Abstract
Significance: Nuclear factor E2-related factor 2 (Nrf2) is a transcription factor that coordinates the basal and stress-inducible activation of a vast array of cytoprotective genes. Understanding the regulation of Nrf2 activity and downstream pathways has major implications for human health.
Recent Advances: Nrf2 regulates the transcription of components of the glutathione and thioredoxin antioxidant systems, as well as enzymes involved in phase I and phase II detoxification of exogenous and endogenous products, NADPH regeneration, and heme metabolism. It therefore represents a crucial regulator of the cellular defense mechanisms against xenobiotic and oxidative stress. In addition to antioxidant responses, Nrf2 is involved in other cellular processes, such as autophagy, intermediary metabolism, stem cell quiescence, and unfolded protein response. Given the wide range of processes that Nrf2 controls, its activity is tightly regulated at multiple levels. Here, we review the different modes of regulation of Nrf2 activity and the current knowledge of Nrf2-mediated transcriptional control.
Critical Issues: It is now clear that Nrf2 lies at the center of a complex regulatory network. A full comprehension of the Nrf2 program will require an integrated consideration of all the different factors determining Nrf2 activity.
Future Directions: Additional computational and experimental studies are needed to obtain a more dynamic global view of Nrf2-mediated gene regulation. In particular, studies comparing how the Nrf2-dependent network changes from a physiological to a pathological condition can provide insight into mechanisms of disease and instruct new treatment strategies.
Keywords: : Nrf2, transcription, antioxidant response
Introduction
In our everyday lives, we are continuously exposed to various chemical and physical insults, including drugs, environmental pollutants, food additives, ultraviolet and ionizing radiation. In addition to these external stresses, free radicals and reactive oxygen species (ROS) are produced as byproducts of both physiological and pathological cellular processes occurring in the mitochondria, peroxisomes, and endoplasmic reticulum (ER). At high levels, these toxicants can cause damage to cellular components, including proteins, lipids, and DNA (133). Cells normally counteract the detrimental effects of ROS and electrophiles through the activation of nuclear factor E2-related factor 2 (Nrf2; Nfe2l2 gene name) (42). Initially identified through cloning experiments as a factor that is able to bind the nuclear factor, erythroid-derived 2/activator protein 1 (NF-E2/AP1) repeat of the beta-globin gene, Nrf2 soon became the subject of extensive research for its role in regulating the expression of many antioxidant and detoxification enzymes (58, 108, 164). Nrf2 activity and abundance are tightly regulated at the transcriptional, post-transcriptional, and post-translational level (49). In response to different activating stimuli, Nrf2 is stabilized and translocates to the nucleus, where it activates the transcription of its downstream targets (49, 117). In this review, we discuss the different modes of regulation of the Nrf2 network and highlight new emerging aspects.
The Nrf2 Regulatory Network
Structure of the Nrf2 protein
Nrf2 (59, 108) belongs to the cap “n” collar (CNC) subfamily of basic-region leucine zipper (bZIP) transcription factors together with Nrf1 (18), Nrf3 (79), NF-E2 p45 subunit (7), as well as the more distantly related factors BTB domain and CNC homolog 1 and 2 (Bach1 and Bach2) (127). Nrf2 is a modular protein with seven Nrf2-ECH homology domains (Neh1–7), each of which fulfills distinct functions (Fig. 1) (49). The Neh1 domain comprises the CNC-bZIP region that is necessary for DNA binding and association with Nrf2 dimerization partners, the small masculoaponeurotic fibrosarcoma (sMaf) proteins (110). Of note, the amino acid sequence of this domain, in particular the basic region, is highly conserved across a wide range of species, underlining how crucial the transcriptional activity is for Nrf2 function (38). The Neh2 domain contains two highly conserved amino acid stretches, the DLG and ETGE motifs, which mediate the interaction with Nrf2 negative regulator Kelch-like ECH-associated protein 1 (Keap1) and seven lysine residues targeted for ubiquitylation and subsequent proteasomal degradation of Nrf2 (60, 102, 166). The C-terminal Neh3 domain harbors transactivation activity and functions in concert with the Neh4 and Neh5 domains to activate transcription of Nrf2 target genes (71, 120, 143). The Neh6 domain is a serine-rich region that is involved in the negative regulation of Nrf2 stability independent of Keap1. It contains two conserved peptide motifs, DSGIS and DSAPGS, which are recognized by β-transducing repeat-containing protein (β-TrCP) (24). β-TrCP binds more efficiently to the Neh6 domain after glycogen synthase kinase-3β (Gsk-3β)-mediated phosphorylation of the DSGIS motif and promotes the recruitment of Skp1-Cul1-F-box protein (SCF) ubiquitin ligase complex and consequent proteasomal degradation of Nrf2 (131, 132, 158, 183). The Neh7 domain is involved in the repression of Nrf2 transcriptional activity by the retinoid X receptor α through a physical association between the two proteins (175).
FIG. 1.
Structure of the human Nrf2 protein. The Nrf2 protein comprises seven Neh domains. The Neh1 CNC-bZIP domain is responsible for DNA binding and dimerization with the small Maf proteins; the Neh2 domain mediates the interaction with Keap1 through the DLG and ETGE motifs and contains seven lysine residues that are targets of ubiquitylation; the Neh3, Neh4 and Neh5 domains are transactivation domains; the Neh6 domain is a serine-rich region that regulates Nrf2 stability; and the Neh7 domain is involved in RXRα binding. bZIP, basic-region leucine zipper; CNC, cap “n” collar; Keap1, Kelch-like ECH-associated protein 1; Neh, Nrf2-ECH homology; Nrf2, nuclear factor E2-related factor 2; RXRα, retinoid X receptor α.
The Nrf2-Keap1-ARE stress–response pathway
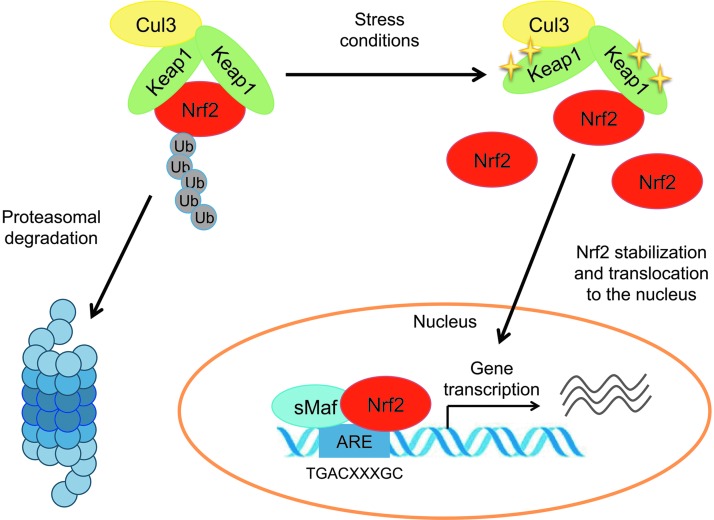
Nrf2 abundance within the cell is tightly regulated by Keap1, a redox-sensitive E3 ubiquitin ligase substrate adaptor (60). Keap1 was initially identified in a yeast two-hybrid screen by using the Neh2 domain of Nrf2 as bait and was confirmed as an Nrf2 repressor by showing that mouse embryonic fibroblasts (MEFs) and livers from Keap1 knockout mice express constitutively high levels of Nrf2 and Nrf2 target genes (60, 171). Under homeostatic conditions, two molecules of Keap1 are bound to the Neh2 domain of Nrf2 at the ETGE and DLG motifs (via their Kelch-repeat domain) (Fig. 2) (166). Keap1 functions as an adaptor protein for the Cul3 E3 ubiquitin ligase, which is responsible for the continuous ubiquitylation and degradation of Nrf2 (28, 37, 80, 192). Under unperturbed conditions, Nrf2 has a short half-life of approximately 10–30 min; therefore, Keap1-mediated high turnover of Nrf2 keeps Nrf2 basal levels extremely low (118, 156). In response to oxidative stress, Keap1 is oxidized at reactive cysteine residues, resulting in Keap1 inactivation, Nrf2 stabilization and translocation into the nucleus (10, 32, 61, 92). Here, Nrf2 heterodimerizes with members of the sMaf protein family (MafF, MafG, and MafK) (112). Genetic evidence for the essential role of sMaf proteins as Nrf2 binding partners came from crossing sMaf knockout mice to Keap1 knockout mice. Keap1-null animals show postnatal lethality due to hyperactivation of Nrf2, which results in aberrant proliferation of keratinocytes in the esophagus and forestomach (171). This phenotype is reversed not only by the concomitant disruption of Nfe2l2 (171) but also by the simultaneous deletion of both MafG and MafF, thus identifying the sMafs as essential Nrf2 dimerization partners (110).
FIG. 2.
The classical view of Nrf2 activation and response. Under unstressed conditions, Nrf2 is bound to Keap1, constantly ubiquitylated by the Cul3 E3 ubiquitin ligase and subsequently degraded by the proteasome. In response to stress, Keap1 is inactivated, resulting in Nrf2 stabilization. Nrf2 translocates to the nucleus where it heterodimerizes with the small Maf proteins, binds to the ARE and activates the transcription of its target genes. ARE, antioxidant response element.
The Nrf2-sMaf complex binds, in a sequence-specific manner, to the antioxidant response element (ARE 5′-TGACXXXGC-3′) in the promoter region of Nrf2 target genes. The AREs were initially identified as cis-regulatory elements for NADPH quinone dehydrogenase 1 (Nqo1) and glutathione S-transferase (Gst) genes (36, 136, 161). Subsequent studies expanded the list of proteins that are encoded by the ARE gene battery including genes involved in drug detoxification, antioxidant responses, NADPH regeneration and regulation of metabolism (49). Nfe2l2 knockout mice provided in vivo evidence that Nrf2 regulates the expression of these antioxidant and cytoprotective genes (58, 164).
Nrf2-mediated response to xenobiotic and oxidative stress
Nrf2 controls the expression of key components of the glutathione (GSH) and thioredoxin (TXN) antioxidant system, as well as enzymes involved in NADPH regeneration, ROS and xenobiotic detoxification, heme metabolism, thus playing a fundamental role in maintaining the redox homeostasis of the cell (Fig. 3) (42). Nrf2 tightly regulates GSH levels by directly controlling the expression of the two subunits that constitute the glutamate-cysteine ligase (Gcl) complex: the catalytic subunit (Gclc) and the modifier subunit (Gclm) (109, 180). Gcl catalyzes the reaction of glutamate with cysteine, the rate-limiting step in the synthesis of GSH. Cysteine is generated from the reduction of cystine, which is imported into the cell by the system xc− (11, 22). Nrf2 increases the supply of cysteine by directly activating Slc7a11, the gene encoding the xCT subunit of system xc− (141). In addition to GSH synthesis, Nrf2 plays a role in GSH maintenance. Nrf2 regulates the transcription of numerous ROS-detoxifying enzymes such as glutathione peroxidase 2 (Gpx2) and several glutathione S-transferases (Gsts) (Gsta1, Gsta2, Gsta3, Gsta5, Gstm1, Gstm2, Gstm3 and Gstp1) (19, 164). These enzymes use GSH to inactivate ROS, generating oxidized glutathione (GSSG). GSSG is reduced back to GSH by glutathione reductase 1 (Gsr1), another Nrf2 target, in an NADPH-dependent manner (46). Through the coordinated activation of GSH production, utilization, and regeneration, Nrf2 ensures that intracellular levels of reduced GSH are maintained. In addition to the regulation of GSH levels within the cells, Nrf2 controls the thioredoxin (TXN)-based antioxidant system. Nrf2 regulates the expression of TXN (48), thioredoxin reductase 1 (Txnrd1) (139, 170), and sulfiredoxin (Srxn1) (1), which are essential for the reduction of oxidized protein thiols (48).
FIG. 3.
The Nrf2-regulated cytoprotective defense system. Through the coordinated regulation of GSH and TXN production, utilization and regeneration, NADPH regeneration, heme and iron metabolism, ROS and xenobiotic detoxification, Nrf2 provides the main cytoprotective defense system in the cell. GSH, glutathione; HMOX1, heme oxygenase 1; Idh1, isocitrate dehydrogenase 1; NAPDH, nicotinamide adenine dinucleotide phosphate; Nqo1, NADPH quinone dehydrogenase 1; Pgd, 6-phosphogluconate dehydrogenase; ROS, reactive oxygen species; TXN, thioredoxin.
NADPH is an obligatory cofactor for many drug-metabolizing enzymes and antioxidant systems, such as cytochromes p450 (Cyp) enzymes and the Nrf2 target Nqo1 (49). Nrf2 supports NADPH production through the positive regulation of the principal NADPH-generating enzymes: glucose-6-phosphate dehydrogenase (G6pd), 6-phosphogluconate dehydrogenase (Pgd), isocitrate dehydrogenase 1 (Idh1), and malic enzyme 1 (Me1), as shown in primary cortical astrocytes (91), lung cancer cells (107), mouse small intestine (164), and mouse liver (184). Another important cytoprotective enzyme regulated by Nrf2 is heme oxygenase (Hmox1), which catalyzes the breakdown of heme molecules (5). Heme degradation results in the release of free Fe2+. Fe2+ catalyzes the Fenton reaction, which describes the conversion of H2O2 to the highly damaging OH• radical (43). To prevent OH• formation, in conjunction with Hmox1 upregulation, Nrf2 induces the expression of the genes encoding the components of the ferritin complex: the ferritin light polypeptides (Ftl) and heavy polypeptides (Fth) (23, 184). The ferritin complex oxidizes Fe2+ to Fe3+ and stores it within its own structure, thus making it unavailable for the Fenton reaction (126). Additionally, Nrf2 can also influence cellular elimination of xenobiotics by controlling the expression of many phase I and phase II drug-metabolizing enzymes (49), as well as the multi-drug-resistance-associated transporters (Mrps) (99, 188). In summary, Nrf2 increases the cellular defense mechanisms against xenobiotic and oxidative stress through the coordinated expression of numerous antioxidant and detoxification genes. The Nrf2 cytoprotective response has been elucidated in various mammalian tissues and cultured cells. In addition, other model organisms, such as Danio rerio, Drosophila melanogaster, and Caenorhabditis elegans, have been shown to possess similar anti-stress systems to mammals, suggesting that the Nrf2 antioxidant system represents an evolutionary conserved defense mechanism (38). Of note, C. elegans does not have an authentic ortholog of Keap1 (14). Skinhead-1 (Skn-1), the Nrf2 ortholog, seems to be regulated at the protein level; however, the mechanism is unclear (38). This suggests that the redox-sensing function of Keap1 might have been acquired later during evolution.
Emerging functions of Nrf2
In recent years, additional functions of Nrf2 have been discovered that go beyond the classical view of Nrf2 as a master regulator of antioxidant responses. For example, Nrf2 has been shown to regulate mitochondrial bioenergetics (54). In murine neurons and embryonic fibroblasts, loss of Nrf2 decreases the mitochondrial membrane potential, ATP production and respiration (54). In agreement with this observation, mitochondrial oxidation of the long-chain palmitic acid and the short-chain hexanoic acid is diminished in Nfe2l2 knockout MEFs (96). In addition, Nrf2 has been shown to be involved in the unfolded protein response (UPR), which is triggered by the accumulation of misfolded proteins in the ER lumen (104, 176). In the absence of Nrf2, several UPR-associated proteins show reduced expression in the liver of mice on a high-fat diet (104). Furthermore, Nrf2 can promote the removal of damaged or misfolded proteins by regulating proteasome activity (69, 87, 88). Multiple proteasome subunits are upregulated after treatment with Nrf2 inducers both in mouse tissues and in human fibroblasts (69, 87, 88). Nrf2 can therefore prevent the accumulation of abnormal proteins that might otherwise interfere with cellular functions (69, 87, 88, 104).
Moreover, Nrf2 has been shown to regulate intermediary metabolism (29, 107). In human lung cancer cells, NRF2 regulates serine biosynthesis via activating transcription factor 4 (ATF4) and phosphoglycerate dehydrogenase (PHGDH) to support GSH and nucleotide production and coordinately activates the pentose phosphate pathway (PPP) to supply ribose for nucleic acid biosynthesis (29, 107). In proliferating cells, the oxidative PPP and serine-driven one-carbon metabolism are the main contributors to cytosolic NADPH production (35). By controlling the PPP and thus the fluctuations in NADPH levels that affect the oxidation of peroxiredoxin, Nrf2 has been shown to influence the transcriptional oscillations of the circadian genes in human cells, mouse tissues and living flies (134). This provides an example of how Nrf2 can influence other cellular processes indirectly, through the regulation of redox homeostasis. In normal stem cells, Nrf2-mediated redox control plays an important role in maintaining stem cell quiescence (53, 63, 130, 167). In Drosophila intestinal stem cells (ISCs), constitutive activation of cap ‘n’ collar isoform C (CncC), a homolog of Nrf2, sustains quiescence by maintaining low intracellular ROS levels (53). In response to paraquat-induced stress, Keap1-mediated repression of CncC results in the accumulation of ROS that promotes ISCs proliferation and accelerates age-related degeneration of the intestinal epithelium (53). In ISCs, the regulation of CncC works in the opposite way compared to differentiated cells: CncC activity is inhibited in response to stress in ISCs, whereas it is induced in differentiated cells. The mechanism of CncC repression in ISCs is still unclear, but it involves Keap1 (53). Similar to ISCs, low intracellular ROS levels are required for the maintenance of quiescence in mouse and human airway basal stem cells (ABSCs) (130). After polidocanol-mediated injury of the mouse tracheobronchial epithelium, changes in ROS levels from low to moderate activate Nrf2, which induces the Notch pathway to stimulate stem cell self-renewal and proliferation for repair (130). In parallel, Nrf2 induces antioxidant genes that return overall ROS levels to a low state and this inhibits ABSCs proliferation, thus preventing hyperproliferation that would be detrimental for the repairing tissue (130). Nrf2 also regulates proliferation and differentiation of mouse hematopoietic stem cells under physiological conditions (167). Nrf2 controls the expansion of the stem and progenitor cells and supports their efficient homing by positively regulating C-X-C chemokine receptor type 4 (Cxcr4) (167). In addition to stem cell self-renewal, NRF2 participates in human embryonic stem cells differentiation into neuroectoderm (63). These studies provide examples of how redox control by Nrf2 can affect different physiological processes through the regulation of ROS levels. It is however unclear which are the targets of ROS cytotoxicity that would explain the observed phenotypes. In this regard, some light has been shed in pancreatic cancer, where Nrf2 antioxidant activity is known to be important for tumor initiation and progression (30). Cysteine residues contain highly reactive thiol groups that render them sensitive to changes in intracellular ROS levels. Alterations in the cellular redox levels are therefore likely to affect the oxidation status of reactive cysteine-containing proteins. The development of a highly sensitive proteomic method to quantify changes in the cysteine proteome showed that Nrf2-antioxidant activity promotes pancreatic tumor maintenance by preventing cysteine oxidation of the mRNA translational machinery to support efficient protein synthesis (21). This cysteine proteomic approach can now be applied to investigate the impact of changes in ROS levels on additional cellular processes.
Modes of Regulation of Nrf2 Activity
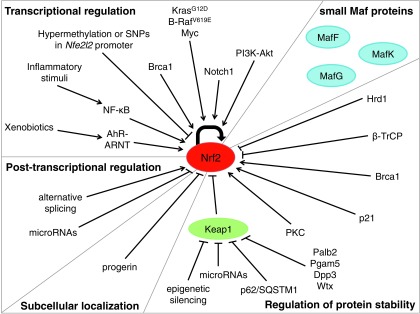
Given the vast array of stimuli that activate Nrf2 and the diverse cellular processes that it controls, the regulation of Nrf2 activity is complex and multifactorial. Indeed, Nrf2 activation can be controlled at the transcriptional and post-transcriptional level, through the regulation of protein stability, post-transcriptional modifications, and the availability of binding partners (Fig. 4).
FIG. 4.
Mechanisms of regulation of Nrf2 activity. The mechanisms of modulation of Nrf2 activity include the regulation of transcription, mRNA processing, translation, subcellular localization, protein stability, and availability of binding partners. Arrows indicate activating regulation, and bars indicate inhibitory regulation. β-TrCP, β-transducing repeat-containing protein; AhR, aryl hydrocarbon receptor; BRCA1, breast cancer susceptibility 1; NF-κB, nuclear factor-κB; PI3K, phopshoinositide 3-kinase; PKC, protein kinase C.
Transcription-associated regulation of Nrf2
Nfe2l2 transcription is regulated by several transcription factors. Nrf2 is induced by aryl hydrocarbon receptor (AhR) in response to polycyclic aromatic hydrocarbon exposure (106). AhR binds as a heterodimer with AhR nuclear translocator (Arnt) to the xenobiotic response element-like sequences in the promoter of Nfe2l2 and transactivates its transcription (106). Thus, xenobiotic ligands are able to activate the Nrf2 pathway by inducing AhR. In addition, the NFE2L2 promoter contains a binding site for nuclear factor (NF)-κB, which allows it to be induced by inflammatory stimuli (138). NFE2L2 transcription is indeed activated by lipopolysaccharide (LPS) treatment in human monocytes (137). High basal NRF2 activity in acute myeloid leukemia (AML) has been attributed to constitutive NF-κB-mediated upregulation of the NFE2L2 gene and is believed to be one cause of resistance to chemotherapy in AML cells (138). In addition, NFE2L2 is induced by breast cancer susceptibility 1 (BRCA1) in human MCF10A mammary epithelial cells on xenobiotic stress (67). Oncogenic Kras and B-Raf, Myc (30), the phosphoinositide 3-kinase (PI3K)-Akt pathway (107), and the Notch signaling pathway (173) have also been reported to augment Nfe2l2 transcription, thus suggesting a possible mechanism for the increased expression of Nrf2 in tumor cells. Of note, the Nfe2l2 gene promoter contains ARE-like sequences, providing a positive feedback mechanism to amplify Nrf2 effects (86). Indeed, in murine keratinocytes, Nrf2 has been shown to bind these sequences and overexpression of the wild-type but not a truncated form of Nrf2 lacking the N-terminal region (amino acids 1–368, including the transactivation domains) induces the activity of the isolated promoter-proximal region of the Nfe2l2 gene in a luciferase reporter assay (86). Additional evidence for the transcriptional regulation of Nrf2 came from the observation that modifications of the Nfe2l2 promoter region such as hypermethylation or single nucleotide polymorphisms (SNPs) result in decreased Nrf2 expression; however, the significance of these alterations awaits further study (101, 159, 191).
Post-transcriptional regulation of Nrf2
microRNAs (miRNAs) are short (20–22 nucleotides long), single-stranded, noncoding RNAs that regulate gene expression by sequence-specific binding with mRNA molecules and consequent inhibition of translation or degradation of the targets (181). miR-144 was the first miRNA identified as an NRF2 negative regulator in reticulocytes of patients with homozygous sickle cell disease (HbSS) (140). A subset of HbSS patients with more severe anemia shows higher erythrocytic miR-144 expression. Increased miR-144 is associated with reduced NRF2 levels, decreased GSH regeneration and impaired oxidative stress tolerance, thereby providing a possible mechanism for the increased anemia severity seen in these patients (140). Subsequently, other miRNAs have been identified to control Nrf2 levels in the cell. Ectopic expression of miR-28 in human MCF7 breast cancer cells (187); miR-27a, miR142-5p, miR-144, and miR-153 in human SH-SY5Y neuroblastoma cells (116); and miR-93 in human MCF10A mammary epithelial cells and T47D breast cancer cells decreased NRF2 mRNA and protein levels (151). However, validation in physiological conditions is still lacking.
Nrf2 can also be regulated through alternative splicing. In lung and head and neck cancers, aberrant NFE2L2 transcript variants missing exon 2, or exons 2 and 3, have been observed (40). The NRF2 protein isoforms encoded by these splice variants lack the KEAP1 interaction domain, thus resulting in NRF2 stabilization and induction of the NRF2 program (40). The impact of NFE2L2 exon skipping on tumorigenesis remains to be evaluated. However, silencing of NRF2 in a hepatocellular carcinoma cell line with heterozygous skip of NFE2L2 exon 2 results in decreased cell viability (40).
Regulation of Nrf2 protein stability
Under unstressed conditions, Nrf2 is constantly targeted for proteasomal degradation by the Keap1/Cul3 E3 ubiquitin ligase complex (37, 60, 80, 192). The high turnover of Nrf2 provides a readily available pool of newly translated protein that can be rapidly stabilized in response to stress by inhibiting ubiquitylation and proteasomal degradation. A decrease in the amount of Keap1 results in Nrf2 accumulation, as evidenced by deletion of Keap1 in the mouse and knockdown of Keap1 in human cells (31, 171). Epigenetic silencing of KEAP1 by hypermethylation of its promoter causes an increase in Nrf2 expression levels in lung (114, 177), prostate (194), colorectal cancers (45) and gliomas (113), conferring a growth advantage to the cancer cells. In addition, KEAP1 is negatively regulated by miR-200a in breast cancer cells, leading to increased NRF2 stabilization (33).
The interaction between NRF2 and KEAP1 can also be disrupted by somatic mutations of the NFE2L2 and KEAP1 genes, as observed in carcinomas of the lung (123, 147, 150, 153, 190), gallbladder (146), ovary (84), breast (119, 152), stomach, liver (190), skin, larynx, and esophagus (77). Moreover, several cytoplasmic proteins that interfere with Keap1-Nrf2 interaction have been identified. These include p62, also known as sequestosome 1 (SQSTM1), a ubiquitin-binding protein that targets protein aggregates for degradation via the autophagic pathway (26, 76, 83, 90). The STGE motif of p62 is similar to the ETGE motif of Nrf2 and therefore p62 competes with Nrf2 for the binding with Keap1. When autophagy is impaired, p62 levels are elevated, leading to degradation of Keap1 and consequent Nrf2 stabilization (26, 83, 90). Of note, p62 is an Nrf2 target gene, thus creating another positive feedback loop (62). In addition to p62, other proteins that bind Keap1 and interfere with Keap1-Nrf2 interaction include partner and localizer of Brca2 (PALB2) (97), phosphoglycerate mutase 5 (PGAM5) (95), dipeptidyl-peptidase 3 (DPP3) (47), and Wilms tumor gene on X chromosome (WTX) (16), among others. Competing proteins that disrupt the Keap1-Nrf2 association by physically interacting with Nrf2 have also been identified. The cyclin-dependent kinase inhibitor p21 is induced by p53 in response to oxidative stress and competes with Keap1 for the binding to the DLG motif of Nrf2, thus compromising Nrf2 ubiquitylation and promoting the Nrf2-dependent antioxidant response (20, 34). The DNA repair protein Brca1 can also induce Nrf2 stabilization by binding Nrf2 and preventing Keap1-mediated inhibition (41). Loss of Brca1 in mouse premalignant mammary epithelial cells results in reduced expression of Nrf2 and Nrf2-regulated antioxidant enzymes, leading to accumulation of ROS (41).
Additionally, Keap1-mediated regulation of Nrf2 activity can be modulated by post-translational modifications of Nrf2. Protein kinase C (PKC) phosphorylates Ser40 in the Neh2 domain of NRF2, disrupting KEAP1-NRF2 association and thus promoting NRF2 activation (55). This allows Nrf2 activity to be induced by signals that induce PKC, such as oxidative stress (155).
Keap1 is not the only negative regulator of Nrf2. Keap1-independent degradation of Nrf2 was first noted when it was observed that deletion of the ETGE and/or DLG motifs in the Neh2 domain results only in a modest increase in its stability under unstressed conditions (103). Further examination of the Nrf2 protein sequence led to the identification of two highly conserved regions within the Neh6 domain of Nrf2, deletion of either of which increases the half-life of Nrf2 mutants that lack the Neh2 domain in Keap1-null MEFs (24). These regions contain two peptide sequences, the DSGIS and the DSAPGS motifs, which are recognized by β-TrCP. β-TrCP binding to Nrf2 follows Gsk-3β-mediated phosphorylation of the Neh6 domain of Nrf2. β-TrCP functions as an adaptor for the SCF E3 ubiquitin ligase complex to regulate proteasomal degradation of Nrf2 (131, 132). Thus, Nrf2 activity could potentially be enhanced by some kinases, such as extracellular signal-regulated kinase (ERK), p38 MAP kinase (MAPK), PI3K and PKC, through the inhibition of Gsk-3β (66). Another ubiquitin-dependent system responsible for Nrf2 degradation involves the E3 ubiquitin ligase synoviolin (Hrd1) (185). During ER stress in the context of liver cirrhosis induced experimentally by administration of CCl4, the Nrf2 antioxidant activity is repressed by Hrd1. Hrd1 interacts with the Neh4 and Neh5 domains of Nrf2 though its C-terminal domain and causes Nrf2 ubiquitylation and subsequent degradation. Hrd1-mediated regulation of Nrf2 is independent of both Keap1 and β-TrCP and prevents Nrf2 from activating the antioxidant response and therefore counteracting the high levels of ROS produced during cirrhosis. Thus, pharmacological inhibition of Hrd1 may represent a potential therapeutic strategy for mitigating liver cirrhosis (185).
Regulation of Nrf2 subcellular localization
Hutchinson-Gilford progeria syndrome (HGPS) is a rare, invariably fatal genetic condition characterized by premature aging beginning in childhood (169). The disease is caused by constitutive production of progerin, a mutant form of the nuclear architectural protein lamin A (39). HGPS cells present a variety of cellular defects including nuclear distortion, loss of heterochromatin structure, altered patterns of histone modifications and increased levels of persistent DNA damage (39). Until a few years ago, the mechanism through which progerin caused these morphological and epigenetic alterations was unclear. Recently, it was shown that progerin traps NRF2 at the nuclear periphery, thus impairing its activity (85). NRF2 sequestration by progerin results in chronic oxidative stress and contributes to HGPS aging defects, which can be reverted by the reactivation of NRF2. Thus, impairment of NRF2 activity is a driver mechanism of HGPS and restoration of its function may represent a therapeutic opportunity for HGPS patients (85).
Small Maf proteins
The regulation of Nrf2 activity is not limited to the control of its abundance but can also be modulated by the availability of its binding partners. As mentioned earlier, Nrf2 forms heterodimers with the sMaf proteins and recognizes the AREs in the genome (72). The sMafs (MafF, MafG and MafK) are members of the bZIP family of transcription factors: the basic domain binds DNA, whereas the leucine zipper mediates homo- or hetero-dimerization with CNC proteins and Bach proteins (68). The CNC and Bach proteins cannot bind DNA on their own and require sMafs as obligatory dimerization partners to exert their role as transcriptional regulators (112, 127). The sMaf proteins lack the transactivation domain and sMaf homodimers have been shown to act as transcriptional repressors in overexpression experiments (56, 78, 115). Thus, the sMaf proteins and their binding partners form a complex network of interacting transcription factors. Changes in the abundance or activity of the participant molecules of the network can lead to major changes in the regulation of gene expression (111).
sMaf genes expression is detected in various tissues, but each sMaf gene has a distinct expression profile (124, 165). In human adult tissues, MAFK mRNA levels are high in heart, skeletal muscle, and placenta; whereas MAFG mRNA is abundant in skeletal muscle and is moderately expressed in heart and brain. Both are expressed in all hematopoietic cell lines, including erythroid and megakaryocytic lineages (165). In the mouse, MafK and MafG are expressed in most tissues, albeit at different levels, whereas MafF gene expression is restricted to the lung (124). The sMaf proteins show a high degree of conservation among vertebrates, including human, mouse, rat, chicken, and zebrafish, and a significant similarity in the primary structure to each other (68, 74). The high degree of similarity suggests that the sMaf proteins have redundant activity and that the specificity is determined by their expression pattern.
To dissect in vivo the specific functions of the different sMafs, MafF, MafG, and MafK knockout mice were generated by deleting the entire coding sequence (124, 125, 145). Gene targeting of MafF and MafK does not cause any apparent phenotype (124, 125), while MafG-null mice exhibit abnormal megakaryocyte differentiation and thrombocytopenia accompanied by a late-onset neurological disorder (145). MafK and MafG double knockout mice survive embryogenesis, but they die postnatally (125). These mutant mice develop more severe deficiencies in megakaryopoiesis compared with MafG-null mice, specifically in proplatelet formation, resulting in profound thrombocytopenia (125). In addition, they present severe anemia accompanied by abnormal erythrocyte morphology and develop severe neurological disorders (73, 125). These observations indicate that MafG and MafK have redundant functions, although MafG is preponderant (73, 125). Mice deficient for Nfe2, another member of the CNC subfamily of bZIP transcription factors (7), exhibit impaired megakaryopoiesis, suggesting that the NF-E2 p45-sMafG heterodimer is necessary for the production of platelets from megakaryocytes (149). Mice carrying a central nervous system-specific deletion of Nfe2l1, the gene encoding Nrf1, display similar neurological disorders as the ones observed in sMaf-deficient mice, indicating that Nrf1 and the sMaf proteins likely collaborate in maintaining neuronal homeostasis (81).
MafF, MafG, and MafK triple knockout embryos develop normally until embryonic day 9.5 (E9.5), then show severe growth retardation and liver hypoplasia and die around E13.5 (186). Basal expression of ARE-dependent genes is unaffected in E10.5 triple knockout embryos compared to wild-type embryos but is significantly reduced in the livers of E13.5 mutant embryos in concomitance with the severe liver hypoplasia observed in these embryos (186). Importantly, the embryonic lethality and liver hypoplasia could be completely rescued by transgenic expression of exogenous MafG (186). Basal expression of cytoprotective genes is severely compromised in sMaf triple knockout fibroblasts prepared from E11 or E13.5 embryos, confirming that the sMafs are essential for the expression of ARE-regulated genes (72). MafG is sufficient to rescue the inducible expression of cytoprotective genes in MEFs (72).
In summary, the investigation of sMaf knockout mice showed that the sMaf proteins are functionally redundant and indispensable for supporting Nrf2-mediated transcriptional activity (72, 186).
Mechanisms of Nrf2-Mediated Gene Transactivation
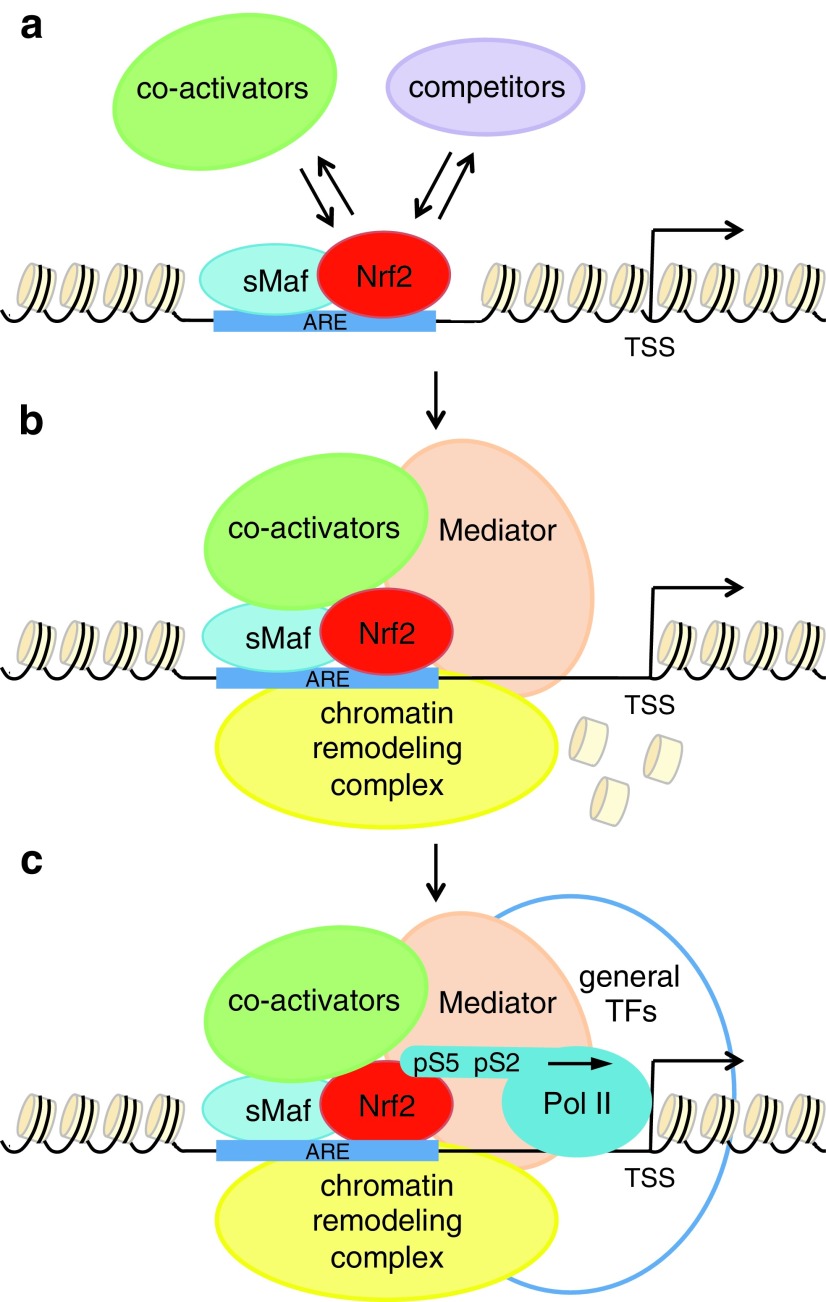
Nrf2 activity is tightly regulated (160). Once Nrf2 is activated, it translocates to the nucleus where it binds to target sequences in association with the sMaf proteins (Fig. 5) (49). The investigation of Nrf2-DNA interactions in a genome-wide manner through chromatin immunoprecipitation followed by sequencing (ChIP-Seq) in MEFs from Keap1-null mice showed that the ARE is strongly enriched within Nrf2 binding sites (100). Subsequent ChIP-Seq analyses of Nrf2 binding sites in human lymphoblastoid cells treated with the dietary isothiocyanate sulforaphane (SFN) and in the mouse hepatoma cell line Hepa1c1c7 treated with the electrophilic agent diethyl maleate (DEM) showed preferential binding of Nrf2-sMaf heterodimer to 5′-TGACTCAGC-3′ (23, 52). In addition, these studies showed that a small fraction of Nrf2 binding sites did not contain an ARE, indicating that Nrf2 probably interacts with other DNA-binding proteins (23, 52, 100). However, the functional relevance of ARE-independent binding requires further investigation.
FIG. 5.
Nrf2-mediated induction of gene expression. (a) Nrf2 selects the genes to be activated by binding as a heterodimer with sMaf to the ARE in promoter regions of the target genes. The recognition of the ARE can be influenced by the cooperation or competition with other activators or repressors. (b) Nrf2 recruits co-activators, components of the transcription machinery and nucleosome-remodeling complexes through protein–protein interactions to make the chromatin structure accessible to the Pol II machinery. (c) Together, co-activators and chromatin remodelers favor the recruitment of Pol II and the general transcription factors to form the PIC. Following phosphorylation of Ser2 and Ser5 in Pol II CTD transcription starts. CTD, carboxy-terminal domain; PIC, pre-initiation complex; Pol II, RNA polymerase II; sMaf, small masculoaponeurotic fibrosarcoma.
The extent of Nrf2 transactivation depends on the levels of Nrf2 protein, as shown in a gene dose response study analyzing expression changes in livers from Nfe2l2-null, wild-type, Keap1 knockdown and Keap1 knockout mice (184). Genes involved in the antioxidant response, GSH and xenobiotic metabolism show a graded activation by Nrf2, suggesting that the Nrf2-regulated cytoprotective response can be tuned to the intensity of the stress by varying Nrf2 levels (184).
The integrated analysis of Nrf2 binding and transcription profiles showed that not all genes in the vicinity of bound Nrf2 are transcriptionally regulated as a result of Nrf2 binding (23, 52, 100). These genes may require the recruitment of specific cofactors for a complete activation. Motif analysis of Nrf2 binding sites identified the consensus motifs for other transcription factors, such as Fos, Mafb, Lhx3 and MEF2A; however, further experiments are required to evaluate their cooperation with Nrf2 to induce gene transactivation (100). In addition, ARE-like sequences are recognized by other CNC transcription factors and the members of the AP1 complex, such as Jun, Fos, Atf and Maf proteins in electrophoretic mobility shift assays (4) and NRF2 was reported to form heterodimers with ATF4 (50). Full understanding of Nrf2-mediated gene transactivation requires taking into account the cooperation or competition with other transcription factors at the available binding sites.
Once the decision to activate a gene is made, Nrf2 recruits co-activators and components of the transcription machinery through protein–protein interactions to initiate transcription. One of the first co-activator identified to interact with Nrf2 is CREB binding protein (CBP) (25, 71). Nrf2 binds CBP through its Neh4 and Neh5 domains and together they activate transcription via the AREs (71). CBP can stimulate gene transcription through its histone acetyltransferase activity or by functioning as a scaffold to stabilize additional components of the general transcriptional machinery (12, 17). However, the precise mechanism of how Nrf2 and CBP cooperate to transduce the ARE gene battery remains to be elucidated (71). In addition to CBP, NRF2 interacts with its close homologue p300 (143). As CBP, p300 acetylates histones to facilitate chromatin decondensation and the recruitment of the transcription machinery (122). In addition, p300/CBP have been reported to associate with NRF2 in response to arsenite-induced stress and acetylate a number of lysine residues within the Neh1 DNA binding domain of NRF2 (157). Mutation of 18 lysine sites to arginine does not affect NRF2 protein stability but does compromise the DNA-binding ability of NRF2 and consequently the transcription of NQO1, TXNRD1 and GCLM, but not HMOX1 in HCT116 cells (157). Together with histone-modifying enzymes, Nrf2 recruits Mediator, a multi-subunit protein complex that communicates the activation signals from a DNA-bound transcription factor to RNA polymerase II (Pol II) (6, 143). The subunit composition of Mediator can change: subunits can be lost or added to affect its biological function (6). NRF2 has been shown to interact directly with the MED16 subunit of Mediator though the Neh1, Neh4, and Neh5 domains (143). MED16 bridges the interaction between NRF2 and the Mediator complex and its depletion specifically reduces the transcription of several NRF2 target genes in response to electrophilic stress, without affecting hypoxia-induced gene expression. MED16 knockout does not impact cell proliferation but renders the cells more sensitive to cytotoxicity induced by menadione (143). In addition to histone-modifying enzymes and the Mediator complex, NRF2 recruits ATP-dependent nucleosome-remodeling complexes. BRG1, the central ATPase subunit of the SWI/SNF chromatin-remodeling complex, has been shown to interact with NRF2 and to selectively influence the transcription of NRF2 target genes with Z-DNA formation (193). Moreover, NRF2 associates with other co-activators of the transcription machinery, such as chromodomain helicase DNA-binding protein 6 (CHD6) (120), receptor-associated co-activator 3 (RAC3) (75), and NAD+-dependent histone deacetylase sirtuin 6 (SIRT6) (128). However, the functional significance of these interactions has not been extensively elucidated.
Together, co-activators and chromatin remodelers promote the recruitment of Pol II and the components of the general transcription machinery to form the pre-initiation complex (PIC) at the promoters of target genes. Following PIC assembly, the carboxy-terminal domain (CTD) of Pol II has to be phosphorylated at Ser5 to initiate transcription of the gene and subsequently at Ser2 to promote productive transcription elongation (179). In Drosophila, cyclin-dependent kinase 12 (Cdk12) has been identified as the Pol II kinase responsible for the phosphorylation of Ser2 in Pol II CTD at Nrf2 target genes on exposure to oxidative stress. Under unstressed conditions, knockdown of Cdk12 in cell culture and in vivo does not affect cell viability or the expression of genes involved in basic housekeeping processes; however, in response to the oxidative stress inducer paraquat, it specifically impairs the expression of Nrf2 target genes and decreases the survival of the flies (93). In human cells, CDK12 has been identified as an essential regulator for the transcription of various DNA damage response and DNA repair genes, increasing the interest in the development of pharmacological inhibitors of CDK12 to act as sensitizers to chemotherapeutic agents (64, 94). If Cdk12 gene selectivity for Nrf2 antioxidant targets is conserved in humans, the mechanism of action of Cdk12 inhibitors may not be limited to the repression of the DNA damage response and DNA repair pathway, but may also involve the suppression of Nrf2 cytoprotective response.
Deciphering the Nrf2-Regulated Network
Mapping of the Nrf2 transcriptional program
Continuous efforts have been made to identify the genes regulated by Nrf2 and thus the function of Nrf2 in a given cellular context. These studies have utilized pharmacological activation of Nrf2, Nfe2l2-deficient or Keap1-deficient mice to define the Nrf2 responsive genes. The first of these studies performed gene expression profiling by microarray of the small intestine from wild-type and Nfe2l2 knockout mice treated with vehicle or the Nrf2 inducer SFN and identified that the basal and inducible expression of several genes involved in ROS detoxification, GSH synthesis and NADPH regeneration is dependent on Nrf2 (164).
Transcriptional profiles of Nfe2l2 knockout mice showed that Nrf2 is not just involved in inducible gene expression in response to an activating agent but also involved in the constitutive expression of several antioxidant and detoxification genes in the absence of external stresses (13, 100, 164). Under normal conditions, Nrf2 is a very unstable protein with a short half-life; therefore, it was surprising to observe Nrf2-mediated regulation of gene expression in the absence of exogenous stress stimuli (118, 156). Since ROS and other endogenous reactive molecules are constantly generated from physiological cellular processes, it is possible that Keap1 activity is slightly impaired under unperturbed conditions. As discussed above, Nrf2 can also be regulated at the transcriptional level (49). Moderate Nrf2 activation in the absence of external stresses is likely the result of equilibrium between Nfe2l2 transcription and protein stability.
These gene expression analyses were performed in several tissues and used different agents to induce Nrf2, including naturally occurring chemopreventive drugs, such as SFN, soy isoflavone and triterpenoids, and toxicants, such as hypochlorous acid and inorganic arsenite, and showed that Nrf2 regulates a common set of genes irrespective of the type of stimulus and the cellular context (13, 27, 164, 182, 188). This “default Nrf2 program” includes genes such as Nqo1, Gclc, Gclm, and Txnrd1 and is in part conserved from Drosophila to humans, constituting an ancient Nrf2 regulatory network (89). Together with the “default Nrf2 program,” Nrf2 selectively activates other genes that are specific to the cell type and the nature of the inducing agent (2, 13, 82, 89, 107, 164, 182, 188).
The current studies of the Nrf2-dependent program are limited to the investigation of the response to pharmacological activation of Nrf2 or to the deletion of either Keap1 or Nfe2l2 in different cellular contexts. Studies comparing how the Nrf2-dependent network changes from a physiological to a pathological condition are still lacking. The only exception is represented by a work on the mechanism of Nrf2-mediated oncogenicity in lung cancer (142). Transcriptional profile in normal lung tissue and urethane-induced tumors from wild-type and Nfe2l2 knockout mice showed induced expression of genes involved in cell growth and proliferation, Wnt/β-catenin signaling, and Notch signaling in an Nrf2-dependent manner, suggesting a possible mechanism for the contribution of Nrf2 to cancer progression (142).
Nrf2-mediated gene repression
These large-scale studies indicated that Nrf2 not only activates but also suppresses the expression of a wide range of targets. However, the mechanisms underlying transcriptional repression by Nrf2 are still unclear. Gene expression profiling by microarray in mouse liver identified genes that were repressed by H-1,2-dithiole-3-thione (D3T) in wild-type mice, but not in Nfe2l2-knockout mice, compared with vehicle-treated wild-type mice. Of note, these downregulated genes were not detected at the early time point (6 h) but only at the late time point (24 h), suggesting that the repression observed might be indirect (88). In line with this observation, the integrated analysis of ChIP-Seq and RNA-Seq data showed that the majority of Nrf2 direct targets are upregulated rather than repressed, indicating that Nrf2 is primarily an activator and blocks gene expression indirectly (23, 52) (C.T., I.I.C.C. and D.A.T. unpublished observations). These data contradicted previous studies, which reported that several transcription factors could form inhibitory complexes with Nrf2 and bind the promoters of Nrf2 target genes, thus causing their transcriptional repression (8, 15, 57). Nrf2 may repress transcription indirectly by inducing the expression of transcriptional repressors or miRNAs (144). NRF2 ChIP-Seq in lymphoblastoid cell lines identified several NRF2 binding sites in the vicinity of multiple miRNAs, the most notable being the miR-365-1/miR-193b cluster and miR-29b-1, which were previously linked to cancer progression and oxidative stress, respectively (23). An additional example of Nrf2-controlled miRNA is miR-125-b1, which is upregulated after activation of Nrf2 in the kidney of mice treated with oltipraz (65).
Nrf2 distal binding sites
The global mapping of Nrf2 binding sites indicated that the majority of Nrf2 binding sites lie outside the promoter-proximal region (23, 52). Nrf2 function at distal genomic sites is still unclear. Some of these binding sites could be located in the promoter region of long non-coding RNAs (lncRNAs), which were beginning to be annotated when Nrf2 binding sites were first profiled (44). To date, few Nrf2-regulated lncRNAs have been described. NRF2 was shown to activate SCAL1 lncRNA in response to cigarette smoke in lung cancer cell lines (162) and to repress the pluripotency lncRNA ROR in human MCF10A mammary epithelial cells (195). Furthermore, a recent study identified additional NRF2-regulated lncRNAs by transcriptomic analysis of tumors with activating mutations in NFE2L2 and validated LINC00942 as a new Nrf2 direct target involved in modulating GCLC expression through an unclear mechanism (9). The profiling of Nrf2-dependent genes has been mainly performed by microarray analysis, which usually does not allow the detection of lncRNAs. With the transition to deep sequencing approaches for measuring gene expression, the list of Nrf2-regulated lncRNAs will continue to grow.
In addition, distal Nrf2 binding sites could be located at enhancers. Enhancers are genomic domains that regulate transcription by functioning as binding platforms for transcription factors and are characterized by specific chromatin signatures of histone methylation and acetylation (135). The generation of chromatin state maps by profiling combinations of epigenetic marks, in addition to Nrf2 in a given cell type will allow the identification of noncoding regulatory elements bound by Nrf2 and provide unique insights into Nrf2-mediated transcriptional regulation. The investigation of the relationship between Nrf2 binding and the dynamics of the local chromatin environment will also inform on Nrf2 activity at those sites.
The systematic characterization of NRF2-bound regulatory elements becomes particularly relevant in light of evidence for positive selection of SNPs at specific NRF2 binding sites that could influence gene expression and, ultimately, disease risk (105, 178).
Crosstalk between Nrf2 and other signaling pathways
Analyses of transcriptional responses have revealed complex interactions between the Nrf2 regulatory network and other signaling cascades. As previously discussed, Nrf2 activity can be modulated in multiple ways by several signaling pathways, thereby affecting the expression of Nrf2 target genes. Conversely, Nrf2 can both positively and negatively influence the downstream pathways of other transcription factors and these interconnections can occur in various forms. For example, Nrf2 can regulate the expression or the stability of other transcriptional regulators. Gene expression microarray analyses comparing Nfe2l2-null and wild-type MEFs showed that expression of Notch1 and AhR, as well as their target genes, is decreased in Nfe2l2-depleted cells (148, 172). In addition, a study on Nfe2l2-null MEFs revealed enhanced IκB kinase β activity, which phosphorylates IκB, the negative regulator of NF-κB, thus inducing IκB degradation and NF-κB activation (163).
Moreover, several Nrf2 inducing agents can concomitantly trigger the activation of other transcriptional regulators, thus creating complex interconnections between their signaling pathways (3, 51, 70, 154). For example, Nrf2 and AhR have been shown to collaborate to mediate the response to 2,3,7,8-tetrachlorodibenzo-p-dioxin, 3-methylchoranthrene, butylated hydroxyanisole, and phenobarbital (98, 121, 189). In addition, disruption of Nrf2 delayed liver regeneration after partial hepatectomy and this phenotype was rescued by expression of the Notch1 intracellular domain, suggesting a functional crosstalk between the Nrf2 and Notch1 pathways (172). It is noteworthy that AhR, Notch, and NF-κB can regulate Nrf2 expression, indicating bidirectional interactions between these pathways (138, 148, 173).
Finally, Nrf2 activity can be modulated through cross-binding with other transcription factors. In cancer cells, mutant p53 has been shown to piggyback on NRF2 to regulate the expression of proteasome genes, leading to resistance to the proteasome inhibitor carfilzomib (174).
A more comprehensive elucidation of the crosstalk between Nrf2 and other signaling pathways will help to decipher the complexity of Nrf2-driven cellular processes. In an attempt to dissect the Nrf2 interactome and regulome, Korcsmáros and colleagues developed NRF2-ome, an integrated web resource containing information on Nrf2 interacting factors, target genes, regulating transcription factors, and miRNAs (129, 168). It will be important to extend the current computational and experimental approaches to obtain a more dynamic global view of Nrf2-mediated gene regulation that integrates all the factors that influence the final transcriptional output.
Conclusions and Perspectives
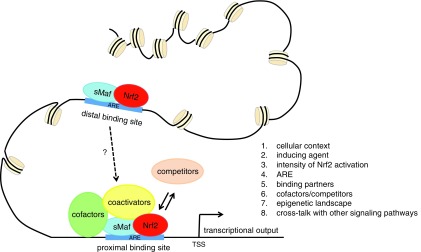
Since the discovery of Nrf2 in 1994 (108), our understanding of its biology has continued to grow. Nrf2 has been implicated in different cellular processes, such as the response to oxidative and xenobiotic stress, mitochondrial respiration, stem cell quiescence, mRNA translation, autophagy and UPR. Significant advances have been made in understanding the regulation of Nrf2 activity, downstream pathways and implications for the development of disease. It is now clear that Nrf2 lies at the center of a complex regulatory network. Nrf2-mediated transcriptional regulation is determined by the cellular context, the activating stimulus, the recognition of the ARE, the availability of binding partners, the competition or cooperation with other activators and repressors, the crosstalk with other signaling pathways and the epigenetic landscape of the target gene promoters, among others (Fig. 6). A complete appreciation of the Nrf2 program will require an integrated consideration of all these factors, which will allow these efforts to have the most profound benefits to human health.
FIG. 6.
Dissecting the Nrf2 network. Nrf2 lies at the center of a complex regulatory network. Nrf2-mediated regulation of gene expression depends on the cellular context, the inducing agent, the levels of Nrf2 activation, the recognition of the ARE, binding partners, cofactors and competitors, the crosstalk with other signaling pathways, and the epigenetic landscape of the target gene promoter, among others.
Abbreviations Used
- β-TrCP
β-transducing repeat-containing protein
- ABSC
airway basal stem cell
- AhR
aryl hydrocarbon receptor
- AML
acute myeloid leukemia
- AP1
activator protein 1
- ARE
antioxidant response element
- ATF4
activating transcription factor 4
- Bach1
BTB domain and CNC homolog 1
- Bach2
BTB domain and CNC homolog 2
- bZIP
basic-region leucine zipper
- BRCA1
breast cancer susceptibility 1
- CBP
CREB binding protein
- Cdk
cyclin-dependent kinase
- ChIP-Seq
chromatin immunoprecipitation followed by sequencing
- CNC
cap ‘n’ collar
- CncC
cap ‘n’ collar isoform C
- CTD
carboxy-terminal domain
- ER
endoplasmic reticulum
- Gcl
glutamate-cysteine ligase
- Gclc
glutamate-cysteine ligase catalytic subunit
- Gclm
glutamate-cysteine ligase modifier subunit
- Gpx2
glutathione peroxidase 2
- GSH
glutathione
- Gsk-3β
glycogen synthase kinase-3β
- GSSG
oxidized glutathione
- Gst
glutathione S-transferase
- HbSS
homozygous sickle cell disease
- HGPS
Hutchinson-Gilford progeria syndrome
- Hmox1
heme oxygenase 1
- Hrd1
synoviolin
- Idh1
isocitrate dehydrogenase 1
- ISC
intestinal stem cell
- Keap1
Kelch-like ECH-associated protein 1
- lncRNAs
long non-coding RNAs
- MEF
mouse embryonic fibroblast
- miRNA
microRNA
- NADPH
nicotinamide adenine dinucleotide phosphate
- Neh
Nrf2-ECH homology
- NF-E2
nuclear factor, erythroid-derived 2
- NF-κB
nuclear factor-κB
- Nqo1
NADPH quinone dehydrogenase 1
- Nrf2
nuclear factor E2-related factor 2
- Pgd
6-phosphogluconate dehydrogenase
- PI3K
phosphoinositide 3-kinase
- PIC
pre-initiation complex
- PKC
protein kinase C
- Pol II
RNA polymerase II
- PPP
pentose phosphate pathway
- ROS
reactive oxygen species
- RXRα
retinoid X receptor α
- SCF
Skp1-Cul1-F-box
- SFN
sulforaphane
- sMaf
small masculoaponeurotic fibrosarcoma
- SNP
single nucleotide polymorphism
- TXN
thioredoxin
- Txnrd1
thioredoxin reductase 1
- UPR
unfolded protein response
Acknowledgments
D.A.T. is a distinguished scholar of the Lustgarten Foundation (LF) and director of the LF-designated Laboratory of Pancreatic Cancer Research and of Cold Spring Harbor Laboratory (CSHL) Cancer Center. D.A.T. is supported by the CSHL Association; the NIH (5P30CA45508-26, 5P50CA101955-07, 1U10CA180944-03, 5U01CA168409-5, 1R01CA190092-03; 1R01CA188134-01A1); the V Foundation; the Thompson Family Foundation; Stand Up to Cancer/KWF; the STARR Foundation (I7-A718), and the DOD (W81XWH-13-PRCRP-IA); and the Precision Medicine Research Associates. We are also grateful for support from the following: American Italian Cancer Foundation (Muzinich Family fellow to C.T.), the Damon Runyon Cancer Research Foundation (Shirley Stein fellow DRG-2165-13 to I.I.C.C.), and the Human Frontier Science Program (LT000190/2013 to I.I.C.C.).
References
- 1.Abbas K, Breton J, Planson AG, Bouton C, Bignon J, Seguin C, Riquier S, Toledano MB, and Drapier JC. Nitric oxide activates an Nrf2/sulfiredoxin antioxidant pathway in macrophages. Free Radic Biol Med 51: 107–114, 2011 [DOI] [PubMed] [Google Scholar]
- 2.Agyeman AS, Chaerkady R, Shaw PG, Davidson NE, Visvanathan K, Pandey A, and Kensler TW. Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Res Treat 132: 175–187, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad R, Raina D, Meyer C, Kharbanda S, and Kufe D. Triterpenoid CDDO-Me blocks the NF-kappaB pathway by direct inhibition of IKKbeta on Cys-179. J Biol Chem 281: 35764–35769, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Alam J, Killeen E, Gong P, Naquin R, Hu B, Stewart D, Ingelfinger JR, and Nath KA. Heme activates the heme oxygenase-1 gene in renal epithelial cells by stabilizing Nrf2. Am J Physiol Renal Physiol 284: F743–F752, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, and Cook JL. Nrf2, a Cap ‘n’ Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem 274: 26071–26078, 1999 [DOI] [PubMed] [Google Scholar]
- 6.Allen BL. and Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 16: 155–166, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, and Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362: 722–728, 1993 [DOI] [PubMed] [Google Scholar]
- 8.Ansell PJ, Lo SC, Newton LG, Espinosa-Nicholas C, Zhang DD, Liu JH, Hannink M, and Lubahn DB. Repression of cancer protective genes by 17beta-estradiol: ligand-dependent interaction between human Nrf2 and estrogen receptor alpha. Mol Cell Endocrinol 243: 27–34, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Ashouri A, Sayin VI, Van den Eynden J, Singh SX, Papagiannakopoulos T, and Larsson E. Pan-cancer transcriptomic analysis associates long non-coding RNAs with key mutational driver events. Nat Commun 7, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baird L, Swift S, Lleres D, and Dinkova-Kostova AT. Monitoring Keap1-Nrf2 interactions in single live cells. Biotechnol Adv 32: 1133–1144, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bannai S. and Kitamura E. Transport interaction of L-cystine and L-glutamate in human diploid fibroblasts in culture. J Biol Chem 255: 2372–2376, 1980 [PubMed] [Google Scholar]
- 12.Bannister AJ. and Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature 384: 641–643, 1996 [DOI] [PubMed] [Google Scholar]
- 13.Barve A, Khor TO, Nair S, Lin W, Yu S, Jain MR, Chan JY, and Kong AN. Pharmacogenomic profile of soy isoflavone concentrate in the prostate of Nrf2 deficient and wild-type mice. J Pharm Sci 97: 4528–4545, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, and Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med 88: 290–301, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown SL, Sekhar KR, Rachakonda G, Sasi S, and Freeman ML. Activating transcription factor 3 is a novel repressor of the nuclear factor erythroid-derived 2-related factor 2 (Nrf2)-regulated stress pathway. Cancer Res 68: 364–368, 2008 [DOI] [PubMed] [Google Scholar]
- 16.Camp ND, James RG, Dawson DW, Yan F, Davison JM, Houck SA, Tang X, Zheng N, Major MB, and Moon RT. Wilms tumor gene on X chromosome (WTX) inhibits degradation of NRF2 protein through competitive binding to KEAP1 protein. J Biol Chem 287: 6539–6550, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, and Evans RM. Role of CBP/P300 in nuclear receptor signalling. Nature 383: 99–103, 1996 [DOI] [PubMed] [Google Scholar]
- 18.Chan JY, Han XL, and Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci U S A 90: 11371–11375, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chanas SA, Jiang Q, McMahon M, McWalter GK, McLellan LI, Elcombe CR, Henderson CJ, Wolf CR, Moffat GJ, Itoh K, Yamamoto M, and Hayes JD. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem J 365: 405–416, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, and Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell 34: 663–673, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chio II, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Ohlund D, Wright K, Filippini D, Lee EJ, Da Silva B, Schoepfer C, Wilkinson JE, Buscaglia JM, DeNicola GM, Tiriac H, Hammell M, Crawford HC, Schmidt EE, Thompson CB, Pappin DJ, Sonenberg N, and Tuveson DA. NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell 166: 963–976, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho Y. and Bannai S. Uptake of glutamate and cysteine in C-6 glioma cells and in cultured astrocytes. J Neurochem 55: 2091–2097, 1990 [DOI] [PubMed] [Google Scholar]
- 23.Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, and Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res 40: 7416–7429, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, and Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32: 3765–3781, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, and Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855–859, 1993 [DOI] [PubMed] [Google Scholar]
- 26.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, and Park BK. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem 285: 16782–16788, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cordova EJ, Martinez-Hernandez A, Uribe-Figueroa L, Centeno F, Morales-Marin M, Koneru H, Coleman MA, and Orozco L. The NRF2-KEAP1 pathway is an early responsive gene network in arsenic exposed lymphoblastoid cells. PLoS One 9: e88069, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cullinan SB, Gordan JD, Jin J, Harper JW, and Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24: 8477–8486, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, and Cantley LC. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47: 1475–1481, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, and Tuveson DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475: 106–109, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devling TW, Lindsay CD, McLellan LI, McMahon M, and Hayes JD. Utility of siRNA against Keap1 as a strategy to stimulate a cancer chemopreventive phenotype. Proc Natl Acad Sci U S A 102: 7280–7285A, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, and Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A 99: 11908–11913, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eades G, Yang M, Yao Y, Zhang Y, and Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem 286: 40725–40733, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esposito F, Cuccovillo F, Russo L, Casella F, Russo T, and Cimino F. A new p21waf1/cip1 isoform is an early event of cell response to oxidative stress. Cell Death Differ 5: 940–945, 1998 [DOI] [PubMed] [Google Scholar]
- 35.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, and Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510: 298–302, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friling RS, Bergelson S, and Daniel V. Two adjacent AP-1-like binding sites form the electrophile-responsive element of the murine glutathione S-transferase Ya subunit gene. Proc Natl Acad Sci U S A 89: 668–672, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furukawa M. and Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol 25: 162–171, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuse Y. and Kobayashi M. Conservation of the Keap1-Nrf2 system: an evolutionary journey through stressful space and time. Molecules 22, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, and Collins FS. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A 101: 8963–8968, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein LD, Lee J, Gnad F, Klijn C, Schaub A, Reeder J, Daemen A, Bakalarski CE, Holcomb T, Shames DS, Hartmaier RJ, Chmielecki J, Seshagiri S, Gentleman R, and Stokoe D. Recurrent loss of NFE2L2 Exon 2 Is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep 16: 2605–2617, 2016 [DOI] [PubMed] [Google Scholar]
- 41.Gorrini C, Baniasadi PS, Harris IS, Silvester J, Inoue S, Snow B, Joshi PA, Wakeham A, Molyneux SD, Martin B, Bouwman P, Cescon DW, Elia AJ, Winterton-Perks Z, Cruickshank J, Brenner D, Tseng A, Musgrave M, Berman HK, Khokha R, Jonkers J, Mak TW, and Gauthier ML. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med 210: 1529–1544, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorrini C, Harris IS, and Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12: 931–947, 2013 [DOI] [PubMed] [Google Scholar]
- 43.Gozzelino R, Jeney V, and Soares MP. Mechanisms of cell protection by heme oxygenase-1. Annu Rev Pharmacol Toxicol 50: 323–354, 2010 [DOI] [PubMed] [Google Scholar]
- 44.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, and Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, Ishiguro A, Kijima H, Mimura J, Itoh K, Fukuda S, and Saijo Y. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 12: 66, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harvey CJ, Thimmulappa RK, Singh A, Blake DJ, Ling G, Wakabayashi N, Fujii J, Myers A, and Biswal S. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic Biol Med 46: 443–453, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hast BE, Goldfarb D, Mulvaney KM, Hast MA, Siesser PF, Yan F, Hayes DN, and Major MB. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res 73: 2199–2210, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawkes HJ, Karlenius TC, and Tonissen KF. Regulation of the human thioredoxin gene promoter and its key substrates: a study of functional and putative regulatory elements. Biochim Biophys Acta 1840: 303–314, 2014 [DOI] [PubMed] [Google Scholar]
- 49.Hayes JD. and Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39: 199–218, 2014 [DOI] [PubMed] [Google Scholar]
- 50.He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AM, and Alam J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem 276: 20858–20865, 2001 [DOI] [PubMed] [Google Scholar]
- 51.Heiss E, Herhaus C, Klimo K, Bartsch H, and Gerhauser C. Nuclear factor kappa B is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J Biol Chem 276: 32008–32015, 2001 [DOI] [PubMed] [Google Scholar]
- 52.Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, Engel JD, and Yamamoto M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 40: 10228–10239, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hochmuth CE, Biteau B, Bohmann D, and Jasper H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell 8: 188–199, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holmstrom KM, Baird L, Zhang Y, Hargreaves I, Chalasani A, Land JM, Stanyer L, Yamamoto M, Dinkova-Kostova AT, and Abramov AY. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol Open 2: 761–770, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang HC, Nguyen T, and Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem 277: 42769–42774, 2002 [DOI] [PubMed] [Google Scholar]
- 56.Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, and Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367: 568–572, 1994 [DOI] [PubMed] [Google Scholar]
- 57.Ikeda Y, Sugawara A, Taniyama Y, Uruno A, Igarashi K, Arima S, Ito S, and Takeuchi K. Suppression of rat thromboxane synthase gene transcription by peroxisome proliferator-activated receptor gamma in macrophages via an interaction with NRF2. J Biol Chem 275: 33142–33150, 2000 [DOI] [PubMed] [Google Scholar]
- 58.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, and Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236: 313–322, 1997 [DOI] [PubMed] [Google Scholar]
- 59.Itoh K, Igarashi K, Hayashi N, Nishizawa M, and Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol 15: 4184–4193, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, and Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13: 76–86, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, and Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8: 379–391, 2003 [DOI] [PubMed] [Google Scholar]
- 62.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, and Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 285: 22576–22591, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jang J, Wang Y, Lalli MA, Guzman E, Godshalk SE, Zhou H, and Kosik KS. Primary cilium-autophagy-Nrf2 (PAN) axis activation commits human embryonic stem cells to a neuroectoderm fate. Cell 165: 410–420, 2016 [DOI] [PubMed] [Google Scholar]
- 64.Johnson SF, Cruz C, Greifenberg AK, Dust S, Stover DG, Chi D, Primack B, Cao S, Bernhardy AJ, Coulson R, Lazaro JB, Kochupurakkal B, Sun H, Unitt C, Moreau LA, Sarosiek KA, Scaltriti M, Juric D, Baselga J, Richardson AL, Rodig SJ, D'Andrea AD, Balmana J, Johnson N, Geyer M, Serra V, Lim E, and Shapiro GI. CDK12 inhibition reverses de novo and acquired PARP inhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Rep 17: 2367–2381, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Joo MS, Lee CG, Koo JH, and Kim SG. miR-125b transcriptionally increased by Nrf2 inhibits AhR repressor, which protects kidney from cisplatin-induced injury. Cell Death Dis 4: e899, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaidanovich-Beilin O. and Woodgett JR. GSK-3: Functional insights from cell biology and animal models. Front Mol Neurosci 4: 40, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang HJ, Hong YB, Kim HJ, Rodriguez OC, Nath RG, Tilli EM, Albanese C, Chung FL, Kwon SH, and Bae I. Detoxification: a novel function of BRCA1 in tumor suppression? Toxicol Sci 122: 26–37, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kannan MB, Solovieva V, and Blank V. The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochim Biophys Acta 1823: 1841–1846, 2012 [DOI] [PubMed] [Google Scholar]
- 69.Kapeta S, Chondrogianni N, and Gonos ES. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J Biol Chem 285: 8171–8184, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karuri AR, Huang Y, Bodreddigari S, Sutter CH, Roebuck BD, Kensler TW, and Sutter TR. 3H-1,2-dithiole-3-thione targets nuclear factor kappaB to block expression of inducible nitric-oxide synthase, prevents hypotension, and improves survival in endotoxemic rats. J Pharmacol Exp Ther 317: 61–67, 2006 [DOI] [PubMed] [Google Scholar]
- 71.Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, and Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6: 857–868, 2001 [DOI] [PubMed] [Google Scholar]
- 72.Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, and Yamamoto M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol Cell Biol 25: 8044–8051, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katsuoka F, Motohashi H, Tamagawa Y, Kure S, Igarashi K, Engel JD, and Yamamoto M. Small Maf compound mutants display central nervous system neuronal degeneration, aberrant transcription, and Bach protein mislocalization coincident with myoclonus and abnormal startle response. Mol Cell Biol 23: 1163–1174, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katsuoka F. and Yamamoto M. Small Maf proteins (MafF, MafG, MafK): history, structure and function. Gene 586: 197–205, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim JH, Yu S, Chen JD, and Kong AN. The nuclear cofactor RAC3/AIB1/SRC-3 enhances Nrf2 signaling by interacting with transactivation domains. Oncogene 32: 514–527, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim PK, Hailey DW, Mullen RT, and Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A 105: 20567–20574, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, Eom HS, Yoo NJ, and Lee SH. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol 220: 446–451, 2010 [DOI] [PubMed] [Google Scholar]
- 78.Kimura M, Yamamoto T, Zhang J, Itoh K, Kyo M, Kamiya T, Aburatani H, Katsuoka F, Kurokawa H, Tanaka T, Motohashi H, and Yamamoto M. Molecular basis distinguishing the DNA binding profile of Nrf2-Maf heterodimer from that of Maf homodimer. J Biol Chem 282: 33681–33690, 2007 [DOI] [PubMed] [Google Scholar]
- 79.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, and Yamamoto M. Molecular cloning and functional characterization of a new Cap ‘n’ collar family transcription factor Nrf3. J Biol Chem 274: 6443–6452, 1999 [DOI] [PubMed] [Google Scholar]
- 80.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, and Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24: 7130–7139, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kobayashi A, Tsukide T, Miyasaka T, Morita T, Mizoroki T, Saito Y, Ihara Y, Takashima A, Noguchi N, Fukamizu A, Hirotsu Y, Ohtsuji M, Katsuoka F, and Yamamoto M. Central nervous system-specific deletion of transcription factor Nrf1 causes progressive motor neuronal dysfunction. Genes Cells 16: 692–703, 2011 [DOI] [PubMed] [Google Scholar]
- 82.Kobayashi EH, Suzuki T, Funayama R, Nagashima T, Hayashi M, Sekine H, Tanaka N, Moriguchi T, Motohashi H, Nakayama K, and Yamamoto M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun 7: 11624, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, and Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223, 2010 [DOI] [PubMed] [Google Scholar]
- 84.Konstantinopoulos PA, Spentzos D, Fountzilas E, Francoeur N, Sanisetty S, Grammatikos AP, Hecht JL, and Cannistra SA. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res 71: 5081–5089, 2011 [DOI] [PubMed] [Google Scholar]
- 85.Kubben N, Zhang W, Wang L, Voss TC, Yang J, Qu J, Liu GH, and Misteli T. Repression of the antioxidant NRF2 pathway in premature aging. Cell 165: 1361–1374, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kwak MK, Itoh K, Yamamoto M, and Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol 22: 2883–2892, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, and Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol 23: 8786–8794, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, and Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem 278: 8135–8145, 2003 [DOI] [PubMed] [Google Scholar]
- 89.Lacher SE, Lee JS, Wang X, Campbell MR, Bell DA, and Slattery M. Beyond antioxidant genes in the ancient Nrf2 regulatory network. Free Radic Biol Med 88: 452–465, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, and Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 30: 3275–3285, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee JM, Calkins MJ, Chan K, Kan YW, and Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem 278: 12029–12038, 2003 [DOI] [PubMed] [Google Scholar]
- 92.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, and Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J 378: 373–382, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li X, Chatterjee N, Spirohn K, Boutros M, and Bohmann D. Cdk12 is a gene-selective RNA polymerase II kinase that regulates a subset of the transcriptome, including Nrf2 target genes. Sci Rep 6: 21455, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liang K, Gao X, Gilmore JM, Florens L, Washburn MP, Smith E, and Shilatifard A. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol Cell Biol 35: 928–938, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lo SC. and Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res 314: 1789–1803, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ludtmann MH, Angelova PR, Zhang Y, Abramov AY, and Dinkova-Kostova AT. Nrf2 affects the efficiency of mitochondrial fatty acid oxidation. Biochem J 457: 415–424, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma J, Cai H, Wu T, Sobhian B, Huo Y, Alcivar A, Mehta M, Cheung KL, Ganesan S, Kong AN, Zhang DD, and Xia B. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol Cell Biol 32: 1506–1517, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma Q, Kinneer K, Bi Y, Chan JY, and Kan YW. Induction of murine NAD(P)H:quinone oxidoreductase by 2,3,7,8-tetrachlorodibenzo-p-dioxin requires the CNC (cap ‘n’ collar) basic leucine zipper transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2): cross-interaction between AhR (aryl hydrocarbon receptor) and Nrf2 signal transduction. Biochem J 377: 205–213, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, Dalton TP, Yamamoto M, and Klaassen CD. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 46: 1597–1610, 2007 [DOI] [PubMed] [Google Scholar]
- 100.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, and Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res 38: 5718–5734, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, and Kleeberger SR. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J 21: 2237–2246, 2007 [DOI] [PubMed] [Google Scholar]
- 102.McMahon M, Itoh K, Yamamoto M, and Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem 278: 21592–21600, 2003 [DOI] [PubMed] [Google Scholar]
- 103.McMahon M, Thomas N, Itoh K, Yamamoto M, and Hayes JD. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem 279: 31556–31567, 2004 [DOI] [PubMed] [Google Scholar]
- 104.Meakin PJ, Chowdhry S, Sharma RS, Ashford FB, Walsh SV, McCrimmon RJ, Dinkova-Kostova AT, Dillon JF, Hayes JD, and Ashford ML. Susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis upon consumption of a high-fat diet is associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes but not with insulin resistance. Mol Cell Biol 34: 3305–3320, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Melton C, Reuter JA, Spacek DV, and Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet 47: 710–716, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miao W, Hu L, Scrivens PJ, and Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem 280: 20340–20348, 2005 [DOI] [PubMed] [Google Scholar]
- 107.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, and Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22: 66–79, 2012 [DOI] [PubMed] [Google Scholar]
- 108.Moi P, Chan K, Asunis I, Cao A, and Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A 91: 9926–9930, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moinova HR. and Mulcahy RT. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem Biophys Res Commun 261: 661–668, 1999 [DOI] [PubMed] [Google Scholar]
- 110.Motohashi H, Katsuoka F, Engel JD, and Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci U S A 101: 6379–6384, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Motohashi H, Katsuoka F, Shavit JA, Engel JD, and Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell 103: 865–875, 2000 [DOI] [PubMed] [Google Scholar]
- 112.Motohashi H, O'Connor T, Katsuoka F, Engel JD, and Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294: 1–12, 2002 [DOI] [PubMed] [Google Scholar]
- 113.Muscarella LA, Barbano R, D'Angelo V, Copetti M, Coco M, Balsamo T, la Torre A, Notarangelo A, Troiano M, Parisi S, Icolaro N, Catapano D, Valori VM, Pellegrini F, Merla G, Carella M, Fazio VM, and Parrella P. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient's outcome. Epigenetics 6: 317–325, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muscarella LA, Parrella P, D'Alessandro V, la Torre A, Barbano R, Fontana A, Tancredi A, Guarnieri V, Balsamo T, Coco M, Copetti M, Pellegrini F, De Bonis P, Bisceglia M, Scaramuzzi G, Maiello E, Valori VM, Merla G, Vendemiale G, and Fazio VM. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics 6: 710–719, 2011 [DOI] [PubMed] [Google Scholar]
- 115.Nagai T, Igarashi K, Akasaka J, Furuyama K, Fujita H, Hayashi N, Yamamoto M, and Sassa S. Regulation of NF-E2 activity in erythroleukemia cell differentiation. J Biol Chem 273: 5358–5365, 1998 [DOI] [PubMed] [Google Scholar]
- 116.Narasimhan M, Patel D, Vedpathak D, Rathinam M, Henderson G, and Mahimainathan L. Identification of novel microRNAs in post-transcriptional control of Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells. PLoS One 7: e51111, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nguyen T, Nioi P, and Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284: 13291–13295, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nguyen T, Sherratt PJ, Huang HC, Yang CS, and Pickett CB. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J Biol Chem 278: 4536–4541, 2003 [DOI] [PubMed] [Google Scholar]
- 119.Nioi P. and Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun 362: 816–821, 2007 [DOI] [PubMed] [Google Scholar]
- 120.Nioi P, Nguyen T, Sherratt PJ, and Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol 25: 10895–10906, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Noda S, Harada N, Hida A, Fujii-Kuriyama Y, Motohashi H, and Yamamoto M. Gene expression of detoxifying enzymes in AhR and Nrf2 compound null mutant mouse. Biochem Biophys Res Commun 303: 105–111, 2003 [DOI] [PubMed] [Google Scholar]
- 122.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, and Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87: 953–959, 1996 [DOI] [PubMed] [Google Scholar]
- 123.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, and Hirohashi S. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res 68: 1303–1309, 2008 [DOI] [PubMed] [Google Scholar]
- 124.Onodera K, Shavit JA, Motohashi H, Katsuoka F, Akasaka JE, Engel JD, and Yamamoto M. Characterization of the murine mafF gene. J Biol Chem 274: 21162–21169, 1999 [DOI] [PubMed] [Google Scholar]
- 125.Onodera K, Shavit JA, Motohashi H, Yamamoto M, and Engel JD. Perinatal synthetic lethality and hematopoietic defects in compound mafG::mafK mutant mice. EMBO J 19: 1335–1345, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Orino K, Lehman L, Tsuji Y, Ayaki H, Torti SV, and Torti FM. Ferritin and the response to oxidative stress. Biochem J 357: 241–247, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, and Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol 16: 6083–6095, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pan H, Guan D, Liu X, Li J, Wang L, Wu J, Zhou J, Zhang W, Ren R, Zhang W, Li Y, Yang J, Hao Y, Yuan T, Yuan G, Wang H, Ju Z, Mao Z, Li J, Qu J, Tang F, and Liu GH. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res 26: 190–205, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Papp D, Lenti K, Modos D, Fazekas D, Dul Z, Turei D, Foldvari-Nagy L, Nussinov R, Csermely P, and Korcsmaros T. The NRF2-related interactome and regulome contain multifunctional proteins and fine-tuned autoregulatory loops. FEBS Lett 586: 1795–1802, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Paul MK, Bisht B, Darmawan DO, Chiou R, Ha VL, Wallace WD, Chon AT, Hegab AE, Grogan T, Elashoff DA, Alva-Ornelas JA, and Gomperts BN. Dynamic changes in intracellular ROS levels regulate airway basal stem cell homeostasis through Nrf2-dependent Notch signaling. Cell Stem Cell 15: 199–214, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, and Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31: 1121–1133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rada P, Rojo AI, Evrard-Todeschi N, Innamorato NG, Cotte A, Jaworski T, Tobon-Velasco JC, Devijver H, Garcia-Mayoral MF, Van Leuven F, Hayes JD, Bertho G, and Cuadrado A. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol Cell Biol 32: 3486–3499, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rahal A, Kumar A, Singh V, Yadav B, Tiwari R, Chakraborty S, and Dhama K. Oxidative stress, prooxidants, and antioxidants: the interplay. Biomed Res Int 2014: 761264, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rey G, Valekunja UK, Feeney KA, Wulund L, Milev NB, Stangherlin A, Ansel-Bollepalli L, Velagapudi V, O'Neill JS, and Reddy AB. The pentose phosphate pathway regulates the circadian clock. Cell Metab 24: 462–473, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rivera CM. and Ren B. Mapping human epigenomes. Cell 155: 39–55, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rushmore TH, Morton MR, and Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem 266: 11632–11639, 1991 [PubMed] [Google Scholar]
- 137.Rushworth SA, MacEwan DJ, and O'Connell MA. Lipopolysaccharide-induced expression of NAD(P)H:quinone oxidoreductase 1 and heme oxygenase-1 protects against excessive inflammatory responses in human monocytes. J Immunol 181: 6730–6737, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rushworth SA, Zaitseva L, Murray MY, Shah NM, Bowles KM, and MacEwan DJ. The high Nrf2 expression in human acute myeloid leukemia is driven by NF-kappaB and underlies its chemo-resistance. Blood 120: 5188–5198, 2012 [DOI] [PubMed] [Google Scholar]
- 139.Sakurai A, Nishimoto M, Himeno S, Imura N, Tsujimoto M, Kunimoto M, and Hara S. Transcriptional regulation of thioredoxin reductase 1 expression by cadmium in vascular endothelial cells: role of NF-E2-related factor-2. J Cell Physiol 203: 529–537, 2005 [DOI] [PubMed] [Google Scholar]
- 140.Sangokoya C, Telen MJ, and Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood 116: 4338–4348, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sasaki H, Sato H, Kuriyama-Matsumura K, Sato K, Maebara K, Wang H, Tamba M, Itoh K, Yamamoto M, and Bannai S. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J Biol Chem 277: 44765–44771, 2002 [DOI] [PubMed] [Google Scholar]
- 142.Satoh H, Moriguchi T, Takai J, Ebina M, and Yamamoto M. Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res 73: 4158–4168, 2013 [DOI] [PubMed] [Google Scholar]
- 143.Sekine H, Okazaki K, Ota N, Shima H, Katoh Y, Suzuki N, Igarashi K, Ito M, Motohashi H, and Yamamoto M. The mediator subunit MED16 transduces NRF2-activating signals into antioxidant gene expression. Mol Cell Biol 36: 407–420, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shah NM, Rushworth SA, Murray MY, Bowles KM, and MacEwan DJ. Understanding the role of NRF2-regulated miRNAs in human malignancies. Oncotarget 4: 1130–1142, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shavit JA, Motohashi H, Onodera K, Akasaka J, Yamamoto M, and Engel JD. Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev 12: 2164–2174, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, and Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135: 1358–1368, 1368 e 1–e4, 2008 [DOI] [PubMed] [Google Scholar]
- 147.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, and Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A 105: 13568–13573, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, Agoston ES, Yamamoto M, and Kensler TW. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol 27: 7188–7197, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shivdasani RA, Rosenblatt MF, Zucker-Franklin D, Jackson CW, Hunt P, Saris CJ, and Orkin SH. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell 81: 695–704, 1995 [DOI] [PubMed] [Google Scholar]
- 150.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, and Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 3: e420, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Singh B, Ronghe AM, Chatterjee A, Bhat NK, and Bhat HK. MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis. Carcinogenesis 34: 1165–1172, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, and Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274, 2006 [DOI] [PubMed] [Google Scholar]
- 153.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, Corvalan AH, Biswal S, Swisher SG, Bekele BN, Minna JD, Stewart DJ, and Wistuba II. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 16: 3743–3753, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sriram N, Kalayarasan S, and Sudhandiran G. Epigallocatechin-3-gallate augments antioxidant activities and inhibits inflammation during bleomycin-induced experimental pulmonary fibrosis through Nrf2-Keap1 signaling. Pulm Pharmacol Ther 22: 221–236, 2009 [DOI] [PubMed] [Google Scholar]
- 155.Steinberg SF. Mechanisms for redox-regulation of protein kinase C. Front Pharmacol 6: 128, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stewart D, Killeen E, Naquin R, Alam S, and Alam J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J Biol Chem 278: 2396–2402, 2003 [DOI] [PubMed] [Google Scholar]
- 157.Sun Z, Chin YE, and Zhang DD. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol Cell Biol 29: 2658–2672, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Suzuki H, Chiba T, Suzuki T, Fujita T, Ikenoue T, Omata M, Furuichi K, Shikama H, and Tanaka K. Homodimer of two F-box proteins betaTrCP1 or betaTrCP2 binds to IkappaBalpha for signal-dependent ubiquitination. J Biol Chem 275: 2877–2884, 2000 [DOI] [PubMed] [Google Scholar]
- 159.Suzuki T, Shibata T, Takaya K, Shiraishi K, Kohno T, Kunitoh H, Tsuta K, Furuta K, Goto K, Hosoda F, Sakamoto H, Motohashi H, and Yamamoto M. Regulatory nexus of synthesis and degradation deciphers cellular Nrf2 expression levels. Mol Cell Biol 33: 2402–2412, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tebay LE, Robertson H, Durant ST, Vitale SR, Penning TM, Dinkova-Kostova AT, and Hayes JD. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med 88: 108–146, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Telakowski-Hopkins CA, King RG, and Pickett CB. Glutathione S-transferase Ya subunit gene: identification of regulatory elements required for basal level and inducible expression. Proc Natl Acad Sci U S A 85: 1000–1004, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Thai P, Statt S, Chen CH, Liang E, Campbell C, and Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol 49: 204–211, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW, and Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest 116: 984–995, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, and Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res 62: 5196–5203, 2002 [PubMed] [Google Scholar]
- 165.Toki T, Itoh J, Kitazawa J, Arai K, Hatakeyama K, Akasaka J, Igarashi K, Nomura N, Yokoyama M, Yamamoto M, and Ito E. Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif. Oncogene 14: 1901–1910, 1997 [DOI] [PubMed] [Google Scholar]
- 166.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, and Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol 26: 2887–2900, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tsai JJ, Dudakov JA, Takahashi K, Shieh JH, Velardi E, Holland AM, Singer NV, West ML, Smith OM, Young LF, Shono Y, Ghosh A, Hanash AM, Tran HT, Moore MA, and van den Brink MR. Nrf2 regulates haematopoietic stem cell function. Nat Cell Biol 15: 309–316, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Turei D, Papp D, Fazekas D, Foldvari-Nagy L, Modos D, Lenti K, Csermely P, and Korcsmaros T. NRF2-ome: an integrated web resource to discover protein interaction and regulatory networks of NRF2. Oxid Med Cell Longev 2013: 737591, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ullrich NJ. and Gordon LB. Hutchinson-Gilford progeria syndrome. Handb Clin Neurol 132: 249–264, 2015 [DOI] [PubMed] [Google Scholar]
- 170.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, and Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A 101: 2040–2045, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, and Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet 35: 238–245, 2003 [DOI] [PubMed] [Google Scholar]
- 172.Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak MK, Misra V, Biswal S, Yamamoto M, and Kensler TW. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal 3: ra52, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wakabayashi N, Skoko JJ, Chartoumpekis DV, Kimura S, Slocum SL, Noda K, Palliyaguru DL, Fujimuro M, Boley PA, Tanaka Y, Shigemura N, Biswal S, Yamamoto M, and Kensler TW. Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol Cell Biol 34: 653–663, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Walerych D, Lisek K, Sommaggio R, Piazza S, Ciani Y, Dalla E, Rajkowska K, Gaweda-Walerych K, Ingallina E, Tonelli C, Morelli MJ, Amato A, Eterno V, Zambelli A, Rosato A, Amati B, Wisniewski JR, Del and Sal G. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol 18: 897–909, 2016 [DOI] [PubMed] [Google Scholar]
- 175.Wang H, Liu K, Geng M, Gao P, Wu X, Hai Y, Li Y, Li Y, Luo L, Hayes JD, Wang XJ, and Tang X. RXRalpha inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res 73: 3097–3108, 2013 [DOI] [PubMed] [Google Scholar]
- 176.Wang M. and Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer 14: 581–597, 2014 [DOI] [PubMed] [Google Scholar]
- 177.Wang R, An J, Ji F, Jiao H, Sun H, and Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun 373: 151–154, 2008 [DOI] [PubMed] [Google Scholar]
- 178.Wang X, Campbell MR, Lacher SE, Cho HY, Wan M, Crowl CL, Chorley BN, Bond GL, Kleeberger SR, Slattery M, and Bell DA. A polymorphic antioxidant response element links NRF2/sMAF binding to enhanced MAPT expression and reduced risk of Parkinsonian disorders. Cell Rep 2016. DOI: 10.1016/j.celrep.2016.03.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Weake VM. and Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet 11: 426–437, 2010 [DOI] [PubMed] [Google Scholar]
- 180.Wild AC, Moinova HR, and Mulcahy RT. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J Biol Chem 274: 33627–33636, 1999 [DOI] [PubMed] [Google Scholar]
- 181.Winter J, Jung S, Keller S, Gregory RI, and Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234, 2009 [DOI] [PubMed] [Google Scholar]
- 182.Woods CG, Fu J, Xue P, Hou Y, Pluta LJ, Yang L, Zhang Q, Thomas RS, Andersen ME, and Pi J. Dose-dependent transitions in Nrf2-mediated adaptive response and related stress responses to hypochlorous acid in mouse macrophages. Toxicol Appl Pharmacol 238: 27–36, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, and Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell 11: 1445–1456, 2003 [DOI] [PubMed] [Google Scholar]
- 184.Wu KC, Cui JY, and Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci 123: 590–600, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wu T, Zhao F, Gao B, Tan C, Yagishita N, Nakajima T, Wong PK, Chapman E, Fang D, and Zhang DD. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev 28: 708–722, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yamazaki H, Katsuoka F, Motohashi H, Engel JD, and Yamamoto M. Embryonic lethality and fetal liver apoptosis in mice lacking all three small Maf proteins. Mol Cell Biol 32: 808–816, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yang M, Yao Y, Eades G, Zhang Y, and Zhou Q. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res Treat 129: 983–991, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yates MS, Tran QT, Dolan PM, Osburn WO, Shin S, McCulloch CC, Silkworth JB, Taguchi K, Yamamoto M, Williams CR, Liby KT, Sporn MB, Sutter TR, and Kensler TW. Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis 30: 1024–1031, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yeager RL, Reisman SA, Aleksunes LM, and Klaassen CD. Introducing the “TCDD-inducible AhR-Nrf2 gene battery.” Toxicol Sci 111: 238–246, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yoo NJ, Kim HR, Kim YR, An CH, and Lee SH. Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology 60: 943–952, 2012 [DOI] [PubMed] [Google Scholar]
- 191.Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, Foster BA, Kan YW, and Kong AN. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS One 5: e8579, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhang DD, Lo SC, Cross JV, Templeton DJ, and Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24: 10941–10953, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang J, Ohta T, Maruyama A, Hosoya T, Nishikawa K, Maher JM, Shibahara S, Itoh K, and Yamamoto M. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol Cell Biol 26: 7942–7952, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, and Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther 9: 336–346, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang Y, Xia J, Li Q, Yao Y, Eades G, Gernapudi R, Duru N, Kensler TW, and Zhou Q. NRF2/long noncoding RNA ROR signaling regulates mammary stem cell expansion and protects against estrogen genotoxicity. J Biol Chem 289: 31310–31318, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]