A heterozygous or wild-type mice, with tumor incidence rates of 76%, 11%, and 10%, respectively [78]. In vitro experiments demonstrated that overexpression of GPRC5A inhibits cell viability and colony-formation and enhances apoptosis in NSCLC cell lines [31, 56, 66, 78]. Similar results were found in another study, which reported that lung epithelial cells from GPRC5A wild-type mice have worse viability and colony-formation ability than lung cells from GPRC5A knockout mice [56]. Importantly, the effect of GPRC5A knockout on lung tumorigenesis can be strengthened by tobacco-specific carcinogen nicotine-derived nitrosamine ketone (NNK). The NNK-treated group developed lung adenocarcinoma sooner than the saline-treated control group, an effect which was most likely enhanced by mutations in multiple genes, such as those for ATM, histone methyltransferase 2D (KMT2D), neurofibromatosis type 1 (NF1), transformation related protein 53 (Trp53), MET, and enhancer of zeste homolog 2 (Ezh2) [79, 80]. Several parallel studies showed that GPRC5A exerts its tumor suppressive effect by regulating the NF-κB and EGFR/STAT3 signaling pathways. Compared to wild-type cells, the NF-κB signaling pathway is activated in GPRC5A knockout cells, which contributes to lung inflammation and tumorigenesis. These effects can be reversed by silencing of the P65 subunit of NF-κB [79, 81]. Additionally, GPRC5A knockout enhances the transformed phenotype in normal and tumor cells through the aberrant activation of the EGFR/STAT3 signaling pathway [66]. Interestingly, there is a mutual effect between EGFR and GPRC5A. On the one hand, EGF induces TYR phosphorylation on the C terminal of GPRC5A, resulting in the suppression of GPRC5A-mediated inhibition of cell invasion and anchorage-independent growth of NSCLCs [45]. On the other hand, GPRC5A interacts with EGFR through its 7TM domains, leading to the activation of EGFR/STAT3 signaling pathway and its downstream target genes, preventing spontaneous and ionizing radiation-induced lung tumorigenesis [82].
5.2. GPRC5A and Breast Cancer
Elevated GPRC5A mRNA expression has been observed in breast cancer cell lines and clinical tumor tissues (25 primary breast cancer tissues), and GPRC5A knockdown leads to inhibition of cell growth in cell lines MCF7 and T47D [83]. Similar results have been obtained in 293 cells (HEK-293 F cells) which exhibited augmented anchorage-independent growth ability upon GPRC5A ectopic expression [30]. Furthermore, GPRC5A together with FXYD domain-containing ion transport regulator 3 (FXYD3) and PYCARD have been reported as potential predictors of pathological grading of breast cancer and might benefit the management of clinical treatments [84]. However, immunohistochemical (IHC) analysis of a tissue microarray consisting of 147 invasive breast cancer samples and 44 normal breast tissue samples showed that GPRC5A is abundantly expressed in breast cancers, whereas no association was discovered between GPRC5A expression and clinicopathological characteristics [85]. Additionally, knockout of GPRC5A results in reduced cell adhesion and spreading ability, via deregulation of ITGB1 expression and suppression of FAK/Src signaling [75]. All these results reveal a tumor-promoting role of GPRC5A in breast cancer. However, one early study suggested that GPRC5A exhibits a tumor-suppressive role in EGFR-expressing MDA-MB-231 cells and that GPRC5A knockdown promotes colony formation, cell growth, cell migration and invasion capacities in this cell line, but has no such effect in EGFR-negative MCF7 cells. Specifically, GPRC5A knockdown augmented EGF signaling, an effect which can be reversed by inhibiting EGFR phosphorylation [86].
5.3. GPRC5A and Colorectal Cancer
GPRC5A is highly expressed in colorectal cancer (CRC), and elevated GPRC5A expression is significantly associated with inferior prognosis [87, 88]. In addition, liquid chromatography analysis demonstrates that GPRC5A expression is lower in polyps than in metastatic and non-metastatic CRC samples, suggesting that GPRC5A may serve as a biomarker to differentiate CRC from normal tissues [89]. What is more, GPRC5A deficiency reduces cell proliferation and promotes cell apoptosis in vitro and inhibits tumorigenesis of a colitis-associated cancer model in vivo. Furthermore, GPRC5A can be induced by hypoxia, regulates the NF-κB-mediated expression of Vanin-1 (a key enzyme of cysteamine generation), and influences the reactive oxygen levels contributing to tumor progression [88, 90].
5.4. GPRC5A and GC
GPRC5A is expressed in the membrane of cells in gastric tissues. Compared to normal gastric tissues, GPRC5A mRNA and protein expression levels are significantly elevated in GC tissues [91]. Increased GPRC5A expression is significantly related to aggressive clinical parameters (larger tumor size, diffuse type, serosal invasion, and lymph node metastasis) and shorter overall survival (OS) [92].
5.5. GPRC5A and Hepatocellular Carcinoma
Conflicting information exists concerning GPRC5A's expression status in hepatocellular carcinoma (HCC). Lower GPRC5A mRNA levels have been reported in seven cell lines established from patients-derived tumor xenografts [93]. Conversely, several studies found that GPRC5A expression is elevated in HCC compared to in paratumor and normal liver tissues, and high GPRC5A expression is related to advanced clinical stage, high serum alpha-fetoprotein (AFP), vascular invasion, tumor recurrence, and worse prognosis (OS and disease-free survival) [94, 95].
5.6. GPRC5A and Pancreatic Carcinoma
GPRC5A expression is generally low in normal pancreatic ductal cells but is dramatically increased in pancreatic ductal cells of primary and metastatic tumor samples [37, 96]. Knockdown of GPRC5A with siRNAs leads to morphological changes in pancreatic tumor cells AsPc-1 [30]. Suppression of GPRC5A impaired the cell growth, proliferation, colony formation, and migration ability of pancreatic ductal adenocarcinoma cells [37, 68, 96].
5.7. GPRC5A in Other Cancers
In intrahepatic cholangiocarcinoma, GPRC5A is up-regulated compared to normal controls [95]. In oral squamous cell carcinoma (OSCC), GPRC5A is downregulated compared to normal oral epithelium, and this downregulation is associated with poorly differentiated OSCCs. Consistently, GPRC5A overexpression reversed the malignant phenotype of OSCC cell lines, implying that GPRC5A may serve as a powerful biomarker for malignant OSCCs [97]. HNSCC is associated with suppressed expression of GPRC5A, which is positively associated with tumor grade, along with the activation of STAT3. Overexpression of GPRC5A suppressed interleukin (IL)-6-induced STAT3 signaling pathway activation and inhibited colony-formation in HNSCC cells [67].
6. Clinical Application Value of GPRC5A
As described above, GPRC5A is dysregulated in a broad range of cancers, which indicates that it can potentially be used as a diagnostic candidate, especially in lung cancer. Further large-scale studies are therefore warranted to evaluate its diagnostic sensitivity and specificity in different cancer types. Moreover, GPRC5A, as a member of the largest family of protein targets for approved drugs (GPCRs) [98], is also a potential therapeutic target in patients with elevated GPRC5A levels. Until now, only tretinoin (ATRA, DB00755) has been demonstrated to be related to GPRC5A, whereas its role in antitumor therapy remains unknown. Future studies are therefore urgently warranted. Notably, GPRC5A also has great values in the optimization of clinical medication. In pancreatic cancer, suppression of GPRC5A was found to increase the cell sensitivity to multiple chemotherapeutic drugs, including gemcitabine, oxaliplatin, and fluorouracil [37, 96]. Additionally, EGFR inhibitors have been shown to be more effective in GPRC5A knockout lung cancer cells than in GPRC5A wild-type lung cancer cells, indicating that they are more suitable for lung cancer patients with lower GPRC5A expression [76]. Therefore, despite few studies having focused on its clinical application, GPRC5A's importance is clear as it could benefit accurate diagnosis and it should be taken in consideration for targeted-therapies and optimizing clinical medications.
7. Conclusions
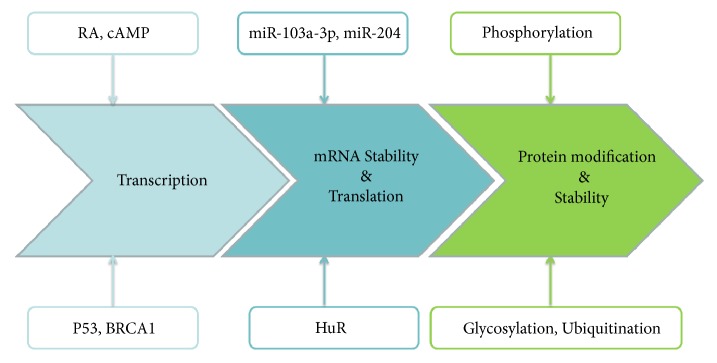
The lack of effective biomarkers for early diagnosis and lack of valid therapeutic methods for the treatment of aggressive cancers are the most intractable issues in clinical cancer management. GPRC5A is a member of orphan class C of the GPCR superfamily and was originally identified as a tumor suppressor playing an important role in lung tumor development. The GPRC5A gene contains many binding sites for transcription factors: this allows the regulation of GPRC5A expression by RA, cAMP, BRCA1, and many others. Additionally, GPRC5A expression is regulated posttranscriptionally and posttranslationally (Figure 2). Accumulating studies have demonstrated GPRC5A dysregulation in various human cancers, although its expression status differs among different cancer types. Aberrant GPRC5A expression induces the deregulation of signaling pathways such as cAMP, NF-κB, STAT3, and FAK/Src signaling and is related to prognosis. Especially, GPRC5A expression is associated with a poor response rate to chemotherapy. These data suggest that GPRC5A can be regarded as a potent biomarker for accurate diagnosis, prognosis prediction, and personalized treatment for patients with cancer. However, current knowledge of the exact mechanism of these processes is limited. Further studies focused on the cellular and molecular mechanisms will reveal novel insights into the details of its intricate function in cancer.
Figure 2.
The regulation of GPRC5A expression in cancer cells. GPRC5A is subjected to multiple levels of regulation from transcription to translation as detailed in the text.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (nos. 81201089 and 81272676) and by a grant from the National Science and Technology Major Project of the Ministry of Science and Technology of China (no. 2013ZX09506015).
Disclosure
Xiaoxia Jiang and Xin Xu are cofirst authors.
Conflicts of Interest
The authors report no conflicts of interest in this work.
References
- 1.Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pu X., Ye Y., Wu X. Development and validation of risk models and molecular diagnostics to permit personalized management of cancer. Cancer. 2014;120(1):11–19. doi: 10.1002/cncr.28393. [DOI] [PubMed] [Google Scholar]
- 3.de Castro D. G., Clarke P. A., Al-Lazikani B., Workman P. Personalized cancer medicine: molecular diagnostics, predictive biomarkers, and drug resistance. Clinical Pharmacology & Therapeutics. 2013;93(3):252–259. doi: 10.1038/clpt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho W. C. Molecular diagnostics for monitoring and predicting therapeutic effect in cancer. Expert Review of Molecular Diagnostics. 2011;11(1):9–12. doi: 10.1586/erm.10.111. [DOI] [PubMed] [Google Scholar]
- 5.De Castro D. G. Challenges for the implementation of routine molecular diagnostics in cancer care. Expert Review of Molecular Diagnostics. 2011;11(6):549–551. doi: 10.1586/erm.11.37. [DOI] [PubMed] [Google Scholar]
- 6.Takeda S., Kadowaki S., Haga T., Takaesu H., Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Letters. 2002;50(1-3):97–101. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- 7.Zhang D., Zhao Q., Wu B. Structural studies of G protein-coupled receptors. Molecules and Cells. 2015;38(10):836–842. doi: 10.14348/molcells.2015.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shonberg J., Kling R. C., Gmeiner P., Löber S. GPCR crystal structures: Medicinal chemistry in the pocket. Bioorganic & Medicinal Chemistry. 2015;23(14):3880–3906. doi: 10.1016/j.bmc.2014.12.034. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., An S., Ward R., et al. G protein-coupled receptors as promising cancer targets. Cancer Letters. 2016;376(2):226–239. doi: 10.1016/j.canlet.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 10.Gutkind J. S. Cell growth control by G protein-coupled receptors: from signal transduction to signal integration. Oncogene. 1998;17(11):1331–1342. doi: 10.1038/sj.onc.1202186. [DOI] [PubMed] [Google Scholar]
- 11.Moore B. B., Keane M. P., Addison C. L., Arenberg D. A., Strieter R. M. CXC chemokine modulation of angiogenesis: the importance of balance between angiogenic and angiostatic members of the family. Journal of Investigative Medicine. 1998;46(4):113–120. [PubMed] [Google Scholar]
- 12.Richard D. E., Vouret-Craviari V., Pouysségur J. Angiogenesis and G-protein-coupled receptors: Signals that bridge the gap. Oncogene. 2001;20(13):1556–1562. doi: 10.1038/sj.onc.1204193. [DOI] [PubMed] [Google Scholar]
- 13.Rozengurt E. Neuropeptides as growth factors for normal and cancerous cells. Trends in Endocrinology & Metabolism. 2002;13(3):128–134. doi: 10.1016/S1043-2760(01)00544-6. [DOI] [PubMed] [Google Scholar]
- 14.Dorsam R. T., Gutkind J. S. G-protein-coupled receptors and cancer. Nature Reviews Cancer. 2007;7(2):79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 15.Spiegelberg B. D., Hamm H. E. Roles of G-protein-coupled receptor signaling in cancer biology and gene transcription. Current Opinion in Genetics & Development. 2007;17(1):40–44. doi: 10.1016/j.gde.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Cheng Y., Lotan R. Molecular cloning and characterization of a novel retinoic acid- inducible gene that encodes a putative G protein-coupled receptor. The Journal of Biological Chemistry. 1998;273(52):35008–35015. doi: 10.1074/jbc.273.52.35008. [DOI] [PubMed] [Google Scholar]
- 17.Rask-Andersen M., Almén M. S., Schiöth H. B. Trends in the exploitation of novel drug targets. Nature Reviews Drug Discovery. 2011;10(8):579–590. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- 18.Gurbel P. A., Kuliopulos A., Tantry U. S. G-Protein-Coupled receptors signaling pathways in new antiplatelet drug development. Arteriosclerosis, Thrombosis, and Vascular Biology. 2015;35(3):500–512. doi: 10.1161/ATVBAHA.114.303412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou H., Rigoutsos I. The emerging roles of GPRC5A in diseases. Oncoscience. 2014;1(12):765–776. doi: 10.18632/oncoscience.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bräuner-Osborne H., Krogsgaard-Larsen P. Sequence and expression pattern of a novel human orphan G-protein- coupled receptor, GPRC5B, a family C receptor with a short amino-terminal domain. Genomics. 2000;65(2):121–128. doi: 10.1006/geno.2000.6164. [DOI] [PubMed] [Google Scholar]
- 21.Robbins M. J., Michalovich D., Hill J., et al. Molecular cloning and characterization of two novel retinoic acid- inducible orphan G-protein-coupled receptors (GPRC5B and GPRC5C) Genomics. 2000;67(1):8–18. doi: 10.1006/geno.2000.6226. [DOI] [PubMed] [Google Scholar]
- 22.Bräuner-Osborne H., Jensen A. A., Sheppard P. O., Brodin B., Krogsgaard-Larsen P., O'Hara P. Cloning and characterization of a human orphan family C G-protein coupled receptor GPRC5D. Biochimica et Biophysica Acta—Gene Structure and Expression. 2001;1518(3):237–248. doi: 10.1016/S0167-4781(01)00197-X. [DOI] [PubMed] [Google Scholar]
- 23.Fredriksson R., Lagerström M. C., Lundin L.-G., Schiöth H. B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Molecular Pharmacology. 2003;63(6):1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 24.Fagerberg L., Hallstrom B. M., Oksvold P., et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & Cellular Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.m113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutierrez-Mazariegos J., Schubert M., Laudet V. Evolution of retinoic acid receptors and retinoic acid signaling. Subcellular Biochemistry. 2014;70:55–73. doi: 10.1007/978-94-017-9050-5_4. [DOI] [PubMed] [Google Scholar]
- 26.Di Masi A., Leboffe L., De Marinis E., et al. Retinoic acid receptors: From molecular mechanisms to cancer therapy. Molecular Aspects of Medicine. 2015;41:1–115. doi: 10.1016/j.mam.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Allenby G., Bocquel M.-T., Saunders M., et al. Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acids. Proceedings of the National Acadamy of Sciences of the United States of America. 1993;90(1):30–34. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye X., Tao Q., Wang Y., Cheng Y., Lotan R. Mechanisms underlying the induction of the putative human tumor suppressor GPRC5A by retinoic acid. Cancer Biology & Therapy. 2009;8(10):951–962. doi: 10.4161/cbt.8.10.8244. [DOI] [PubMed] [Google Scholar]
- 29.Hirano M., Zang L., Oka T., et al. Novel reciprocal regulation of cAMP signaling and apoptosis by orphan G-protein-coupled receptor GPRC5A gene expression. Biochemical and Biophysical Research Communications. 2006;351(1):185–191. doi: 10.1016/j.bbrc.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Wu Q., Ding W., Mirza A., et al. Integrative genomics revealed RAI3 is a cell growth-promoting gene and a novel P53 transcriptional target. The Journal of Biological Chemistry. 2005;280(13):12935–12943. doi: 10.1074/jbc.M409901200. [DOI] [PubMed] [Google Scholar]
- 31.Jin E., Wang W., Fang M., et al. Lung cancer suppressor gene GPRC5A mediates p53 activity in non-small cell lung cancer cells in vitro. Molecular Medicine Reports. 2017;16(5):6382–6388. doi: 10.3892/mmr.2017.7343. [DOI] [PubMed] [Google Scholar]
- 32.Sokolenko A. P., Bulanova D. R., Iyevleva A. G., et al. High prevalence of GPRC5A germline mutations in BRCA1-mutant breast cancer patients. International Journal of Cancer. 2014;134(10):2352–2358. doi: 10.1002/ijc.28569. [DOI] [PubMed] [Google Scholar]
- 33.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 34.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou H., Rigoutsos I. MiR-103a-3p targets the 5′ UTR of GPRC5A in pancreatic cells. RNA. 2014;20(9):1431–1439. doi: 10.1261/rna.045757.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrestha S., Yang C.-D., Hong H.-C., et al. Integrated microRNA–mRNA analysis reveals miR-204 inhibits cell proliferation in gastric cancer by targeting CKS1B, CXCL1 and GPRC5A. International Journal of Molecular Sciences. 2018;19(1) doi: 10.3390/ijms19010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H., Telonis A. G., Jing Y., et al. GPRC5A is a potential oncogene in pancreatic ductal adenocarcinoma cells that is upregulated by gemcitabine with help from HuR. Cell death & disease. 2016;7:p. e2294. doi: 10.1038/cddis.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennan C. M., Steitz J. A. HuR and mRNA stability. Cellular and Molecular Life Sciences. 2001;58(2):266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nature Reviews Molecular Cell Biology. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon J. H., Abdelmohsen K., Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. Journal of Molecular Biology. 2013;425(19):3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnan J., Mishra R. K. Emerging trends of long non-coding RNAs in gene activation. FEBS Journal. 2014;281(1):34–45. doi: 10.1111/febs.12578. [DOI] [PubMed] [Google Scholar]
- 42.Dephoure N., Zhou C., Villén J., et al. A quantitative atlas of mitotic phosphorylation. Proceedings of the National Acadamy of Sciences of the United States of America. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen J. V., Vermeulen M., Santamaria A. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Science Signaling. 2010;3(104, article ra3) doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 44.Zhou H., Di Palma S., Preisinger C., et al. Toward a comprehensive characterization of a human cancer cell phosphoproteome. Journal of Proteome Research. 2013;12(1):260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 45.Lin X., Zhong S., Ye X., et al. EGFR phosphorylates and inhibits lung tumor suppressor GPRC5A in lung cancer. Molecular Cancer. 2014;13(1) doi: 10.1186/1476-4598-13-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meierhofer D., Wang X., Huang L., Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. Journal of Proteome Research. 2008;7(10):4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danielsen J. M. R., Sylvestersen K. B., Bekker-Jensen S., et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Molecular & Cellular Proteomics. 2011;10(3) doi: 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emanuele M. J., Elia A. E. H., Xu Q., et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147(2):459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim W., Bennett E. J., Huttlin E. L., et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Molecular Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee K. A., Hammerle L. P., Andrews P. S., et al. Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. The Journal of Biological Chemistry. 2011;286(48):41530–41538. doi: 10.1074/jbc.M111.248856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner S. A., Beli P., Weinert B. T., et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Molecular & Cellular Proteomics. 2011;10(10) doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Povlsen L. K., Beli P., Wagner S. A., et al. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nature Cell Biology. 2012;14(10):1089–1098. doi: 10.1038/ncb2579. [DOI] [PubMed] [Google Scholar]
- 53.Thompson J. W., Nagel J., Hoving S., et al. Quantitative Lys--Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 Ligase HUWE1. The Journal of Biological Chemistry. 2014;289(42):28942–28955. doi: 10.1074/jbc.M114.573352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vallabhapurapu S., Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annual Review of Immunology. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 55.Hallam S., Escorcio-Correia M., Soper R., Schultheiss A., Hagemann T. Activated macrophages in the tumour microenvironment - Dancing to the tune of TLR and NF-κB. The Journal of Pathology. 2009;219(2):143–152. doi: 10.1002/path.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng J., Fujimoto J., Ye X.-F., et al. Knockout of the tumor suppressor gene Gprc5a in mice leads to NF-κB activation in airway epithelium and promotes lung inflammation and tumorigenesis. Cancer Prevention Research. 2010;3(4):424–437. doi: 10.1158/1940-6207.CAPR-10-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y., Song H., Xu D., et al. Gprc5a-deficiency confers susceptibility to endotoxin-induced acute lung injury via NF-kappaB pathway. Cell Cycle. 2015;14(9):1403–1412. doi: 10.1080/15384101.2015.1006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bowman T., Garcia R., Turkson J., Jove R. STATs in oncogenesis. Oncogene. 2000;19(21):2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 59.Bourguignon L. Y. W., Earle C., Wong G., Spevak C. C., Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31(2):149–160. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alexandrow M. G., Song L. J., Altiok S., Gray J., Haura E. B., Kumar N. B. Curcumin: a novel Stat3 pathway inhibitor for chemoprevention of lung cancer. European Journal of Cancer Prevention. 2012;21(5):407–412. doi: 10.1097/CEJ.0b013e32834ef194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anglesio M. S., George J., Kulbe H., et al. IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clinical Cancer Research. 2011;17(8):2538–2548. doi: 10.1158/1078-0432.CCR-10-3314. [DOI] [PubMed] [Google Scholar]
- 62.Takemoto S., Ushijima K., Kawano K., et al. Expression of activated signal transducer and activator of transcription-3 predicts poor prognosis in cervical squamous-cell carcinoma. British Journal of Cancer. 2009;101(6):967–972. doi: 10.1038/sj.bjc.6605212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo C., Yang G., Khun K., et al. Activation of Stat3 in renal tumors. American Journal of Translational Research. 2009;1(3):283–290. [PMC free article] [PubMed] [Google Scholar]
- 64.Sheen-Chen S.-M., Huang C.-C., Tang R.-P., Chou F.-F., Eng H.-L. Prognostic value of signal transducers and activators of transcription 3 in breast cancer. Cancer Epidemiology, Biomarkers & Prevention. 2008;17(9):2286–2290. doi: 10.1158/1055-9965.EPI-08-0089. [DOI] [PubMed] [Google Scholar]
- 65.Mora L. B., Buettner R., Seigne J., et al. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Research. 2002;62(22):6659–6666. [PubMed] [Google Scholar]
- 66.Chen Y., Deng J., Fujimoto J., et al. Gprc5a deletion enhances the transformed phenotype in normal and malignant lung epithelial cells by eliciting persistent Stat3 signaling induced by autocrine leukemia inhibitory factor. Cancer Research. 2010;70(21):8917–8926. doi: 10.1158/0008-5472.CAN-10-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu S., Ye D., Wang T., et al. Repression of GPRC5A is associated with activated STAT3, which contributes to tumor progression of head and neck squamous cell carcinoma. Cancer Cell International. 2017;17(1) doi: 10.1186/s12935-017-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jahny E., Yang H., Liu B., et al. The G protein-coupled receptor RAI3 is an independent prognostic factor for pancreatic cancer survival and regulates proliferation via STAT3 phosphorylation. PLoS ONE. 2017;12(1) doi: 10.1371/journal.pone.0170390.e0170390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reddig P. J., Juliano R. L. Clinging to life: cell to matrix adhesion and cell survival. Cancer and Metastasis Reviews. 2005;24(3):425–439. doi: 10.1007/s10555-005-5134-3. [DOI] [PubMed] [Google Scholar]
- 70.Makrilia N., Kollias A., Manolopoulos L., Syrigos K. Cell adhesion molecules: role and clinical significance in cancer. Cancer Investigation. 2009;27(10):1023–1037. doi: 10.3109/07357900902769749. [DOI] [PubMed] [Google Scholar]
- 71.Berrier A. L., Yamada K. M. Cell-matrix adhesion. Journal of Cellular Physiology. 2007;213(3):565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 72.Desgrosellier J. S., Cheresh D. A. Integrins in cancer: biological implications and therapeutic opportunities. Nature Reviews Cancer. 2010;10(1):9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hamadi A., Bouali M., Dontenwill M., Stoeckel H., Takeda K., Rondé P. Regulation of focal adhesion dynamics and disassembly by phosphorylation of FAK at tyrosine 397. Journal of Cell Science. 2005;118(19):4415–4425. doi: 10.1242/jcs.02565. [DOI] [PubMed] [Google Scholar]
- 74.Geiger B., Spatz J. P., Bershadsky A. D. Environmental sensing through focal adhesions. Nature Reviews Molecular Cell Biology. 2009;10(1):21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 75.Bulanova D. R., Akimov Y. A., Rokka A., et al. Orphan G protein-coupled receptor GPRC5A modulates integrin β1-mediated epithelial cell adhesion. Cell Adhesion & Migration. 2017;11(5-6):434–446. doi: 10.1080/19336918.2016.1245264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong S., Yin H., Liao Y., et al. Lung tumor suppressor GPRC5A binds EGFR and restrains its effector signaling. Cancer Research. 2015;75(9):1801–1814. doi: 10.1158/0008-5472.CAN-14-2005. [DOI] [PubMed] [Google Scholar]
- 77.Fujimoto J., Kadara H., Garcia M. M., et al. G-protein coupled receptor family C, group 5, member A (gprc5a) expression is decreased in the adjacent field and normal bronchial epithelia of patients with chronic obstructive pulmonary disease and non-small-cell lung cancer. Journal of Thoracic Oncology. 2012;7(12):1747–1754. doi: 10.1097/JTO.0b013e31826bb1ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tao Q., Fujimoto J., Men T., et al. Identification of the retinoic acid-inducible Gprc5a as a new lung tumor suppressor gene. Journal of the National Cancer Institute. 2007;99(22):1668–1682. doi: 10.1093/jnci/djm208. [DOI] [PubMed] [Google Scholar]
- 79.Fujimoto J., Kadara H., Men T., van Pelt C., Lotan D., Lotan R. Comparative functional genomics analysis of NNK tobacco-carcinogen induced lung adenocarcinoma development in Gprc5a-knockout mice. PLoS ONE. 2010;5(7) doi: 10.1371/journal.pone.0011847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fujimoto J., Nunomura-Nakamura S., Liu Y., et al. Development of Kras mutant lung adenocarcinoma in mice with knockout of the airway lineage-specific gene Gprc5a. International Journal of Cancer. 2017;141(8):1589–1599. doi: 10.1002/ijc.30851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barta P., Van Pelt C., Men T., Dickey B. F., Lotan R., Moghaddam S. J. Enhancement of lung tumorigenesis in a Gprc5a Knockout mouse by chronic extrinsic airway inflammation. Molecular Cancer. 2012;11, article no. 4 doi: 10.1186/1476-4598-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J., Farris A. B., Xu K., et al. GPRC5A suppresses protein synthesis at the endoplasmic reticulum to prevent radiation-induced lung tumorigenesis. Nature Communications. 2016;7 doi: 10.1038/ncomms11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nagahata T., Sato T., Tomura A., Onda M., Nishikawa K., Emi M. Identification of RAI3 as a therapeutic target for breast cancer. Endocrine-Related Cancer. 2005;12(1):65–73. doi: 10.1677/erc.1.00890. [DOI] [PubMed] [Google Scholar]
- 84.Dairkee S. H., Sayeed A., Luciani G., et al. Immutable functional attributes of histologic grade revealed by context-independent gene expression in primary breast cancer cells. Cancer Research. 2009;69(19):7826–7834. doi: 10.1158/0008-5472.CAN-09-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jörißen H., Bektas N., Dahl E., et al. Production and characterisation of monoclonal antibodies against RAI3 and its expression in human breast cancer. BMC Cancer. 2009;9 doi: 10.1186/1471-2407-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang L., Ma T., Zhang J. GPRC5A exerts its tumor-suppressive effects in breast cancer cells by inhibiting EGFR and its downstream pathway. Oncology Reports. 2016;36(5):2983–2990. doi: 10.3892/or.2016.5062. [DOI] [PubMed] [Google Scholar]
- 87.Zougman A., Hutchins G. G., Cairns D. A., et al. Retinoic acid-induced protein 3: Identification and characterisation of a novel prognostic colon cancer biomarker. European Journal of Cancer. 2013;49(2):531–539. doi: 10.1016/j.ejca.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 88.Greenhough A., Bagley C., Heesom K. J., et al. Cancer cell adaptation to hypoxia involves a HIF‐GPRC5A‐YAP axis. EMBO Molecular Medicine. 2018 doi: 10.15252/emmm.201708699.e8699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kume H., Muraoka S., Kuga T., et al. Discovery of colorectal cancer biomarker candidates by membrane proteomic analysis and subsequent verification using Selected Reaction Monitoring (SRM) and Tissue Microarray (TMA) Analysis. Molecular & Cellular Proteomics. 2014;13(6):1471–1484. doi: 10.1074/mcp.M113.037093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang L., Li L., Gao G., et al. Elevation of GPRC5A expression in colorectal cancer promotes tumor progression through VNN-1 induced oxidative stress. International Journal of Cancer. 2017;140(12):2734–2747. doi: 10.1002/ijc.30698. [DOI] [PubMed] [Google Scholar]
- 91.Cheng L., Yang S., Yang Y., et al. Global gene expression and functional network analysis of gastric cancer identify extended pathway maps and GPRC5A as a potential biomarker. Cancer Letters. 2012;326(1):105–113. doi: 10.1016/j.canlet.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 92.Liu H., Zhang Y., Hao X., et al. GPRC5A overexpression predicted advanced biological behaviors and poor prognosis in patients with gastric cancer. Tumor Biology. 2016;37(1):503–510. doi: 10.1007/s13277-015-3817-0. [DOI] [PubMed] [Google Scholar]
- 93.Xin H., Wang K., Hu G., et al. Establishment and characterization of 7 novel hepatocellular carcinoma cell lines from patient-derived tumor xenografts. PLoS ONE. 2014;9(1):p. e85308. doi: 10.1371/journal.pone.0085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng J., Guo X., Gao X., Liu H., Tu Y., Zhang Y. Overexpression of retinoic acid-induced protein 3 predicts poor prognosis for hepatocellular carcinoma. Clinical & Translational Oncology: Official Publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2014;16(1):57–63. doi: 10.1007/s12094-013-1040-2. [DOI] [PubMed] [Google Scholar]
- 95.Subrungruang I., Thawornkuno C., Porntip C.-P., Pairojkul C., Wongkham S., Petmitr S. Gene expression profiling of intrahepatic cholangiocarcinoma. Asian Pacific Journal of Cancer Prevention. 2013;14(1):557–563. doi: 10.7314/APJCP.2013.14.1.557. [DOI] [PubMed] [Google Scholar]
- 96.Liu B., Yang H., Pilarsky C., Weber G. The effect of GPRC5a on the proliferation, migration ability, chemotherapy resistance, and phosphorylation of GSK-3β in pancreatic cancer. International Journal of Molecular Sciences. 2018;19(7):p. 1870. doi: 10.3390/ijms19071870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu S. L., Zhong S. S., Ye D. X., Chen W. T., Zhang Z. Y., Deng J. Repression of G protein-coupled receptor family C group 5 member A is associated with pathologic differentiation grade of oral squamous cell carcinoma. Journal of Oral Pathology & Medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2013;42:761–768. doi: 10.1111/jop.12077. [DOI] [PubMed] [Google Scholar]
- 98.Insel P. A., Sriram K., Wiley S. Z., et al. GPCRomics: GPCR expression in cancer cells and tumors identifies new, potential biomarkers and therapeutic targets. Frontiers in Pharmacology. 2018;9 doi: 10.3389/fphar.2018.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]