Abstract
The arylamine N-acetyltransferase (NAT) nomenclature committee assigns functional phenotypes for human NAT1 alleles in those instances in which the committee determined a consensus has been achieved in the scientific literature. In the most recent nomenclature update, the committee announced that functional phenotypes for NAT1*10 and NAT1*11 alleles were not provided due to lack of consensus. Phenotypic inconsistencies observed among various studies for NAT1*10 and NAT1*11 may be due to variable allelic expression among different tissues, the limitations of the genotyping assays (which mostly relied on techniques not involving direct DNA sequencing), the differences in recombinant protein expression systems used (bacteria, yeast, mammalian cell lines) and/or the known inherent instability of human NAT1 protein which requires very careful handling of native and recombinant cell lysates. Three recent studies provide consistent evidence of the mechanistic basis underlying the functional phenotype of NAT1*10 and NAT1*11 as “increased-activity” alleles. Some NAT1 variants (e.g. NAT1*14, NAT1*17, and NAT1*22) may be designated as “decreased-activity” alleles and other NAT1 variants (e.g., NAT1*15 and NAT1*19) may be designated as “no-activity” alleles compared to the NAT1*4 reference allele. We propose that phenotypic designations as “rapid” and “slow” acetylator should be discontinued for NAT1 alleles, although these designations remain very appropriate for N-acetyltransferase 2 (NAT2) alleles.
Genetic variants of the arylamine N-acetyltransferases are expressed in human populations and a consensus nomenclature for arylamine N-acetyltransferase 1 (NAT1) and 2 (NAT2) alleles or haplotypes was initially published in Pharmacogenetics over 20 years ago [1]. Subsequently, additional NAT1 and NAT2 alleles appeared in the scientific literature. In order to achieve consensus for identification and naming of new NAT1 and NAT2 alleles, an arylamine N-acetyltransferase nomenclature committee was initiated [2] to establish, publish and maintain consensus listings of NAT1 and NAT2 alleles on a website originally housed at the University of Louisville and presently housed at Democritus University of Thrace (http://nat.mbg.duth.gr). As globally the most common functional allele, NAT1*4 has been assigned reference allelic status [1]. Therefore, studies comparing the sequence and phenotypic impact of other NAT1 alleles use NAT1*4 as a reference for comparison.
Updates and discussions of N-acetyltransferase gene nomenclature have been conducted at each of the seven N-acetyltransferase workshops held at three year intervals. At the 4th N-acetyltransferase workshop held in Alexandroupolis, Greece [3], the N-acetyltransferase nomenclature committee was asked to post functional phenotypes for the human NAT1 and NAT2 alleles in those instances in which the committee determined a consensus has been achieved in the scientific literature. In the most recent N-acetyltransferase nomenclature update [4], the committee announced that functional phenotypes for NAT1 and NAT2 alleles were provided where consensus is evident in the scientific literature, but that functional phenotypes for some alleles such as NAT1*10 and NAT1*11 were not provided due to lack of consensus.
NAT1*10 allele
The NAT1*10 allele was first described by Vatsis and Weber [5] and is defined by two single nucleotide polymorphisms (SNPs) in the 3’-untranslated region (UTR) of the NAT1 gene, namely 1088T>A (c.*215T>A, rs1057126) and 1095C>A (c.*222C>A, rs15561) (Table 1). It is the most common NAT1 variant allele, with an average global population frequency of about 35–40%. Its allelic prevalence is highest (~53%) in East Asian populations and lowest (15–25%) in Europeans [6, 7] (also see the dbSNP database for rs1057126). The 3’-UTR polymorphisms cause no amino acid changes, but SNP 1088T>A (c.*215T>A) causes a change in polyA-1 (AATAAA→AAAAAA), one of multiple active consensus polyadenylation signals of human NAT1 gene [8–10]. NAT1*10 has been reported to be associated with slightly elevated NAT1 activity levels in human bladder [11, 12], colon [12], liver [9, 13], and white blood cells [9, 14]. Some studies also detected higher levels of carcinogen-DNA adducts in bladder and breast tissue of individuals carrying the NAT1*10 allele [11, 15]. As described by Hein et al. [16], urinary metabolites were measured in 547 healthy individuals administered caffeine. Probit plots of the caffeine urinary metabolites 5-acetylamino-6-formylamino-3-methyluracil/1-methylxanthine (AFMU/1X) are normally used to separate rapid from slow acetylator phenotypes for NAT2 using a cut-point of 0.6, with the remaining activity being attributed to NAT1 [17]. This attribute was used to plot probits of AFMU/1X according to NAT1*4 homozygous, NAT1*10 homozygous and NAT1*4/*10 heterozygous genotypes. Presence of the NAT1*10 allele resulted in a gene-dose increase in acetylation in vivo with NAT1*10/*10 > NAT1*4/*10 > NAT1*4/*4 [16]. This trend could, however, be at least partially due to the reported linkage disequilibrium between NAT1*10 and NAT2*4 alleles [18, 19], with the observed gene-dose increase in acetylation potentially attributed to the “rapid acetylator” NAT2*4 allele frequently co-localizing with NAT1*10 on the same haplotype. In contrast, in other studies, NAT1*10 did not confer higher N-acetylation in blood cells [20–24] or healthy tissue of bladder [25] and breast [26]. Similar observations were made in vivo [23, 25, 27] and when measuring carcinogen-hemoglobin [28] or carcinogen-DNA [26, 29] adduct formation. Furthermore, transfection of NAT1*10 did not increase acetylation activity in COS-1 cells [22, 30]. Consequently, numerous reviews [31–37] conclude that these inconsistent findings reflect lack of consensus regarding NAT1*10 phenotype.
Table 1:
Description of NAT1 haplotypes *4, *10 and *11A-C. The current NAT nomenclature is aligned to the consensus Human Genome Variation Society (HGVS) nomenclature used by public electronic databases to localize SNPs in the human genome1–3.
| Homo sapiens chromosome 8 GRCh38.p12 primary assembly4, GeneID: 95, HGNC:76456, official gene symbol: NAT1 (arylamine N-acetyltransferase 1)6 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Description of nucleotide change (Current NAT nomenclature)7 | −344C>T | −40A>T | 445G>A | 459G>A | 640T>G | Δ9 between 1065–1090 | 1088T>A | 1095C>A | |
| Description of nucleotide change (HGVS nomenclature)8 | c.−344C>T | c.−40A>T | c.445G>A | c.459G>A | c.640T>G | c.*192_c.*217del AATAATAAT | c.*215T>A | c.*222C>A | |
| Reference (Allele)3 | NAT1*4 (AJ307007.1) | C | A | G | G | T | No deletion | T | C |
| Variant 1 | NAT1*10 | C | A | G | G | T | No deletion | A | A |
| Variant 2 | NAT1*11A | T | T | A | A | G | Deletion | T | A |
| Variant 3 | NAT1*11B | T | T | A | A | G | Deletion | T | C |
| Variant 4 | NAT1*11C | T | T | G | A | G | Deletion | T | A |
| Location in Gene | Intron 8, upstream of exon 9 (coding) | Intron 8, upstream of exon 9 (coding) | Exon 9 (coding) | Exon 9 (coding) | Exon 9 (coding) | 3’-untranslated region (polyA-1) | 3’-untranslated region (polyA-1) | 3’-untranslated region | |
| Description of amino acid change (Current NAT nomenclature)7 | N/A | N/A | V149I | T153T | S214A | N/A | N/A | N/A | |
| Description of amino acid change (HGVS nomenclature)8 | N/A | N/A | p.Val149Ile | p.Thr153= | p.Ser214Ala | N/A | N/A | N/A | |
| Reference (SNP)9 | rs4986988 | rs4986989 | rs4987076 | rs4986990 | rs4986783 | rs367921464 | rs1057126 | rs15561 | |
| Reference (Genomic)10 | NC_000008.11 | g.18221704C>T | g.18222008A>T | g.18222492G>A | g.18222506G>A | g.18222687T>G | g.18223127_18223135delAATAATAAA | g.18223135A>T | g.18223142A>C |
| Reference (Gene)11 | NG_012245.2 | g.56243C>T | g.56547A>T | g.57031G>A | g.57045G>A | g.57226T>G | g.57666_57674delAATAATAAA | g.57674A>T | g.57681A>C |
| Reference (Transcript)12 | NM_000662.7 (Exons 4,8,9) | c.−6–338C>T | c.−6–34A>T | c.445G>A | c.459G>A | c.640T>G | c.*207_*215delAATAATAAA | c.*215A>T | c.*222A>C |
| Reference (Protein)13 | NP_000653.3 | N/A | N/A | p.Val149Ile | p.Thr153= | p.Ser214Ala | N/A | N/A | N/A |
| Minor Allele Frequency/Minor Allele Count (1000 Genomes)14 | T=0.0168/84 | T=0.0168/84 | A=0.0170/85 | A=0.0170/85 | G=0.0170/85 | −=0.0170/85 | A=0.4006/2006 | A=0.4373/2190 | |
Harmonization of allele nomenclature was recently proposed for all human genes relevant to pharmacogenomics research and clinical practice, and designations should follow the consensus HGVS nomenclature [62].
All websites/databases were accessed on May 14, 2018.
Note that all sequences derived from the Human Genome Reference Consortium (see footnotes 9–12 below) correspond to NAT1 gene variant NAT1*10, instead of the most common functional NAT1 allele found in global populations, designated as NAT1*4 (Nucleotide ID: AJ307007.1) and considered as the reference allele for human NAT1 (see NAT website, http://nat.mbg.duth.gr/background_2013.html).
Genome Reference Consortium Human Build 38 patch release 12 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.38/#/st).
NCBI Gene Database page for human NAT1 gene (https://www.ncbi.nlm.nih.gov/gene?cmd=Retrieve&dopt=Graphics&list_uids=9).
HUGO Gene Nomenclature Committee (HGNC) page for human NAT1 gene (https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:7645).
NAT database page for human NAT1 alleles (http://nat.mbg.duth.gr/).
Sequence variant nomenclature according to current recommendations of the Human Genome Variation Society (HGVS) (http://varnomen.hgvs.org/).
Reference SNP (rs) numbers available from the NCBI dbSNP Database (https://www.ncbi.nlm.nih.gov/snp/?term=).
Homo sapiens chromosome 8 NCBI reference sequence NC_000008.11 (GRCh38.p12) (https://www.ncbi.nlm.nih.gov/nuccore/NC_000008.11).
Homo sapiens NAT1 NCBI RefSeqGene NG_012245.2 (https://www.ncbi.nlm.nih.gov/nuccore/NG_012245.2).
Homo sapiens NAT1 major transcript comprising non-coding exons 4 and 8, together with coding exon 9 [8, 43, 44] (NCBI RefSeq NM_000662.7, https://www.ncbi.nlm.nih.gov/nuccore/NM_000662.7).
Homo sapiens NAT1 protein, allozyme NAT1_4 expressed from reference allele NAT1*4 (NCBI RefSeq NP_000653.3 (https://www.ncbi.nlm.nih.gov/protein/NP_000653.3).
Reported by the NCBI dbSNP Database in the context of allele frequencies determined by the 1000 Genomes project.
NAT1*11 allele
A similar lack of consensus exists for the NAT1*11 allelic group comprising haplotypes NAT1*11A, *11B and *11C [5, 38, 39]. Those three related but distinct haplotypes bear combinations of the following variations: c.−344C>T (rs4986988), c.−40A>T (rs4986989), c.445G>A (p.Val149Ile, rs4987076; not present in NAT1*11C), c.459G>A (p.Thr153=, rs4986990), c.640T>G (p.Ser214Ala, rs4986783), 1095C>A (c.*222C>A, rs15561; not present in NAT1*11B), and a 9 bp deletion between nucleotide positions 1065–1090 (c.*192-c.*217, rs367921464) affecting a stretch of eight TAA repeats adjacent to polyadenylation signal polyA-1 [8] (Table 1). Those allelic variants are rare with an average global population frequency of about 1.8% and higher prevalence observed in Eurasian populations [6] (also see the dbSNP database for rs367921464). Previous studies with recombinant NAT1 variants bearing only the coding SNPs of NAT1*11 alleles did not show any substantial effects on NAT1 enzymatic function when expression took place in bacterial or yeast cells [23, 38, 40, 41]. However, the results were inconsistent between studies when the same variants were expressed in mammalian COS-1 cells [22, 42] or when genotype-phenotype correlation was undertaken for NAT1*11 in blood cells [14, 21]. Inclusion of the 3’-UTR SNPs in the NAT1*11 recombinant constructs expressed in yeast cells did not resolve those ambiguities [22, 30]. Consequently, the functional phenotype for NAT1*11 alleles has remained elusive [35] and no designation is presently posted on the N-acetyltransferase nomenclature committee database (http://nat.mbg.duth.gr).
Insights into the mechanistic basis of NAT1*10 and NAT1*11 allelic function
The aforementioned studies have attributed the phenotypic inconsistencies observed for NAT1*10 and NAT1*11 to the possible variable allelic expression among different tissues, the limitations of the genotyping assays (which mostly relied on techniques not involving direct DNA sequencing), the differences in recombinant protein expression systems used (bacteria, yeast, mammalian cell lines) and/or the known inherent instability of human NAT1 protein which requires very careful handling of native and recombinant cell lysates.
More recent studies may provide a mechanistic basis to identify NAT1*10 and NAT1*11 allelic function. Although the open reading frame of human NAT1 gene is contained in a single 873 bp exon, the gene is transcribed into mRNAs with variable 5’- and 3’-UTRs formed via alternative splicing of eight upstream non-coding exons and differential utilization of at least three downstream polyadenylation signals [8–10, 43, 44] (Figure 1). Using recombinant constructs expressing the sequence of the major transcript of human NAT1 gene (comprising upstream non-coding exons 4 and 8, as well as the coding exon and an adjacent 888 bp portion encompassing the 3’-UTR), Millner and colleagues [10] studied the effects of NAT1*10 polymorphisms relative to NAT1*4 reference allele in mammalian CHO cells subjected to transient or stable transfection. Although no differences between NAT1*4 and NAT1*10 polyadenylation pattern and no differences in mRNA stability were observed, nevertheless cells transfected with NAT1*10 haplotype expressed higher N- and O- acetylation activity, NAT1 mRNA, and immunoreactive protein compared to cells transfected with NAT1*4. Incubation of these cells with the arylamine carcinogen 4-aminobiphenyl showed higher DNA adducts and mutants in cells transfected with NAT1*10 compared to NAT1*4. Those effects were more pronounced in cells transfected with a third variant (named NAT1*10B) combining NAT1*10 SNPs at positions 1088 (c.*215) and 1095 (c.*222) with additional downstream SNPs linked together in high allelic frequencies according to current population data. Such polymorphisms have not been examined by previous genotyping studies and could explain the inconsistencies reported for NAT1*10 phenotype in different studies [10].
Figure 1:
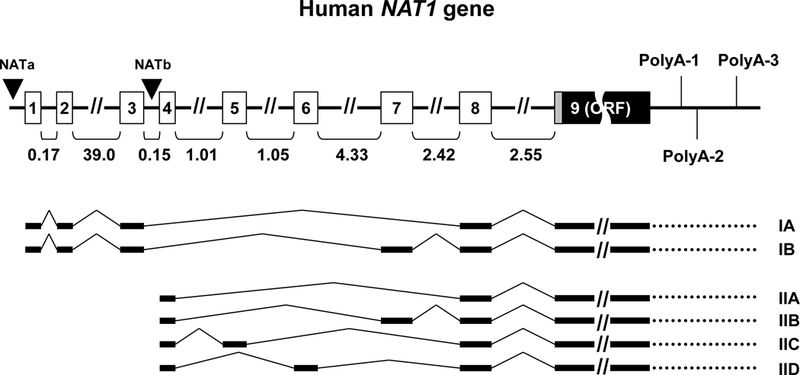
Overview of NAT1 gene structure and main alternative transcripts. The gene comprises 8 non-coding exons differentially spliced to generate alternative transcripts with variable 5’-UTRs. Exon 9 contains the entire open reading frame (ORF) of the gene, as well as the adjacent 3’-UTR terminated after three differentially utilized polyadenylation signals located at 213 (polyA-1), 331 (polyA-2; major) and 734 (polyA-3) nucleotides downstream of the coding exon. Type I transcripts (A,B) are initiated by promoter NATa located upstream of non-coding exon 1. Type II transcripts (A-D) are initiated by promoter NATb (major) located upstream of non-coding exon 4. The size of introns (in kilobases) is indicated below the gene. The figure was compiled from information previously published [7–9, 12] (figure not drawn to scale).
In another study, Wang and colleagues [9] measured transcription and translation of NAT1*10 and NAT1*11 alleles, assessing the influence of various transcription start sites, alternative splicing of 5’-UTR exons and differential usage of polyadenylation sites, employing liver biopsies, B-lymphocyte samples and transfected mammalian cell lines (HepG2 and HEK293). The determined allele frequencies in clinical samples were 19% for NAT1*10 and 2.4% for NAT1*11. These alleles did not significantly affect total levels of NAT1 mRNA in either tissue compared to the NAT1*4 allele. The relative abundance of alternative transcripts, i.e. NAT1 transcripts bearing variable 5’-UTR sequences (Figure 1), was also similar for all three alleles. The two NAT1*11 polymorphisms found upstream of the gene coding region (c.−344C>T and c.−40A>T) had no effect on the transcription initiation site or the splicing pattern of the 5’-UTR. Moreover, no effect was evident for NAT1*11 coding SNPs c.445G>A (p.Val149Ile) and c.640T>G (p.Ser214Ala) on mRNA or enzymatic activity levels, consistent with earlier studies outlined above.
The investigators then turned their attention to the region downstream of NAT1 coding exon, focusing on the effects of NAT1*10 and NAT1*11 SNPs located within the 3’-UTR of the gene [9]. First, they undertook quantification of NAT1*4 transcripts terminated after three active polyadenylation signals, located at 213 (polyA-1), 331 (polyA-2) and 734 (polyA-3) nucleotides downstream of the coding exon (Figure 1), and determined their relative amount to be 30, 60 and 10%, respectively, in both livers and B-lymphocytes. However, using a computational algorithm, an effect was predicted on transcriptional strength of polyA-1 signal due to the adjacent 9 bp deletion of the NAT1*11 allele, unlike NAT1*10 which was predicted to have no such effect. Consistent with these predictions, in ten NAT1*4/*11 heterozygous samples (6 liver and 4 B-lymphocytes), allelic mRNA analyses showed NAT1*11 to increase the amount of transcript terminated after polyadenylation signals polyA-2 (major) and potentially polyA-3, at the expense of the shorter transcript terminated after polyA-1, but without apparent change in the total NAT1 mRNA levels expressed. Further luciferase reporter gene assays demonstrated that NAT1*11 enhances translation by favoring formation of transcripts with intermediate or long 3’-UTRs, additionally implicating three NAT1*11-linked SNPs downstream of polyA-2 signal.
Undertaking a similar investigation for the NAT1*10 allele [9], no apparent differences were observed between NAT1*10 and NAT1*4 allelic transcripts expressed, in terms of both the total amount of mRNA measured and the relative abundance of generated transcripts with variable 3’-UTR lengths. However, compared with NAT1*4, reporter gene assays produced higher levels of luciferase activity with NAT1*10 3’-UTR constructs (irrespectively of their length), suggesting some enhancing effect on protein translation efficiency. The above findings were further corroborated by measurement of NAT1 protein/enzymatic activity in liver and B-cell samples genotyped as NAT1*4/*4, NAT1*4/*10, NAT1*10/*10 and NAT1*4/*11. An increase was evident for samples carrying the NAT1*10 allele, and this increase was even higher for carriers of the NAT1*11 allele.
In a more recent study, Mascarenhas and colleagues [45] undertook allele-selective whole-transcriptome analysis to assess which allelic variants of genes are likely to be recruited more efficiently by the polysomes. Levels of polysome-bound mRNA (translatome) are better correlated with levels of expressed proteins, allowing more comprehensive insight into the possible effects of SNPs located within the 5’- and 3’-UTR of transcriptionally active genes. In the course of validating their methodology, the investigators undertook allelic RNA ratio analysis to compare total cellular to polysomal RNA using a heterozygous NAT1*4/*10 lymphoblast cell line as model. No apparent differences were observed when measuring the cytoplasmic mRNA ratio for the two alleles, suggesting that NAT1*10 has no significant effect on expression and processing of NAT1 transcripts. However, when the analysis was focused on polysomal mRNA, it became evident that NAT1*10 increased protein translation by enhancement of mRNA loading to the translational apparatus of cells [45]. This is a very significant finding, as it provides a mechanism by which the NAT1*10 allele may enhance protein expression without affecting transcription of the NAT1 gene [9].
Concluding remarks
Despite some minor differences, the three studies above [9, 10, 45] succeed to reach a consensus about the mechanistic basis underlying the functional phenotype of NAT1*10 and NAT1*11 as “increased-activity” alleles compared to the NAT1*4 reference function allele. Some NAT1 variants (e.g. NAT1*14, NAT1*17 and NAT1*22) may be designated as “decreased-activity” alleles and other NAT1 variants (e.g. NAT1*15 and NAT1*19) may be designated as “no-activity” alleles compared to NAT1*4.
Designation of variants as “increased-function”, “decreased-function” and “no-function” alleles is well established for CYP2D6 gene [46]. We consider phenotypic designations described as “increased-activity”, “decreased-activity” and “no-activity” to be most suitable for NAT1 alleles. We also propose that phenotypic designations as “rapid” and “slow” acetylator should be discontinued for NAT1 alleles, as they have been used inconsistently and in different contexts in the literature, often causing confusion. For instance, some investigators have used the designation “rapid” allele to describe NAT1*4, while others have used the same designation to describe NAT1*10. Similarly, the designation “slow” allele has been used to describe low activity alleles (like NAT1*14, NAT1*17 and NAT1*22), but also prematurely terminated “null” alleles (like NAT1*15 and NAT1*19). Moreover, we consider the term “ultra-rapid” allele to be inappropriate for NAT1*10 and NAT1*11, in view of their apparently modest increases in activity. We thus propose that NAT1 variants be grouped as “increased-activity”, “decreased-activity” or “no-activity” alleles, with NAT1*4 as the “reference” allele. These designations would sufficiently incorporate the different mechanisms by which various NAT1 alleles may exert their phenotypic effects (e.g. via changes in transcription or translation, protein integrity or turnover, enzymatic activity etc.). However, “rapid” and “slow” acetylator remain very appropriate phenotypic designations for N-acetyltransferase 2 (NAT2) alleles, where genotype-phenotype correlations are much more straightforward.
Whether or not the presence of NAT1*10 and/or NAT1*11 increased-activity alleles is sufficient to modify disease risk (particularly cancer) is subject to ongoing investigations discussed by several recent reviews and meta-analyses [37, 47–51]. The NAT1 isoenzyme is expressed in many tissues, where it is likely to compete with other xenobiotic metabolizing enzymes [52]. Therefore, it is difficult to predict how toxicity of xenobiotic compounds may be modulated by NAT1*10 and NAT1*11, as the moderate phenotypic impact of those alleles is likely to be influenced by a range of other factors. Furthermore, current evidence implicates NAT1 in carcinogenesis via mechanisms not directly relevant to allelic variation [37, 53–61].
Acknowledgements
The authors gratefully acknowledge the contribution of the Erasmus+ International Credit Mobility programme (2016–2017 and 2017–2018) for faculty and student exchanges funded by the European Union. DWH is supported by United States Public Health Service grants R25-CA134283, P20-GM113226 and P42-ES023716.
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- [1].Vatsis KP, Weber WW, Bell DA, Dupret JM, Evans DA, Grant DM, et al. Nomenclature for N-acetyltransferases. Pharmacogenetics 1995;5:1–17. [DOI] [PubMed] [Google Scholar]
- [2].Hein DW, Grant DM, Sim E. Update on consensus arylamine N-acetyltransferase gene nomenclature. Pharmacogenetics 2000;10:291–2. [DOI] [PubMed] [Google Scholar]
- [3].Boukouvala S, Westwood IM, Butcher NJ, Fakis G. Current trends in N-acetyltransferase research arising from the 2007 International NAT Workshop. Pharmacogenomics 2008;9:765–71. [DOI] [PubMed] [Google Scholar]
- [4].Hein DW, Boukouvala S, Grant DM, Minchin RF, Sim E. Changes in consensus arylamine N-acetyltransferase gene nomenclature. Pharmacogenet Genomics 2008;18:367–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vatsis KP, Weber WW. Structural heterogeneity of Caucasian N-acetyltransferase at the NAT1 gene locus. Arch Biochem Biophys 1993;301:71–6. [DOI] [PubMed] [Google Scholar]
- [6].Patin E, Barreiro LB, Sabeti PC, Austerlitz F, Luca F, Sajantila A, et al. Deciphering the ancient and complex evolutionary history of human arylamine N-acetyltransferase genes. Am J Hum Genet 2006;78:423–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mortensen HM, Froment A, Lema G, Bodo JM, Ibrahim M, Nyambo TB, et al. Characterization of genetic variation and natural selection at the arylamine N-acetyltransferase genes in global human populations. Pharmacogenomics 2011;12:1545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Boukouvala S, Sim E. Structural analysis of the genes for human arylamine N-acetyltransferases and characterisation of alternative transcripts. Basic Clin Pharmacol Toxicol 2005;96:343–51. [DOI] [PubMed] [Google Scholar]
- [9].Wang D, Para MF, Koletar SL, Sadee W. Human N-acetyltransferase 1 *10 and *11 alleles increase protein expression through distinct mechanisms and associate with sulfamethoxazole-induced hypersensitivity. Pharmacogenet Genomics 2011;21:652–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Millner LM, Doll MA, Stepp MW, States JC, Hein DW. Functional analysis of arylamine N-acetyltransferase 1 (NAT1) NAT1*10 haplotypes in a complete NATb mRNA construct. Carcinogenesis 2012;33:348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Badawi AF, Hirvonen A, Bell DA, Lang NP, Kadlubar FF. Role of aromatic amine acetyltransferases, NAT1 and NAT2, in carcinogen-DNA adduct formation in the human urinary bladder. Cancer Res 1995;55:5230–7. [PubMed] [Google Scholar]
- [12].Bell DA, Badawi AF, Lang NP, Ilett KF, Kadlubar FF, Hirvonen A. Polymorphism in the N-acetyltransferase 1 (NAT1) polyadenylation signal: association of NAT1*10 allele with higher N-acetylation activity in bladder and colon tissue. Cancer Res 1995;55:5226–9. [PubMed] [Google Scholar]
- [13].Zenser TV, Lakshmi VM, Rustan TD, Doll MA, Deitz AC, Davis BB, et al. Human N-acetylation of benzidine: role of NAT1 and NAT2. Cancer Res 1996;56:3941–7. [PubMed] [Google Scholar]
- [14].Zhangwei X, Jianming X, Qiao M, Xinhua X. N-Acetyltransferase-1 gene polymorphisms and correlation between genotype and its activity in a central Chinese Han population. Clin Chim Acta 2006;371:85–91. [DOI] [PubMed] [Google Scholar]
- [15].Ambrosone CB, Abrams SM, Gorlewska-Roberts K, Kadlubar FF. Hair dye use, meat intake, and tobacco exposure and presence of carcinogen-DNA adducts in exfoliated breast ductal epithelial cells. Arch Biochem Biophys 2007;464:169–75. [DOI] [PubMed] [Google Scholar]
- [16].Hein DW, McQueen CA, Grant DM, Goodfellow GH, Kadlubar FF, Weber WW. Pharmacogenetics of the arylamine N-acetyltransferases: a symposium in honor of Wendell W. Weber. Drug Metab Dispos 2000;28:1425–32. [PubMed] [Google Scholar]
- [17].Cribb AE, Isbrucker R, Levatte T, Tsui B, Gillespie CT, Renton KW. Acetylator phenotyping: the urinary caffeine metabolite ratio in slow acetylators correlates with a marker of systemic NAT1 activity. Pharmacogenetics 1994;4:166–70. [PubMed] [Google Scholar]
- [18].Smelt VA, Mardon HJ, Sim E. Placental expression of arylamine N-acetyltransferases: Evidence for linkage disequilibrium between NAT1*10 and NAT2*4 alleles of the two human arylamine N-acetyltransferase loci NAT1 and NAT2. Pharmacol Toxicol 1998;83:149–57. [DOI] [PubMed] [Google Scholar]
- [19].Cascorbi I, Brockmöller J, Mrozikiewicz PM, Müller A, Roots I. Arylamine N-acetyltransferase activity in man. Drug Metab Rev 1999;31:489–502. [DOI] [PubMed] [Google Scholar]
- [20].Payton MA, Sim E. Genotyping human arylamine N-acetyltransferase type 1 (NAT1): the identification of two novel allelic variants. Biochem Pharmacol 1998;55:361–6. [DOI] [PubMed] [Google Scholar]
- [21].Bruhn C, Brockmoller J, Cascorbi I, Roots I, Borchert HH. Correlation between genotype and phenotype of the human arylamine N-acetyltransferase type 1 (NAT1). Biochem Pharmacol 1999;58:1759–64. [DOI] [PubMed] [Google Scholar]
- [22].de Leon JH, Vatsis KP, Weber WW. Characterization of naturally occurring and recombinant human N-acetyltransferase variants encoded by NAT1. Mol Pharmacol 2000;58:288–99. [DOI] [PubMed] [Google Scholar]
- [23].Hughes NC, Janezic SA, McQueen KL, Jewett MA, Castranio T, Bell DA, et al. Identification and characterization of variant alleles of human acetyltransferase NAT1 with defective function using p-aminosalicylate as an in-vivo and in-vitro probe. Pharmacogenetics 1998;8:55–66. [DOI] [PubMed] [Google Scholar]
- [24].Kukongviriyapan V, Prawan A, Warasiha B, Tassaneyakul W, Aiemsa-ard J. Polymorphism of N-acetyltransferase 1 and correlation between genotype and phenotype in a Thai population. Eur J Clin Pharmacol 2003;59:277–81. [DOI] [PubMed] [Google Scholar]
- [25].Vaziri SA, Hughes NC, Sampson H, Darlington G, Jewett MA, Grant DM. Variation in enzymes of arylamine procarcinogen biotransformation among bladder cancer patients and control subjects. Pharmacogenetics 2001;11:7–20. [DOI] [PubMed] [Google Scholar]
- [26].Williams JA, Stone EM, Fakis G, Johnson N, Cordell JA, Meinl W, et al. N-Acetyltransferases, sulfotransferases and heterocyclic amine activation in the breast. Pharmacogenetics 2001;11:373–88. [DOI] [PubMed] [Google Scholar]
- [27].Sy SK, de Kock L, Diacon AH, Werely CJ, Xia H, Rosenkranz B, et al. N-acetyltransferase genotypes and the pharmacokinetics and tolerability of para-aminosalicylic acid in patients with drug-resistant pulmonary tuberculosis. Antimicrob Agents Chemother 2015;59:4129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Probst-Hensch NM, Bell DA, Watson MA, Skipper PL, Tannenbaum SR, Chan KK, et al. N-acetyltransferase 2 phenotype but not NAT1*10 genotype affects aminobiphenyl-hemoglobin adduct levels. Cancer Epidemiol Biomarkers Prev 2000;9:619–23. [PubMed] [Google Scholar]
- [29].Pfau W, Stone EM, Brockstedt U, Carmichael PL, Marquardt H, Phillips DH. DNA adducts in human breast tissue: association with N-acetyltransferase-2 (NAT2) and NAT1 genotypes. Cancer Epidemiol Biomarkers Prev 1998;7:1019–25. [PubMed] [Google Scholar]
- [30].Zhu Y, States JC, Wang Y, Hein DW. Functional effects of genetic polymorphisms in the N-acetyltransferase 1 coding and 3’ untranslated regions. Birth Defects Res A Clin Mol Teratol 2011;91:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Butcher NJ, Boukouvala S, Sim E, Minchin RF. Pharmacogenetics of the arylamine N-acetyltransferases. Pharmacogenomics J 2002;2:30–42. [DOI] [PubMed] [Google Scholar]
- [32].Boukouvala S, Fakis G. Arylamine N-acetyltransferases: what we learn from genes and genomes. Drug Metab Rev 2005;37:511–64. [DOI] [PubMed] [Google Scholar]
- [33].Sim E, Westwood I, Fullam E. Arylamine N-acetyltransferases. Expert Opin Drug Metab Toxicol 2007;3:169–84. [DOI] [PubMed] [Google Scholar]
- [34].Sim E, Fakis G, Laurieri N, Boukouvala S. Arylamine N-acetyltransferases--from drug metabolism and pharmacogenetics to identification of novel targets for pharmacological intervention. Adv Pharmacol 2012;63:169–205. [DOI] [PubMed] [Google Scholar]
- [35].Hein DW. N-acetyltransferase SNPs: emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Expert Opin Drug Metab Toxicol 2009;5:353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Walker K, Ginsberg G, Hattis D, Johns DO, Guyton KZ, Sonawane B. Genetic polymorphism in N-Acetyltransferase (NAT): Population distribution of NAT1 and NAT2 activity. J Toxicol Environ Health B Crit Rev 2009;12:440–72. [DOI] [PubMed] [Google Scholar]
- [37].Butcher NJ, Minchin RF. Arylamine N-acetyltransferase 1: a novel drug target in cancer development. Pharmacol Rev 2012;64:147–65. [DOI] [PubMed] [Google Scholar]
- [38].Doll MA, Jiang W, Deitz AC, Rustan TD, Hein DW. Identification of a novel allele at the human NAT1 acetyltransferase locus. Biochem Biophys Res Commun 1997;233:584–91. [DOI] [PubMed] [Google Scholar]
- [39].Johnson N, Bell P, Jonovska V, Budge M, Sim E. NAT gene polymorphisms and susceptibility to Alzheimer’s disease: identification of a novel NAT1 allelic variant. BMC Med Genet 2004;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fretland AJ, Doll MA, Leff MA, Hein DW. Functional characterization of nucleotide polymorphisms in the coding region of N-acetyltransferase 1. Pharmacogenetics 2001;11:511–20. [DOI] [PubMed] [Google Scholar]
- [41].Fretland AJ, Doll MA, Zhu Y, Smith L, Leff MA, Hein DW. Effect of nucleotide substitutions in N-acetyltransferase-1 on N-acetylation (deactivation) and O-acetylation (activation) of arylamine carcinogens: implications for cancer predisposition. Cancer Detect Prev 2002;26:10–4. [DOI] [PubMed] [Google Scholar]
- [42].Zhu Y, Hein DW. Functional effects of single nucleotide polymorphisms in the coding region of human N-acetyltransferase 1. Pharmacogenomics J 2008;8:339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Husain A, Barker DF, States JC, Doll MA, Hein DW. Identification of the major promoter and non-coding exons of the human arylamine N-acetyltransferase 1 gene (NAT1). Pharmacogenetics 2004;14:397–406. [DOI] [PubMed] [Google Scholar]
- [44].Butcher NJ, Arulpragasam A, Goh HL, Davey T, Minchin RF. Genomic organization of human arylamine N-acetyltransferase Type I reveals alternative promoters that generate different 5’-UTR splice variants with altered translational activities. Biochem J 2005;387:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mascarenhas R, Pietrzak M, Smith RM, Webb A, Wang D, Papp AC, et al. Allele-Selective Transcriptome Recruitment to Polysomes Primed for Translation: Protein-Coding and Noncoding RNAs, and RNA Isoforms. PLoS One 2015;10:e0136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med 2017;19:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dhaini HR, El Hafi B, Khamis AM. NAT1 genotypic and phenotypic contribution to urinary bladder cancer risk: a systematic review and meta-analysis. Drug Metab Rev 2018;50:208–19. [DOI] [PubMed] [Google Scholar]
- [48].Xu Z, Li X, Qin Z, Xue J, Wang J, Liu Z, et al. Association of N-acetyltransferase 1 polymorphism and bladder cancer risk: an updated meta-analysis and trial sequential analysis. Int J Biol Markers 2017;32:e297–e304. [DOI] [PubMed] [Google Scholar]
- [49].Zhang K, Gao L, Wu Y, Chen J, Lin C, Liang S, et al. NAT1 polymorphisms and cancer risk: a systematic review and meta-analysis. Int J Clin Exp Med 2015;8:9177–91. [PMC free article] [PubMed] [Google Scholar]
- [50].Wu K, Wang X, Xie Z, Liu Z, Lu Y. N-acetyltransferase 1 polymorphism and bladder cancer susceptibility: a meta-analysis of epidemiological studies. J Int Med Res 2013;41:31–7. [DOI] [PubMed] [Google Scholar]
- [51].Cai J, Zhao Y, Zhu CL, Li J, Huang ZH. The association of NAT1 polymorphisms and colorectal carcinoma risk: evidence from 20,000 subjects. Mol Biol Rep 2012;39:7497–503. [DOI] [PubMed] [Google Scholar]
- [52].Grant DM, Hughes NC, Janezic SA, Goodfellow GH, Chen HJ, Gaedigk A, et al. Human acetyltransferase polymorphisms. Mutat Res 1997;376:61–70. [DOI] [PubMed] [Google Scholar]
- [53].Tiang JM, Butcher NJ, Minchin RF. Small molecule inhibition of arylamine N-acetyltransferase Type I inhibits proliferation and invasiveness of MDA-MB-231 breast cancer cells. Biochem Biophys Res Commun 2010;393:95–100. [DOI] [PubMed] [Google Scholar]
- [54].Tiang JM, Butcher NJ, Cullinane C, Humbert PO, Minchin RF. RNAi-mediated knock-down of arylamine N-acetyltransferase-1 expression induces E-cadherin up-regulation and cell-cell contact growth inhibition. PLoS One 2011;6:e17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tiang JM, Butcher NJ, Minchin RF. Effects of human arylamine N-acetyltransferase I knockdown in triple-negative breast cancer cell lines. Cancer Med 2015;4:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Stepp MW, Doll MA, Samuelson DJ, Sanders MA, States JC, Hein DW. Congenic rats with higher arylamine N-acetyltransferase 2 activity exhibit greater carcinogen-induced mammary tumor susceptibility independent of carcinogen metabolism. BMC Cancer 2017;17:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Stepp MW, Doll MA, Carlisle SM, States JC, Hein DW. Genetic and small molecule inhibition of arylamine N-acetyltransferase 1 reduces anchorage-independent growth in human breast cancer cell line MDA-MB-231. Mol Carcinog 2018;57:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Johansson I, Nilsson C, Berglund P, Lauss M, Ringnér M, Olsson H, et al. Gene expression profiling of primary male breast cancers reveals two unique subgroups and identifies N-acetyltransferase-1 (NAT1) as a novel prognostic biomarker. Breast Cancer Res 2012;14:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Endo Y, Yamashita H, Takahashi S, Sato S, Yoshimoto N, Asano T, et al. Immunohistochemical determination of the miR-1290 target arylamine N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast cancer. BMC Cancer 2014;14:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Minchin RF, Butcher NJ. Trimodal distribution of arylamine N-acetyltransferase 1 mRNA in breast cancer tumors: association with overall survival and drug resistance. BMC Genomics 2018;19(1):513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Carlisle SM, Hein DW. Retrospective analysis of estrogen receptor 1 and N-acetyltransferase gene expression in normal breast tissue, primary breast tumors, and established breast cancer cell lines. Int J Oncol 2018;53:694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kalman LV, Agúndez J, Appell ML, Black JL, Bell GC, Boukouvala S, et al. Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin Pharmacol Ther 2016;99:172–85. [DOI] [PMC free article] [PubMed] [Google Scholar]


