SUMMARY
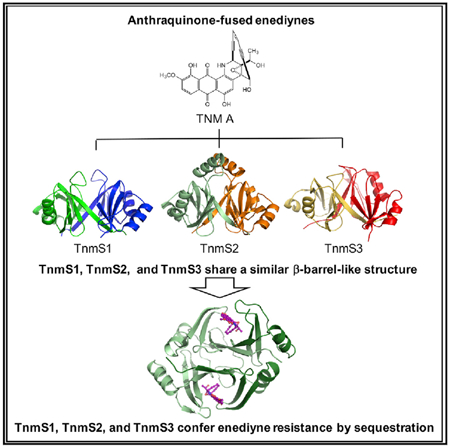
The enediynes, microbial natural products with extraordinary cytotoxicities, have been translated into clinical drugs. Two self-resistance mechanisms are known in the enediyne producers—apoproteins for the nine-membered enediynes and self-sacrifice proteins for the ten-membered enediyne calicheamicin. Here we show that: (1) tnmS1, tnmS2, and tnmS3 encode tiancimycin (TNM) resistance in its producer Streptomyces sp. CB03234, (2) tnmS1, tnmS2, and tnmS3 homologs are found in all anthraquinone-fused enediyne producers, (3) TnmS1, TnmS2, and TnmS3 share a similar β barrel-like structure, bind TNMs with nanomolar KD values, and confer resistance by sequestration, and (4) TnmS1, TnmS2, and TnmS3 homologs are widespread in nature, including in the human microbiome. These findings unveil an unprecedented resistance mechanism for the enediynes. Mechanisms of self-resistance in producers serve as models to predict and combat future drug resistance in clinical settings. Enediyne-based chemotherapies should now consider the fact that the human microbiome harbors genes encoding enediyne resistance.
Graphical Abstract
In Brief
TnmS1, TnmS2, and TnmS3 confer tiancimycin resistance by sequestration. Their homologs are found in all anthraquinone-fused enediyne producers and are widespread in nature, including in the human microbiome. These findings unveil an unprecedented resistance mechanism for the enediynes and should be considered in future efforts to develop enediyne-based chemotherapies.
INTRODUCTION
The enediynes represent one of the most fascinating families of natural products for their unprecedented molecular architecture and extraordinary cytotoxicity against a broad panel of cancer cell lines (Van Lanen and Shen, 2008). Since the determination of the enediyne chromophore structure of neocarzinostatin (NCS) in 1985 (Edo et al., 1985), 13 enediynes have been isolated and structurally characterized (Yan et al., 2016, 2017). All enediynes contain an enediyne core consisting of two acetylenic groups conjugated to a double bond or incipient double bond within a nine- or ten-membered carbocycle (Galm et al., 2005). Members of the nine-membered enediyne core subcategory include NCS, C-1027, kedarcidin, maduropeptin, and N1999A2. Members of the ten-membered enediyne core subcategory include calicheamicin (CAL), esperamicin, namenamicin, shishijimicin, and the anthraquinone-fused enediynes dynemicin (DYN), uncialamycin (UCM), tiancimycin (TNM), and yangpumicin (YPM). As a consequence of this structural feature, the enediynes share a common mode of action—electronic rearrangement of the nine- or ten-membered enediynes core to produce a transient benzenoid diradical. When positioned within the minor groove of DNA, the diradical abstracts hydrogen atoms from the deoxyribose backbone of DNA. The DNA-centered radicals can then cause interstrand crosslinks, react with molecular oxygen leading ultimately to DNA double-strand breaks, or both, endowing the enediyne natural products with an extraordinary cytotoxicity (Kennedy et al., 2007).
The enediynes have been successfully translated into clinical drugs by targeted delivery. Poly(styrene-co-maleic acid)-conjugated NCS (SMANCS) has been marketed since 1994 for the treatment of hepatocellular carcinoma (Abe and Otsuki, 2002). A CD33 monoclonal antibody-CAL conjugate, gemtuzumab ozogamicin (Mylotarg), and a CD22 monoclonal antibody-CAL conjugate, inotuzumab ozogamicin (Besponsa), have been approved since 2000 and 2017, respectively, for the treatment of acute myeloid leukemia (Pfizer voluntarily withdrew Mylotarg from the market in 2010, and the US Food and Drug Administration reapproved Mylotarg in 2017). In addition, C-1027, also known as lidamycin (Chen et al., 2015), and UCM as payloads of antibody-drug conjugates for cancers have been actively pursued in several preclinical studies (Li et al., 2016; Shen et al., 2015).
Self-resistance mechanisms are essential for antibiotic-producing microbes to protect themselves from the lethal effects of their own cytotoxic molecules. The three major self-resistance mechanisms in antibiotic-producing bacteria are: (1) modification or degradation of the antibiotics to render them biologically inactive, (2) modification of the antibiotic targets to render them insensitive, and (3) effluxion of the antibiotics out of the cell to keep them under the lethal level (Blair et al., 2015). In addition to these common resistance mechanisms, antibiotic sequestration, target overproduction, and metabolic pathway modification are also known (Blair et al., 2015). Self-resistance determinants in the producers represent a major reservoir for the development of drug resistance in the clinical setting by horizontal gene transfer. Although direct horizontal gene transfer has only been shown in a few cases, the evidence is now clear that the environment, including antibiotic producers, is the single largest reservoir of resistance (Surette and Wright, 2017). Mechanisms of self-resistance in antibiotic producers, therefore, serve as models to predict and combat future drug resistance in clinical settings.
Two mechanisms of self-resistance are known for the enediyne producers. Members of the nine-membered enediyne core subcategory are produced as chromoproteins, employing apoproteins to stabilize the otherwise unstable enediyne chromophores and facilitate their transport (Liu and Shen, 2000). For the ten-membered enediyne core subcategory, only CAL has a reported mechanism—the self-sacrifice proteins, encoded by calC, calU16, and calU19 in the cal biosynthetic gene cluster, which trigger aromatization of the enediyne core via radical-dependent protein cleavage (Biggins et al., 2003; Elshahawi et al., 2014). The anthraquinone-fused enediynes are a subclass of ten-membered enediyne core subcategory that includes DYN (Konishi et al., 1989), UCM (Davies et al., 2005), TNM (Yan et al., 2016), and YPM (Yan et al., 2017) (Figure 1A). The gene cluster for DYN biosynthesis has been cloned previously (Gao and Thor-son, 2008). We recently cloned and characterized the gene clusters for TNM (Yan et al., 2016), UCM (Yan et al., 2016), and YPM (Yan et al., 2017) biosynthesis. We subsequently constructed an enediyne genome neighborhood network (GNN), which allowed prediction of new structural features and biosynthetic insights that could be exploited for enediyne discovery (Rudolf et al., 2016; Yan et al., 2016, 2017). However, the mechanism of self-resistance for the anthraquinone-fused enediynes remains unknown.
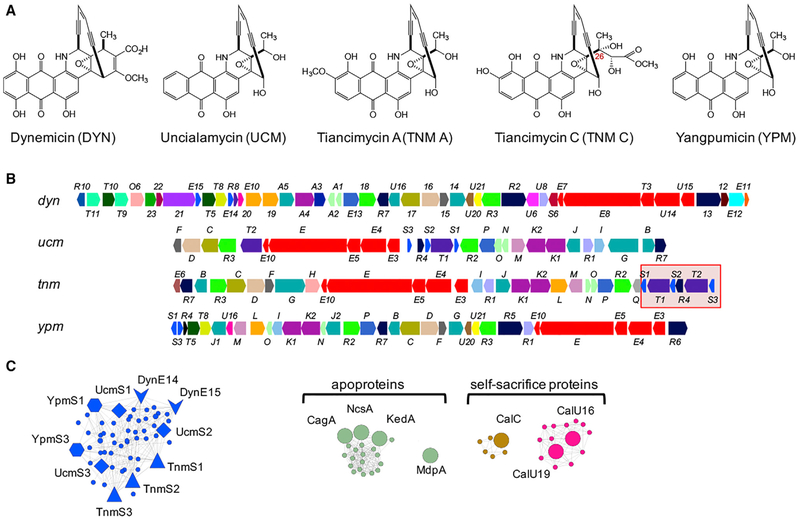
Figure 1. Bioinformatics Analysis Revealing Candidates for Self-Resistance Genes in the Biosynthetic Gene Clusters of Anthraquinone-Fused Enediynes.
(A) Structures of representative anthraquinone-fused enediynes DYN, UCM, TNM A and TNM C, and YPM.
(B) The biosynthetic gene clusters of DYN, UCM, TNM, and YPM. Genes are color-coded based on GNN annotation (see Figure S1 for details). The 5.5-kb fragment harboring tnmS1 to tnmS3 in the tnm biosynthetic gene cluster is highlighted by a red box.
(C) Selected clusters from the enediyne GNN (Figure S1) that highlight apoproteins and self-sacrifice proteins conferring enediyne resistance by the two known mechanisms and reveal new candidates conferring resistance to anthraquinone-fused enediynes by an unprecedented mechanism. The genes in the same color are homologous proteins.
Here we report (1) the discovery and characterization of sequestration as an unprecedented mechanism of self-resistance for the anthraquinone-fused enediynes and (2) the wide distribution of these resistance elements in nature, including in the human microbiome. These findings should now be considered in current and future efforts to develop enediyne-based chemotherapies.
RESULTS AND DISCUSSION
Bioinformatics Analysis Revealing Candidates for the Self-Resistance Genes in the Biosynthetic Gene Clusters of Anthraquinone-Fused Enediynes
We first constructed an enediyne GNN, including all predicted and experimentally confirmed enediyne biosynthetic gene clusters, to search for candidates conferring self-resistance to the anthraquinone-fused enediynes. The GNN clearly revealed additional homologs of the apoproteins, known for the nine-membered enediyne core subcategory, and the self-sacrifice proteins, known for the ten-membered enediyne CAL, suggesting that both mechanisms are likely shared by many other enediynes; these findings also demonstrated the utility of GNN in identifying candidates conferring enediyne resistance by the known mechanisms (Figures 1C and S1). Strikingly, GNN analysis failed to identify any candidates for the two known resistance mechanisms from the gene clusters encoding the biosyn-thesis of the four anthraquinone-fused enediynes, DYN, UCM, TNM, and YPM (Figure 1A). Since these producers must have evolved self-resistance elements, close examination of the GNN revealed a distinct family of proteins, consisting of TnmS1, TnmS2, TnmS3, UcmS1, UcmS2, UcmS3, DynE14, DynE15, YpmS1, and YpmS3, from the TNM, UCM, DYN, and YPM biosynthetic machineries, that share 31%–90% amino acid sequence identities (Figures 1B, 1C, and S1; Data S1A), suggesting an unprecedented mechanism of self-resistance for the anthraquinone-fused enediynes. BLASTP analysis revealed that the proteins in this family belong to the glyoxalase super-family, which are known to perform versatile functions, varying from isomerization (Cameron et al., 1997), oxidative cleavage of C-C bonds (Cho et al., 2010), epimerization (McCarthy et al., 2001), and epoxide hydrolysis (Thompson et al., 2013). Selected members in this superfamily are also known to render antibiotics ineffective, such as fosfomycin epoxide hydrolases and thiol transferases (Fillgrove et al., 2003; Thompson et al., 2013, 2014), and bleomycin (BLM) (Dumas et al., 1994) and mitomycin (Martin et al., 2002) binding proteins. With the exception of the antibiotic binding proteins, proteins in this family often possess the conserved amino acid residues His, Glu, and Asp for metal chelating. Since TnmS1, TnmS2, TnmS3, UcmS1, UcmS2, UcmS3, DynE14, DynE15, YpmS1, and YpmS3 lacked these conserved residues, we hypothesized, in the absence of alternative proposals for these proteins to play a role in biosynthesis of the anthraquinone-fused enediynes, that they might be antibiotic binding proteins that confer DYN, UCM, TNM, and YPM resistance by sequestration.
Deletion of the tnmS1, tnmS2, and tnmS3 Genes in Streptomyces sp. CB03234 Confirming their Necessity in Encoding TNM Resistance
We next decided to use Streptomyces sp. CB03234, the wild-type producer of TNMs, as a model to investigate the self-resistance mechanisms for the anthraquinone-fused enediynes in vivo, confirming that tnmS1, tnmS2, and tnmS3 are necessary to endow Streptomyces sp. CB03234 with high resistance to TNMs. We deleted a 5.5-kb DNA fragment, harboring tnmS1, tnmS2, and tnmS3, as well as three additional genes, tnmT1 and tnmT2 (encoding two putative transporters) and tnmR4 (encoding a putative regulator), which were interspersed among the three predicted resistance genes (Figure 1B), in Streptomyces sp. CB03234 to generate the ΔtnmS1-tnmS3 mutant strain SB20003. Its sensitivity to TNMs was first tested by a disk diffusion assay, with the wild-type Streptomyces sp. CB03234 as a positive control. While CB03234 showed high resistance, SB20003 was remarkably sensitive to TNM A, and obvious inhibition zones were observed for SB20003 even at the lowest amount of TNM A (1 ng) tested (Figure 2A). We further determined the minimal inhibitory concentrations (MICs) of TNM A against CB03234 and SB20003 by a plate growth assay, with Streptomyces lividans TK24, a naive strain that is sensitive to TNM A, as a negative control. SB20003 was shown to be extremely sensitive to TNM A, with an MIC of 4 ng/mL, which was the same as the MIC for S. lividans TK24 (4 ng/mL) and was minimally 125-fold more sensitive than the MIC for CB03234 (500 ng/mL) (Table 1; Figure S2A). These results unambiguously establish that the inherent self-resistant elements for TNMs in Streptomyces sp. CB03234 reside within this six-gene fragment and that there are no other genes, outside the tnm biosynthetic gene cluster, that encode TNM resistance.
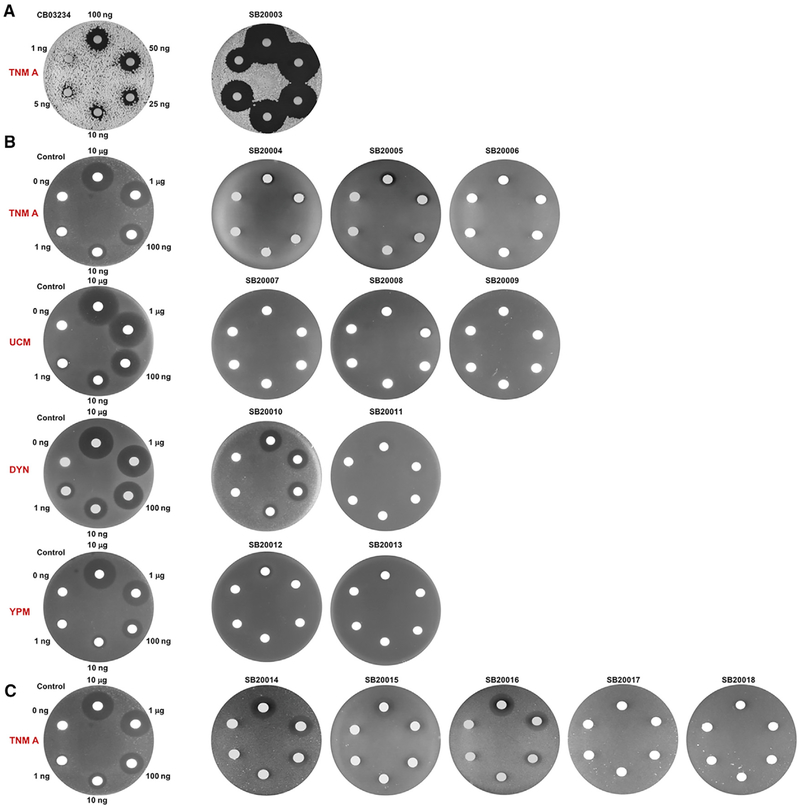
Figure 2. The tnmS1, tnmS2, tnmS3 Genes, and their Homologs Encoding Resistance to Anthraquinone-Fused Enediynes as Demonstrated by the Disk Diffusion Assays.
(A) The ΔtnmS1-tnmS3 mutant strain SB20003 is sensitive to TNM A, in comparison with the wild-type strain CB03234 as a positive control, establishing that TnmS1, TnmS2, and TnmS3 are necessary and sufficient to confer TNM resistance in its producer.
(B) E. coli recombinant strains SB20004 to SB20013, expressing tnmS1, tnmS2, tnmS3, and their homologs from the UCM, DYN, and YPM producers, are resistant to TNM A, UCM, DYN, and YPM, respectively, in comparison with E. coli BL21(DE3)/pET28a as a negative control, demonstrating that each of these gene products is sufficient to confer resistance to anthraquinone-fused enediynes in their respective producers.
(C) E. coli recombinant strains SB20015 to SB20018, expressing the five homologs from the human microbiome, show resistance to TNM A, in comparison withE. coli BL21(DE3)/pET28a as a negative control, revealing that the human microbiome might already possess resistant elements to anthraquinone-fused enediynes. Depicted next to the disks are the amounts of DYN, UCM, TNM A, and YMP, respectively, loaded to each set of the diffusion assays.
Table 1.
Sensitivity of Selected Streptomyces and E. coli Strains against TNM A as Determined by the MICs Values
| Strain | MIC (ng/mL) |
|---|---|
| Streptomyces strains | |
| CB03234 | 500 |
| SB20003 | 4 |
| S. lividans TK24 | 4 |
| E. coli strains | |
| BL21(DE3)/pET28a | 2.5 |
| SB20004 (TnmS1) | 2,560 |
| SB20005 (TnmS2) | 2,560 |
| SB20006 (TnmS3) | 20,480 |
| SB20014 (HMPREF1015_01744) | 40 |
| SB20015 (HMPREF0765_3744) | 40 |
| SB20016 (HMPREF1004_01465) | 10 |
| SB20017 (HMPREF1211_07544) | 20,480 |
| SB20018 (CBFG_04671) | 1,280 |
The tnmS1, tnmS2, and tnmS3 Genes and their Homologs Encoding Self-Resistance for All Anthraquinone-Fused Enediynes, Show Cross-Resistance
We then focused on tnmS1, tnmS2, and tnmS3 genes from the tnm cluster, as well as their homologs ucmS1, ucmS2, and ucmS3 genes from the ucm cluster, dynE14 and dynE15 genes from the dyn cluster, and ypmS1and ypmS3 genes from the ypm cluster for comparative studies (Figure 1), and revealed that they encoded self-resistance to the four anthraquinone-fused enediynes TNM, UCM, DYN, and YPM, respectively, capable of conferring cross-resistance. We adopted an in vivo approach to screen if tnmS1, tnmS2, tnmS3, or their homologs could confer resistance to TNMs or the other anthraquinone-fused enediynes UCM, DYN, and YPM, respectively, in E. coli BL21(DE3), which is known to be sensitive to these enediyne natural products. Each of the candidate-resistant genes was cloned into pET28a, affording expression plasmids pBS20007 to pBS20016, and transformed into E. coli BL21(DE3), yielding recombinant strains SB20004 to SB20013 (Table S2). Disk diffusion assays revealed that SB20004[C0]SB20013 are significantly more resistant to the corresponding anthraquinone-fused enediynes, in comparison with the negative control E. coli BL21(DE3)/pET28a (Figure 2B). These results provided direct evidence, supporting that tnmS1, tnmS2, tnmS3, or their homologs, individually, was sufficient to endow E. coli BL21(DE3) with resistance to their respective anthraquinone-fused enediynes in vivo. We subsequently determined the MICs of TNM A against SB20004, SB20005, and SB20006 by the plate growth assay for quantitative comparison. Expression of tnmS1, tnmS2, or tnmS3 inE. coli BL21(DE3) revealed a similar level for protein overproduction (Figure S2B), and individually endowed E. coli BL21(DE3) with >1,000-fold increase in cellular resistance to TNM A as exemplified by the MICs for SB20004 (2,560 ng/mL), SB20005 (2,560 ng/mL), and SB20006 (20,480 ng/mL) in comparison with the E. coli BL21(DE3) (2.5 ng/mL) (Table 1; Figure S2C). Furthermore, SB20004, SB20005, and SB20006 showed cross-resistance to UCM, DYN, and YPM (Figure S3A). These results support a common self-resistance mechanism for the anthraqui-none-fused enediynes.
TnmS1, TnmS2, and TnmS3 Binding TNMs with Nanomolar Affinity
We subsequently characterized the TnmS1, TnmS2, and TnmS3 proteins in vitro, revealing that they bind TNMs with nanomolar affinity. Recombinant TnmS1, TnmS2, and TnmS3 proteins were produced in E. coli and purified to homogeneity. At high concentration (50 mg/mL), the purified TnmS1, TnmS2, and TnmS3 appeared light yellow. To identify the yellow pigment co-purified with TnmS1, TnmS2, and TnmS3, these proteins were denatured by treating with trifluoroacetic acid. The released yellow pigment, which was isolated and subjected to LC-MS analysis, was identified to be riboflavin in comparison with an authentic standard. Given the low occupancy (<20%) and micromolar KD values of riboflavin to TnmS1, TnmS2, and TnmS3 (Figure S4; Table 2), we concluded that the co-purified riboflavin resulted from adventitious binding of riboflavin to these proteins upon their overproduction in E. coli and would not interfere with TnmS1, TnmS2, and TnmS3 binding to TNMs, thereby their ability to confer TNM resistance.
Table 2.
Binding of TnmS1, TnmS2, TnmS3 and the Selected Homologs from the Human Microbiome to TNM A, TNM C, and Riboflavin as Determined by the KD Values
| Protein | Ligand | KD (nM) |
|---|---|---|
| TnmS1 | TNM A | 163 ± 17 |
| TNMC | 350 ± 41 | |
| riboflavin | (84.1 ± 13.5) × 103 | |
| TnmS2 | TNM A | 246 ± 36 |
| TNM C | 361 ± 65 | |
| riboflavin | (59.5 ± 8.6) × 103 | |
| TnmS3 | TNM A | 108 ± 13 |
| TNM C | 215 ± 27 | |
| riboflavin | (1.1 ± 0.1) × 103 | |
| HMPREF1211_07544 | TNM A | 164 ± 23 |
| CBFG_04671 | TNM A | 855 ± 66 |
To directly measure the binding of TNMs to TnmS1, TnmS2, and TnmS3, we first removed the partially bound riboflavin from TnmS1, TnmS2, and TnmS3 by treatment with activated charcoal. TNM A and TNM C (Figure 1A) were used as representative anthraquinone-fused enediynes to measure their direct binding to TnmS1, TnmS2, and TnmS3 in vitro. The purified TnmS1, TnmS2, and TnmS3 were each found to be dimeric in solution upon analysis by size-exclusion chromatography (Figure S5A). Fluorescence-quenching analysis revealed that TNM A and TNM C were tightly bound by TnmS1, TnmS2, and TnmS3 with nanomolar KD values (Figure S4; Table 2). TNM A, upon recovery from binding to TnmS1, TnmS2, and TnmS3, showed no modification or degradation, further demonstrating the non-covalent, non-destructive, and reversible binding between these proteins and TNMs.
Crystal Structures of TnmS1, TnmS2, and TnmS3 Revealing the Molecular Details for Resistance to the Anthraquinone-Fused Enediynes by Sequestration
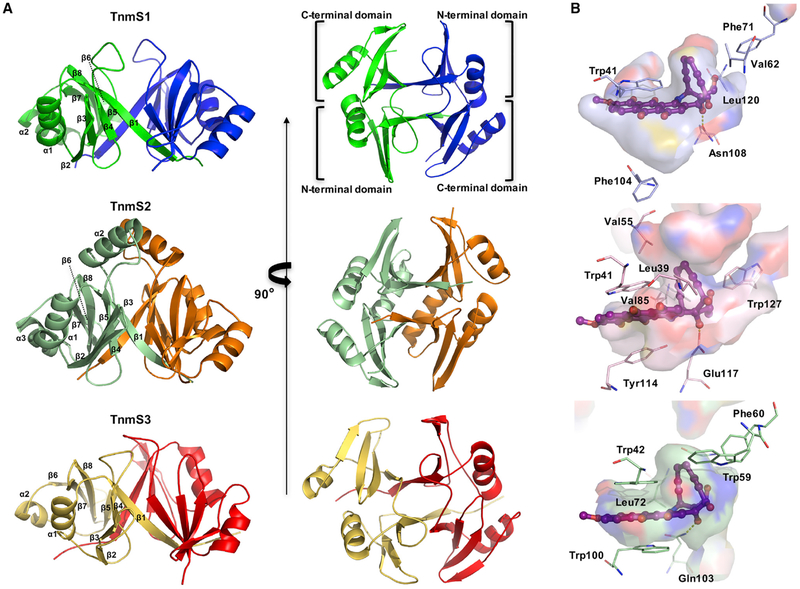
We solved the crystal structures of TnmS1, TnmS2, and TnmS3 to understand the molecular details of TNM binding, revealing sequestration as an unprecedented self-resistance mechanism for the anthraquinone-fused enediynes. TnmS1, TnmS2, and TnmS3 are packed as homodimers in crystal structures, consistent with their oligomeric structures determined upon size-exclusion chromatography. Despite their low amino acid sequence identities, ranging from 16% to 29%, TnmS1, TnmS2, and TnmS3 were found to share similar structural folds with known antibiotic binding proteins, such as BLM (PDB: 1EWJ, 1QTO, and 1BYL) (Maruyama et al., 2001; Kawano et al., 2000; Dumas et al., 1994) and mitomycin (PDB: 1KLL) (Martin et al., 2002) (Data S1). Each monomer of TnmS1, TnmS2, and TnmS3 contains an N- and a C-terminal domain with a β-α-β-β-β topology (Figure 3A). The strand β1 in the N-terminal domain contributes to dimerization by alternate arm exchange between the two monomers to form a β barrel-like structure and the two large concavities (Figure 3A). The amino acid sequence of TnmS2 is 9 and 11 residues longer than that of TnmS1 and TnmS3, respectively. Structural comparison of TnmS1, TnmS2, and TnmS3 revealed an α helix (α2) comprising 12 amino acids in TnmS2, instead of a loop in TnmS1 or TnmS3, which connects the N- and C-terminal domains (Figure 3A; Data S1B).
Figure 3. The Crystal Structures of TnmS1, TnmS2, and TnmS3 Revealing a β Barrel-like Structure that Forms Two Large Concavities to Accommodate TNMs.
(A) The overall structures of TnmS1, TnmS2, and TnmS3 shown in ribbon diagram. The two chains of TnmS1, TnmS2, and TnmS3 are shown by green/blue, pale green/orange, and yellow/red, respectively.
(B) Solvent accessible surfaces of the binding cavities of TnmS1, TnmS2, and TnmS3, highlighting a common mode for enediyne binding. The binding modes of TNM A in TnmS1 and TnmS2 were modeled based on the structure of TnmS3 in complex with TNM A.
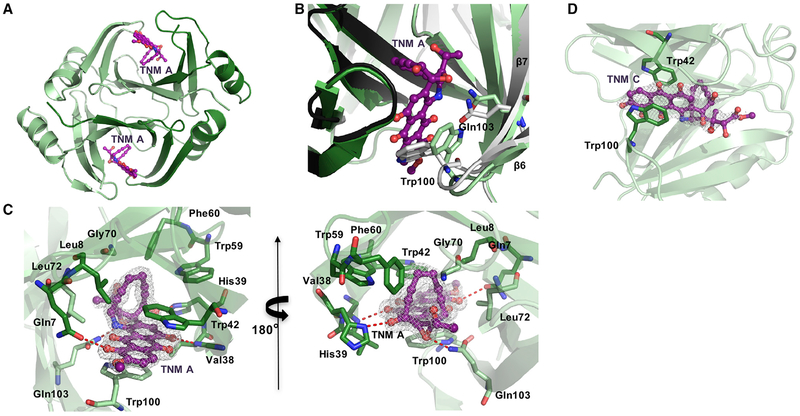
The structure of TnmS3 in complex with TNM A shows that each cavity houses one TNM A molecule (Figure 4A). TnmS3 undergoes a slight conformational change in β6-loop-β7 upon TNM A binding (Figure 4B). The loop between β6 and β7 shifts ~1.5Å upon TNM A binding, and the side chains of Trp100 and Gln103 flip into new conformations to interact with TNM A (Figure 4B). The structure of TnmS3 in complex with TNM C, a biosynthetic intermediate of TNM A accumulated by the ΔtnmH mutant strain Streptomyces sp. SB20002 (Yan et al., 2016, 2017), shows a similar binding environment as TNM A in TnmS3 (Figures 4C and 4D). The side chain at C-26 of TNM C reveals no strong interaction with TnmS3 (Figures 1A and 4D). The complex structures of TnmS3 with TNM A and TNM C, therefore, provide direct evidence and reveal the molecular details of how TnmS3 sequesters TNMs. Thus, TNMs are sequestered in the cavity of the β barrel-like structure by three main interactions (Figure 4C):(1) the flat anthraquinone moiety of TNM A is sandwiched between the side chains of Trp100A and Trp42B via electrostatic interactions of the aromatic rings; (2) the side chains of Gln103A, Gln7A, His39B, and the main chain amine group of Val38B provide hydrogen bond interactions with the hydroxyl and epoxide groups of TNM A; and (3) Gly70A and Leu72A and Leu8B, Trp42B, Trp59B, and Phe60B create a hydrophobic environment to accommodate the enediyne core.
Figure 4. The Crystal Structure of TnmS3 in Complex with TNMs Revealing the Molecular Details for Resistance to the Anthraquinone-Fused Enediynes by Sequestration.
(A) The overall structure of TnmS3 in complex with TNM A shown in ribbon diagram. Two molecules of TNM A are bound by one TnmS3 dimer.
(B) Local view showing that TnmS3 undergoes a slight conformational change upon TNM A binding, as depicted by the side chains of Trp100 and Gln103 flipping into new conformations to interact with TNM A, with the apo-TnmS3 structure shown in gray.
(C) Local view of the enediyne binding cavity highlighting the residues involved in interactions between TnmS3 and TNM A. Hydrogen bonds are shown in red dashed lines.
(D) The local view of the enediyne binding cavity highlighting the residues involved in interactions between TnmS3 and TNM C. While TNM A and TNM C share many of the same interactions with TnmS3, the side chain at C-26 of TNM C reveals no major interaction. The 2Fo–Fc electron density maps are contoured at 1s shown as white mesh. The σ-A-weighted difference (mFo–DFc) omit maps of TnmS3 in complex with TNM A and TNM C, respectively, are shown in Figure S5.
By superposition of TnmS1 and TnmS2 with the structure of TnmS3 in complex with TNM A, the binding modes of TNM A in TnmS1 and TnmS2 were mapped. The models of TnmS1 and TnmS2 in complex with TNM A revealed that the cavities of the β barrel-like structures provide sufficient space to accommodate TNM A, the binding residues of which are proposed (Figure 3B): (1) the anthraquinone moiety interacts electrostatically with Trp41 and Phe104 in TnmS1 and Trp41 and Tyr114 in TnmS2, (2) the epoxide ring is hydrogen bonded by Asn108 in TnmS1 and Glu117 in TnmS2, and (3) the enediyne core interacts hydrophobically with Trp41, Val62, Phe71, and Leu120 and Leu39 in TnmS1, and Trp41, Val55, Val85, and Trp127 in TnmS2.
Taken together, both the anthraquinone moiety and the enediyne core contribute to the major interactions between the enediynes and the binding proteins, accounting for the findings that TnmS1, TnmS2, and TnmS3 can confer resistance to varying members of the TNMs and cross-resistance to other members of the anthraquinone-fused enediynes (Figures 1A and S3A). It is fascinating to note that each of the anthraquinone-fused enediyne producers harbors multiple genes encoding proteins with the same mechanism of self-resistance (Figures 1B and 1C). A similar phenomenon has been observed for the CAL producer that encodes three self-sacrifice proteins to confer CAL resistance (Big-gins et al., 2003; Elshahawi et al., 2014). Multiple resistance genes may ensure that sufficient resistance proteins are available to accommodate the extreme toxicity of the enediynes at all times during their biosynthesis. In addition to the final anthraquinone-fused enediyne natural products, biosynthetic intermediates and shunt metabolites have also been isolated from these producers (Konishi et al., 1989; Davies et al., 2005; Gao and Thorson, 2008; Yan et al., 2016), indicative that multiple cytotoxic molecules may be accumulated during their biosynthesis. Multiple resistance genes would allow the producers to evolve proteins with varying substrate specificity to account for enediyne structural diversity during biosynthesis, a hypothesis that would be consistent with the findings that TnmS1, TnmS2, and TnmS3 bind TNM A and TNM C with varying KD values (Table 2; Figure S4A).
We also solved the structure of TnmS3 in complex with riboflavin (Figure S5C), and mapped riboflavin binding in TnmS2 (Figure S5B), revealing that riboflavin binds TnmS2 and TnmS3 at the same cavity as TNMs. In analogy to the anthraquinone moiety of TNMs, the isoalloxazine ring of riboflavin is sandwiched between the side chains of Trp41 and Tyr 114 in TnmS2 and Trp42 and Trp100 in TnmS3 via electrostatic interactions (Figures S5B and S5C), accounting for the adventitious binding of riboflavin upon production of these proteins in E. coli. In contrast, despite the fact that TnmS1, TnmS2, and TnmS3 share the same overall structural folds as several antibiotic binding proteins, including the BLM and mitomycin binding proteins, they feature distinct ligand binding cavities (Figure S6) (Maruyama et al., 2001; Martin et al., 2002). This is consistent with the findings that TnmS1, TnmS2, and TnmS3 displayed no cross-resistance to BLM or mitomycin, respectively, upon overproduction in E. coli (Figures S3B and S3C).
Homologs of TnmS1, TnmS2, and TnmS3 from the Human Microbiome Conferring Resistance to TNM A
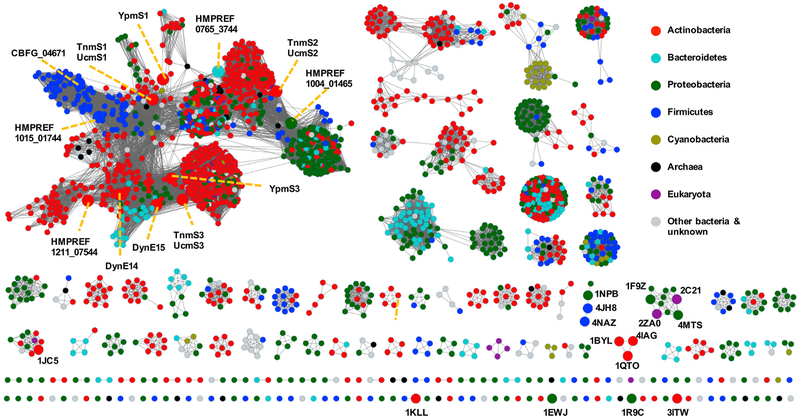
Homologs of TnmS1, TnmS2, and TnmS3 are widely distributed in nature, including the human microbiome, and confer resistance to anthraquinone-fused enediynes. Inspired by the unprecedented resistance mechanism to the anthraqui-none-fused enediynes, we constructed a sequence similarity network (Zhao et al., 2014) of 205,686 proteins from the glyoxalase family (InterPro family IPR029068, release 65.0, as of October 30, 2017) to search for homologs of TnmS1, TnmS2, and TnmS3 from the InterPro database. When an e value threshold of 10−20 (~30% sequence identity over 80 amino acids) was applied, 4,095 homologs of TnmS1, TnmS2, and TnmS3 were grouped in a large cluster. These homologs are from a broad range of bacteria, with Actinobacteria (~65%) and Proteobacteria (~22%) as the major sources, together with 19 proteins from the Archaea domain and one protein from the Eukarya domain (Figure 5; Data S2A). Strikingly, 89 of the 4,095 homologs are from the human micro-biome at different body sites. As the human microbiome has been reported as a reservoir of antibiotic resistance genes (Sommer et al., 2009), characterization of selected proteins from the 89 homologs will provide direct evidence whether these proteins could also confer resistance to the anthraqui-none-fused enediynes.
Figure 5. Sequence Similarity Network of Proteins from the InterPro Family IPR029068 Revealing Homologs of TnmS1, TnmS2, and TnmS3 Are Widely Distributed in Nature, Including the Human Microbiome.
The SSN of the IPR029068 family was generated using the online Enzyme Function Initiative-Enzyme Similarity Tool (Gerlt et al., 2015) and visualized with Cytoscape with an e value threshold of 10−20 (~30% sequence identity over 80 amino acids). Each node represents proteins with above 75% sequence identities. The nodes are colored by the taxonomy of the sources of these proteins. The TnmS1, TnmS2, TnmS3, and their homologs characterized in this study are highlighted in larger circles and labeled with orange dashed lines. Other characterized proteins from the IPR029068 family are also highlighted in large circles and labeled with their respective PDB numbers. PDB: 1NPB, 4JH8, and 4NAZ are thiol transferases; PDB: 1F9Z, 2C21, 2ZA0, and 4MTS are glyoxalases-I; PDB: 1BYL, 4IAG, 1QTO, and 1EWJ are BLM binding proteins; PDB: 1JC5 is epimerase; PDB: 1KLL is mitomycin binding protein; PDB: 1R9C is epoxide hydrolase; and PDB: 3ITW is thiocoraline binding protein.
We selected five representative proteins from the human microbiota, HMPREF0765_3744 from Sphingobacterium spiritivorum ATCC 33300 (isolated from the urogenital tract), HMPREF1004_01465 from Ralstonia sp. 5_7_47FAA, HMPREF1015_01744 from Bacillus smithii 7_3_47FAA, HMPREF1211_07544 from Streptomyces sp. HGB0020, and CBFG_04671 from Clostridiales bacterium 1_7_47FAA (all isolated from the gastrointestinal tract), none of which possesses the gene clusters encoding enediyne biosynthesis (Data S2B). The five representative proteins, selected from the 89 homologs, share 29%–48% amino acid sequence identities to TnmS1, TnmS2, and TnmS3, from the 89 homologs (Data S1A). We directly tested, by both in vivo and in vitro methods, their ability to confer resistance to TNM A as a representative of anthraquinone-fused enediynes. Thus, the genes encoding these five proteins were synthesized and cloned into pET28a, affording expression plasmids pBS20017 to pBS20021, and transformed into E. coli BL21(DE3), yielding recombinant strains SB20014 to SB20018 (Table S2). SDS-PAGE analysis confirmed that these strains produced similar levels of the five representative proteins (Figure S2B). Disk diffusion assay revealed that all five homologs confer some degree of resistance to TNM A (Figure 2C), with MICs, determined by the plate growth assay, ranging from 10 to 20,480 ng/mL (Table 1; Figure S2C). It is remarkable that HMPREF1211_07544 and CBFG_04671 confer TNM A resistance at the same level as the TnmS1, TnmS2, and TnmS3, displaying similar nanomolar KD values (Table 2; Figure S4A). Like TnmS1, TnmS2, and TnmS3, these homologs conferred resistance specific to TNM A, and no cross-resistance to BLM or mitomycin was observed (Figure S3). Sequence alignment of TnmS1, TnmS2, TnmS3, with the five selected homologs from the human microbiome, revealed that the residues for electrostatic interactions are highly conserved in each of these proteins, while the residues for hydrogen bond and hydrophobic interactions are diverse (Data S1D).
Little is known how the human microbiome acquires its immense diversity of drug-resistance genes. Although the human microbiota has been suggested to acquire resistance genes from pathogens resistant to clinical antibiotics (Penders et al., 2013), we present here that the human microbiome already possesses the resistant elements to anthraquinone-fused enediynes, which have yet to be developed and used clinically. It has been recently reported that intratumor bacteria could produce enzymes that metabolize the anti-cancer drug into its inactive form, thereby influencing the efficacy of cancer chemotherapy (Geller et al., 2017). Although it is far from certain if these alleles are actual components of the human microbiome, rather than environmental contaminants, our findings raise the possibility that the human microbiome might impact the efficacy of anthraquinone-fused enediynes and should now be taken into consideration in current and future efforts to develop enediyne-based chemotherapies.
STAR+METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Streptomyces lividans TK324 | Hopwood et al., 1983 | N/A |
| Micromonospora chersina ATCC53710 | Gao and Thorson, 2008 | N/A |
| Streptomyces sp. CB03234 | Yan et al., 2016 | N/A |
| Streptomyces uncialis DCA2648 | Yan et al., 2016 | N/A |
| Micromonospora yangpuensis DSM45577 | Yan et al., 2017 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Selenomethionine | Acros | Cat#AC259960025 |
| Bleomycin | bioWORLD | Cat#40210016-2 |
| Mitomycin | RPI | Cat#M92010 |
| Deposited Data | ||
| Crystal structure of member of Glyoxalase/bleomycin resistance protein/dioxygenase superfamily from Sphaerobacter thermophilus DSM 20745 | Protein Data Bank | PDB: 4HC5 https://www.rcsb.org/structure/4hc5 |
| TnmS1, TnmS2, and TnmS3 from S. sp. CB03234 | GenBank | Accession No.: AME18020 AME18022 AME18026 |
| UcmS1, UcmS2, and UcmS3 from S. uncialis DCA2648 | GenBank | Accession No.: AMK92568 AMK92566 AMK92564 |
| DynE14 and DynE15 from M. chersina ATCC53710 | GenBank | Accession No.: WP_091316676 WP_091316667 |
| YpmS1 and YpmS3 from M. yangpuensis DSM45577 | GenBank | Accession No.: WP_091439392 WP_091439387 |
| The selected five homologues of TnmS1, TnmS2, and TnmS3 from the human microbiome: HMPREF1015_01744, HMPREF0765_3744, HMPREF1004_01465, HMPREF1211_07544, and CBFG_04671 | GenBank | Accession No.: WP_003353922 WP_003002659 WP_004633594 WP_016437401 WP_008724152 |
| Oligonucleotides | ||
| hmpref1015_01744 gene | Genewiz | N/A |
| hmpref0765_3744 gene | Genewiz | N/A |
| hmpref1004_01465 gene | Genewiz | N/A |
| hmpref1211_07544 gene | Genewiz | N/A |
| cbfg04671 gene | Genewiz | N/A |
| Recombinant DNA | ||
| pBS8007 | Tao et al., 2007 | N/A |
| pBS20002 | Yan et al., 2016 | N/A |
| Software and Algorithms | ||
| MOLREP | Vagin and Teplyakov, 2010 | N/A |
| AutoSol | Terwilliger et al., 2009 | N/A |
| HKL-3000 | Minor et al., 2006 | N/A |
| REFMAC | Murshudov et al., 1997 | N/A |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ben Shen (shenb@scripps.edu).
EXPERIMENTAL MODEL AND SUBJECTE DETAILS
E. coli BL21(DE3) was used for protein production. Micromonospora chersina ATCC53710, Streptomyces sp. CB03234, Streptomyces uncialis DCA2648, and Micromonospora yangpuensis DSM45577 were used for production of DYN, TNMs, UCM, and YPM, respectively. Culture conditions followed literature procedures, and references are provided in Method Details.
METHOD DETAILS
General Materials
Primers, plasmids, and strains used in this study are summarized in Tables S1 and S2.
Gene Cloning and Protein Production and Purification
The tnmS1, tnmS2, and tnmS3 genes were amplified by PCR from the genomic DNA of the TNM producer S. sp. strain CB03234 using primers summarized in Table S1. The PCR products were cloned into pET28a to afford the expression plasmids pBS20007, pBS20008, and pBS20009, which were transformed into E. coli BL21(DE3) to yield recombinant strains SB20004, SB20005, and SB20006. The latter strains were grown at 37°C, 200 rpm in 1 L LB medium with 50 μg/mL kanamycin until it reached an OD600 =0.5. To produce the selenomethionyl (SeMet)-labeled TnmS2 protein, SB20005 was grown at 37°C, 200 rpm in 1 L of enriched M9 medium with 50 μg/mL kanamycin until it reached an OD600=1.0. After air-cooling at 4°C for 60 min, methionine biosynthesis inhibitory amino acids (25 mg each/L of L-valine, L-isoleucine, L-leucine, L-lysine, L-threonine, L-phenylalanine) and 90 mg/L of selenomethionine were added. Protein overproduction was induced by 0.5 mM isopropyl-β-D-thiogalactoside (IPTG). The cells were incubated overnight at 18°C and were subsequently harvested and resuspended in lysis buffer (500 mM NaCl, 20 mM Tris, pH 8.0). Cells were disrupted by lysozyme treatment (1 mg/mL) and sonication, and the insoluble cellular material was removed by centrifugation. TnmS1, TnmS2 or SeMet-labeled SnmS2, and TnmS3 were purified using Ni-NTA affinity chromatography. The column was washed sequentially with 20 mL of lysis buffer and 12 mL of washing buffer (lysis buffer containing 50 mM imidazole) and eluted with 10 mL of elution buffer (lysis buffer containing 300 mM imidazole). The eluted protein was further purified by size exclusion chromatography using a Superdex 200 16/200 column under 200 mM NaCl and 50 mM Tris buffer, pH 8.0. The buffer was exchanged with 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (20 mM, pH 7.0) using Amicon Ultra-15 concentrators (Millipore). Protein purity was examined by SDS-PAGE protein electrophoresis. Protein concentrations were determined from the absorbance at 280 nm using a calculated molar absorption coefficient (ε280 = 8,480, 18,450, and 17,990 M−1 cm−1 for TnmS1, TnmS2, and TnmS3, respectively). The concentration of TnmS1, TnmS2, and TnmS3 used for crystallization were 15, 20, and 30 mg/mL, respectively.
The ucmS1, ucmS2, and ucmS3 genes from the UCM producer Streptomyces uncialis DCA2648, dynE14 and dynE15 genes from the DYN producer Micromonospora chersina ATCC53710, and ypmS1 and ypmS3 genes from the YPM producer Micromonospora yangpuensis DSM45577 were amplified by PCR from the corresponding genomic DNAs using primers summarized in Table S1. The genes encoding the five homologues of TnmS1, TnmS2, and TnmS3, hmpref1015_01744, hmpref0765_3744, hmpref1004_01465, hmpref1211_07544, and cbfg04671, were synthesized with their codons optimized for E. coli expression (Gouy and Gautier, 1982). The targeted genes were cloned into pET28a to afford the expression plasmids pBS20010 to pBS20021, which were transformed into E. coli BL21(DE3) to yield recombinant strains SB20007 to SB20018. Gene expression and protein production and purification of HMPREF1015_01744, HMPREF 0765_3744, HMPREF 1004_01465, HMPREF 1211_07544, and CBFG04671 followed the same procedure described above for TnmS1, TnmS2, and TnmS3. Protein concentrations were determined from the absorbance at 280 nm using a calculated molar absorption coefficient (ε280 = 23,960, 12,950, 16,960, 12,490, and 22,460 M−1 cm−1 for HMPREF1015_01744, HMPREF 0765_3744, HMPREF 1004_01465, HMPREF 1211_07544, and CBFG04671, respectively).
Construction of the ΔtnmS1–tnmS3 Mutant Strain SB20003
The ΔtnmS1–tnmS3 mutant strain SB20003 was constructed in S. sp. CB03234 by gene replacement via homologous recombination. The 1.7-kb ermE-neo resistance cassette was amplified by PCR from pBS8007 (Tao et al., 2007) using primers tnmS1-S3tgtF and tnmS1-S3tgtR (Table S1) and used to replace the 5.5-kb tnmS1–tnmS3 fragment in pBS20002 (Yan et al., 2016) via λ-RED-mediated PCR targeting mutagenesis (Gust et al., 2003) to generate pBS20006. The pBS20006 plasmid was then introduced into S. sp. CB03234 by E. coli–Streptomyces intergeneric conjugation, and kanamycin-resistant and apramycin-sensitive colonies were isolated and named SB20003, the genotype of which was confirmed by Southern analysis.
Production and Isolation of the Anthraquinone-Fused Enediynes TNM A, TNM C, UCM, DYN, and YPM
Production and isolation of TNM A, TNM C, UCM, DYN, and YPM from S. sp. CB03234, S. uncialis DCA2648, M. chersina ATCC53710, and M. yangpuensis DSM45577, respectively, followed literature procedures (Yan et al., 2016; Davies et al., 2005; Konishi et al., 1989). The authenticity of TNM A, TNM C, UCM, DYN, and YPM was validated by high resolution MS and 1H and 13C NMR analysis.
Protein Crystallization
The purified proteins were crystallized using the hanging drop vapor-diffusion method. TnmS1 was crystallized in 50 mM ammonium sulfate, 50 mM bis-Tris propane at pH 6.5, and 30% v/v pentaerythriol ethoxylate (15/4 EO/OH). SeMet-labeled TnmS2 was crystallized in 50 mM magnesium sulfate hydrate, 50 mM HEPES sodium at pH 7.0, and 1.6 M lithium sulfate monohydrate. TnmS3 was crystallized in 100 mM ammonium acetate, 100 mM bis-Tris at pH 5.5, and 17% w/v polyethylene glycol 3350. The crystals of TnmS3 in complex with riboflavin were obtained by soaking TnmS3 crystals with 1 μL of 20 μM riboflavin dissolved in the crystalized condition for 12 h. The crystals of TnmS3 in complex with TNM A or TNM C were obtained by soaking TnmS3 crystals with 0.1 μL of 5 mM TNM A or TNM C, respectively, which were dissolved in the crystalized condition with 50% DMSO, every 48 h until the color of the crystals became dark purple (~10 days).
Data Collection, Structural Determination, and Refinement
The protein crystals were transferred to a cryoprotectant solution containing glycerol (30% v/v) prior to X-ray diffraction. The diffraction data of TnmS1, TnmS2, and TnmS3 were collected at 100 K at the 19-ID beamline of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory (Rosenbaum et al., 2006). The data were recorded on Q315r (ADSC) for TnmS1 and TnmS3 and on Pilatus 6M (DECTRIS) for TnmS2. The diffraction data of TnmS3 complexed with TNM A or TNM C were collected at 100 K by an in-house diffraction facility equipped with Micromax-007 HEM (Rigaku). The data were recorded on Mar345dtb (Mar research). Data collection strategy, integration, and scaling were performed with the HKL3000 program package (Minor et al., 2006) for the crystals of TnmS1, TnmS2, and TnmS3 and the MOSFLM program (Vonrhein et al., 2011; Leslie, 2006) for the crystals of TnmS3 in complex with TNM A or TNM C.
The crystal structures of TnmS1 and TnmS3 were determined by the molecular replacement method of MOLREP (CCP4) (Vagin and Teplyakov, 2010) using the structure of a glyoxalase superfamily protein (PDB entry: 4HC5) as a search model. The crystal structures of TnmS3 complexed with TNM A, TNM C, or riboflavin were determined using the structure of TnmS3 as a search model. The crystal structure of TnmS2 was determined by the multi-wavelength anomalous dispersion (MAD) method. AutoSol (PHENIX) (Terwilliger et al., 2009) was used to find the phase solution and build the initial model. Extensive manual model building was performed using COOT (Emsley and Cowtan, 2004). The models were refined with REFMAC (Murshudov et al., 1997). The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 5UMQ for TnmS1, 5UMW for TnmS2, 5UMP for TnmS3, and 5UMY, 6BBX, and 5UMX for TnmS3 in complex with TNM A, TNM C, and riboflavin, respectively. A summary of the crystallographic data and the refinement statistics are given in Table S3
Disk Diffusion Assay
For S. sp. CB03234, SB20003, and S. lividans TK24, disk diffusion assay was carried out using 105 spores for each strain in molten tryptic soy agar. Spores of the Streptomyces strains were mixed well with molten tryptic soy agar and poured into Petri dishes. The 7 mm disks (Whatman) were placed onto the agar and loaded with varying quantities of TNM A, ranging from 0 to 100 ng in 10 μL DMSO (Figure 2A). The inhibition zones were observed after 48 h at 28°C. For recombinant E. coli strains SB20004 to SB20018, as well as E. coli BL21(DE3)/pET-28a as a negative control, disk diffusion assays were carried out using overnight cultures diluted to OD600 = 0.2 in molten LB agar with 50 μg/mL kanamycin and 200 μM IPTG. The resultant E. coli cultures, kanamycin, and IPTG were mixed well with molten LB agar and poured into Petri dishes. The 7 mm disks (Whatman) were placed onto the agar and loaded with varying quantities of TNM A, UCM, DYN, YPM, ranging from 0 to 10 μg, BLM, ranging from 0 to 30 μg, and mitomycin, ranging from 0 to 5 μg, all in 10 μL DMSO (Figures 2B, 2C, and S3). The inhibition zones were observed after 24 h at 37°C.
Determination of MICs
For S. sp. CB03234, SB20003, and S. lividans TK24, 104 spores of each strain were added into 96 wells, each containing 200 μL TSB, and treated with varying concentrations of TNM A, ranging from 2.56 × 10−4 ng/mL to 2,500 ng/mL. The 96-well plates were incubated for 48 h at 28°C. For E. coli strains SB20004 to SB20006 and SB20014 to SB20018, as well as E. coli BL21(DE3)/pET-28a as a negative control, they were grown overnight at 37°C in LB medium with 50 μg/mL kanamycin. Each of the overnight cultures was then diluted to OD600 = 0.1 aliquot in 96 wells and was subsequently treated with 200 mM IPTG and varying concentrations of TNM A, ranging from 0.625 ng/mL–40,960 ng/mL. The 96-well plates were incubated for 24 h at 37°C (Figures S1A and S2C).
Production of TnmS1, TnmS2, TnmS3, and their homologues in E. coli was examined to confirm that varying values of MICs were not due to varying expression levels. Strains SB20004-SB20006 and SB20014-SB20018, as well as BL21(DE3)/pET-28a as a negative control, were grown overnight at 37°C in LB medium with 50 μg/mL kanamycin. Each of the overnight cultures was then added to 5 mL of LB to reach OD600 = 0.1, induced with 200 μM IPTG, incubated for 16 h at 37°C, and harvested by centrifugation at 4000 × g for 15 min at 4°C. The pellets were resuspended in 500 μL of 50 mM Tris, pH 8.0, containing 100 mM NaCl. The cells were lysed by sonication, the cells debris were removed by centrifugation at 15,000 × g for 15 min at 4°C, and 10 μL of the supernatant were finally subjected to SDS-PAGE analysis (Figure S2B).
Measurement of Dissociation Constants of KDs
Dissociation constant KDs were determined according to literature procedure using fluorescence-quenching analysis (Sugiyama et al., 2002). Adventitiously bound riboflavin was first removed from TnmS1, TnmS2, and TnmS3 by treatment with activated charcoal. Proteins and ligands (i.e., TNM A, TNM C, or riboflavin) were mixed in 96-well plates, and the fluorescence intensity was recorded at 28°C on an Infinite M1000pro Microplate Reader (Tecan) under the condition of λ excitation = 280 nm, λ emission = 340 nm, 50 number of flashes, 20 μs integration time, and the slit-width = 5 nm. The proteins were diluted with MOPS buffer (20 mM, pH 7.0) to a final concentration of 2 μM for 180 μL in each 96-well. TNM A, TNM C, or riboflavin, with varying concentrations in 20 μL of DMSO, was then added to the protein solution. The fluorescent intensity of the resultant protein solutions was progressively quenched by the increasing concentrations of TNM A, TNM C, or riboflavin. All reactions were carried out in triplicate.
Recovery of TNM A from Its TnmS1, TnmS2, and TnmS3 Complexes
To form the protein-TNM A complexes, 2 μL of TNM A (2 mM in methanol) was mixed with 98 μL of TnmS1, TnmS2, or TnmS3 (1 mM in 20 mM MOPS buffer, pH 7.0), respectively, and incubated at room temperature for 10 min. The resultant protein-TNM A complexes were concentrated with Microcon-3 kDa centrifugal filter (Sigma-Aldrich), washed three times with 100 μL of MOPS buffer (20 mM, pH 7.0), and recovered by dissolving in 100 μL of MOPS buffer (20 mM, pH 7.0). TNM A was recovered by extracting the protein-TNM A complexes three times with equal volume of ethyl acetate. The ethyl acetate extracts were combined, dried in vacuo, and the residues were dissolved in 100 μL methanol and subjected to LC-MS analysis. The identity and integrity of TNM A recovered was confirmed by comparison with an authentic TNM A standard, and the percentage of recovered TNM A from each of the complexes was >90%.
QUANTIFICATION AND STATISTICAL ANALYSIS
For disk diffusion assay and MICs determination, each sample was repeated two times. For binding assay, three independent assays of each sample were run. X-ray data collected were from single crystals.
DATA AND SOFTWARE AVAILABILITY
The crystal structures have been deposited in Protein Data Bank (http://www.rcsb.org/pdb) under ID codes 5UMQ, 5UMW, 5UMP, 5UMX, 5UMY, and 6BBX for TnmS1, TnmS2, TnmS3, TnmS3–RBF, TnmS3–TNM A, and TnmS3–TNM C, respectively.
Supplementary Material
Highlights.
The tnmS1, tnmS2, and tnmS3 genes encode tiancimycin resistance in its producer
Their homologs are found in all anthraquinone-fused enediyne producers
TnmS1, TnmS2, and TnmS3 bind tiancimycins, conferring resistance by sequestration
TnmS1, TnmS2, and TnmS3 homologs are widespread, including in the human microbiome
SIGNIFICANCE.
The enediynes represent one of the most fascinating families of natural products for their unprecedented molecular architecture and extraordinary cytotoxicity and have been successfully translated into clinical drugs. Two self-resistance mechanisms are known for the enediyne producers—apoproteins for the nine-membered enediynes and self-sacrifice proteins for the ten-membered enediyne calicheamicin. Here we unveil an unprecedented resistance mechanism for the enediynes by sequestration. We show that tnmS1, tnmS2, and tnmS3 encode tiancimycin resistance in its producer, and that their homologs are found in all known anthraquinone-fused enediyne producers. TnmS1, TnmS2, and TnmS3 share a similar β barrel-like structure, bind tiancimycins with nano-molar KD values, and confer resistance by a common mechanism. TnmS1, TnmS2, and TnmS3 homologs are widespread in nature, including the human microbiome. Mechanisms of self-resistance in producers serve as models to predict and combat future drug resistance in clinical settings. Little is known how the human microbiome acquires its immense diversity of drug-resistance genes. Although the human micro-biota has been suggested to acquire resistance genes from pathogens resistant to clinical antibiotics, we present here that the human microbiome already possesses the resistant elements to anthraquinone-fused enediynes, which have yet to be developed and used clinically. While it is far from certain if these alleles are actual components of the human micro-biome, rather than environmental contaminants, our findings raise the possibility that the human microbiome might impact the efficacy of anthraquinone-fused enediynes and should now be taken into consideration in current and future efforts to develop enediyne-based chemotherapies.
ACKNOWLEDGMENTS
This work is supported in part by NIH grants GM098248, GM109456, GM121060 (to G.N.P.), GM094585 (to A.J.), CA078747, GM115575, and CA204484 (to B.S.). The use of Structural Biology Center beamlines at the Advanced Photon Source was supported in part by the U.S. Department of Energy, Office of Biological and Environmental Research, grant DE-AC02–06CH11357 (to M.E.C. and A.J.). We thank the John Innes Center, Norwich, UK, for providing the REDIRECT technology kit. C.-Y.C., I.C., and J.D.R. are supported in part by postdoctoral fellowships from the Academia Sinica – The Scripps Research Institute Talent Development Program, the German Research Foundation, and the Arnold and Mabel Beckman Foundation, respectively. This is manuscript no. 29647 from The Scripps Research Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures, three tables, and two data files and can be found with this article online at https://doi.org/10.1016/j.chembiol. 2018.05.012.
DECLARATION OF INTERESTS
B.S. has filed a patent (US, 2018/0009823 A1). The other authors declare no competing interests.
REFERENCES
- Abe S, and Otsuki M (2002). Styrene maleic acid neocarzinostatin treatment for hepatocellular carcinoma. Curr. Med. Chem. Anticancer Agents 2, 715–726. [DOI] [PubMed] [Google Scholar]
- Biggins JB, Onwueme KC, and Thorson JS (2003). Resistance to enediyne antitumor antibiotics by CalC self-sacrifice. Science 301, 1537–1541. [DOI] [PubMed] [Google Scholar]
- Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, and Piddock LJV (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol 13, 42–51. [DOI] [PubMed] [Google Scholar]
- Cameron AD, Olin B, Ridderstrom M, Mannervik B, and Jones TA (1997). Crystal structure of human glyoxalase I-evidence for gene duplication and 3D domain swapping. EMBO J 16, 3386–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yu D, Zhang C, Shang B, He H, Chen J, Zhang H, Zhao W, Wang Z, Xu X, et al. (2015). Lidamycin inhibits tumor initiating cells of hepatocellular carcinoma Huh7 through GSK3b/b-catenin pathway. Mol. Carcinog 54, 1–8. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Kim K, Sohn SY, Cho HY, Kim KJ, Kim MH, Kim D, Kim E, and Kang BS (2010). Substrate binding mechanism of a type I extradiol dioxygenase. J. Biol. Chem 285, 34643–34652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Wang H, Taylor T, Warabi K, Huang XH, and Andersen RJ (2005). Uncialamycin, a new enediyne antibiotic. Org. Lett 7, 5233–5236. [DOI] [PubMed] [Google Scholar]
- Dumas P, Bergdoll M, Cagnon C, and Masson JM (1994). Crystal structure and site-directed mutagenesis of a bleomycin resistance protein and their significance for drug sequestering. EMBO J 13, 2483–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edo K, Mizugaki M, Koide Y, Seto H, Furihata K, Otake N, and Ishida N (1985). The structure of neocarzinostatin chromophore possessing a novel bicyclo-[7,3,0]dodecadiyne system. Tetrahedron Lett 26, 331–334. [Google Scholar]
- Elshahawi SI, Ramelot TA, Seetharaman J, Chen J, Singh S, Yang Y, Pederson K, Kharel MK, Xiao R, Lew S, et al. (2014). Structure-guided functional characterization of enediyne self-sacrifice resistance proteins, CalU16 and CalU19. ACS Chem. Biol 9, 2347–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fillgrove KL, Pakhomova S, Newcomer ME, and Armstrong RN (2003). Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms.J. Am. Chem. Soc 125, 15730–15731. [DOI] [PubMed] [Google Scholar]
- Galm U, Hager MH, Van Lanen SG, Ju J, Thorson JS, and Shen B (2005). Antitumor antibiotics: bleomycin, enediynes, and mitomycin. Chem. Rev 105, 739–758. [DOI] [PubMed] [Google Scholar]
- Gao Q, and Thorson JS (2008). The biosynthetic genes encoding for the production of the dynemicin enediyne core in Micromonospora chersina ATCC53710. FEMS Microbiol. Lett 282, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. (2017). Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, and Whalen KL (2015). Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, and Gautier C (1982). Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res 10, 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B, Challis GL, Fowler K, Kieser T, and Chater KF (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100, 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA, Kieser T, Wright H, and Bibb MJ (1983). Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J. Gen. Microbiol 129, 2257–2269. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kumagai T, Muta K, Matoba Y, Davies J, and Sugiyama M (2000). The 1.5Å crystal structure of a bleomycin resistance determinant from belomycin-producing Streptomyces verticillus. J. Mol. Biol 28, 915–925. [DOI] [PubMed] [Google Scholar]
- Kennedy DR, Ju J, Shen B, and Beerman TA (2007). Designer enediynes generate DNA breaks interstrand cross-links, or both, with concomitant changes in the regulation of DNA damage responses. Proc. Natl. Acad. Sci. USA 104, 17632–17637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Ohkuma H, Matsumoto K, Tsuno T, Kamei H, Miyaki T, Oki T, Kawaguchi H, VanDuyne GD, and Clardy J (1989). Dynemicin A, a novel antibiotic with the anthraquinone and 1,5-diyn-3-ene subunit. J. Antibiot. (Tokyo) 42, 1449–1452. [DOI] [PubMed] [Google Scholar]
- Leslie AG (2006). The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr 62, 48–57. [DOI] [PubMed] [Google Scholar]
- Li W, Li X, Huang T, Teng Q, Crnovcic I, Rader C, and Shen B (2016). Engineered production of cancer targeting peptide (CTP)-containing C-1027 in Streptomyces globisporus and biological evaluation. Bioorg. Med. Chem 24, 3887–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, and Shen B (2000). Genes for production of the enediyne antitumor antibiotic C-1027 in Streptomyces globisporus are clustered with the cagA gene that encodes the C-1027 apoprotein. Antimicrob. Agents Chemother 44, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TW, Dauter Z, Devedjiev Y, Sheffield P, Jelen F, He M, Sherman DH, Otlewski K, Derewenda ZS, and Derewenda U (2002). Molecular basis of mitomycin C resistance in Streptomyces: structure and function of the MRD protein. Structure 10, 933–942. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Kumagai T, Matoba Y, Hayashida M, Fujii T, Hata Y, and Sugiyama M (2001). Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.J. Biol. Chem 276, 9992–9999. [DOI] [PubMed] [Google Scholar]
- McCarthy AA, Baker HM, Shewry SC, Patchett ML, and Baker EN (2001). Crystal structure of methylmalonyl-coenzyme A epimerase fromP. shermanii: a novel enzymatic function on an ancient metal binding scaffold. Structure 9, 637–646. [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, and Chruszcz M (2006). HKL-3000: the integration of data reduction and structure solution – from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr 62, 859–866. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, and Dodson EJ (1997). Refinement of macro-molecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Penders J, Stobberingh EE, Savelkoul PHM, and Wolffs PFG (2013). The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol 4, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang RG, Ginell SL, Duke N, Naday I, Lazarz J, et al. (2006). The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J. Synchrotron Radiat 13, 30–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf JD, Yan X, and Shen B (2016). Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and priori-tization for discovery. J. Ind. Microbiol. Biotechnol 43, 261–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Hindra, Yan X, Huang T, Ge H, Yang D, Tang D, Rudolf JD, and Lohman JR (2015). Enediyne: exploration of microbial genomics to discover new anticancer drug leads. Bioorg. Med. Chem. Lett 25, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer MOA, Dantas G, and Church GM (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Kumagai T, Hayashida M, Maruyama M, and Matoba Y (2002). The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus. J. Biol. Chem 277, 2311–2320. [DOI] [PubMed] [Google Scholar]
- Surette MD, and Wright GD (2017). Lessons from the environmental antibiotic resistome. Annu. Rev. Microbiol 71, 309–329. [DOI] [PubMed] [Google Scholar]
- Tao M, Wang L, Wendt-Pienkowski E, George NP, Galm U, Zhang G, Coughlin JM, and Shen B (2007). The tallysomycin biosynthetic gene cluster from Streptoalloteichus hindustanus E465–94 ATCC 31158 unveiling new insights into the biosynthesis of the bleomycin family of antitumor antibiotics. Mol. Biosyst 3, 60–74. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, and Hung LW (2009). Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr 65, 582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MK, Keithly ME, Goodman MC, Hammer ND, Cook PD, Jagessar KL, Harp J, Skaar EP, and Armstrong RN (2014). Structure and function of the genomically encoded fosfomycin resistance enzyme, FosB, from Staphylococcus aureus. Biochemistry 53, 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MK, Keithly ME, Harp J, Cook PD, Jagessar KL, Sulikowski GA, and Armstrong RN (2013). Structural and chemical aspects of resistance to the antibiotic fosfomycin conferred by FosB from Bacillus cereus. Biochemistry 52, 7350–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, and Teplyakov A (2010). Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr 66, 22–25. [DOI] [PubMed] [Google Scholar]
- Van Lanen SG, and Shen B (2008). Biosynthesis of enediyne antitumor antibiotics. Curr. Top. Med. Chem 8, 448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, and Bricogne G (2011). Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr 67, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Chen JJ, Adhikari A, Yang D, Crnovcic I, Wang N, Chang CY, Rader C, and Shen B (2017). Genome mining of Micromonospora yangpuensis DSM 45577 as a producer of an anthraquinone-fused enediyne. Org. Lett 19, 6192–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Ge H, Huang T, Hindra, Yang D, Teng Q, Crnovcic I, Li X, Rudolf JD, Lohman JR, et al. (2016). Strain prioritization and genome mining for enediyne natural products. MBio 7, e02104–e02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Sakai A, Zhang X, Vetting MW, Kumar R, Hillerich B, San Francisco B, Solbiati J, Steves A, Brown S, et al. (2014). Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks. Elife 3, e03275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.