Abstract
Background
Insects share similar fundamental molecular principles with mammals in innate immunity. For modulating normal gut microbiota, insects produce phenoloxidase (PO), which is absent in all vertebrates, and reactive nitrogen species (ROS) and antimicrobial proteins (AMPs). However, reports on insect gut phagocytosis are very few. Furthermore, most previous studies measure gene expression at the transcription level. In this study, we provided proteomic evidence on gut modulation of normal microorganisms by investigating the anal droplets from a weevil, Cryptorhynchus lapathi.
Results
The results showed that the anal droplets contained diverse proteins related to physical barriers, epithelium renewal, pattern recognition, phenoloxidase activation, oxidative defense and phagocytosis, but AMPs were not detected. According to annotations, Scarb1, integrin βν, Dscam, spondin or Thbs2s might mediate phagocytosis. As a possible integrin βν pathway, βν activates Rho by an unknown mechanism, and Rho induces accumulation of mDia, which then promotes actin polymerization.
Conclusions
Our results well demonstrated that insect anal droplets can be used as materials to investigate the defense of a host to gut microorganisms and supported to the hypothesis that gut phagocytosis occurs in insects.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5182-z) contains supplementary material, which is available to authorized users.
Keywords: Anal droplet, Intestine, Innate immunity, Phagocytosis, Cryptorhynchus lapathi
Background
Essentially, gut microbiota are composed of both commensal and pathogenic microbes, and they may be benign, beneficial or pathogenic in basal conditions [1, 2]. For multicellular organisms, nonself recognition is particularly important to protect an individual from potential environmental pathogens. Additionally, it is now firmly established that the animal gut immune system not only eliminates microbial pathogens but also maintains an adequate level of commensal microbiota [1, 3, 4].
The molecular basis of the interactions between the host gut epithelium and microorganisms has been investigated extensively in mammals and Drosophila. Mammal intestinal epithelial cells (IECs) keep the microbes at bay by secreting highly glycosylated mucins, reactive oxygen species (ROS) and antimicrobial proteins (AMPs) and by establishing a physical and biochemical barrier to microbial contact with the epithelial surface and underlying immune cells [4]. Specialized mammal IECs, called microfold cells (M cells), are capable of both specific receptor-mediated microbial uptake and nonspecific antigen uptake from the intestinal lumen [4]. Insects and other invertebrates share similar fundamental molecular principles with mammals in innate immunity [5]. In addition to ROS and AMPs, invertebrates also produce phenoloxidase (PO), which is absent in all vertebrates [3, 6]. Many insect guts also have a peritrophic envelope preventing microorganisms from direct contact with the epithelial cells [6]. In contrast to many reports on gut AMPs and ROS, reports on insect gut phagocytosis are very few. Whether any immune cells are associated with the Drosophila intestine remains an open question [7], although cells able to phagocytose bacteria and dying cells are present in the Drosophila proventriculus (PV) [8].
To study insect gut immunity, most previous studies measure gene expression at the transcriptional level [9]. In this study, we provided proteomics data from the anal droplets of a weevil, Cryptorhynchus lapathi (L.) (Coleoptera: Curculionidae). Insect feces are anal excretions either in liquid form or packaged in pellets and known as frass. Malpighian tubules initiate the insect excretory process. The primary urine from the Malpighian tubules mixed with the end products of digestion from the midgut are modified by continued resorption by rectal glands to produce a secondary urine that is then expelled through the anus. The liquid form of insect feces is called honeydew, anal secretion or anal droplet. Most insects produce either frass or anal droplets, whereas few insects produce both. The poplar-and-willow borer, C. lapathi, is a wood-boring pest of economic importance throughout Europe, China, Japan, the United States and Canada [10]. When disturbed, the larvae produce anal droplets. As the larvae spend all their life within a tree stem, this phenomenon has never been previously reported.
Previous studies revealed the chemical composition of hemipteran honeydew, which primarily contains sugar, amino acids and other chemicals [11]. Unfortunately, the proteins in insect anal droplets were not reported until small AMPs were detected in the anal secretions of Nicrophorus (burying beetles) [12]. Additionally, very recently, proteomic investigation of aphid honeydew revealed an unexpected diversity of proteins [11]. Thus, the gut immune system can possibly be reconstructed by investigation of an anal droplet proteome.
In this paper, a proteome investigation of the larval weevil showed many proteins related to physical barriers, epithelium renewal, pattern recognition, phenoloxidase activation, oxidative defense and phagocytosis, but no AMPs were detected.
Materials and methods
Anal droplet collection
Larval weevils were collected in the wild. Anal droplets were collected directly onto foils placed against the anal areas of each weevil, while gently squeezing their abdomens. Samples on the foil were then moved into a mini glass bottle with a pipette and stored at − 20 °C when not immediately used.
Q-TOF MS sequencing
The anal droplets were first separated on SDS-PAGE before LC-MS analysis. Ten microliters of pooled sample was resuspended in 50 μL of Laemmli sample buffer supplemented with 2% β-mercaptoethanol and heated at 95 °C for 5 min. After electrophoresis, the gel was rinsed with three changes of Nanopure water, stained for 20 min with Bio-safe TM Coomassie and destained with three changes of Nanopure water. The gel lane was carefully cut into eleven pieces (Fig. 1), placed into Eppendorf tubes and rinsed twice for 10 min with 1 mL of MilliQ water. After destained with freshly prepared destaining solution (25 mM (NH4)HCO3, 50% acetonitrile), the gel pieces were dehydrated until they shrank and became white (approximately 2 min) with 25 mM (NH4)HCO3 with 50% acetonitrile and then once more for 30 s in 100% acetonitrile. The gel pieces were then rehydrated in freshly prepared 10 mM Dithiothreitol for 1 h at 56 °C (water bath) and were alkylated with freshly prepared 55 mM Iodoacetamide for 1 h at room temperature in the dark. Subsequently, the gel pieces were washed with 25 mM (NH4)HCO3 twice for 10 min and destained to become white as before. Trypsin digestion was performed overnight with trypsin work solution (1 μg/uL stock solution was diluted 15-fold with 25 mM (NH4)HCO3) at 37 °C. Digested proteins were extracted 4 times with 50 μL of 50 mM (NH4)HCO3, 50 μL of 0.1%(V/V)FA in water, 50 μL of 0.1%(V/V)FA in acetonitrile and 50 μL of acetonitrile. All extracts were pooled, freeze-dried at − 20 °C and resuspended in 0.1% FA for sequencing.
Fig. 1.
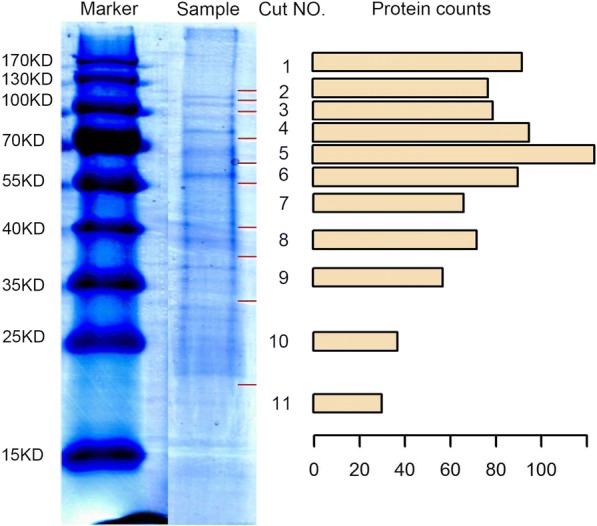
SDS-PAGE of the anal droplets. The gel was cut into 11 segments at the red line for Q-TOF. The counts of identified protein from each segment are shown in a barplot
The resuspended peptides were fractionated using reversed-phase high-pressure liquid chromatography (HPLC; prominence nano 2D, Shimazu, Kyoto, Japan), and the gradient-eluted peptides were analyzed using a MicrOTOF-QII (Bruker Daltonics, Billerica, MA, USA). The liquid chromatography columns were packed in-house with C18 (5 μm, 150 Å; Downers Grove, IL, USA). LC-MS conditions were as follow: Mobile phase: (A) 100% H2O with 0.1% FA and (B) 100% acetonitrile with 0.1% formic acid; Gradient: 0–4 min, 5–5% B; 4–30 min, 5–40% B; 30–35 min, 40–80% B; 35–45 min, 80–80% B; 45–45.1 min, 80–5% B; 45.1–60 min, 5–5%; Flow rate, 400 nL/min; Drying gas temperature, 150 °C; Capillary voltage, 1.5 kV; Collision gas, argon. The results were exported as. MGF file for X!Tandem [13] analysis.
Database searching and protein identification
For protein identification, the peak list data from MS were searched against a protein database. Instead of downloading insect proteins from a public database (for example, National Center for Biotechnology Information (NCBI)), we constructed a protein database of C. lapathi itself. A transcriptomic database of C. lapathi was first constructed by de novo assembly (Trinity software [14]) of the sequences from an Illumina sequencing platform (Illumina HiSeq2500) based on pooled RNAs of the larvae, pupae and adults and was then clustered by CD-HIT software (http://weizhongli-lab.org) to obtain unigenes. The unigenes were subsequently mapped to the proteome of Dendroctonus ponderosae with a cutoff E-value of 10− 4 using BLASTx to obtain a proteomic database of C. lapathi. The proteome of D. ponderosae was downloaded from UniProtKB (http://www.uniprot.org/). The database searches were performed by an R package, rTANDEM [13]. The proteins were identified from at least one peptide and with an X!tandem [15] score corresponding to an expected value of better than 0.05.
We performed BLASTP against the Antimicrobial Peptide Database (APD) [16] to identify AMPs from the anal droplet. For identification of other enzymes, local databases were set up and BLASTP was conducted.
Assignment of proteins to Kyoto encyclopedia of genes and genomes (KEGG) pathways
The IDs of proteins identified by X!tandem were labeled with their homologue gene IDs (UniProtKB protein entry, http://www.uniprot.org) of D. ponderosae, and then KEGG IDs were obtained from UniProtKB using the ID Mapping function. Enrichment analyses of KEGG were conducted by an R package, GOstats [17].
Results
Proteins identified by LC-MS
The BLASTX results showed that 26,685 unigenes of C. lapathi were aligned to the proteome of D. ponderosae. The deduced amino acid sequences of these unigenes were used for the database search by X!tandem. In total, 819 proteins homologous to D. ponderosae were identified (Fig. 1), and those related to gut homeostasis are summarized in Table 1.
Table 1.
Identified proteins with putative defensive roles from anal droplets of Cryptorhynchus lapathi
Proteins functioning as a physical barrier
Two proteins, mucin and ZO-1 (Zonula occludens-1), were identified from the anal droplet (Additional file 1: Table S1; Fig. 2). Insects have a mucus layer that lines enterocytes along the midgut, and the production of mucin is regulated by infection [1]. Tight junctions connect adjacent IECs and could be important targets for increasing the integrity of the intestinal barrier [4]. Occluding junctions play crucial roles in epithelial barrier function. The occluding junctions found in vertebrates are called tight junctions; whereas invertebrate species have a different type of occluding junction, the septate junction (SJ). Arthropods contain two types of SJs, the pleated SJ (pSJ) and the smooth SJ (sSJ). The pSJ exists in ectodermally derived epithelial cells and glial cells, whereas the sSJ is found in endodermally derived epithelia, such as in the midgut of insects. Malpighian tubules of insects also have sSJs. Together, Tsp2A (tetraspanin 2A), Ssk (snakeskin) and Mesh form a protein complex, and all three proteins are required for sSJ formation and for intestinal barrier function in Drosophila [18]. From the anal droplet, none of these proteins were detected; whereas a tight junction–associated protein, ZO-1, was detected (Additional file 1: Table S1). ZO-1 and other homologues of vertebrate tight junction proteins do not localize to the septate junction but instead are found in adherens junctions or the marginal zone (reviewed in [19]). In insect epithelial cells, adherens junctions are at the apical apex and septate junctions localize more to the basal domains; therefore, ZO-1 perhaps plays the role of physical barrier similar to that in humans and mammals. In humans, normal gut microbes induce the expression and redistribution of ZO, suggesting the weevil gut microbes also regulate adherens junctions [20].
Fig. 2.
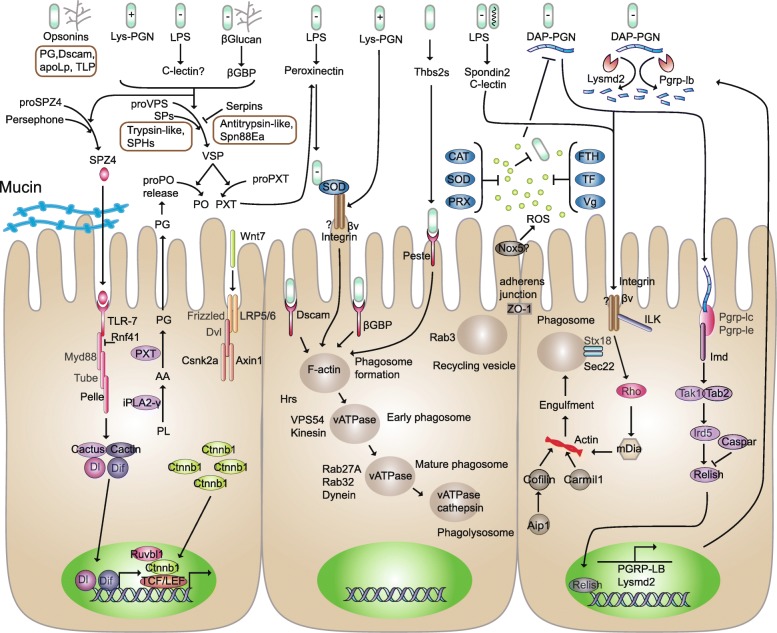
Anal droplet-derived proteins and possible pathways in innate immune. Proteins of which labels in grey were not detected in the anal droplets. ApoLp, apolipophorins; CAT, catalase; CTLs, C-type lectins; DAP, meso-diaminopimelic acid; FTH ferritin; LPS, lipopolysaccharide; PGN, Peptidoglycan; PGRPs, peptidoglycan recognition proteins; PO, phenoloxidase; proPO, prophenoloxidase; PRX, peroxiredoxin; PXT, peroxinectin; RNS, Reactive nitrogen species; ROS, reactive oxygen species; SOD, superoxide dismutase; TF, transferrin; TLPs, Thaumatin-like proteins; Vg, Vitellogenin; VSP, venom serine protease; ZO-1, Zonula occludens-1; βGBP, beta-1,3-glucan-binding protein
Proteins involved in gut epithelium renewal
Gut epithelium renewal is considered a tolerance mechanism of insects to increase the capacity to endure infection. This response depends on induction of the Wingless, JAK-STAT and Epidermal growth factor receptor (Egfr) pathways in progenitor cells, which stimulates their proliferation and increases their differentiation into enterocytes (reviewed in [1]). Egfr ligands are required to activate the Egfr pathway. Drosophila Egfr and its activity are modulated by five ligands. The TGF-α-like molecules, Gurken (Grk), Spitz (Spi), and Keren, and the neuregulin-like molecule Vein (Vn) function as receptor activators. The fifth Egfr ligand, Argos (Aos), is a receptor antagonist (reviewed in [21]). We did not find any coleopteran Grk homologs by BLASTP against the Nr database, whereas the other four Egfs were found in the weevil transcriptome. However, none of them were detected from the anal droplet. From the anal droplet, only one enzyme (Sos) for JAK-STAT pathways was identified, suggesting that the JAK-STAT pathway was not activated in the basal condition in gut epithelial cells. By contrast, seven proteins involved in the Wnt signaling pathway were identified, which were Ctnnb1, Ruvbl1, Wnt7, Siah1, Axin1, Plcb and Csnk2a (Additional file 2: Table S2; Fig. 2), suggesting a canonical Wnt pathway was activated. These results are consistent with the conclusion that the gut microbiota also promote epithelial renewal in basal conditions [1].
Proteins involved in pattern recognition
When microbial lipopolysaccharides (LPSs), glucans, or peptidoglycans are present, the coagulation cascade is triggered and/or changes to Ca2+ concentration or pH occur that activate a series of serine proteases to activate prophenoloxidase (proPO) in the hemolymph [22]. From the anal droplet, one beta-1,3-glucan-binding protein (βGBP, also known as GNBP), two C-type lectins (CTLs), one spondin 2 and one peroxinectin were detected (Additional file 1: Table S1; Fig. 2). βGBP binds to beta-1,3-glucan to initiate activation of cell-free prophenoloxidase [23], phagocytosis or Toll signaling [24]. CTLs are involved in opsonization, nodule formation, agglutination, encapsulation, melanization, and prophenoloxidase activation, in addition to maintaining gut microbiome homeostasis [25]. Binding to mannose, fucose, and glucose, among others, C-type lectin binds to β-integrin to promote hemocyte phagocytosis in an invertebrate [26, 27]. C-type lectins are classified into seventeen subgroups. It is difficult to classify the two C-type lectins from the anal droplet because of the very low similarity to the known CTLs. The extracellular matrix protein mindin (spondin) is a pattern-recognition molecule binding LPSs and an integrin ligand and is critical for initiating innate immune responses to both bacterial and viral pathogens (reviewed in [28]). Peroxinectin is a cell adhesive protein functioning in degranulation, encapsulation enhancement and with opsonin and peroxidase (reviewed in [29]). Insect phagocytic receptors include scavenger receptors, the nimrod receptor superfamily, peptidoglycan-recognition receptors, integrins, Dscam, and complement-like molecules [30] and can be divided into nonopsonic or opsonic receptors [31]. From the anal droplet, neither nimrod receptor superfamily receptors nor TEPs were identified, whereas one scavenger receptor class B member 1 (Scarb1, also known as peste in Drosophila), one integrin (beta-nu-like, also known as βν, betaInt-nu, βInt-ν, Βν, Dmel\CG1762, Fly betanu, Itgbetanu), one Dscam, one spondin-2 (also known as mindin) and one cartilage oligomeric matrix protein (Comp, also known as Thbs2s, thrombospondin-5) were detected in the anal droplet (Additional file 1: Table S1; Fig. 2). SR-I1 binds to both gram-negative and gram-positive bacteria [32] and is required for the efficient phagocytosis of two intracellular bacterial pathogens in Drosophila [33]. Dscam contributes to phagocytosis in Drosophila, mosquito and crayfish [34] and is considered a phagocytic receptor or opsonin for bacteria, which is conserved in invertebrates [30]. Integrin beta-nu transcripts are found exclusively in the larval midgut [35]. This protein binds to peptidoglycan to mediate phagocytosis of apoptotic cells and bacteria in Drosophila [30, 36]. An integrin from the oyster Crassostrea gigas that mediates the phagocytosis toward Vibrio splendidus through LPS binding activity is also documented [37]. The thrombospondin-coated apoptotic cells are tethered to the macrophage by CD36, and the vitronectin receptor signals the initiation of phagocytosis (reviewed in [38]). The extracellular matrix protein mindin (spondin-2) is a pattern-recognition molecule binding LPSs and an integrin ligand and is critical for initiating innate immune responses to both bacterial and viral pathogens [28].
Apolipophorin (ApoLp)-III binds to microbial cell wall components, such as gram-negative bacteria LPSs, gram-positive bacteria lipoteichoic acids, and fungal β-1,3-glucans. These binding properties impart to ApoLp-III a role pertaining to the pathogen recognition receptor (PRR) (reviewed in [39]). Thaumatin-like proteins (TLPs) can bind to β-1,3-glucans [40], indicating a potential function in pattern recognition in insect immunity. From the droplets, both ApoLp and TLP were detected (Additional file 1: Table S1; Fig. 2).
Proteins involved in phenoloxidase activation
Insect peroxinectin plays a role in prostaglandin (PG) biosynthesis [41], and an iPLA2-γ that can promote prostaglandin production has also been identified [42]. This particular phospholipase specifically recognizes the sn-2 acyl bond of phospholipids and catalytically hydrolyzes the bond releasing arachidonic acid and lysophosphatidic acid. Upon downstream modification by cyclooxygenases, arachidonic acid is modified into active compounds called eicosanoids [43]. PGs mediate the release of proPO, which may occur via cell surface GPCRs [44, 45].
A cascade of proteinases triggers activation of the proform of prophenoloxidase-activating enzyme (ppA) into active ppA, which cleaves proPO into active PO and results in melanin production. In insects, venom serine protease (VSP) acts as an arthropod prophenoloxidase-activating factor (PPAF), thereby triggering the phenoloxidase (PO) cascade [46]. The active ppA directly, or through an intermediate proteinase, activates properoxinectin (proPXT), a myeloperoxidase homologue, into peroxinectin (PXT). Serpins negatively regulate both PXT and ppA production. ProPXT lacks the cell adhesion, opsonin and encapsulation-promoting activities of PXT. However, both proPXT and PXT possess peroxidase activity. PXT is associated with an extracellular superoxide dismutase (SOD) and with an integrin. An attractive hypothesis is that an active microbe-PXT-dismutase complex is phagocytosed and results in intracellular killing (insert) through the generation of reactive oxygen intermediates (HOCL) by NADPH oxidase (reviewed in [47]). From the anal droplet, one proPO2, two Serpins (Antitrypsin-like, Spn88Ea), 14 trypsin-like serine proteases (some of them orthologous to H2, P127, P27, P40, P80, P128, H129, P155, and H146 of Tribolium castanum [48]; Additional file 1: Table S1, Additional file 3: Table S3), one VSP34-like protein and one VSP-like protein were identified (Additional file 1: Table S1; Fig. 2).
Proteins involved in oxidative defense
ROS in the gut have a bactericidal role and also act as both intra- and intercellular signaling molecules to induce repair responses or other homeostatic pathways [1, 49]. ROS can be produced within mitochondria or generated by Nox enzymes, and most cellular ROS have a mitochondrial origin [50]. Nox is a protein family composed of seven members. The Drosophila genome encodes one Nox5 and one Duox, and the Duox, but not the Nox5, is responsible for the extracellular microbicidal activity in gut mucosal immunity [51]. In Dendroctonus ponderosae, two isoforms of Nox5 (XP_019771050.1, XP_019771051.1) and one Duox (XP_019766183.1) are encoded in the genome. From the anal droplet, only one weevil-derived Nox5 was detected. However, seven weevil-derived enzymes involved in scavenging of ROS were detected, including two copper/zinc SODs, two peroxiredoxins (PRX) (reduces H2O2 in the presence of dithiothreitol [52]), one catalase (CAT), one ferritin (FTH) [53], one transferring (TF) [53] and one Vitellogenin (Vg) [54] (Additional file 1: Table S1; Fig. 2). These results indicated that the excessive ROS was cleared to maintain a balanced defense.
Insect AMPs from the anal droplet
Drosophila AMPs in the gut are apparently required to maintain a healthy gut microbiota [55]. From the anal droplet, only two AMPs were identified. A plant-originated thaumatin-like protein was one of the AMPs, which has antifungal activity against filamentous fungi Beauveria bassiana and Fusarium culmorum [56] (Additional file 4: Table S4). Thaumatin-like proteins are type 5 pathogenesis-related proteins, which are detected in high levels in plants upon biotic (including insect stress) or abiotic stress [57]. However, in insects, the function of thaumatin-like proteins is not well studied yet. A very recent study showed that in the cereal weevil Sitophilus oryzae, the expression of thaumatin drastically decreases during the population burst of midgut endosymbiont Sodalis pierantonius [58]. Thus, the discrimination of thaumatin as an AMP might be premature. The other AMP was c-type lysozyme 3 (Additional file 1: Table S1, Additional file 4: Table S4), which is expressed in the gut of Drosophila and is suggested to play a role in nutrition by lysis of the microbial cells rather than in immunity (reviewed in [5, 59]). These results are consistent with those in Drosophila, i.e., normal gut residents do not incite AMP production [60].
Anal droplet proteins involved in immune pathways
Two major signaling pathways, Imd and Toll, control the expression of AMPs in the body cavity, but in the case of the midgut, AMP production is induced in response to Imd signaling (reviewed in [1]). In mammalian cells, activation of Tak1 requires an upstream kinase complex consisting of Tak1 and two specific Tak1-binding proteins, Tab1 and Tab2. Tab1 functions as an activator of Tak1, and Tab2 functions as an adaptor protein that links Tak1 to the upstream regulator Traf6 in the IL-1 signaling pathway [61]. The Drosophila Imd pathway bifurcates into the JNK pathway at the level of Tak1 and Tab2 (reviewed in [62]). From the anal droplet, proteins Imd, Tab1 and Tab2 were detected (Additional file 1: Table S1, Additional file 2: Table S2; Fig. 2), but Tak1, Ird5 and Relish were not. However, other proteins, for example, Caspar [1], Pgrp-lb [1] and LysM (Lysmd2) [63], which decrease Imd signaling, were also identified from the anal droplet.
Proteins involved in the Toll pathway were also identified from the anal droplet (Additional file 1: Table S1, Additional file 2: Table S2; Fig. 2). Endogenous humoral factors from microbes activate the toll pathway in the hemolymph in which the endogenous ligand Spz is processed to its active form through a proteolytic cascade [64]. Persephone (Psh) is an extracellular Clip-domain serine protease activating Spz through a proteolytic cascade that is partly independent of proteoglycan sensing [64]. Other molecules involved in the toll pathway were also detected, including pelle, spz4, and TLRs (TLR7 and one unclassified TLR). However, Myd88 and Tube were not detected from the anal droplet.
NF-kappa-B inhibitor cactus (IκB homolog) was also detected from the anal droplet. Furthermore, a Cactin (Cactus interactor) was identified that negatively regulates NF-kappaB transcription factor activity [65] and inhibits the promoters of Drosophila and shrimp AMPs [66]. TmCactin may serve as an important regulator of innate immunity, mediating AMP responses against both gram-positive and gram-negative bacteria in Tenebrio molitor [67]. Hun et al. proposed that cactin acts in both Drosophila and T. molitor as a positive regulator of Toll signaling, whereas in humans, cactin functions as a negative regulator [67]. However, no AMPs were detected from the anal droplet, suggesting cactin functions as a negative regulator in the weevil gut. These results suggest that negative regulators of the Imd pathway repress AMPs in the gut to protect the normal microbiota, thereby maintaining the balance between immune tolerance and immune response (reviewed in [68]).
Proteins involved in phagocytosis
The entire process of phagocytosis of microorganisms by phagocytes can be divided into a series of defined steps: recognition, attachment, signaling, engulfment and phagosome maturation. Phagocytosis is a special endocytosis, and the phagosome ultimately fuses with a lysosome. From the anal droplet, 11 proteins (four HSPA1s, HGS, RNF41, IQSEC, SH3GL, RAB11FIP1_2_5, DAB2, EHD1, PSD, RABEP1) involved in endocytosis, 12 proteins (uidA, NAGA, uidA, CTSL, CTSB, MAN2B1, LIPA, CTSL, GGA, MAN2B1, E3.2.1.25, SMPD1) involved in lysosome pathways and 11 proteins (four ACTB_G1s, two CTSLs, SEC22, ITGB1, THBS2S, HGS, ATPeV1C) involving the phagosome pathway were identified by KEGG (Additional file 2: Table S2; Fig. 2).
Proteins involved in microbe internalization and phagosome formation
In addition to the proteins (F-actin, Sec22) identified by KEGG, other proteins involved in phagosome formation were also found in the anal droplet: Cofilin, Aip1, Carmil1, Ilk and Formin (Additional file 1: Table S1; Fig. 2). Cofilin promotes actin cytoskeletal remodeling to form phagocytic cups by accelerating actin turnover and thereby facilitating phagosome formation [69]. Aip1 increases the filament disassembly activity of Cofilin and restricts Cofilin localization to cortical actin patches [70]. By uncapping actin filaments, mammalian Carmil1 increases actin polymerization and plays a role in endocytosis and phagocytosis [71]. Integrin-linked kinase (ILK) couples integrins and cytoskeletal proteins and is an essential link between integrins and uptake of bacterial pathogens by epithelial cells [72]. Rho-GTPases can induce accumulation of mDia1 (mammalian diaphanous related formin 1) and polymerize actin in the phagocytic cup (reviewed in [31]).
Proteins involved in phagosome maturation
In addition to the proteins (vATPase, cathepsin) identified by KEGG, other proteins involved in phagosome maturation were also found in the anal droplet: VPS54, Kinesin, Dynein and Rab GTPases (Additional file 1: Table S1; Fig. 2). The maturation of phagosomes involves interactions with other cellular organelles and includes the stages early phagosome, intermediate phagosome, late phagosome, and phagolysome [30, 31]. GARP (Golgi-associated retrograde protein) complex, functioning in traffic from endosomes to the trans-Golgi network, is composed of VPS51, VPS52, VPS53, and VPS54 [73]. Kinesin is the major microtubule motor involved in trafficking materials required for the early events in phagocytosis [74]. From the anal droplet, two kinesins (Kif23, Kif18a), one trafficking kinesin-binding protein (Milt) and one carboxy-terminal kinesin 2-like protein were detected. Dynein, which travels toward the minus end of microtubules, propels phagosomes centripetally toward the microtubule-organizing center at which late endosomes/lysosomes that are themselves endowed with Rab7, RILP and dynein frequently accumulate [75]. Rab GTPases are at the central node of the machinery that regulates the dynamic process of interactions between phagosomes and intracellular compartments [76]. From the anal droplet, four proteins (Rab-3, Rab-14, Rab-27A, Rab-32) involved in the network of phagosomal Rab GTPases were identified of which Rab-3 and Rab-27A are the less common Rab proteins, whereas Rab-14 and Rab-32 are the most common ones [76].
The final step in the maturation process is the formation of the phagolysosome (pH ~ 4.5). Key cofactors of bacterial housekeeping enzymes are removed from the phagosomal lumen to prevent bacterial growth. ROS and reactive nitrogen species (RNS) attack bacterial DNA, proteins, and lipids to destroy the pathogen [30]. To date, however, no reports show that phagocytic hemocytes in Drosophila have a functional phagocytic respiratory burst pathway. Additionally, no reports show whether Drosophila epithelial cells are capable of microbial phagocytosis or that Drosophila Duox is involved in such a mechanism (for review, see [51]). In vertebrates, phagocytic respiratory burst (PRB) is generated by Nox2 [51], which is not encoded by insect genomes. Thus, in insects, the production of PRB remains to be elucidated.
Proteins involved in phagocytosis regulation
Phagocytosis crucially depends on remodeling of the actin cytoskeleton and on membrane dynamics. Distinct Rho GTPases regulate actin polymerization during phagocytosis and actually define several modes of phagocytosis that are phenotypically different (reviewed in [77]). Members of the Rho-GTPases coordinate actin dynamics, functioning as molecular switches that alternate between active (GTP-bound) and inactive (GDP-bound) states [30]. They are activated by guanine nucleotide exchange factors (GEFs) and inactivated by GTPase-activating proteins (GAPs). From the anal droplet, no Rho-GTPases or downstream elements including WASp, Scar/WAVE and Arp2/3 were detected, whereas two GEFs (Arhgef5, Dock9) and three GAPs (Srgap1, Arhgap39, Abr) were detected (Additional file 1: Table S1; Fig. 2). Arhgef5, a member of the Dbl family of Rho GEFs, strongly activates rhoA and plays a crucial role in regulating cytoskeletal remodeling linked to cell migration and invasion [78]. Dock 9 (dedicator of cytokinesis protein 9) belongs to the Zizimin subfamily (Class D) and is a specific GEF for Cdc42 [79]. Both Srgap1 and Arhgap39 impair the ability of cells to ingest IgG-opsonized targets [80, 81]. Srgap1 inhibits Rho A, and Arhgap39 inhibits Rac1 [80, 81], whereas Abr is specific for Rac and Cdc42 [82].
None of the GEFs for Rab were detected from the anal droplet, whereas three GAPs were detected (Tbc1d9, Tbc1d13, SGSM3) (Additional file 1: Table S1; Fig. 2). TBC1 domain family member 9 (Tbc1d9), a rab GAP gene, regulates the late-endosome trafficking pathway [83]. TBC1 domain family member 13 (Tbc1d13) acts as a GTPase-activating protein for Rab35 [84]. Small G protein signaling modulator 3 (SGSM3) is a specific cellular GAP for Rab5 [85].
Proteins involved in defense against viruses
The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster [86]. Drosophila Dicer-2 has an RNA interference-independent function that modulates Toll immune signaling, which defends against gram-positive bacteria, fungi, and some viruses [87]. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission [88]. From the anal droplet, both Argonaute-2 and Dicer-2 were identified (Additional file 1: Table S1, Additional file 3: Table S3), suggesting the weevil uses RNAi to manipulate some microbes, particularly viruses.
Other proteins related to immune function
Four chitinases were detected from the anal droplet (Additional file 1: Table S1). These chitinases were Group IV chitinases, containing a GH18 domain and a CBM14 domain but lacking an STL region. Group IV chitinases apparently have functions in the intestinal system because they are only expressed in different parts of the gut. This feature suggests that they are involved in the degradation of chitinous material either assimilated with food or as part of the PM. Some of these gut-specific chitinases may also have immune functions. Some are suspected to have antifungal activity, as fungal cell walls also consist of chitin. A SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 homolog (Etl1, also known as CG5899) that showed defensive response to a gram-negative bacterium [89] was identified from the anal droplet (Additional file 1: Table S1).
Discussion
Gut microbiota are essential for the health of humans and animals. These microbes increase resistance to infection, stimulate mucosal immune defenses, synthesize essential vitamins and promote caloric uptake by hydrolyzing complex carbohydrates [90]. Similar to pathogens, the native gut flora induces immune responses in the insect host [91]. To date, no specific mechanism to recognize commensal bacteria has been identified in D. melanogaster [1]. However, obviously, insects evolved regulatory mechanisms that prevent a deleterious induction of the immune response under basal conditions but allow a rapid elimination of microorganisms on pathogenic infection [1]. First, new epithelial cells are constantly generated at the level of intestinal crypts, and a tight balance is maintained between the self-renewal of cells and their elimination that is crucial to homeostasis and epithelium integrity [92]. Our results showed that epithelium renewal occurred in basal conditions, perhaps via the Wingless pathway. Additionally, a mucus layer and adherens junctions provided physical barriers against gut microbes. Second, the insect gut produces AMPs, ROS (by Duox), lysozymes and PO to defend against pathogens [1, 3]. Our results showed that in basal conditions, lysozymes, PO and ROS functioned in the weevil gut, whereas the Imd pathway and AMP production were repressed.
Lysozymes are considered to play a role in nutrition rather than in immune function. The PO cascade is an extracellular pathway in insects that is triggered by certain pathogen-associated molecular patterns and leads to the melanization of pathogens and damaged tissues [3]. Prophenoloxidase activation in hemolymph is required to survive microbial infections in Drosophila [93]. The PPOs in insect molting fluids can be activated by a virulence factor from a fungal spore and effectively melanize and inhibit fungal spore germination [94]. However, the activity of POs in the gut environment is poorly understood in arthropods, although PO secretion is demonstrated in the foregut and hindgut (reviewed in [9]). Our study showed that the PPO activating pathway in the gut was the same as that in hemolymph. PO activity contributes to wound healing by forming a scab at the epithelial injury site. The major function of gut POs is supposed as phenol-polymerization [95], which has been demonstrated in insects [96]. However, melanization induced by microbial infection has been observed in the enterocytes of the hindgut of mutant Drosophila, which does not require the recruitment of hemocytes [97, 98], suggesting that the weevil gut PO plays a role in immune function.
For gram-negative bacteria, the insect Duox system produces ROS, which in turn sterilize infected microbes [99]. In the intestine of Drosophila, the production of ROS was not observed after gram-positive bacteria S. aureus ingestion [99]. Apparently only pathogenic microbes secrete uracil that would activate the Duox pathway [99]. Our study showed that in basal conditions, AMP production was most likely not induced, whereas ROS might be released, which is consistent with reports in Drosophila [100]. These results suggest that in basal conditions, gut microbiota include pathogenic microbes (perhaps few in number). Additionally, the animals remained healthy because their defensive system cleared pathogens. Previous studies also show that the Imd pathway does not affect ROS-dependent gut immunity, and ROS-resistant bacteria remained controlled by local AMP expression [99], indicating that ROS are the primary defense, whereas AMPs are secondary. Although Duox was not found in the anal droplet, ROS were supposed in the gut. Antioxidant enzymes including SOD, CAT and TPX1 and nonenzymatic iron-binding proteins including ferritin, transferrin and vitellogenin were all detected, suggesting excessive ROS were cleared. Additionally, the titer of the microbial community associated with increased Imd pathway activation increased during aging of a fly [101], also suggesting that AMP production can be shut down in basal conditions.
From the anal droplet, enzymes involved in the Toll signal pathway were detected, indicating the same activating pathway as that in hemolymph. The Toll pathway is considered to be nonfunctional in midgut AMP production [1], although it is present in the foregut and hindgut (reviewed in [102]), and epithelial Toll-like receptors (TLRs) are activated by symbiotic bacteria in the gut in response to certain pathogens such as Salmonella enterica (reviewed in [7]). Very recently, the role of the Toll pathway in D. melanogaster gut was demonstrated to be required and sufficient to survive oral infection with Drosophila C virus (DCV) [103]. In humans, functions of Toll receptors are supposed to be related to metabolism [104]. These results indicate that the function of the gut Toll pathway is not clear and requires further study.
In humans and mammals, bacteria that do manage to cross the intestinal epithelial cell barrier are phagocytosed by macrophages in the lamina propria. Whereas in the insect gut, cells able to phagocytose bacteria and dying cells are only observed in the Drosophila proventriculus (PV) [8]. Our results provide support for this observation. From the anal droplet, many proteins involved in the processes of microbe recognition, internalization, and phagosome maturation and regulation were detected, suggesting phagocytes, perhaps nonprofessional phagocytes, were in the weevil gut. The signaling cascades in phagocytic internalization are known in great detail for the Fc receptors and the complement receptors (e.g., integrin CR3). However, none of the Fc receptors were detected from the anal droplet. Based on the proteins from the anal droplet, we propose that an integrin βν-mediated pathway might be involved in gut phagocytosis, i.e., integrin βν activates Rho by an unknown mechanism, and Rho induces accumulation of mDia, which then promotes actin polymerization [31, 105] (Fig. 2). Although none of the Rho-GTPases were identified from the anal droplet, both GEFs and GAPs for Rho were identified, suggesting Rho was in the gut. Phagocytosis is effective in eliminating bacteria. When phagocytosis is blocked, a systemic immune response ensues (reviewed in [7]). In basal conditions, when phagocytosis is sufficient for a few pathogenic microbes, activation of the systemic immune response (for example, AMPs) is not required. This property might be the reason why phagocytosis is presented, whereas the Imd pathway is repressed in the gut of the weevil.
Insects express high levels of ferritin messages in the hind end of the midgut and the yellow region of the Malpighian tubules that associate primarily with the vacuolar system and serve as iron transporters. The opposite is observed for the mammalian ferritins, which are primarily cytoplasmic and serve as iron storage proteins (reviewed in [106, 107]). The anal droplet ferritins had two possible origins: sloughed cells and/or secreted from the gut. In most insects, intracellular holoferritin is rarely observed in the cytosol or the nuclear compartment but is consistently observed in the secretory pathway [106]. Although the proteomic analysis in this study did not distinguish holoferritin and apoferritin, Calpodes larvae secret holoferritin into the posterior midgut lumen, presumably for excretion in the feces [106], suggesting a mechanism for combating iron overload [108]. The exocytosis of iron-loaded ferritin by the midgut is most likely the primary route for iron excretion by insects [109]. Thus, gut ferritin might not be involved in immune defense.
The expression of insect transferrin is up regulated to levels that are similar to those of the antimicrobial peptides after bacterial challenge, and transferrin shows antimicrobial activity (reviewed in [110]). Using immunoblot analysis, the midgut fluid transferrin of Manduca sexta has a similar concentration to that of transferrin in hemolymph [111]. However, no reports identify whether the transferrin is holo- or apo-transferrin, which determines whether it has an antibacterial role. Excess iron in the diet down regulates the transferrin, which is rationalized as a failure of the iron withholding strategy [108]. Thus, gut transferrrin may not have an antibacterial role but function as an antioxidant in stress responses such as heat shock, fungal challenge, and H2O2 exposure [112].
Conclusions
Our results demonstrated that insect anal droplets contain diverse proteins related to gut homeostasis, suggesting that droplets can be used as materials to investigate the defense of a host against gut microorganisms. Compared with materials such as the gut and frass, anal droplets also have many advantages: animals are not killed, DNA or proteins do not have to be extracted, samples are easily collected, repeated measurement experiments can be designed, etc.
Additional files
Table S1. Identified proteins by rtandem package and BlastP. Proteins identified by LC-MS were annotated by BLASTP against proteome of Dendroctonus ponderosae and NCBI CDD database. This table contains the results of rtandem and the outputs of two local BLASTPs. (XLS 59 kb)
Table S2. KEGG enrichment analysis based on the homologous genes of Dendroctonus ponderosae. KEGG enrichment analysis was used to annotate identified proteins by LC-MS. The table only contains those pathways related to innate immunity. (XLS 16 kb)
Table S3. Results of blastP against Tribolium castanum proteins presented in ref. [48]. To annotate the identified proteins, BLASTP was carried out against immune proteins of Tribolium castanum presented in ref. [48]. This file contains the standard outputs of a local BLASTP. Names of putative Drosophila melanogaster orthologs are also presented. (XLS 22 kb)
Table S4. Results of blastP against Antimicrobial Peptide Database (APD). This table shows a standard output of a local BLASTP against Antimicrobial Peptide Database. (XLS 15 kb)
Acknowledgements
We acknowledge the assistance of the graduate student Xiaojie Dou and the undergraduate student Yi Che in anal droplet sample collections.
Funding
National Natural Science Foundation of China (Grant No. 31370591), Fundamental Research Funds for the Central Universities (Grant No. DL13CA07), Postdoctoral Science Start-up Foundation of the Heilongjiang Province (Grant No. LBH-Q14012).
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Abbreviations
- AMPs
Antimicrobial proteins
- ApoLp
Apolipophorins
- CAT
Catalase
- CTLs
C-type lectins
- DAP
meso-diaminopimelic acid
- Egfr
Epidermal growth factor receptor
- FTH
Ferritin
- GAPs
GTPase-activating proteins
- GEFs
Guanine nucleotide exchange factors
- IECs
Intestinal epithelial cells
- LPS
Lipopolysaccharide
- M cells
Microfold cells
- PGN
Peptidoglycan
- PGRPs
Peptidoglycan recognition proteins
- PO
Phenoloxidase
- ppA
Prophenoloxidase-activating enzyme
- PRB
Phagocytic respiratory burst
- proPO
Prophenoloxidase
- PRX
Peroxiredoxin
- PXT
Peroxinectin
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
- SOD
Superoxide dismutase
- TF
Transferring
- TLPs
Thaumatin-like proteins
- Vg
Vitellogenin
- VSP
Venom serine protease
- ZO-1
Zonula occludens-1
- βGBP
Beta-1,3-glucan-binding protein
Authors’ contributions
TJ, and Z.W. designed the experiments. FW, FQ and TJ collected and analysed the data. TJ wrote the paper. All authors have read and approved the manuscript.
Ethics approval and consent to participate
The animals used in this study are forest insect pests. No ethics approval is needed.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tianzhong Jing, Email: jingtianzhong@yahoo.com.
Fuxiao Wang, Email: wang6676516@qq.com.
Fenghui Qi, Email: 147635828@qq.com.
Zhiying Wang, Email: tianzhongj@yahoo.com.
References
- 1.Buchon N, Broderick NA, Lemaitre B. Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat Rev Microbiol. 2013;11:615–626. doi: 10.1038/nrmicro3074. [DOI] [PubMed] [Google Scholar]
- 2.Thakur A, Dhammi P, Saini HS, Kaur S. Pathogenicity of bacteria isolated from gut of Spodoptera litura (Lepidoptera: Noctuidae) and fitness costs of insect associated with consumption of bacteria. J Invertebr Pathol. 2015;127:38–46. doi: 10.1016/j.jip.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Garcia E, Galindo-Villegas J, Mulero V. Mucosal immunity in the gut: the non-vertebrate perspective. Dev Comp Immunol. 2013;40:278–288. doi: 10.1016/j.dci.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 5.Douglas AE. The molecular basis of bacterial-insect symbiosis. J Mol Biol. 2014;426:3830–3837. doi: 10.1016/j.jmb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang JH, Jing X, Douglas AE. The multi-tasking gut epithelium of insects. Insect Biochem Mol Biol. 2015;67:15–20. doi: 10.1016/j.ibmb.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mistry R, Kounatidis I, Ligoxygakis P. Exploring interactions between pathogens and the Drosophila gut. Dev Comp Immunol. 2016;64:3–10. doi: 10.1016/j.dci.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Zaidman-Remy A, Regan JC, Brandao AS, Jacinto A. The Drosophila larva as a tool to study gut-associated macrophages: PI3K regulates a discrete hemocyte population at the proventriculus. Dev Comp Immunol. 2012;36:638–647. doi: 10.1016/j.dci.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Wu K, Yang B, Huang W, Dobens L, Song H, Ling E. Gut immunity in lepidopteran insects. Dev Comp Immunol. 2016;64:65–74. doi: 10.1016/j.dci.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Nef L, Menu JF. Internationally important damaging poplar insects. Meeting I.P.C. 1994. p. 12. [Google Scholar]
- 11.Sabri A, Vandermoten S, Leroy PD, Haubruge E, Hance T, Thonart P, De Pauw E, F F. Proteomic investigation of aphid honeydew reveals an unexpected diversity of proteins. PLoS One. 2013;8:e74656. doi: 10.1371/journal.pone.0074656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall CL, Wadsworth NK, Howard DR, Jennings EM, Farrell LD, Magnuson TS, Smith RJ. Inhibition of microorganisms on a carrion breeding resource: the antimicrobial peptide activity of burying beetle (Coleoptera: Silphidae) Oral and anal secretions. Environ Entomol. 2011;40:669–678. doi: 10.1603/EN10137. [DOI] [PubMed] [Google Scholar]
- 13.Fournier F, Beauparlant CJ, Paradis R, Droit A. rTANDEM: interfaces the tandem protein identification algorithm in R. 2014. [Google Scholar]
- 14.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 16.Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:1087–1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 18.Furuse M, Izumi Y. Molecular dissection of smooth septate junctions: understanding their roles in arthropod physiology. Ann N Y Acad Sci. 2017;1397:17–24. doi: 10.1111/nyas.13366. [DOI] [PubMed] [Google Scholar]
- 19.Wu VM, Schulte J, Hirschi A, Tepass U, Beitel GJ. Sinuous is a <em>Drosophila</em> claudin required for septate junction organization and epithelial tube size control. J Cell Biol. 2004;164:313. doi: 10.1083/jcb.200309134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011;141:769–776. doi: 10.3945/jn.110.135657. [DOI] [PubMed] [Google Scholar]
- 21.Donaldson T, Wang SH, Jacobsen TL, Schnepp B, Price J, Simcox A. Regulation of the Drosophila epidermal growth factor-ligand vein is mediated by multiple domains. Genetics. 2004;167:687–698. doi: 10.1534/genetics.103.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mak TW, Saunders ME. The Immune Response. Burlington: Academic Press; 2006. 21 - Comparative Immunology; pp. 611–637. [Google Scholar]
- 23.Marmaras VJ, Lampropoulou M. Regulators and signalling in insect haemocyte immunity. Cell Signal. 2009;21:186–195. doi: 10.1016/j.cellsig.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Brown GD, Gordon S. Immune recognition of fungal β-glucans. Cell Microbiol. 2005;7:471–479. doi: 10.1111/j.1462-5822.2005.00505.x. [DOI] [PubMed] [Google Scholar]
- 25.Xia X, You M, Rao XJYXQ. Insect C-type lectins in innate immunity. Dev Comp Immunol. 2017;83:70–79. doi: 10.1016/j.dci.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Wang XW, Zhao XF, Wang JX. C-type lectin binds to î2-integrin to promote hemocytic phagocytosis in an invertebrate. J Biol Chem. 2014;289:2405–2414. doi: 10.1074/jbc.M113.528885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerrigan AM, Brown GD. C-type lectins and phagocytosis. Immunobiology. 2009;214:562–575. doi: 10.1016/j.imbio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Cao C, Jia W, Yu L, Mo M, Wang Q, Huang Y, Lim JM, Ishihara M, Wells L, et al. Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule. EMBO J. 2009;28:286–297. doi: 10.1038/emboj.2008.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu CH, Cheng W, Chen JC. The peroxinectin of white shrimp Litopenaeus vannamei is synthesised in the semi-granular and granular cells, and its transcription is up-regulated with Vibrio alginolyticus infection. Fish Shellfish Immunol. 2005;18:431–444. doi: 10.1016/j.fsi.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Nazario-Toole AE, Wu LP. Phagocytosis in insect immunity. Advances in Insect Physiology. 2017;52:35–82. doi: 10.1016/bs.aiip.2016.12.001. [DOI] [Google Scholar]
- 31.Rosales C, Uribe-Querol E. Phagocytosis: a fundamental process in immunity. Biomed Res Int. 2017;2017:9042851. doi: 10.1155/2017/9042851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zani AI, Stephen LS, Mughal AN, Russell D, Homer-Vanniasinkam S, Wheatcroft BS, Ponnambalam S. Scavenger receptor structure and function in health and disease. Cell. 2015;4:178–201. doi: 10.3390/cells4020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulvila J, Vanha-Aho LM, Ramet M. Drosophila phagocytosis- still many unknowns under the surface. APMIS. 2011;119:651–662. doi: 10.1111/j.1600-0463.2011.02792.x. [DOI] [PubMed] [Google Scholar]
- 34.Ng TH, Chiang YA, Yeh YC, Wang HC. Review of Dscam-mediated immunity in shrimp and other arthropods. Dev Comp Immunol. 2014;46:129–138. doi: 10.1016/j.dci.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Yee GH, Hynes RO. A novel, tissue-specific integrin subunit, beta nu, expressed in the midgut of Drosophila melanogaster. Development. 1993;118:845–858. doi: 10.1242/dev.118.3.845. [DOI] [PubMed] [Google Scholar]
- 36.Nonaka S, Nagaosa K, Mori T, Shiratsuchi A, Nakanishi Y. Integrin ?PS3/??-mediated phagocytosis of apoptotic cells and bacteria in Drosophila. J Biol Chem. 2013;288:10374–10380. doi: 10.1074/jbc.M113.451427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia Z, Zhang T, Jiang S, Wang M, Cheng Q, Sun M, Wang L, Song L. An integrin from oyster Crassostrea gigas mediates the phagocytosis toward Vibrio splendidus through LPS binding activity. Dev Comp Immunol. 2015;53:253–264. doi: 10.1016/j.dci.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Lim JJ, Grinstein S, Roth Z. Diversity and versatility of phagocytosis: roles in innate immunity, tissue remodeling, and homeostasis. Front Cell Infect Microbiol. 2017;7:191. doi: 10.3389/fcimb.2017.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhawan R, Gupta K, Kajla M, Kakani P, Choudhury TP, Kumar S, Kumar V, Gupta L. Apolipophorin-III acts as a positive regulator of Plasmodium development in Anopheles stephensi. Front Physiol. 2017;8:185. doi: 10.3389/fphys.2017.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trudel J, Grenier J, Potvin C, Asselin A. Several thaumatin-like proteins bind to β-1,3-glucans. Plant Physiol. 1998;118:1431–1438. doi: 10.1104/pp.118.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park J, Stanley D, Kim Y. Roles of peroxinectin in PGE2-mediated cellular immunity in Spodoptera exigua. PLoS One. 2014;9:e105717. doi: 10.1371/journal.pone.0105717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami M, Masuda S, Ueda-Semmyo K, Yoda E, Kuwata H, Takanezawa Y, Aoki J, Arai H, Sumimoto H, Ishikawa Y, et al. Group VIB Ca2+-independent phospholipase A2Î3 promotes cellular membrane hydrolysis and prostaglandin production in a manner distinct from other intracellular phospholipases A2. J Biol Chem. 2005;280:14028–14041. doi: 10.1074/jbc.M413766200. [DOI] [PubMed] [Google Scholar]
- 43.Dennis EA. Diversity of group types, regulation, and function of phospholipase A2. J Biol Chem. 1994;269:13057–13060. [PubMed] [Google Scholar]
- 44.Stanley D, Zhang L, Kim Y. Insect anti-viral immunity: roles of prostaglandins and other eicosanoids. Invertebrate Surviv J. 2015;12:166–169. [Google Scholar]
- 45.Stanley D. Prostaglandins and their receptors in insect biology. Front Endocrinol. 2011;2:105. doi: 10.3389/fendo.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choo YM, Lee KS, Yoon HJ, Kim BY, Sohn MR, Roh JY, Je YH, Kim NJ, Kim I, Woo SD, et al. Dual function of a bee venom serine protease: Prophenoloxidase-activating factor in arthropods and fibrin(ogen)olytic enzyme in mammals. PLoS One. 2010;5:e10393. doi: 10.1371/journal.pone.0010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerenius L, Lee BL, Soderhall K. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–271. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones RM, Mercante JW, Neish AS. Reactive oxygen production induced by the gut microbiota: Pharmacotherapeutic implications. Curr Med Chem. 2012;19:1519–1529. doi: 10.2174/092986712799828283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Yang P, Huang S, Yan X, Huang G, Dong X, Zheng T, Yuan D, Wang R, Li R, Tan Y, et al. Origin of the phagocytic respiratory burst and its role in gut epithelial phagocytosis in a basal chordate. Free Radic Biol Med. 2014;70:54–67. doi: 10.1016/j.freeradbiomed.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 52.Radyuk SN, Klichko VI, Spinola B, Sohal RS, Orr WC. The peroxiredoxin gene family in drosophila melanogaster. Free Radic Biol Med. 2001;31:1090–1100. doi: 10.1016/S0891-5849(01)00692-X. [DOI] [PubMed] [Google Scholar]
- 53.Felton GW, Summers CB. Antioxidant systems in insects. Arch Insect Biochem Physiol. 1995;29:187–197. doi: 10.1002/arch.940290208. [DOI] [PubMed] [Google Scholar]
- 54.Seehuus SC, Norberg K, Gimsa U, Krekling T, Amdam GV. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc Natl Acad Sci U S A. 2006;103:962–967. doi: 10.1073/pnas.0502681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 56.Altincicek B, Knorr E, Vilcinskas A. Beetle immunity: identification of immune-inducible genes from the model insect Tribolium castaneum. Dev Comp Immunol. 2008;32:585–595. doi: 10.1016/j.dci.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 57.Sinha M, Singh RP, Kushwaha GS, Iqbal N, Singh A, Kaushik S, Kaur P, Sharma S, Singh TP. Current overview of allergens of plant pathogenesis related protein families. Sci World J. 2014;2014:543195. doi: 10.1155/2014/543195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masson F, Mone Y, Vigneron A, Vallier A, Parisot N, Vincent-Monegat C, Balmand S, Carpentier MC, Zaidman-Remy A, Heddi A. Weevil endosymbiont dynamics is associated with a clamping of immunity. BMC Genomics. 2015;16:819. doi: 10.1186/s12864-015-2048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 60.Rajagopal R. Beneficial interactions between insects and gut bacteria. Indian J Microbiol. 2009;49:114–119. doi: 10.1007/s12088-009-0023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhuang ZH, Sun L, Kong L, Hu JH, Yu MC, Reinach P, Zang JW, Ge BX. Drosophila TAB2 is required for the immune activation of JNK and NF-kappaB. Cell Signal. 2006;18:964–970. doi: 10.1016/j.cellsig.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 62.Myllymaki H, Valanne S, Ramet M. The Drosophila Imd signaling pathway. J Immunol. 2014;192:3455–3462. doi: 10.4049/jimmunol.1303309. [DOI] [PubMed] [Google Scholar]
- 63.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 64.Ming M, Obata F, Kuranaga E, Miura M. Persephone/Spatzle pathogen sensors mediate the activation of toll receptor signaling in response to endogenous danger signals in apoptosis-deficient Drosophila. J Biol Chem. 2014;298:7558–7568. doi: 10.1074/jbc.M113.543884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin PH, Huang LH, Steward R. Cactin, a conserved protein that interacts with the Drosophila IκB protein Cactus and modulates its function. Mech Dev. 2000;94:57–65. doi: 10.1016/S0925-4773(00)00314-2. [DOI] [PubMed] [Google Scholar]
- 66.Zhang S, Shi L, Yang QH, Dong XH, Chi SY, Liu HY, Tan BP. Molecular characterization and functional analysis of Cactin gene from Litopenaeus vannamei. Fish Shellfish Immunol. 2014;41:608–617. doi: 10.1016/j.fsi.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 67.Hun Jo Y, Jung Kim Y, Beom Park K, Hwan Seong J, Gon Kim S, Park S, Young Noh M, Lee YS, Han Y. TmCactin plays an important role in gram-negative and -positive bacterial infection by regulating expression of 7 AMP genes in Tenebrio molitor. Sci Rep. 2017;7:46459. doi: 10.1038/srep46459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zug R Hammerstein P. Wolbachia and the insect immune system: what reactive oxygen species can tell us about the mechanisms of Wolbachia-“host interactions. Front Microbiol 2015;6:1201. [DOI] [PMC free article] [PubMed]
- 69.Lu Y, Cao L, Egami Y, Kawai K, Araki N. Cofilin contributes to phagocytosis of IgG-opsonized particles but not non-opsonized particles in RAW264 macrophages. Microscopy (Oxf) 2016;65:233–242. doi: 10.1093/jmicro/dfv376. [DOI] [PubMed] [Google Scholar]
- 70.Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang C, Pring M, Wear MA, Huang M, Cooper JA, Svitkina TM, Zigmond SH. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell. 2005;9:209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang B, Yurecko RS, Dedhar S, Cleary PP. Integrin-linked kinase is an essential link between integrins and uptake of bacterial pathogens by epithelial cells. Cell Microbiol. 2006;8:257–266. doi: 10.1111/j.1462-5822.2005.00618.x. [DOI] [PubMed] [Google Scholar]
- 73.Li Z, Blissard G. The vacuolar protein sorting genes in insects: a comparative genome view. Insect Biochem Mol Biol. 2015;62:211–225. doi: 10.1016/j.ibmb.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 74.Silver KE, Harrison RE. Kinesin 5B is necessary for delivery of membrane and receptors during Fcγ R-mediated phagocytosis. J Immunol. 2011;186:816. doi: 10.4049/jimmunol.1002161. [DOI] [PubMed] [Google Scholar]
- 75.Botelho RJ, Grinstein S. Phagocytosis. Curr Biol. 2011;21:R533–R538. doi: 10.1016/j.cub.2011.05.053. [DOI] [PubMed] [Google Scholar]
- 76.Gutierrez MG. Functional role(s) of phagosomal Rab GTPases. Small GTPases. 2013;4:148–158. doi: 10.4161/sgtp.25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dupuy AG, Caron E. Integrin-dependent phagocytosis--spreading from microadhesion to new concepts. J Cell Sci. 2008;121:1773–1783. doi: 10.1242/jcs.018036. [DOI] [PubMed] [Google Scholar]
- 78.Komiya Y, Onodera Y, Kuroiwa M, Nomimura S, Kubo Y, Nam JM, Kajiwara K, Nada S, Oneyama C, Sabe H, et al. The rho guanine nucleotide exchange factor ARHGEF5 promotes tumor malignancy via epithelial-mesenchymal transition. Oncogene. 2016;5:e258. doi: 10.1038/oncsis.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kulkarni K, Yang J, Zhang Z, Barford D. Multiple factors confer specific Cdc42 and Rac protein activation by dedicator of cytokinesis (DOCK) nucleotide exchange factors. J Biol Chem. 2011;286:25341–25351. doi: 10.1074/jbc.M111.236455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chung J, Serezani CH, Huang SK, Stern JNH, Keskin DB, Jagirdar R, Brock TG, Aronoff DM, Peters-Golden M. Rap1 activation is required for Fcã receptor-dependent phagocytosis. J Immunol. 2008;181:5501–5509. doi: 10.4049/jimmunol.181.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duman JG, Mulherkar S, Tu YK, Cheng X, Tolias KF. Mechanisms for spatiotemporal regulation of rho-GTPase signaling at synapses. Neurosci Lett. 2015;601:4–10. doi: 10.1016/j.neulet.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cho YJ, Cunnick JM, Yi SJ, Kaartinen V, Groffen J, Heisterkamp N. Abr and Bcr, two homologous Rac GTPase-activating proteins, control multiple cellular functions of murine macrophages. Mol Cell Biol. 2007;27:899–911. doi: 10.1128/MCB.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gallo LI, Liao Y, Ruiz WG, Clayton DR, Li M, Liu YJ, Jiang Y, Fukuda M, Apodaca G, Yin XM. TBC1D9B functions as a GTPase-activating protein for Rab11a in polarized MDCK cells. Mol Biol Cell. 2014;25:3779–3797. doi: 10.1091/mbc.e13-10-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davey JR, Humphrey SJ, Junutula JR, Mishra AK, Lambright DG, James DE Stockli J. TBC1D13 is a RAB35 specific GAP that plays an important role in GLUT4 trafficking in adipocytes. Traffic. 2012;13:1429–1441. doi: 10.1111/j.1600-0854.2012.01397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- 86.van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Z, Wu D, Liu Y, Xia X, Gong W, Qiu Y, Yang J, Zheng Y, Li J, Wang YF, et al. Drosophila Dicer-2 has an RNA interference-independent function that modulates toll immune signaling. Sci Adv. 2015;1:e1500228. doi: 10.1126/sciadv.1500228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blair CD. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011;6:265–277. doi: 10.2217/fmb.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.AkS K, Qin W. Recent Developments in Using Advanced Sequencing Technologies for the Genomic Studies of Lignin and Cellulose Degrading Microorganisms. Int J Biol Sci. 2016;12:156–171. doi: 10.7150/ijbs.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Godon JJ, Arulazhagan P, Steyer JP, Hamelin J. Vertebrate bacterial gut diversity: size also matters. BMC Ecol. 2016;16:1–9. doi: 10.1186/s12898-016-0071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kwong Waldan K., Mancenido Amanda L., Moran Nancy A. Immune system stimulation by the native gut microbiota of honey bees. Royal Society Open Science. 2017;4(2):170003. doi: 10.1098/rsos.170003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ribet D, Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015;17:173–183. doi: 10.1016/j.micinf.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 93.Binggeli O, Neyen C, Poidevin M, Lemaitre B. Prophenoloxidase activation is required for survival to microbial infections in Drosophila. PLoS Pathog. 2014;10:e1004067. doi: 10.1371/journal.ppat.1004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang J, Huang W, Yuan C, Lu Y, Yang B, Wang CY, Zhang P, Dobens L, Zou Z, Wang C, et al. Prophenoloxidase-mediated ex vivo immunity to delay fungal infection after insect ecdysis. Front Immunol. 2017;8:1445. doi: 10.3389/fimmu.2017.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mattila JM, Zimmer M, Vesakoski O, Jormalainen V. Habitat-specific gut microbiota of the marine herbivore Idotea balthica (isopoda) J Exp Marine Biol Ecol. 2014;455:22–28. doi: 10.1016/j.jembe.2014.02.010. [DOI] [Google Scholar]
- 96.Wu K, Zhang J, Zhang Q, Zhu S, Shao Q, Clark KD, Liu Y, Ling E. Plant phenolics are detoxified by prophenoloxidase in the insect gut. Sci Rep. 2015;5:16823. doi: 10.1038/srep16823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seisenbacher G, Hafen E, Stocker H. MK2-dependent p38b signalling protects drosophila hindgut enterocytes against JNK-induced apoptosis under chronic stress. PLoS Genet. 2011;7:e1002168. doi: 10.1371/journal.pgen.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen J, Xie C, Tian L, Hong L, Wu X, Han J. Participation of the p38 pathway in Drosophila host defense against pathogenic bacteria and fungi. Proc Natl Acad Sci U S A. 2010;107:20774–20779. doi: 10.1073/pnas.1009223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hori A, Kurata S, Kuraishi T. Unexpected role of the IMD pathway in Drosophila gut defense against Staphylococcus aureus. Biochem Bioph Res Co. 2018;495:395–400. doi: 10.1016/j.bbrc.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 100.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 101.Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 102.Bonfini A, Liu X, Buchon N. From pathogens to microbiota: how Drosophila intestinal stem cells react to gut microbes. Dev Comp Immunol. 2016;64:22–38. doi: 10.1016/j.dci.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 103.Ferreira AG, Naylor H, Esteves SS, Pais IS, Martins NE, Teixeira L. The toll-dorsal pathway is required for resistance to viral oral infection in Drosophila. PLoS Pathog. 2014;10:e1004507. doi: 10.1371/journal.ppat.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yiu JHC, Dorweiler B, Woo CW. Interaction between gut microbiota and toll-like receptor: from immunity to metabolism. J Mol Med. 2017;95:13–20. doi: 10.1007/s00109-016-1474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mao Y, Finnemann SC. Regulation of phagocytosis by rho GTPases. Small GTPases. 2015;6:89–99. doi: 10.4161/21541248.2014.989785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pham DQD, Winzerling JJ. Insect ferritins: typical or atypical? Biochim Biophys Acta. 2010;1800:824–833. doi: 10.1016/j.bbagen.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Locke M, Leung H. The induction and distribution of an insect ferritin – A new function for the endoplasmic reticulum. Tissue Cell. 1984;16:739–766. doi: 10.1016/0040-8166(84)90007-7. [DOI] [PubMed] [Google Scholar]
- 108.Law JH. Insects, oxygen, and Iron. Biochem Bioph Res Co. 2002;292:1191–1195. doi: 10.1006/bbrc.2001.2015. [DOI] [PubMed] [Google Scholar]
- 109.Nichol H, Locke M. The localization of ferritin in insects. Tissue Cell. 1990;22:767–777. doi: 10.1016/0040-8166(90)90042-8. [DOI] [Google Scholar]
- 110.Geiser DL, Winzerling JJ. Insect transferrins: multifunctional proteins. BBA-Gen Subjects. 2012;1820:437–451. doi: 10.1016/j.bbagen.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 111.Brummett LM, Kanost MR, Gorman MJ. The immune properties of Manduca sexta transferrin. Insect Biochem Mol Biol. 2017;81:1–9. doi: 10.1016/j.ibmb.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim BY, Lee KS, Choo YM, Kim I, Je YH, Woo SD, Lee SM, Park HC, Sohn HD, Jin BR. Insect transferrin functions as an antioxidant protein in a beetle larva. Comp Biochem Physiol B Biochem Mol Biol. 2008;150:161–169. doi: 10.1016/j.cbpb.2008.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Identified proteins by rtandem package and BlastP. Proteins identified by LC-MS were annotated by BLASTP against proteome of Dendroctonus ponderosae and NCBI CDD database. This table contains the results of rtandem and the outputs of two local BLASTPs. (XLS 59 kb)
Table S2. KEGG enrichment analysis based on the homologous genes of Dendroctonus ponderosae. KEGG enrichment analysis was used to annotate identified proteins by LC-MS. The table only contains those pathways related to innate immunity. (XLS 16 kb)
Table S3. Results of blastP against Tribolium castanum proteins presented in ref. [48]. To annotate the identified proteins, BLASTP was carried out against immune proteins of Tribolium castanum presented in ref. [48]. This file contains the standard outputs of a local BLASTP. Names of putative Drosophila melanogaster orthologs are also presented. (XLS 22 kb)
Table S4. Results of blastP against Antimicrobial Peptide Database (APD). This table shows a standard output of a local BLASTP against Antimicrobial Peptide Database. (XLS 15 kb)
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].