Abstract
Breast cancer is a heterogeneous disease and one of the most common cancers among women. Recently, microRNAs (miRNAs) have been used as biomarkers due to their effective role in cancer diagnosis. This study proposes a support vector machine (SVM)-based classifier SVM-BRC to categorize patients with breast cancer into early and advanced stages. SVM-BRC uses an optimal feature selection method, inheritable bi-objective combinatorial genetic algorithm, to identify a miRNA signature which is a small set of informative miRNAs while maximizing prediction accuracy. MiRNA expression profiles of a 386-patient cohort of breast cancer were retrieved from The Cancer Genome Atlas. SVM-BRC identified 34 of 503 miRNAs as a signature and achieved a 10-fold cross-validation mean accuracy, sensitivity, specificity, and Matthews correlation coefficient of 80.38%, 0.79, 0.81, and 0.60, respectively. Functional enrichment of the 10 highest ranked miRNAs was analysed in terms of Kyoto Encyclopedia of Genes and Genomes and Gene Ontology annotations. Kaplan-Meier survival analysis of the highest ranked miRNAs revealed that four miRNAs, hsa-miR-503, hsa-miR-1307, hsa-miR-212 and hsa-miR-592, were significantly associated with the prognosis of patients with breast cancer.
Introduction
Breast cancer is one of the major leading causes of death among women, and it accounts for 14% of cancer deaths worldwide1,2. There are different types of breast carcinomas depending on the specific cells in the breast that are affected; most breast cancers are a type of adenocarcinoma. According the American Joint Committee on Cancer, the three features used to stage breast cancer are the size of the primary breast tumour (T), the spread of cancer to lymph nodes (N) and distant metastasis (M)3. In the TNM staging system, the T category represents the primary breast tumour and the spread within the tumour. The T category comprises stages T1 to T4 based on the tumour size. T1 tumours are subdivided into T1a, T1b and T1c, and the tumour size is >10 mm and ≤2 cm in dimension. T2 tumours are >2 cm, T3 tumours are >5 cm, and T4 tumours are any size and may spread to the breast skin or chest wall3. The estimated numbers of invasive and in situ breast cancer cases and breast cancer deaths in 2013 in the United States are 32,340, 64,640 and 39,620, respectively4. Approximately 252,710 new cases and 40,610 breast cancer deaths are estimated for US women in 2017 according to the surveillance, epidemiology, and end result programme (SEER 2017) statistics. Breast cancer survival rates are associated with the stage of the cancer. The 5-year survival rates for stages I, II and III are 100%, 93%, and 72%, respectively; unfortunately, the 5-year survival rate for stage IV breast cancer is only 22%5. Despite the advances in the treatment of breast cancer, metastatic breast cancer remains incurable, and mortality rate is still high due to the emergence of therapy-resistant cancer cells6 and limitations in the current treatment strategies. A better understanding of the molecular markers that affect breast tumours at different stages may lead to the development of new therapeutic strategies.
Recent evidence demonstrated that molecular marker-based targeted therapies have potential for the prognosis and diagnosis of various diseases. Molecular target-based studies focused on advances in microRNA (miRNA) expression profiling because of their prominent role in tumour development and metastasis. MiRNAs are small noncoding RNAs that regulate gene expression and are involved in human carcinogenesis7. Over the past few years, many studies reported the significant role of miRNAs in the molecular pathogenesis of breast tumours. MiRNA profiling studies have identified miRNAs that are aberrantly expressed in breast tumours and their functions. For instance, miRNAs such as miR-125b, miR-145, miR-155, and miR-21 are significantly deregulated in breast tumour tissues compared to normal tissue8. Potential association between miRNA and breast neoplasm has been predicted in studies9,10. Functionally, miRNAs are act as tumour suppressor11 and oncogene12 in breast tumour progression and metastasis. Gene expression and miRNA expression profiling has been used to classify different tumour types13,14. However, it has been confirmed that miRNA expression profiles can classify tumour types more accurately than gene expression profiles14.
Machine learning methods have been developed for cancer survival calculation, risk classification and prognosis prediction in various cancers, including breast cancer15–17. Several researchers have used different machine learning models and the Wisconsin breast cancer dataset to categorize benign and malignant breast cancers. For instance, M.F. Akay has used a support vector machine (SVM) combined with feature selection for a medical decision making system to diagnose breast cancer18. Abonyi and Szeifert have used a supervised rule-based fuzzy classifier to categorize benign and malignant breast cancers19. Pena-Reyes and Sipper have utilized a fuzzy-genetic algorithm method to classify benign and malignant breast tumours20. In addition, other well-known machine learning methods, such as the feed forward neural network algorithm21, the C4.5 decision tree method22, the linear discreet analysis method23 and the neuron-fuzzy technique24, have been developed for breast cancer diagnosis. Most machine learning methods developed for breast cancer classification using breast tumor images25 and gene/miRNA expression profiles26 to distinguish molecular subtypes27. Shimomura et al. identified five miRNAs to distinguish the breast cancer from other cancer types28. However, there are few studies of identifying the miRNA signature associated with the breast cancer stage for exploring the molecular level changes at various breast cancer stages.
Although there are methodologies for breast cancer treatment, challenges regarding early stage detection of breast tumours exist. Early stage detection may help to obtain a better treatment diagnosis. Therefore, we explored whether miRNA expression profiling could be used to categorize early stage breast tumours accurately. In this study, we collected the breast cancer data from the cancer genome atlas (TCGA) database and proposed a SVM-based classifier called SVM-BRC to categorize early stage and advanced stage patients with breast cancer using their miRNA expression profiles. SVM-BRC is based on an SVM incorporating an optimal feature selection method referred to as the inheritable bi-objective combinatorial genetic algorithm (IBCGA)29. We retrieved the miRNA expression profile data on 386 patients with breast cancer, with 193 patients in the early stage and the remaining 193 patients at an advanced stage groups. To the best of our knowledge, this is the first study to use miRNA expression profiles to identify the miRNA signature for predicting the breast cancer stage. SVM-BRC identified a signature consisting of 34 of 503 miRNAs that can distinguish early stage breast cancer patients from advanced stage breast cancer patients and achieved a 10-fold cross-validation (10-CV) mean accuracy, sensitivity, specificity, and Matthews correlation coefficient (MCC) of 80.38%, 0.79, 0.81, and 0.60, respectively. Further, we ranked the identified miRNAs based on the MED scores. The 10 highest ranked miRNAs were analysed based on their involvement in breast cancer and other cancer types. Functional enrichment of the 10 highest ranked miRNAs were analysed using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) annotations. Kaplan-Meier survival analysis of the identified miRNAs revealed that four miRNAs among the 10 highest ranked miRNAs, hsa-miR-503, hsa-miR-1307, hsa-miR-212 and hsa-miR-592, were significantly associated with the overall survival of patients with breast cancer.
Results and Discussion
Prediction performance of SVM-BRC
We used a dataset consisting of 386 patients with breast cancer and 503 miRNA expression profiles. The dataset was divided into early stage (Stages I & II) and advanced stage (Stages III & IV) groups. Then, we attempted to categorize the early stage and the advanced stage groups using miRNA expression alone. The proposed SVM-BRC includes the feature selection algorithm IBCGA to select a significant miRNA signature that is associated with the tumour stage of breast cancer patients. SVM-BRC identified a miRNA signature (34 miRNAs) that can classify early stage and advanced stage groups and achieved a 10-CV mean accuracy, sensitivity, specificity, and MCC of 80.38% ± 1.55%, 0.79 ± 2.7, 0.81 ± 2.26, and 0.60 ± 0.03, respectively. SVM-BRC achieved a 10-CV accuracy, sensitivity, specificity, MCC and AUC of 83.16%, 0.84, 0.81, 0.66 and 0.87, respectively (shown in Table 1), and a jackknife test accuracy of 63.89%. The prediction performance of SVM-BRC was evaluated using a receiver operating curve (ROC), as shown in Fig. 1.
Table 1.
Comparison of SVM-BRC with the some classifiers for the 386-patient breast cancer cohort.
| Method | 10-CV accuracy (%) | Sensitivity | Specificity | MCC |
|---|---|---|---|---|
| SVM-BRC-Mean | 80.38 ± 1.55 | 0.79 ± 2.7 | 0.81 ± 2.26 | 0.60 ± 0.03 |
| SVM-BRC-Best | 83.16 | 0.84 | 0.81 | 0.66 |
| Random forest | 66.83 | 0.66 | 0.67 | 0.33 |
| Multilayer perceptron | 57.25 | 0.57 | 0.57 | 0.14 |
| SMO | 62.69 | 0.62 | 0.63 | 0.25 |
| Naïve Bayes | 64.50 | 0.63 | 0.65 | 0.29 |
| Decision tree | 50.25 | 0.50 | 0.50 | 0.01 |
Figure 1.
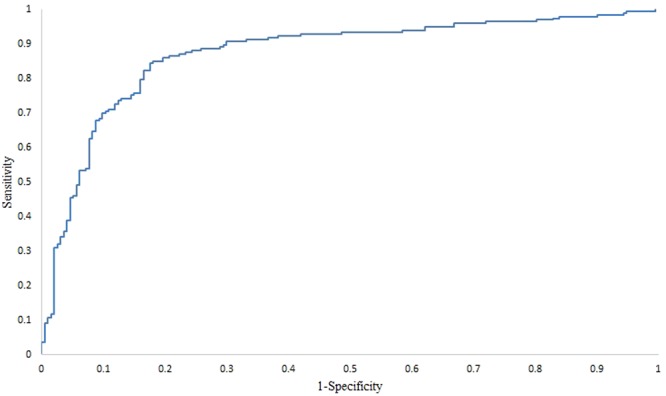
SVM-BRC performance evaluation using the ROC curve. The area under the ROC curve is 0.87 using a 386-patient breast cancer cohort.
We compared SVM-BRC with some machine learning methods of Weka such as Random forest (RF), Multilayer perceptron (MLP), Sequential minimal optimization (SMO), Naïve Bayes, and Decision tree. We used information gain for feature selection and Ranker attribute evaluator method, and obtained 14 miRNAs to distinguish early stage and advanced stage groups. The accuracies of RF, MLP, SMO, Naïve Bayes, and Decision tree methods using the 14 miRNAs with 10-CV were 66.83%, 57.25%, 62.69%, 64.50%, and 50.25% respectively. The results of performance comparison are shown in Table 1. The performance of SVM-BRC is much better than the other machine learning methods in distinguish the early stage and advanced stage groups.
Prioritizing the miRNA signature
We ranked the miRNAs identified by SVM-BRC using main effect difference (MED) analysis30. The 10 highest ranked miRNAs based on their contribution to the prediction accuracy are hsa-miR-200c, hsa-miR-503, hsa-miR-1307, hsa-miR-361, hsa-miR-212, hsa-miR-592, hsa-miR-1185-1, hsa-miR-146b, hsa-miR-1468, and hsa-miR-769. The 10 highest ranked miRNAs and their MED scores are listed in Table 2. The 34 miRNA signature and their rankings are shown in Supplementary Table 1. Further, the significance of the 10 highest ranked miRNAs in breast cancer is discussed.
Table 2.
Ten highest ranked miRNAs and feature knockout analysis of individual miRNAs.
| Rank | miRNA | MED scores | Accuracy difference (%) |
|---|---|---|---|
| 1 | hsa-miR-200c | 69.68 | 20.99 |
| 2 | hsa-miR-503 | 65.02 | 20.73 |
| 3 | hsa-miR-1307 | 48.44 | 21.25 |
| 4 | hsa-miR-361 | 47.92 | 21.25 |
| 5 | hsa-miR-212 | 46.89 | 20.99 |
| 6 | hsa-miR-592 | 46.89 | 19.95 |
| 7 | hsa-miR-1185-1 | 43.26 | 20.73 |
| 8 | hsa-miR-146b | 43.26 | 19.69 |
| 9 | hsa-miR-1468 | 34.45 | 21.25 |
| 10 | hsa-miR-769 | 30.82 | 20.47 |
Hsa-miR-200c
Hsa-miR-200c scored 69.68 and ranked one according to the MED ranking index, which means that the contribution of this miRNA is higher than that of the others. The miR-200 family of miRNAs possesses a unique role in cancer stem cells31, neurogenesis32, and chemosensitivity33. Hsa-miR-200c is aberrantly expressed in several cancers, including breast cancer. A retrospective analysis of 210 breast tumour samples revealed that hsa-miR-200c expression was associated with poor distant relapse-free survival34. A luciferase reporter assay study reported that hsa-miR-200c regulates cancer stem cell functions such as proliferation and self-renewal; miR-200c modulates the expression of the BM1 protein, which is an essential stem cell self-renewal regulator in breast cancer stem cells35. It is also observed that hsa-miR-200c suppresses the tumourigenicity of breast cancer stem cells35. This miRNA targets class III beta tubulin and increases the chemosensitivity in breast tumours33. Hsa-miR-200c is also significantly expressed in several other tumours, such as bladder cancer36, colorectal cancer37 and ovarian cancer38.
Hsa-miR-503
Hsa-miR-503 expression was found to be downregulated in breast cancer cells, and overexpression of this miRNA reduced cell proliferation by targeting CCND139. A quantitative RT-PCR study involving screening a series of 12 inflammatory breast cancer cells showed that hsa-miR-503 was differently expressed and was used as a predictor for an inflammatory breast cancer phenotype40. Recently, overexpression of hsa-miR-503 was found in breast cancer tissue and plasma compared to that in healthy tissue; upregulation of this miRNA in breast cancer cells suppresses the expression of the epithelial-mesenchymal transition-related protein SMAD2 and the epithelial marker protein E-cadherin41. Experimental evidence showed that hsa-miR-503 regulates the oncogene ZNF217 and that higher expression of this miRNA is associated with improved survival in breast cancer42. Hsa-miR-503 acts as a tumour suppressor by targeting DDHD2 in breast cancer cells43.
Hsa-miR-1307
Hsa-miR-1307 was found to be upregulated in breast cancer. Hsa-miR-1307 was differentially expressed with a fold-change of 0.36 between breast cancer and the adjacent normal control tissue44. Hsa-miR-1307 expression was upregulated in BRCA1-associated breast carcinoma compared to that in the normal counterparts45.
Hsa-miR-361
A miRNA expression profiling study of 376 human miRNAs reported that hsa-miR-361 expression was downregulated in MCF-7 docetaxel-resistant breast cancer cells46. A screening study of miRNAs related to different subtypes of breast cancers showed that hsa-miR-361 was upregulated in metastatic breast tumours47. A microarray-based study of 375 breast tumour cases revealed that overexpression of hsa-miR-361 is correlated with the better disease-free survival in patients with breast cancer48. Downregulation of hsa-miR-361 was observed in 60 breast cancer tissues; hsa-miR-361 targets FGFR1 and MMP-1, resulting in inhibition of glycolysis and invasion in breast cancer cells49.
Hsa-miR-212
A case study of patients diagnosed with breast invasive ductal carcinoma reported that hsa-miR-212 was significantly downregulated in breast tumours by 0.328-fold and that this reduced expression was prominent in high grade breast tumours50. Hsa-miR-212 expression was downregulated in 30 paired triple-negative breast cancer samples, and its expression inhibited cell migration and invasion during cancer progression by targeting Prrx251.
Hsa-miR-592
A real-time PCR study of a nonmetastatic breast cancer cell line reported the overexpression of hsa-miR-59252. Antonio Colaprico et al. identified differentially expressed miRNA-regulating pathway crosstalk between breast cancer and healthy samples; hsa-miR-592 expression was approximately twenty-three times higher in breast cancer samples than in healthy samples and regulated the extrinsic prothrombin activation pathway53. Recently, marked downregulation of hsa-miR-592 was observed in a breast cancer cell line compared to that in a normal breast cell line and further, hsa-miR-592 acted as tumour suppressor by targeting the transforming growth factor β-2 in breast cancer54.
Hsa-miR-146b
Hsa-miR-146b was downregulated and negatively regulated nuclear factor-kappaB, resulting in a reduction of the metastatic potential in breast cancer cells55. Higher expression of hsa-miR-146b induced interleukin-6 expression and signal transducer and activator transcription 3 phosphorylation, and this expression was positively correlated with survival in some breast cancer subtypes56. Hsa-miR-146a and hsa-miR-146b were found to be the most expressed in breast cancer metastasis suppressor 1-expressing cells, and upregulation of hsa-miR-146b was observed in the MDA-MB-435 breast cancer cell line57. A reporter assay study of triple negative breast tumours reported that hsa-miR-146b negatively regulates BRCA1 in triple negative sporadic breast cancer58. An RT-PCR study of 120 young women with primary breast tumours and 130 patients with breast fibroadenoma reported that downregulation of hsa-miR-146b expression in breast cancer cells was associated with the development and deterioration of breast cancer59.
Hsa-miR-769
Examination using the Nanostring nCounter assay on 43 miRNAs reported that hsa-miR-769 can inhibit the expression of N-myc downstream-regulated gene 1 upon reoxygenation in the breast adenocarcinoma cell line MCF-7 and that overexpression of hsa-miR-769 significantly enhanced apoptosis60. A study of triple negative breast cancer comparing African-American and non-Hispanic white women reported that 26 miRNAs, including hsa-miR-769, were differentially expressed between these groups61. Hsa-miR-769 found to be upregulated with a log2-fold change of 1.355 between triple negative breast cancer in African-American and non-Hispanic white women61. Differential expression of hsa-miR-769 was also found in male breast cancers62,63.
Our analysis of the 10 highest ranked miRNAs acknowledged that two miRNAs, hsa-miR-1185-1 and hsa-miR-1468, among the 10 highest ranked miRNAs are not directly involved in breast cancer but are implicated in other cancers. For instance, hsa-miR-1185-1 expression was abnormally low in Alzheimer’s disease64 and atherosclerosis65. The expression of hsa-miR-1468 was upregulated in hepatocellular carcinoma tissue66. Dysregulation of hsa-miR-1468 was observed in epithelial ovarian cancer67. Hsa-miR-1468 was significantly associated with the recurrence-free survival in lung adenocarcinoma68. Therefore, these two miRNAs are important molecules to validate further in breast cancer. Eight miRNAs among the 10 highest ranked miRNAs are involved not only in breast cancer but also in several major cancer types.
Additionally, we employed miRNA knockout analysis to observe the difference in the prediction performance by removing one miRNA from the signature. Each miRNA of the 10 highest ranked miRNAs can affect the prediction performance with a mean accuracy difference of 20.73 ± 0.54. We report the results of knockout of the 10 highest ranked miRNAs in Table 2. The accuracy difference after removing each miRNA is depicted in Fig. 2. The accuracy differences obtained from feature knockout analysis for 34 miRNA signature are shown in Supplementary Table 1.
Figure 2.
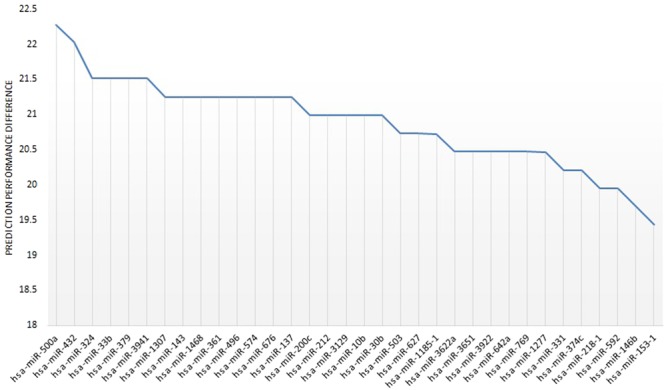
Feature knockout analysis. Prediction performance difference for individual miRNAs using feature knockout analysis.
Difference of expression profiles between early stage and advanced stage groups
We measured expression levels of the 10 highest ranked miRNAs in early stage and advanced stage groups. We observed a slight expression difference between early and advanced stage groups for 10 highest ranked miRNAs. Of the 10 highest ranked miRNAs, the mean expression values of hsa-miR-200c, hsa-miR-503, hsa-miR-1307, hsa-miR-361, hsa-miR-212, hsa-miR-592, hsa-miR-1185-1, hsa-miR-146b, hsa-miR-1468, and hsa-miR-769 are 13.34 ± 0.94, 3.44 ± 1.28, 10.16 ± 1.04, 8.35 ± 0.57, 2.20 ± 0.83, 1.93 ± 1.11, 0.24 ± 0.39, 9.03 ± 0.94, 2.50 ± 1.10, and 4.88 ± 0.70, respectively, in the early stage group, and 13.28 ± 0.77, 3.80 ± 1.39, 9.93 ± 1.12, 8.30 ± 0.55, 2.20 ± 0.80, 1.80 ± 1.11, 0.35 ± 0.39, 9.20 ± 0.96, 2.45 ± 1.06, and 4.75 ± 0.77, respectively, in the advanced stage group. Box-plot representation of expression difference in the early stage and advanced stage groups is given for the signature of 34 miRNAs in Supplementary Fig. 1.
KEGG pathway enrichment analysis
To investigate the functional mechanism of the 10 highest ranked miRNAs, we employed KEGG pathway analysis using the DIANA-mirPath v.3 web server69. The 10 highest ranked miRNAs are significantly enriched in pathways involving fatty acid biosynthesis, fatty acid metabolism, adherens junction, protein processing in endoplasmic reticulum, cytokine-cytokine interaction, bacterial invasion of epithelial cells, spliceosome, and proteoglycans in cancer. The significantly enriched in KEGG pathways for the 10 highest ranked miRNAs and the target genes involved in each pathway are listed in Table 3. The 10 highest ranked miRNAs and the number of targeted genes are shown in Fig. 3. A detailed summary of the 10 highest ranked miRNAs, the enriched KEGG pathways and the number of targeted genes is provided in Supplementary Table 2.
Table 3.
Enriched KEGG pathways and the corresponding target genes for the 10 highest ranked miRNAs.
| KEGG pathway | p-value | Target genes |
|---|---|---|
| Fatty acid biosynthesis (hsa00061) | <1e-325 | FASN |
| Fatty acid metabolism (hsa01212) | <1e-325 | FASN TECR ACOX1 |
| Adherens junction (hsa04520) | 4.47E-06 | TGFBR1, MET, WASL, SMAD2, ACTG1, IQGAP1, IGF1R, VCL, RHOA, TJP1, MLLT4, CDH1, CTNNB1, CTNNA1, WASF2, ACTN4, CREBBP |
| Protein processing in endoplasmic reticulum (hsa04141) | 0.00083483 | HSPA1A, EIF2AK1, SSR1, RAD23B, AMFR, UGGT1, YOD1, SEL1L, HSP90AA1, DNAJC10, UBE2E2, STT3B, HSPH1, PDIA6, RAD23A, PRKCSH, VCP, HSPA8, LMAN1, RPN2, DERL1, HSPA1B |
| Cytokine-cytokine receptor interaction (hsa04060) | 0.002767508 | IL6ST |
| Bacterial invasion of epithelial cells (hsa05100) | 0.01255968 | ARPC5L, MET, ITGB1, WASL, SEPT11, ACTG1, VCL, RHOA, CD2AP, CDH1, CLTA, WASF2, FN1, ARPC2 |
| Spliceosome (hsa03040) | 0.02884541 | RBM25, HSPA1A, HNRNPA1, DDX23, PPIL1, U2SURP, PRPF8, SRSF1, HNRNPM, DHX15, HSPA8, DHX16, SRSF3, HSPA1B, SNRPC, SNRNP200, SRSF8 |
| Proteoglycans in cancer (hsa05205) | 0.03666157 | PDCD4, MET, ITGB1, EZR, ARHGEF12, ACTG1, FRS2, IQGAP1, RHOA, ERBB3, ITGAV, LUM, HOXD10, FN1, MAP2K1, SDC4, TWIST1, VEGFA, MDM2, SMAD2, WNT5A, PPP1CC, ACTG1, TIAM1, IGF1R, AKT2, PTK2, CTNNB1, ITGA2, DDX5, GAB1 |
Figure 3.
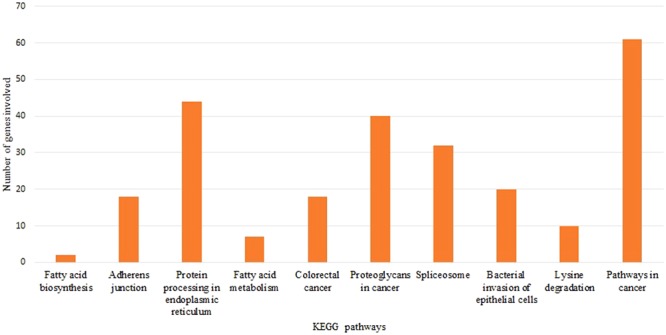
KEGG pathway analysis of the 10 highest ranked miRNAs.
Most of the 34 miRNAs are prevalently involved in the biological pathways. For example, 30 miRNAs of the signature are significantly involved in the RAS signalling pathway, cGMP-signaling pathway, and cancer pathways by targeting 123, 90 and 229 genes, respectively. There are 29 miRNAs significantly involved in focal adhesion, PI3K-Akt signaling pathway, MAPK signaling pathway, and viral carcinogenesis. There are 28 miRNAs in proteoglycans in cancer pathway, ErbB signaling pathway, cAMP signaling pathway, and estrogen signaling pathway to name a few. Details of the miRNA signature involved in biological pathways and their targeted genes are listed in Supplementary Table 3.
Gene ontology analysis
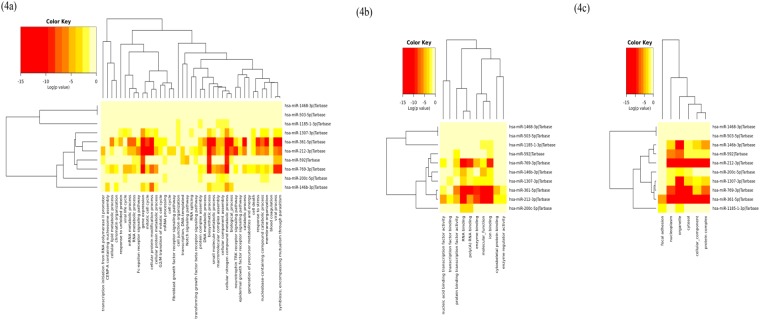
The biological significance of the 10 highest ranked miRNAs was analysed using GO annotations at three levels, includes biological process, molecular functions and cellular component. The 10 highest ranked miRNAs were highly enriched in five biological processes: mitotic cell cycle, cellular protein modification process, viral process, small molecule metabolic process, and symbiosis, encompassing mutualism through parasitism. The 10 highest ranked miRNAs were highly enriched in the molecular functions enzyme binding, RNA binding, and poly(A) RNA binding; the significantly enriched cellular components include protein complex, nucleoplasm, cytosol, organelle and focal adhesion. The enriched biological processes, molecular function and cellular components of the 10 highest ranked miRNAs are shown in Fig. 4(a–c). GO analysis of the 10 highest ranked miRNAs and the targeted genes for biological process, molecular function and cellular component are listed in Supplementary Tables 4, 5 and 6 respectively.
Figure 4.
Gene ontology (GO) annotations for the 10 highest ranked miRNAs. GO enrichment analysis was performed for the 10 highest ranked miRNAs at three levels: biological process (a), molecular functions (b), and cellular component (c).
Survival analysis of the top ranked miRNAs
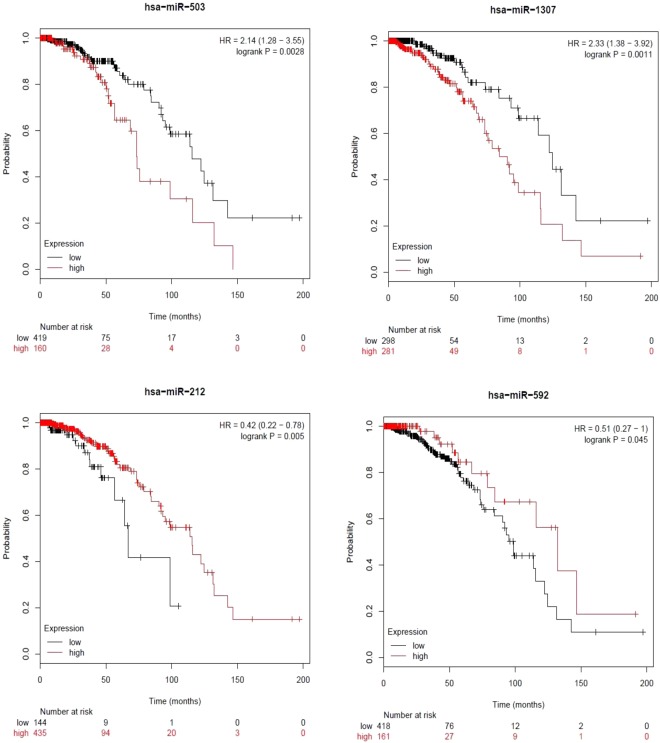
Survival analysis was performed using Kaplan-Meier plotter70 to validate the prognostic value of the top ranked miRNAs. We selected the TCGA dataset and systematically evaluated the patient data using the Kaplan-Meier survival analysis. Four of the 10 highest miRNAs, hsa-miR-503, hsa-miR-1307, hsa-miR-212 and hsa-miR-592, were significantly associated with the prognosis of patients with breast cancer. These four miRNAs, hsa-miR-503, hsa-miR-1307, hsa-miR-212 and hsa-miR-592, obtained P-values of 0.0028, 0.0011, 0.005, and 0.045, respectively, and hazard ratios of 2.14, 2.33, 0.42 and 0.51, respectively, between the high and low expression groups. The Kaplan-Meier survival curves for the four miRNAs are shown in Fig. 5.
Figure 5.
Kaplan-Meier plots of hsa-miR-503, hsa-miR-1307, hsa-miR-212, and hsa-miR-592 for the systemically treated breast cancer cohort.
To confirm the association between the four miRNAs with overall survival, we utilized the METABRIC dataset. Two of the four miRNAs show significant association with prognosis in patients with breast cancer. Two miRNAs, hsa-miR-503 and hsa-miR-1307, obtained P-values of 0.046 and 0.0031, respectively, and hazard ratios of 0.82 and 1.37, respectively, between the high and low expression groups. Whereas another two miRNAs, hsa-miR-212 and hsa-miR-592, obtained P-values of 0.16 and 0.35, respectively, and hazard ratios of 0.87 and 0.9, respectively, between the high and low expression groups.
Another four of the 10 highest miRNAs, hsa-miR-200c, hsa-miR-1185, hsa-miR-146b and hsa-miR-769, were significantly associated with the prognosis of patients with breast cancer. These four miRNAs, hsa-miR-200c, hsa-miR-1185, hsa-miR-146b, and hsa-miR-769, obtained P-values of 0.00017, 1.4e-05, 0.0018, and 0.0078, respectively, and hazard ratios of 1.49, 0.6, 0.73, and 0.76, respectively, between the high and low expression groups. The Kaplan-Meier survival curves for the four miRNAs are shown in Supplementary Fig. 2.
Additionally, we estimated overall survival of the breast cancer patients using Multiple linear regression71, and observed that correlation between these four miRNAs and overall survival is better in the advanced stage group when compared to the early stage group. The correlation coefficient between actual and overall survival in early stage and advanced stage groups is 0.26 and 0.40, respectively. The correlation plots are shown in Supplementary Fig. 3.
Conclusions
The challenges for early stage detection of breast cancer are that breast cancer is a heterogeneous disease with the potential for metastatic spreading at an early stage. Detecting cancer at a treatable stage and removing the lesions can prevent the development of lethal invasive cancers and would prevent death from breast cancer. Currently, it is widely reported that miRNAs can be potential biomarkers for various cancers. Identifying the disease-related miRNAs aids to improve the understanding of pathogenesis and diagnosis. Hence, various potential computational models have been developed to investigate the miRNA disease-association9,72–74. However, only a few studies focused on identifying a miRNA signature for the early stage detection of breast cancer. Accordingly, in this study, we proposed a novel miRNA-based classification method to categorize the early stage and the advanced stages of breast cancer. Recent development of personalized medicine and growing trend in applications of machine learning techniques improved the prognosis and cancer prediction. Various machine learning methods and feature selection algorithms have been widely used to identify the important factors that influence cancer progression, cancer recurrence, and cancer survival. Generally, machine learning based cancer prediction studies used mRNA/miRNA expression profiles, histological variables and clinical factors as input to the cancer prediction procedure75–77. Success in developing computational models for cancer predictions depends on understanding of biological knowledge and limitations of the training data set such as a small set of high-dimensional samples called “curse of dimensionality”78. However, the over-training problem can be coped with proper feature selection and cross-validation methods.
Hence, we proposed an SVM-based classifier called SVM-BRC that incorporated the feature selection method IBCGA to identify a miRNA signature that can distinguish early stage from advanced stage breast cancer. SVM-BRC identified a 34-miRNA signature and obtained a 10-CV accuracy, sensitivity, specificity, MCC and AUC of 83.16% 0.84, 0.81, 0.66 and 0.87, respectively. SVM-BRC obtained an average training accuracy of 80.38% ± 1.55%. Further, we ranked the identified miRNAs using MED scores. The significance of the 10 highest ranked miRNAs was validated using the literature. The importance of the top-10 miRNAs in breast cancer progression and other cancers is discussed. The prediction performance difference was measured for the 10 highest ranked miRNAs using feature knockout analysis. The functional mechanisms of the 10 highest ranked miRNAs were analysed using KEGG pathway enrichment and GO enrichment at three levels, including biological process, molecular functions, and cellular components. Survival analysis of the highest ranked miRNAs in the breast cancer cohort using the Kaplan-Meier curve revealed that four miRNAs, hsa-miR-503, hsa-miR-1307, hsa-miR-212, and hsa-miR-592, among the top-10 miRNAs were significantly (P ≤ 0.05) associated with the prognosis of breast cancer. We hope that our findings will help to improve the early stage detection methodologies by using the miRNA signature as a biomarker of breast cancer.
Materials and Methods
Dataset
The miRNA expression profiles of breast cancer cohort obtained from the Illumina HiSeq 2000 miRNA sequencing platform were obtained from TCGA database. We considered only the patients who underwent radiotherapy or targeted molecular therapy. Further, we divided the patients into early stage and advanced stage based on their pathological condition. After the filtering, the final balanced dataset contained 386 patients, with 193 patients in the early stage group and 193 in the advanced stage group, along with 503 miRNA expression profiles.
SVM-BRC
Support vector machines (SVMs) are based on statistical learning theory79. The main idea of an SVM is to find the optimal hyperplane between the two classes. SVMs have been used to solve biological problems due to their potential discriminating ability. SVMs have been widely used to detect tumour markers80 and to perform cancer predictions81. Thus, we proposed an SVM-based classifier SVM-BRC including the feature selection method IBCGA to categorize early stage and advanced stage groups with breast cancers. The general formulation of the SVM is
| 1 |
where w is vector of the hyperplane, C is the classifier parameter, Si are the variables and n = number of vectors in the training dataset.
Inheritable bi-objective combinatorial genetic algorithm (IBCGA)
To select a small set of miRNAs (signature) from a large number of expression profiles (503 miRNAs) we used a genetic algorithm (GA) based feature selection algorithm IBCGA29. The feature selection algorithm IBCGA uses an intelligent evolutionary algorithm82 to solve the large parameter optimization problem. IBCGA has been successfully applied in several bioinformatics problems, including the prediction of human ubiquitination sites83, the prediction of the regulatory roles of cyclic AMP receptor proteins84 and the estimation of survival time for cancer patients85,86.
In this study, we used IBCGA and identified a miRNA signature (m = 34 miRNAs) from a large number of miRNA expression profiles (n = 503 miRNAs) to distinguish the early stage and advanced stage groups with breast cancer. We used traditional terms of GA, GA-gene and GA-chromosome. The GA-chromosome of IBCGA consists of n binary GA-genes for feature selection and two 4-bit GA-genes for encoding parameters C and γ of SVM. Normalized miRNA expressions of patients with n miRNAs were used as input of IBCGA in designing the SVM-based classifier. The parameter setting of IBCGA was as follows: rstart = 10, rend = 50, Npop = 50, Gmax = 60, and r = rstart. We used the LibSVM package87 to implement SVM-BRC. The steps of IBCGA are as follows.
Step 1: (Initialization) Randomly generate a population of Npop individuals.
Step 2: (Evaluation) Evaluate the fitness value of all individuals using the fitness function that is the prediction accuracy in terms of 10-fold cross-validation (10-CV).
Step 3: (Selection) Use a tournament selection method that selects the winner from two randomly selected individuals to generate a mating pool.
Step 4: (Crossover) Select two parents from the mating pool to perform orthogonal array crossover operation.
Step 5: (Mutation) Apply a conventional mutation operator to the randomly selected individuals in the new population. Mutation is not applied to the best individuals to prevent the best fitness value from deterioration.
Step 6: (Termination test) If the stopping condition for obtaining the solution is satisfied, output the best individual as the solution. Otherwise, go to Step 3.
Step 7: (Inheritance) If r < rend, randomly change one bit in the binary GA-genes for each individual from 0 to 1; increase the number r by one, and go to Step 3. Otherwise, stop the algorithm.
Step 8: (Output) Obtain a set of m miRNAs from the GA-chromosome of the best individual.
Weka classifier
Weka has implementations of all major learning techniques for classification and regression methods. Some methods of Weka data mining software88 were used to compare SVM-BRC such as Random forest (RF), Multilayer perceptron (MLP), Sequential minimal optimization (SMO), Naïve Bayes, and Decision tree for classification to discriminate early stage and advanced stage groups with breast cancer.
We evaluated the prediction performance of SVM-BRC using the prediction accuracy (ACC), sensitivity (Sn), specificity (Sp), Matthews correlation coefficient (MCC), and area under the ROC curve (AUC).
| 2 |
| 3 |
| 4 |
| 5 |
where TP is true positive; TN is true negative; FP is false positive; and FN is false negative.
KEGG and GO term enrichment analysis
We used DIANA-mirPath v3.0 for KEGG pathway analysis. Fisher’s exact t-test was used for enrichment analysis89. GO term analysis was employed to determine the involvement of the 10 highest ranked miRNAs in biological process, molecular functions and cellular components using mirPath v3.0. The DIANA-Tarbase algorithm in the mirPath web server was used to predict the experimentally validated miRNA targets89.
Kaplan-Meier survival analysis
To identify the miRNAs associated with the prognosis of breast cancer patients, we employed Kaplan-Meier survival analysis using the mirPower-Kaplan-Meier plotter web-tool70. We selected TCGA breast cancer dataset, and the analysis was restricted to only patients systemically treated with chemotherapy.
Electronic supplementary material
Acknowledgements
This work was funded by Ministry of Science and Technology ROC under the contract numbers MOST 106-2627-M-009-003-, 106-3114-E-029-001-, 106-2218-E-009-031- and 106-2634-F-075-001 -, and was financially supported by the “Center For Intelligent Drug Systems and Smart Bio-devices (IDS2B)” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Yerukala Sathipati Srinivasulu (Y.S.S.) and Shinn-Ying Ho (S.Y.H.) designed the system and carried out the detailed study. Yerukala Sathipati Srinivasulu (Y.S.S.) participated in the design of the system, implemented programmes, and discussed the results. All authors participated in manuscript preparation and approved the final manuscript.
Data Availability
All the data used in this analysis can be found at TCGA data portal.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34604-3.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, et al. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Edge SB, Compton CC. The American Joint Committee on Cancer: the7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 4.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 5.Altekruse, S. SEER cancer statistics review, 1975–2007, http://seer.cancer. gov/csr/1975_2007/results_merged/sect_13_leukemia.pdf (2009).
- 6.Stockler M, Wilcken NR, Ghersi D, Simes RJ. Systematic reviews of chemotherapy and endocrine therapy in metastatic breast cancer. Cancer Treat Rev. 2000;26:151–168. doi: 10.1053/ctrv.1999.0161. [DOI] [PubMed] [Google Scholar]
- 7.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28:2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 8.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.can-05-1783. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Huang L, Xie D, Zhao Q. EGBMMDA: Extreme Gradient Boosting Machine for MiRNA-Disease Association prediction. Cell Death & Disease. 2018;9:3. doi: 10.1038/s41419-017-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Wang L, Qu J, Guan NN, Li JQ. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
- 11.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–8201. doi: 10.1128/mcb.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong W, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/mcb.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, et al. Identification of combination gene sets for glioma classification. Mol Cancer Ther. 2002;1:1229–1236. [PubMed] [Google Scholar]
- 14.Lu Jun, Getz Gad, Miska Eric A., Alvarez-Saavedra Ezequiel, Lamb Justin, Peck David, Sweet-Cordero Alejandro, Ebert Benjamin L., Mak Raymond H., Ferrando Adolfo A., Downing James R., Jacks Tyler, Horvitz H. Robert, Golub Todd R. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Chen YC, Ke WC, Chiu HW. Risk classification of cancer survival using ANN with gene expression data from multiple laboratories. Comput Biol Med. 2014;48:1–7. doi: 10.1016/j.compbiomed.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Gevaert O, De Smet F, Timmerman D, Moreau Y, De Moor B. Predicting the prognosis of breast cancer by integrating clinical and microarray data with Bayesian networks. Bioinformatics. 2006;22:e184–190. doi: 10.1093/bioinformatics/btl230. [DOI] [PubMed] [Google Scholar]
- 17.De Smet F, et al. Predicting the clinical behavior of ovarian cancer from gene expression profiles. Int J Gynecol Cancer. 2006;16(Suppl 1):147–151. doi: 10.1111/j.1525-1438.2006.00321.x. [DOI] [PubMed] [Google Scholar]
- 18.Akay MF. Support vector machines combined with feature selection for breast cancer diagnosis. Expert systems with applications. 2009;36:3240–3247. doi: 10.1016/j.eswa.2008.01.009. [DOI] [Google Scholar]
- 19.Abonyi J, Szeifert F. Supervised fuzzy clustering for the identification of fuzzy classifiers. Pattern Recognition Letters. 2003;24:2195–2207. doi: 10.1016/S0167-8655(03)00047-3. [DOI] [Google Scholar]
- 20.Pena-Reyes CA, Sipper M. A fuzzy-genetic approach to breast cancer diagnosis. Artif Intell Med. 1999;17:131–155. doi: 10.1016/S0933-3657(99)00019-6. [DOI] [PubMed] [Google Scholar]
- 21.Setiono R. Generating concise and accurate classification rules for breast cancer diagnosis. Artif Intell Med. 2000;18:205–219. doi: 10.1016/S0933-3657(99)00041-X. [DOI] [PubMed] [Google Scholar]
- 22.Quinlan JR. Improved use of continuous attributes in C4. 5. Journal of artificial intelligence research. 1996;4:77–90. doi: 10.1613/jair.279. [DOI] [Google Scholar]
- 23.Šter, B. & Dobnikar, A. In international Conference on Engineering Applications of Neural Networks. 427–430.
- 24.Nauck D, Kruse R. Obtaining interpretable fuzzy classification rules from medical data. Artificial intelligence in medicine. 1999;16:149–169. doi: 10.1016/S0933-3657(98)00070-0. [DOI] [PubMed] [Google Scholar]
- 25.Sewak, M., Vaidya, P., Chan, C.-C. & Duan, Z.-H. In Computer and Computational Sciences, 2007. IMSCCS 2007. Second International Multi-Symposiums on. 32–37 (IEEE).
- 26.Kahraman M, et al. MicroRNA in diagnosis and therapy monitoring of early-stage triple-negative breast cancer. Scientific reports. 2018;8:11584. doi: 10.1038/s41598-018-29917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherafatian, M. Tree-based machine learning algorithms identified minimal set of miRNA biomarkers for breast cancer diagnosis and molecular subtyping. Gene (2018). [DOI] [PubMed]
- 28.Shimomura A, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer science. 2016;107:326–334. doi: 10.1111/cas.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho SY, Chen JH, Huang MH. Inheritable genetic algorithm for biobjective 0/1 combinatorial optimization problems and its applications. IEEE Trans Syst Man Cybern B Cybern. 2004;34:609–620. doi: 10.1109/TSMCB.2003.817090. [DOI] [PubMed] [Google Scholar]
- 30.Tung CW, Ho SY. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinformatics. 2008;9:310. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iliopoulos D, et al. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 33.Cochrane DR, Howe EN, Spoelstra NS, Richer JK. Loss of miR-200c: A Marker of Aggressiveness and Chemoresistance in Female Reproductive Cancers. J Oncol. 2010;2010:821717. doi: 10.1155/2010/821717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korpal Manav, Ell Brian J, Buffa Francesca M, Ibrahim Toni, Blanco Mario A, Celià-Terrassa Toni, Mercatali Laura, Khan Zia, Goodarzi Hani, Hua Yuling, Wei Yong, Hu Guohong, Garcia Benjamin A, Ragoussis Jiannis, Amadori Dino, Harris Adrian L, Kang Yibin. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nature Medicine. 2011;17(9):1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimono Y, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L, et al. miR-200c inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3. J Transl Med. 2014;12:305. doi: 10.1186/s12967-014-0305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen ML, Liang LS, Wang X. K. miR-200c inhibits invasion and migration in human colon cancer cells SW480/620 by targeting ZEB1. Clin Exp Metastasis. 2012;29:457–469. doi: 10.1007/s10585-012-9463-7. [DOI] [PubMed] [Google Scholar]
- 38.Hu X, et al. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Long J, Ou C, Xia H, Zhu Y, Liu D. MiR-503 inhibited cell proliferation of human breast cancer cells by suppressing CCND1 expression. Tumour Biol. 2015;36:8697–8702. doi: 10.1007/s13277-015-3623-8. [DOI] [PubMed] [Google Scholar]
- 40.Lerebours F, et al. miRNA expression profiling of inflammatory breast cancer identifies a 5-miRNA signature predictive of breast tumor aggressiveness. Int J Cancer. 2013;133:1614–1623. doi: 10.1002/ijc.28171. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Z, et al. miR-503-3p promotes epithelial-mesenchymal transition in breast cancer by directly targeting SMAD2 and E-cadherin. J Genet Genomics. 2017;44:75–84. doi: 10.1016/j.jgg.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Baran-Gale J, Purvis JE, Sethupathy P. An integrative transcriptomics approach identifies miR-503 as a candidate master regulator of the estrogen response in MCF-7 breast cancer cells. Rna. 2016;22:1592–1603. doi: 10.1261/rna.056895.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polioudakis D, Abell NS, Iyer VR. miR-503 represses human cell proliferation and directly targets the oncogene DDHD2 by non-canonical target pairing. BMC Genomics. 2015;16:40. doi: 10.1186/s12864-015-1279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CH, et al. MicroRNA-regulated protein-protein interaction networks and their functions in breast cancer. Int J Mol Sci. 2013;14:11560–11606. doi: 10.3390/ijms140611560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vos S, Vesuna F, Raman V, van Diest PJ, van der Groep P. miRNA expression patterns in normal breast tissue and invasive breast cancers of BRCA1 and BRCA2 germ-line mutation carriers. Oncotarget. 2015;6:32115–32137. doi: 10.18632/oncotarget.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kastl L, Brown I, Schofield A. C. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Research and Treatment. 2012;131:445–454. doi: 10.1007/s10549-011-1424-3. [DOI] [PubMed] [Google Scholar]
- 47.Sun EH, et al. Screening miRNAs related to different subtypes of breast cancer with miRNAs microarray. Eur Rev Med Pharmacol Sci. 2014;18:2783–2788. [PubMed] [Google Scholar]
- 48.Cao Z-G, et al. Positive expression of miR-361-5p indicates better prognosis for breast cancer patients. Journal of Thoracic Disease. 2016;8:1772–1779. doi: 10.21037/jtd.2016.06.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma F, et al. MiR-361-5p inhibits glycolytic metabolism, proliferation and invasion of breast cancer by targeting FGFR1 and MMP-1. Journal of Experimental & Clinical Cancer Research: CR. 2017;36:158. doi: 10.1186/s13046-017-0630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Damavandi Z, et al. Aberrant Expression of Breast Development-Related MicroRNAs, miR-22, miR-132, and miR-212, in Breast Tumor Tissues. J Breast Cancer. 2016;19:148–155. doi: 10.4048/jbc.2016.19.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lv ZD, et al. MiR-212-5p Suppresses the Epithelial-Mesenchymal Transition in Triple-Negative Breast Cancer by Targeting Prrx2. Cell Physiol Biochem. 2017;44:1785–1795. doi: 10.1159/000485785. [DOI] [PubMed] [Google Scholar]
- 52.Mohammadi-Yeganeh S, et al. Development of a robust, low cost stem-loop real-time quantification PCR technique for miRNA expression analysis. Molecular Biology Reports. 2013;40:3665–3674. doi: 10.1007/s11033-012-2442-x. [DOI] [PubMed] [Google Scholar]
- 53.Colaprico A, Cava C, Bertoli G, Bontempi G, Castiglioni I. Integrative Analysis with Monte Carlo Cross-Validation Reveals miRNAs Regulating Pathways Cross-Talk in AggressiveBreast Cancer. BioMed Research International. 2015;2015:17. doi: 10.1155/2015/831314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou W, et al. Suppressive role of miR-592 in breast cancer by repressing TGF-beta2. Oncol Rep. 2017;38:3447–3454. doi: 10.3892/or.2017.6029. [DOI] [PubMed] [Google Scholar]
- 55.Bhaumik D, et al. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27:5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiang M, et al. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-kappaB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci Signal. 2014;7:ra11. doi: 10.1126/scisignal.2004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hurst DR, et al. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.can-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia AI, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3:279–290. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Xu Y, Yu C, Zuo W. Associations of miR-146a and miR-146b expression and breast cancer in very young women. Cancer Biomark. 2015;15:881–887. doi: 10.3233/cbm-150532. [DOI] [PubMed] [Google Scholar]
- 60.Luo EC, et al. MicroRNA-769-3p down-regulates NDRG1 and enhances apoptosis in MCF-7 cells during reoxygenation. Sci Rep. 2014;4:5908. doi: 10.1038/srep05908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugita B, et al. Differentially expressed miRNAs in triple negative breast cancer between African-American and non-Hispanic white women. Oncotarget. 2016;7:79274–79291. doi: 10.18632/oncotarget.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fassan M, et al. MicroRNA expression profiling of male breast cancer. Breast Cancer Research: BCR. 2009;11:R58–R58. doi: 10.1186/bcr2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danza K, et al. Combined microRNA and ER expression: a new classifier for familial and sporadic breast cancer patients. Journal of Translational Medicine. 2014;12:319. doi: 10.1186/s12967-014-0319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lau P, et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol Med. 2013;5:1613–1634. doi: 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng H, et al. MicroRNA-1185 Induces Endothelial Cell Apoptosis by Targeting UVRAG and KRIT1. Cell Physiol Biochem. 2017;41:2171–2182. doi: 10.1159/000475571. [DOI] [PubMed] [Google Scholar]
- 66.Liu G, et al. A five-miRNA expression signature predicts survival in hepatocellular carcinoma. Apmis. 2017;125:614–622. doi: 10.1111/apm.12697. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, et al. A Ten-MicroRNA Signature Identified from a Genome-Wide MicroRNA Expression Profiling in Human Epithelial Ovarian Cancer. PLOS ONE. 2014;9:e96472. doi: 10.1371/journal.pone.0096472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin K, et al. MicroRNA expression profiles predict progression and clinical outcome in lung adenocarcinoma. OncoTargets and therapy. 2016;9:5679–5692. doi: 10.2147/OTT.S111241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vlachos IS, et al. DIANA-miRPathv3.0: deciphering microRNA function with experimental support. Nucleic Acids Research. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lanczky A, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 71.Aiken, L. S., West, S. G. & Pitts, S. C. Multiple linear regression. Handbook of psychology, 481–507 (2003).
- 72.Chen X, Huang L. LRSSLMDA: Laplacian Regularized Sparse Subspace Learning for MiRNA-Disease Association prediction. PLoS Comput Biol. 2017;13:e1005912. doi: 10.1371/journal.pcbi.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Xing, Xie Di, Wang Lei, Zhao Qi, You Zhu-Hong, Liu Hongsheng. BNPMDA: Bipartite Network Projection for MiRNA–Disease Association prediction. Bioinformatics. 2018;34(18):3178–3186. doi: 10.1093/bioinformatics/bty333. [DOI] [PubMed] [Google Scholar]
- 74.You ZH, et al. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. PLoS Comput Biol. 2017;13:e1005455. doi: 10.1371/journal.pcbi.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cruz JA, Wishart DS. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. 2007;2:59–77. [PMC free article] [PubMed] [Google Scholar]
- 76.Exarchos KP, Goletsis Y, Fotiadis DI. Multiparametric decision support system for the prediction of oral cancer reoccurrence. IEEE Trans Inf Technol Biomed. 2012;16:1127–1134. doi: 10.1109/titb.2011.2165076. [DOI] [PubMed] [Google Scholar]
- 77.Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med. 2001;23:89–109. doi: 10.1016/S0933-3657(01)00077-X. [DOI] [PubMed] [Google Scholar]
- 78.Bellman, R. E. Adaptive control processes: a guided tour. Vol. 2045 (Princeton university press, 2015).
- 79.Vapnik VN. An overview of statistical learning theory. IEEE transactions on neural networks. 1999;10:988–999. doi: 10.1109/72.788640. [DOI] [PubMed] [Google Scholar]
- 80.Wang H, Huang G. Application of support vector machine in cancer diagnosis. Medical Oncology. 2011;28:613–618. doi: 10.1007/s12032-010-9663-4. [DOI] [PubMed] [Google Scholar]
- 81.Chu F, Wang L. Applications of support vector machines to cancer classification with microarray data. Int J Neural Syst. 2005;15:475–484. doi: 10.1142/s0129065705000396. [DOI] [PubMed] [Google Scholar]
- 82.Ho S-Y, Shu L-S, Chen J-H. Intelligent evolutionary algorithms for large parameter optimization problems. IEEE Transactions on evolutionary computation. 2004;8:522–541. doi: 10.1109/TEVC.2004.835176. [DOI] [Google Scholar]
- 83.Wang J-R, et al. ESA-UbiSite: accurate prediction of human ubiquitination sites by identifying a set of effective negatives. Bioinformatics. 2017;33:661–668. doi: 10.1093/bioinformatics/btw701. [DOI] [PubMed] [Google Scholar]
- 84.Tsai M-J, et al. PredCRP: predicting and analysing the regulatory roles of CRP from its binding sites in Escherichia coli. Scientific Reports. 2018;8:951. doi: 10.1038/s41598-017-18648-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yerukala Sathipati S, Ho S-Y. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Scientific Reports. 2017;7:7507. doi: 10.1038/s41598-017-07739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yerukala Sathipati S, Huang H-L, Ho S-Y. Estimating survival time of patients with glioblastoma multiforme and characterization of the identified microRNA signatures. BMC Genomics. 2016;17:1022. doi: 10.1186/s12864-016-3321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2:1–27. doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- 88.Frank E, Hall M, Trigg L, Holmes G, Witten IH. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 89.Vlachos Ioannis S., Zagganas Konstantinos, Paraskevopoulou Maria D., Georgakilas Georgios, Karagkouni Dimitra, Vergoulis Thanasis, Dalamagas Theodore, Hatzigeorgiou Artemis G. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Research. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in this analysis can be found at TCGA data portal.