Abstract
Breeding vegetative crops (e.g. beets, cabbage, forage grasses) is challenged by two conflicting aims. For field production, flowering must be avoided while flowering and seed set is necessary for breeding and seed production. The biennial species sugar beet makes shoot elongation (‘bolting’) followed by flowering after a long period of cold temperatures. Field production in northern geographical regions starts in spring. A thickened storage root is formed only during vegetative growth. It is expected that winter beets, which are sown before winter would have a much higher yield potential. However, field production was not possible so far due to bolting after winter. We propose a strategy to breed winter beets exploiting haplotype variation at two major bolting time loci, B and B2. Both genes encode transcription factors controlling the expression of two orthologs of the Arabidopsis gene FLOWERING LOCUS T (FT). We detected an epistatic interaction between both genes because F2 plants homozygous for two B/B2 mutant alleles did not bolt even after vernalization. Fluorescence complementation studies revealed that both proteins form a heterodimer in vivo. In non-bolting plants, the bolting activator BvFT2 was completely downregulated whereas the repressor BvFT1 was upregulated which suggests that both genes acquire a CONSTANS (CO) like function in beet. Like CO, B and B2 proteins house CCT and BBX domains which, in contrast to CO are split between the two beet genes. We propose an alternative regulation of FT orthologs in beet that can be exploited to breed winter beets.
Introduction
The transition from the vegetative to the generative phase is of major interest to crop breeders due to its high relevance for yield and quality. Crop plants show great variation regarding their phenological development. If vegetative parts of the plant are harvested (leaves, roots) they must not enter the reproductive phase, a major step in plant development commonly referred to as floral transition. Sugar beet (Beta vulgaris L.) is a typical vegetative crop with a biennial life cycle. After sowing in spring, it produces huge leaf and root mass until harvest in autumn. As a result of secondary thickening, a storage root is produced with sucrose contents between 17–20%1. As a biennial plant it enters the reproductive phase only after exposure to a long period of cold temperatures (<4 °C). Then, the shoot is elongated (‘bolting’) and flowers are produced. Early bolting under field conditions must be strictly avoided because it gives rise to flowering plants with small roots and low sucrose content. For seed production, plants must bolt and flower early after winter. This follows, that conventional sugar beet cannot be cultivated as a vegetative crop over winter, commonly referred to as ‘winter beet’1.
Quantitative trait loci (QTL) and major genes controlling bolting time have been mapped to the nine beet chromosomes2. The bolting time QTL SEASONAL BOLTING- 4 and -9 (SBT-4, SBT-9) accounts for up to 52% of the phenotypic variation3. The phenotypic effect of SBT-4 is likely caused by the major flowering time regulator BvFT2 because they were mapped to the same position on chromosome 4. SBT-9 was precisely mapped to the position of BR1. This QTL was recently fine mapped by a sequencing approach and a gene similar to CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 73-I (CPSF73-I) from Arabidopsis was suggested as a candidate gene for this QTL4.
Sugar beet has two sequences which share high homology to FLOWERING LOCUS T (FT) a major integrator of signals from different regulatory pathways triggering floral transition in Arabidopsis5. BvFT1 is a floral repressor which is transcriptionally active before winter and prevents bolting. In contrast, BvFT2 is a floral inducer which is activated during vernalization. A high BvFT2 activity is indicative for generative (bolting) beet plants5.
Two upstream regulators of the two BvFT orthologs have been cloned. BOLTING TIME CONTROL 1 (BTC1) belongs to the PRR3/7 clade of PSEUDO RESPONSE REGULATOR (PRR) genes that are components of the photoperiod pathway in Arabidopsis6. A dominant allele which is highly abundant in wild beet (B. vulgaris ssp. maritima) populations from the Mediterranean causes early bolting (without vernalization) resulting in an annual life cycle. Another PRR7 homolog, BvPRR7, is a cold responsive gene with a clock function in beets but not involved in bolting time regulation7. The second bolting time gene, BvBBX19 encodes a putative transcription factor with two B-Box zinc finger motifs but lacking a CCT domain8. Recently, haplotype variation of the four major bolting time genes from beet have been studied in wild and cultivated beet accessions9. For BTC1 and BvBBX19, 14 and 7 haplotypes were found, respectively6,9,10. They were classified as annual or biennial bolting time regulators. BTC1 and BvBBX19 share homology with the transcription factor CONSTANS (CO), which regulates floral transition in Arabidopsis in a long day (LD) dependent manner11. It has two consecutive Zn finger domains which are called B-Boxes12. Mutants with amino acid alterations in conserved residues of the B-Boxes are late flowering. At the C-terminus, the CO protein has a CCT (CO, CONSTANS-LIKE, and TIMING OF CAB EXPRESSION1) domain which includes a nuclear import signal. By its CCT domain, CO binds to the ubiquitin ligase COP1 and to the FT promoter by forming complexes with other transcription factors13. This sequence is strictly conserved in proteins which are constituents of the circadian clock12. CDF (CYCLING DOF FACTORS) transcription factors bind to the CO promoter and inhibit its expression during the morning. Later, they are degraded by the proteasome when GIGANTEA (GI) interacts with FLAVIN BINDING, KELCH REPEAT, F-BOX PROTEIN 1 (FKF1) and ZEITLUPE (ZTL) resulting in strong transcriptional upregulation of CO14. The CO protein is stabilized by light and degraded in darkness after ubiquitination and proteolysis by the 26S proteasome15.
In Arabidopsis, apart from CO there are at least 31 genes encoding proteins with B-Box and CCT domains, 16 are CO-Like (COL) proteins with one or two B-Boxes and one CCT domain, the remaining ones are either lacking the CCT domain, or one B-Box and the CCT domain16. BBX19 and CO are forming dimers which jointly regulate FT in an antagonistic way17. BBX32 physically interacts with COL3 to form a dimer which targets the FT promoter15. Interestingly, beet has a large CONSTANS-LIKE gene family but is lacking a functional CO ortholog with both domains18. BTC1 is lacking a B-Box and BvBBX19 is lacking a CCT domain.
The purpose of this work was to understand the genetic and physical interaction between BTC1 and BvBBX19 and to lay the foundations to breed winter beets. We assumed that both proteins work together to acquire a CO-like function. To test our hypothesis, we studied an F2 population segregating for both genes. We found an epistatic interaction between both loci which resulted in three different life cycle regimes. Combining two mutant alleles resulted in plants which completely lost their competence to bolt after vernalization. The genetic data were confirmed by yeast-two-hybrid interaction and in planta bimolecular fluorescence complementation studies. Double mutant plants are proposed as prototypes for winter beet breeding which requires complete bolting control after winter.
Results
The B2 locus is epistatic to B
We produced an F2 population from a cross between two biennial beet genotypes, seed code 093187 (BaBa B2fB2f) and 056822 (BdBd B2hB2h) which differed by their B and B2 alleles. 145 plants were grown under long day conditions together with their parents and the annual and biennial controls. We determined the genotypes of the B and B2 loci for all F2 plants using the markers CAU4234 and CAU4235 (Supplementary Tables 1 and 2). In accordance with their position on different chromosomes, the observed genotypic segregation fitted a random segregation ratio (χ2 = 15.77; α = 0.05) (Supplementary Tables 3 and 4).
The biennial controls bolted within 3–4 weeks after vernalization whereas the annual controls bolted early (4–6 weeks after sowing) without vernalization required (Fig. 1A). Most F2 plants carrying at least one Bd and one B2f allele were lacking a vernalization requirement because they bolted within 114 days after sowing like plants from the annual controls (Fig. 1A). Six F2 plants with the BdBa B2fB2h genotype did not bolt prior to cold treatment. However, these plants were the earliest biennials as they bolted already 9–16 days after vernalization. Neither the F2 genotypes carrying the homozygous Ba allele in combination with B2h or B2f alleles (BaBa BfB2f, BaBa B2fB2h, BaBa B2hB2h) nor those plants carrying the Bd allele (heterozygous or homozygous) in combination with the homozygous B2h allele (BdBd B2hB2h, BdBa B2hB2h) were able to bolt without cold treatment. After vernalization, all F2 plants homozygous or heterozygous for the B2f allele (BaBa B2f Bf, BaBa B2fB2h) started shoot elongation within three weeks which is typical for biennial beets.
Figure 1.
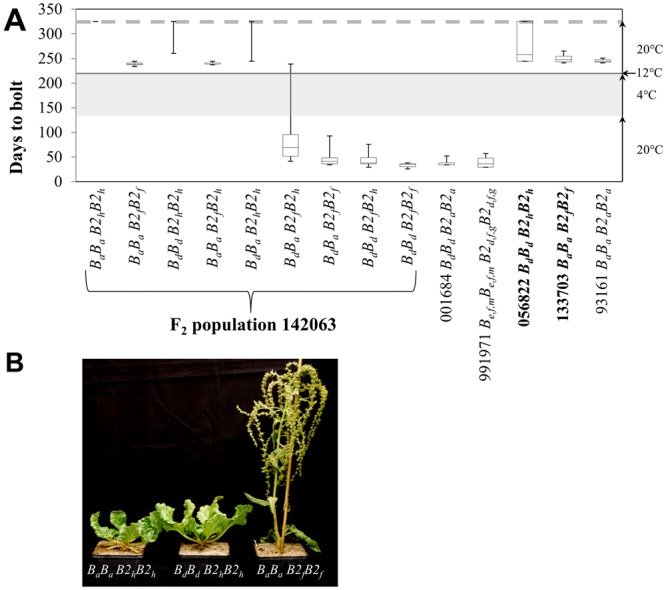
Phenotypic variation for bolting time in the F2 population. Box plots indicate the first and third quartile range of the phenotypic variation for each genotype. The top and bottom whiskers indicate the maximum data value. (A) F2 plants (seed code 142063) and annual (001684, 991971) and biennial (056822, 133703, 930176) controls were grown in a climate chamber under long day conditions (16 h light/8 h dark) at 20 °C. After 135 days, non-bolting plants were kept for 12 weeks at 4 °C. For plants which did not start to bolt until the end of the experiment (grey dotted line), days to bolting was set at 325. The respective genotypes for BTC1 (B) and BvBBX19 (B2) are given according to Höft et al.9. Seed code 056822 and 133703 are the parental genotypes (bold letters). (B) Three F2 plants with their respective genotypes after 12 weeks of cold treatment.
Consistent with our initial hypothesis, the F2 population displayed a third phenotypic class for bolting time because 27 out of 30 F2 plants that carry the homozygous B2h allele in combination with the homozygous and heterozygous Bd allele and all 17 BaBa B2hB2h F2 plants failed to bolt until the end of the experiment (325 days after sowing). The fact that almost all F2 plants carrying the homozygous B2h allele were non-bolting after vernalization irrespective of the B allele indicates that the B2h allele is able to ‘mask’ the phenotypic effect of the Bd or Ba alleles.
How can transgressive variation in the F2 population be explained? We tested two genetic hypotheses to explain the phenotypic segregation observed in this experiment (Supplementary Table 4). Our initial hypothesis follows the assumption that all plants carrying at least one Bd and B2f allele are annual, plants which are homozygous for either the Ba or B2h allele are biennial, and only the double homozygous F2 plants (BaBa B2hB2h) do not bolt after vernalization giving rise to a phenotypic segregation of 9:6:1 (annual: biennial: non-bolting after vernalization). This hypothesis was rejected after a χ² test for goodness of fit to a 9:6:1 ratio (χ2 = 150.03; α = 0.01). The second hypothesis is based on the assumption that the B2h allele acts epistatically over the B locus. In this case, a 9:4:3 phenotypic segregation was to be expected (Supplementary Table 4). As this segregation rate was not rejected (χ2 = 2.24; α = 0.01), we assume that the B2h allele which was derived from an EMS mutagenesis acts epistatically to B resulting in a non-bolting (after vernalization) phenotype (Supplementary Table 4). However, this interaction does not fully explain phenotypic variation because biennial plants were found in the BdB2h parent 056822 and among the corresponding F2 genotypes (Fig. 1A). In conclusion, genetic analyses are clearly pointing at a joint activity of both loci to control the onset of bolting.
The floral promoter BvFT2 is completely downregulated in beets which do not bolt after vernalization
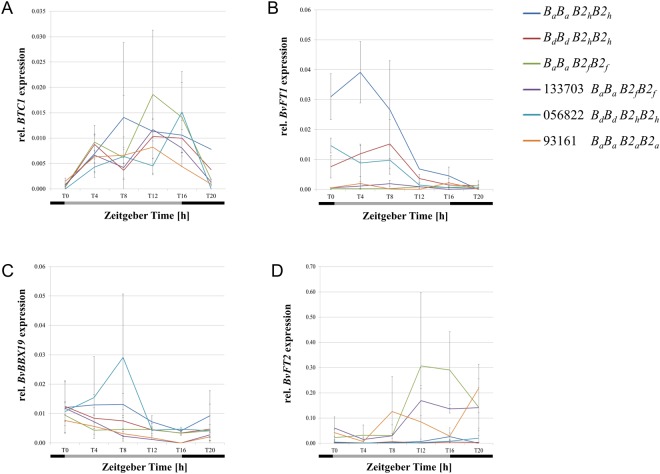
We questioned whether the transcript levels of BTC1 and BvBBX19 differ between F2 plants bolting and non-bolting after vernalization. Therefore, the effect of different Ba/Bd and B2f/B2h allele combinations on their transcriptional activity was investigated using the same F2 plants as for the genetic experiments. Leaves were taken 23 days after cold treatment every 4 hours over 24 hours. We observed no significant differences in the diurnal expression pattern of BTC1 between double homozygous F2 plants (BaBa B2fB2f), F2 plants that are homozygous for the B2h allele and the biennial controls (Fig. 2A) despite of strikingly different life cycle regimes. BTC1 was upregulated during the day and the transcript levels decreased during the night in controls as well as in F2 individuals bolting after vernalization (BaBa B2fB2f) and F2 genotypes non-bolting after vernalization (BdBd B2hB2h and BaBa B2hB2h). For BvBBX19 we detected generally low expression levels during the day with continuously increasing transcript levels during the night in all plants which bolted after vernalization except for the parental genotype 056822 (BdBd B2hB2h). In general, the BvBBX19 transcript levels were increased during the day (ZT8) in all plants which is in accordance with previous data8. Interestingly, upregulation was also observed in F2 genotypes that failed to bolt after the cold treatment (BdBd B2hB2h and BaBa B2hB2h) however at a much lower level when compared to the parental genotype 056822 (Fig. 2C).
Figure 2.
Diurnal expression analysis for BTC1, BvBBX19, BvFT1 and BvFT2 in bolting and non-bolting F2 plants after cold treatment. The expression was measured in bolting (BaBa B2fB2f) or non-bolting (BaBa B2hB2h, BdBd B2hB2h) F2 plants after vernalization. The BvBBX19 mutant parent 056822 (BdBd B2hB2h) and the biennial genotypes 133703 (BaBa B2fB2f) and 930176 (BaBa B2aB2a) were used as controls. Each value is the mean of three biological and three technical replicates, except for the BvFT1 expression of the BaBaB2hB2h genotype where each value is the mean of two biological replicates and three technical replicates. The relative gene expression is given on the vertical axis. Night and day periods are indicated by black and grey bars. Error bars represent the SD of biological replicates.
We reasoned that the BTC1/BvBBX19 genotype impacts the expression of the two FT paralogs BvFT1 and BvFT2. It had been demonstrated that floral transition in beet is promoted through downregulation of the floral repressor BvFT1 and therewith upregulation of the floral inducer BvFT2, which both are downstream targets of BTC1 and BvBBX195,6,8. We observed that the transcriptional activity of BvFT1 and BvFT2 follows the anticipated expression pattern (Fig. 2B,D). As expected, BvFT2 was highly upregulated and BvFT1 completely downregulated after vernalization in the biennial controls and in biennial F2 plants. Interestingly, a contrasting expression pattern was observed in F2 plants which did not bolt after vernalization. The transcriptional activity of BvFT1 was two times higher in non-bolting F2 plants homozygous for Ba/B2h as compared to F2 plants homozygous for Bd/B2h.
BvBBX19 and BTC1 physically interact with each other
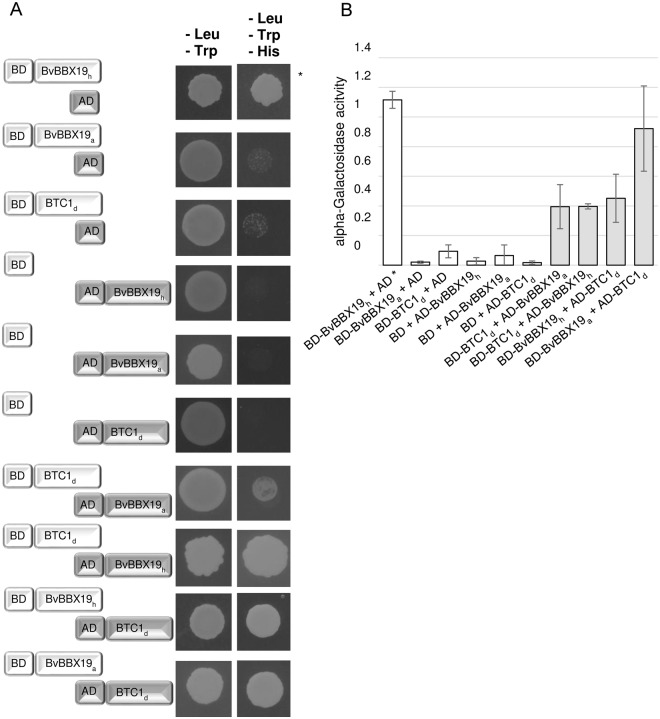
The absence of a gene in sugar beet encoding a canonical CO protein suggested that one or several other proteins fulfill the function of CO in this plant. The most likely candidates are BvBBX19 and BTC1 since they contain two B-Box domains and the CCT domain, respectively resembling the CO domain structure8. One likely scenario how BvBBX19 together with BTC1 can replace CO is direct physical interaction between the two proteins resulting in a functional CO ortholog. To test this hypothesis, we performed yeast-two-hybrid studies. Constructs were made containing the full-length coding regions of BvBBX19 and BTC1 fused to either the GAL4 DNA-binding domain (BD) or the GAL4 activation domain (AD) at the N-terminus of the respective proteins. In addition, we included constructs of the previously identified BvBBX19 mutant (BvBBX19h), which contains a premature stop codon resulting in a BvBBX19 variant with only one B-Box8. Again, AD or BD-domains were fused to the N-terminus of BvBBX19h. Wild type BvBBX19a and BTC1d constructs showed no autoactivation and were thus useful to study interaction between the two proteins. For both combinations of wild type BvBBX19awith BTC1d we observed growth of yeast cells on selective plates (-Leu, -Trp, -His) as well as induction of the α-galactosidase reporter in the quantitative assays (Fig. 3). Thus, BvBBX19a and BTC1d interact. In case of BvBBX19h we observed autoactivation for the BD-BvBBX19h construct. Thus, this construct was not useful for further interaction studies. However, AD-BvBBX19h did not result in autoactivation. In combination with BD-BTC1d, colonies were formed on selective medium and α-galactosidase activity induced. This result implies that the second C-terminally located B-box in BvBBX19a, which is missing in BvBBX19h, is not essential but supportive for the interaction with BTC1d.
Figure 3.
Yeast-2-Hybrid analysis showing interaction of BTC1 with BvBBX19. The proteins were fused at their N-terminus to either the DNA binding domain (BD) or the activation domain (AD) of the GAL4 transcription factor. (A) Yeast cells were transformed with vectors harboring the indicated constructs. Aliquots of overnight cultures were spotted on non-selective (-Leu, -Trp) or selective (-Leu, -Trp, -His) plates and tested for His auxotrophy. Nine clones of each plasmid combination were tested and one representative result is shown. (B) Quantification of BvBBX19/BTC1 interaction using α-galactosidase assay. Means ± SD of three technical replicates are displayed. The BvBBX19 mutant (BvBBX19h) with a premature stop codon after the first B-Box domain was included in these studies. BvBBX19h showed autoactivation (indicated by asterisks).
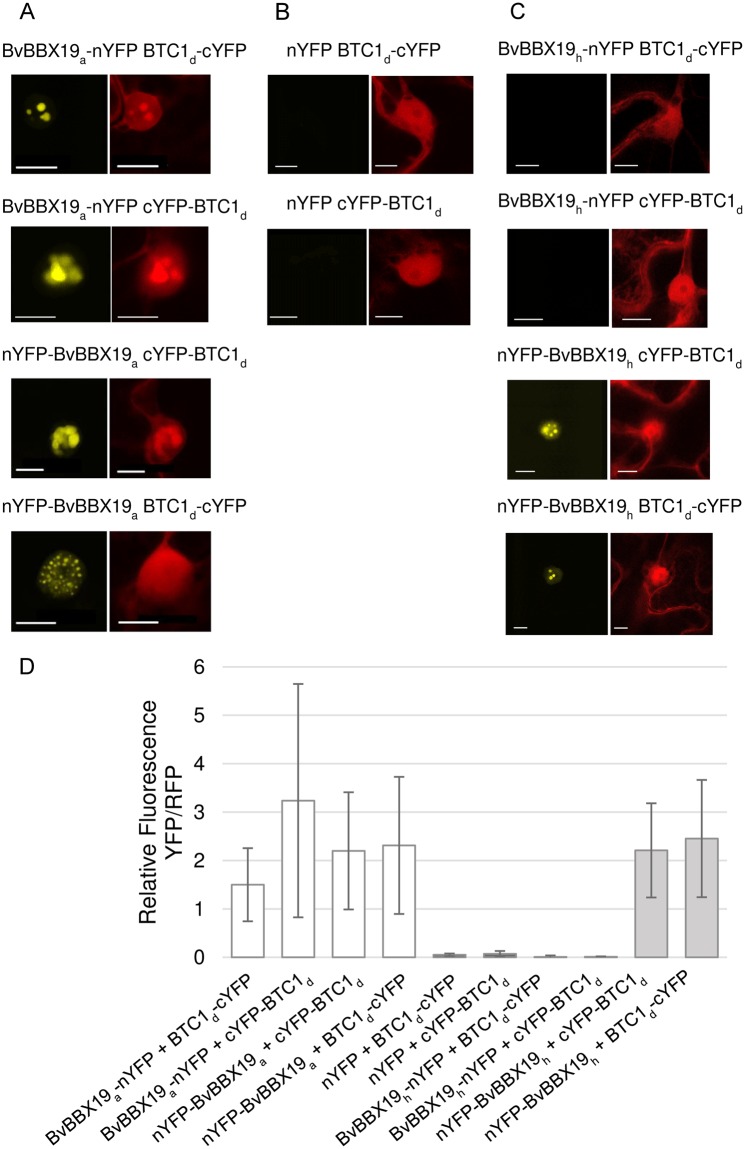
Y2H data strongly suggested direct physical interaction between BvBBX19 and BTC1. For further confirmation, we applied ratiometric bimolecular fluorescence complementation assays (rBiFC). We used constructs where either the 5′- or the 3′ region of BvBBX19a was fused with the 5′-terminal half of YFP (nYFP). Accordingly, BTC1d was fused with the C-terminal part of YFP (cYFP) at its N- or C-terminus. These constructs were co-transfected into Nicotiana benthamiana leaves using Agrobacterium-mediated infiltration. All four combinations resulted in YFP signals (Fig. 4A) in contrast to co-expression of the non-fused nYFP controls with BTC1d fused to cYFP at its N-terminus or C-terminus (Fig. 4B). The truncated BBX19 version nYFP-BvBBX19h co-transfected with BTC1d carrying cYFP at the N-terminus or C-terminus also gave a clear YFP signal in contrast to BvBBX19h constructs carrying nYFP at the C-terminus of BvBBX19h (Fig. 4C). The latter result is expected since BvBBX19h contains a stop codon upstream of the second B-Box and thus does not allow expression of the C-terminal YFP half. Interestingly, in all cases complemented YFP signals were observed in nuclear bodies. Quantification of the YFP against the RFP fluorescence signals from at least 20 images (Fig. 4D) are consistent with the representative pictures presented in Fig. 4A–C.
Figure 4.
Interaction between BvBBX19 and BTC1 analyzed by ratiometric bimolecular fluorescence complementation (rBiFC). (A–C) Confocal pictures of Nicotiana benthamiana leaves three days after Agrobacterium tumefaciens infiltration with the rBiFC constructs. The C-terminal part of YFP (cYFP) or the N-terminal part of YFP (nYFP) was fused to the target proteins (X) at their N-terminus (c/nYFP-X) or at their C-terminus (X-c/nYFP). Fluorescence was detected with a Leica TCS SP5 Confocal Laser Scanning Microscope. YFP was excited with a 488 nm laser and RFP with a 561 nm laser. YFP fluorescence was detected between 535 nm and 560 nm. RFP fluorescence was detected between 600 nm and 625 nm. (A) shows BvBBX19a interaction with BTC1d, (B) negative controls for BTC1d, and (C) interaction of mutant BvBBX19 (BvBBX19h) with BTC1d. Scale bars, 10 µm. (D) Quantification of the mean fluorescence of split-YFP normalized against RFP. Data are means ± SD of at least 20 images selected at random.
Discussion
We have performed a genetic study with all combinations of BTC1 (B) and BvBBX19 (B2) alleles in an F2 population. Bd is a typical annual allele only found in wild beet populations from the Mediterranean. Ba carries six non-synonymous SNPs and a large insertion in the promoter compared to ‘annual’ alleles6. B2f is found in annual beets9 while B2h is a nonsense EMS mutant allele8.
Beet is a typical long day plant. Bolting even in the presence of the early bolting alleles or after vernalization is strongly delayed in short days19. Therefore, all experiments were performed under long day conditions. An annual life cycle requires an annual B allele and a functional B2 allele (Bd/B2f). The competence for early flowering is lost in plants homozygous for the B2 mutant allele BvBBX19h irrespective of the B allele. Likewise, B knockdown (RNAi) plants cannot flower any more even after vernalization6.
Double mutants homozygous for the biennial B and the non-functional B2 allele completely lost their competence to bolt which confirmed our initial hypothesis that B and B2 jointly regulate the onset of bolting in sugar beet. By combining two mutant alleles, we could select plants that did not bolt even after cold treatment. We found an incomplete epistatic interaction between bolting resistant plants among all B2h homozygous genotypes, but the presence of the Bd alleles modified the B2h effect because biennial plants were present in the BdB2h parent and F2 plants. We assume that apart from these two major bolting time regulators, additional genes can modify bolting time. Moreover, the rare occurrence of spontaneously bolting plants in production fields points at environmental factors modifying the activities of B and B2. These factors together may explain the presence of biennial plants in the 056822 parent.
The genus Beta comprises iteroparous perennials with an annually repeated requirement for vernalization20. Future studies with perennial wild beets will resolve the question whether the BTC1/BvBBX19 module and the BR1 QTL4 control the perennial life cycle. However, strictly non-bolting genotypes are likely to be a dead end of evolution because they cannot reproduce sexually in contrast to iteroparous plants which flower and set seeds in subsequent years after winter. Thus, it is no surprise that despite of extensive screenings BTC1/BvBBX19 double mutants have not been found in nature so far.
The B2h genotypes exhibited a strong requirement for vernalization even in the presence of the early bolting allele Bd. This indicates that there are upstream regulators of the BTC1/BvBBX19 module which respond to cold temperatures and to alterations of the B2h protein. This makes B2 a primary target of a putative vernalization regulatory pathway. However, no further mutants have been detected so far. Searching for orthologous genes from Arabidopsis has not been successful and beet lacks a functional ortholog of FLOWERING LOCUS C which is a major integrator of signals from the vernalization pathway in Arabidopsis. Seemingly, divergent vernalization pathways have evolved in both species. Because vernalization has an epigenetic basis, genes responding to methylation might be interesting candidates. Consequently, two genes, SHORT VEGETATIVE PHASE (BvSVP) and BvVIN3 come into focus as upstream regulators of the BTC1/BvBBX19 module because they are hypomethylated and/or differentially expressed after cold exposure21,22.
How can the bolting-resistant phenotype be explained by protein-interaction and expression studies? BTC1 requires a functional BvBBX19 protein consistent with our data that beets carrying the annual Bd allele do not bolt in the presence of the mutant B2h allele. Loss of competence to bolt is due to downregulation of the floral inducer BvFT2 and upregulation of the floral repressor BvFT16. The non-bolting phenotype of the B2 mutant is not caused by a lack of protein-interaction because binding between BvBBX19h and BTC1d was demonstrated (Figs 3 and 4). Y2H data imply that BBX19 and BTC1 do not require another beet protein for their interaction. However, it cannot be excluded that BTC1 and BvBBX19 interact with other proteins as CO does with PHYTOCHROME INTERACTING FACTOR 423. We propose a model where BTC1 and BvBBX19 (mutated and wild type) dimerize to bind to the BvFT2 promoter. Likewise, the interaction between Arabidopsis COL proteins and CO has been demonstrated. B-BOX 32 binds to CONSTANS-LIKE315 and BBX19 binds to CO to suppress its function as an activator of FT17. Consequently, the heterodimer cannot bind to the Box 1 Motif of the FT promoter which is essential for binding of the CO protein. We reason that BTC1 or BvBBX19 alone are not able to bind to the BvFT2 promoter and that the heterodimer of mutant BvBBX19h with the BTC1d protein cannot bind to the BvFT2 promoter. The importance of intact domains for their binding activities was recently reported for Arabidopsis where a truncated COβ variant lacking the CCT domain lost its DNA-binding affinity24. The variant protein results from alternative splicing of the CO mRNA. Moreover, the truncated protein inhibits the function of the full-size COα protein by reducing its protein abundance and preventing its DNA-binding. A similar mechanism of CO-BBX functional interaction has been reported for rice where OsBBX14 activates the CO ortholog Hd1 which is a repressor of the rice FT ortholog Heading date 3a (Hd3a) under LD conditions25. In Arabidopsis, CO was shown to bind to a tandemly repeated sequence element of the FT promoter [consensus TGTG(N2-3)ATG motif]26. A promoter analysis of the beet FT genes revealed that this element is lacking from the 5′ regions of BvFT1 and BvFT2 (Supplementary Table 5). Moreover, overexpression of BvBBX19 and BTC1 in Arabidopsis CO mutants did not accelerate flowering (data not shown). Evidently, plants have evolved different mechanisms to control FT expression. In Arabidopsis and rice, CO-like genes and CO orthologs gained different functions as both activators and suppressors of their downstream target. We propose an alternative mechanism for beet, where two FT paralogs are differentially regulated by two CO-like genes whose function depends on vernalization. The upstream regulators responding to external cues are still unknown. Moreover, the involvement of other homologs of CO binding proteins such as TARGET OF EAT1 (TOE1) or small B-BOX protein (MiP1a and MiP1b)11 from Arabidopsis remains to be demonstrated.
Simon et al.27 suggested, that CO has been derived from COL genes and that the function of the CO protein, which is specific to Brassicaceae species gave Arabidopsis an adaptive advantage during its expansion to northern geographical regions. Also B. maritima spread to northern regions after the last ice age. It is tempting to speculate that the flexible B/B2 module was an important factor for its adoption to winter climates and LD conditions. We reason that BTC1 and BvBBX19 must also perceive signals from the photoperiod and the vernalization signaling pathways because bolting initiation depends on long days and exposure to cold temperatures. A recent study with Arabidopsis demonstrated that the PRR proteins play an important role in stabilizing the CO protein. They suppress the proteasomal degradation of CO and contribute to light-mediated accumulation of CO during the day28. In beet, one member of the PRR clade has been further studied. Interestingly, BvPRR7 is a cold responsive gene with a clock function and caused late flowering after overexpression in Arabidopsis7. Future studies are needed to show if this gene plays a role in beet as an upstream regulator of the BTC1/BvBBX19 module.
This study has importance for breeding vegetative crops which are sown in spring under winter climate conditions. After early sowing, cold temperatures can pose a risk because they cause early bolting which drastically reduces yield (e.g. cabbage, carrots, salad, beet root)29. Therefore, breeders have been selecting for bolting resistant mutants, many of these carry mutations in functional orthologs of FLC (only Brassica species), CO or FT. Breeding winter beets requires full bolting control after winter. In contrast to traditional ‘spring beets’, they must not bolt after winter. This can be achieved by selecting for non-bolting (after vernalization) alleles from the BR1 QTL on chromosome 94,30. Alternatively, we propose a haplotype-based breeding strategy using well defined BTC1 and BvBBX19 alleles. But how can we harvest seeds from the parents if they are already bolting resistant? This problem could be overcome by introducing the early bolting B allele (e.g. Bd) into non-bolting parents turning them into biennials which can flower and set seeds after winter. We propose a haplotype swapping strategy where different B and B2 alleles9 are combined with each other. A second approach relies on conditional bolting of Bh parents. Non-bolting plants can enter the reproductive phase under extreme environments. We have obtained seeds from Bh genotypes after cultivation in a climate chamber under 24 hours light and largely extended vernalization period. As an alternative, the bolting resistance of Bh seed parents could be overcome by field cultivation in southern regions under high temperatures. It was recently shown, that CO expression increases under high temperatures23. Although this was observed under SD conditions, it is tempting to speculate that B/B2 allele combinations display different temperature sensitivity before and after vernalization.
Materials and Methods
Plant material and growth conditions
We performed a cross between two single plants of the biennial beet lines seed code 056822 (plant #15) and 093187 (plant #8). The female parent 056822 carries the BTC1d allele only found in annual beets which confers early bolting without vernalization6 and a mutated BvBBX19 allele8 which we termed BvBBX19h following the haplotype nomenclature described by Höft et al.9. The pollinator parent 093187/8 carries the btc1a allele and the functional BvBBX19f allele. For ease of understanding we will use the allele nomenclature as Bd (haplotype BTC1d), Ba (haplotype btc1a), B2f (haplotype BvBBX19f) and B2h (haplotype BvBBX19h). A single F1 plant (seed code 133580/1, BdBa B2hB2f) was selected and propagated by bag isolation to produce F2 seeds (seed code 142063).
For phenotyping, 145 plants of the F2 population 142063 were grown in a climate chamber under long day conditions (16 h light/8h dark, 320 µmol m−2 s−1) for 325 days. The two parent lines 056822 and 133703 (selfing progeny of 093187), three biennial (seed codes 092492, 930184, 930176) and two annual genotypes (001684, 991971) were grown as controls (five plants per line). Plants were first grown in 9 cm pots for 135 days at 20 °C and then cold treated at 4 °C for 12 weeks, followed by an acclimatization phase at 12 °C for three days. For the rest of the experiment, they grew again in 11 cm pots at 20 °C for another 102 days. Every second day, plants were randomized and the onset of bolting was recorded (BBCH scale code: 51) according to Meier et al.31. Finally, plants were classified as follows: (1) annual plants which bolted within 135 days, (2) biennial plants which bolted only after cold treatment, and (3) plants, which did not bolt until the end of the experiment after 325 days.
DNA techniques
For DNA isolation, leaves were harvested from six-weeks-old F2 plants and freeze dried. Genomic DNA was isolated applying the CTAB method32. A 10-fold dilution of the extracted DNA was later used for PCR using Taq DNA Polymerase (Invitrogen). We used the InDel marker CAU4234 and the CAPS marker CAU4235 for genotyping the BTC1 and BvBBX19 locus, respectively (Supplementary Table 2). PCR products were separated on 1% agarose gels.
Gene expression analysis
We measured the diurnal expression of the four flowering time genes BTC1, BvBBX19, BvFT1 and BvFT2 by qRT-PCR in F2 plants with the BTC1 and BvBBX19 haplotypes BaBa B2fB2f, BaBa B2hB2h and BdBd.B2hB2h, and in the biennial controls 056822 (BdBd B2hB2h), 133703 (BaBa B2fB2f) and 930176 (BaBa B2aB2a). Total RNA was isolated from young leaves that were harvested 23 days after cold treatment in a 4 hours interval over 24 hours (first measurement at ZT 0, the time of lights on). Total RNA was extracted with the peqGOLD Plant RNA Kit (PeqLab) and subsequently treated with DNase. 500 ng of total RNA was reverse transcribed using a First Strand cDNA Synthesis kit (Fermentas). Resulting cDNA was diluted 10-fold and 2 µl of the dilution were used as a template for qRT-PCR. Three independent biological and three technical replicates were analyzed. qRT-PCR was performed with a Platinum SYBR Green Mastermix (Invitrogen) on a CFX96 Real-Time PCR detection system (Bio-Rad) with a final reaction volume of 20 µl and a final primer concentration of 20 pM. The housekeeping gene BvGAPDH was used as a reference. Data were analyzed with the CFX Manager Software v2.1 (Bio-Rad). Expression levels were first calculated with the comparative CT (ΔCT) method and then normalized to the geometric mean of BvGAPDH to calculate the relative expression levels.
Yeast-2-Hybrid assays
Yeast-2-Hybrid experiments were performed using the Matchmaker Gold Yeast-Two-Hybrid System (Clontech). The proteins of interest were fused at their N-terminus to either the DNA binding domain (BD) or the activation domain (AD) of the GAL4 transcription factor by insertion of the full-length coding sequences of BvBBX19a, BvBBX19h (BvBBX19 mutant) and BTC1d into the NcoI and XhoI sites of the vector pACT2 or the NcoI and SalI sites of the vector pAS2-1. Full-length coding sequences of BvBBX19a, BvBBX19h and BTC1d were obtained by PCR with primers listed in Supplementary Table 2. The correctness of the amplified sequences was verified by sequencing. Yeast cells (strain Y2H Gold, Clontech) were transformed according to the supplier’s manual. Screening for histidine auxotrophy was done with nine clones of each transformant which were spread on non-selective (-Leu, -Trp) or selective (-Leu, -Trp, -His) plates and incubated for two days at 28 °C.
Quantification of interaction was determined by the α-galactosidase-assay33 with minor modifications. For this purpose, yeast transformants were cultured overnight in 3 mL selective medium. After measuring OD600 of the overnight culture and pelleting the cells, 200 µL of the overnight medium were mixed with 600 µL assay buffer (0.33 M sodium acetate, pH 4.5, 10 mg mL−1 p-nitrophenyl-alpha-D-galactopyranoside) and incubated at 29 °C. After 21 h of incubation 200 µL stopping buffer (2 M sodium carbonate) were added and OD410 was measured. α-galactosidase activity was calculated as: α-galactosidase units = 1,000 × OD410/(t × V × OD600), where t = time of incubation in min, V = volume of culture, OD410 = absorbance by p-nitrophenol, OD600 = cell density at the beginning.
Ratiometric Bimolecular Fluorescence Complementation (rBiFC) and immunoblots
For rBiFC, BvBBX19a, BvBBX19h and BTC1d were C- or N-terminally fused to either the N-terminal or the C-terminal half of YFP (n/cYFP). Thus, eight different construct combinations were obtained and the unfused N-terminal half of YFP was used as negative control in combination with cYFP-BTC1d or BTC1d -cYFP.
For generation of constructs, the full-length coding sequences of BvBBX19a, BvBBX19h and BTC1d were PCR amplified with att sites allowing recombination into the entry vectors pDONR221-P1P4 and pDONR221-P3P2 followed by recombination into the Gateway vector pBiFCt-2in1 that also provides RFP as an internal standard34. The obtained constructs were transformed into Agrobacterium tumefaciens strain GV3101(pMP90)35. 4-weeks-old Nicotiana benthamiana plants were transiently co-transformed by Agrobacterium infiltration36 with the construct combinations mentioned above and with p19 to suppress gene silencing37. Three days after infiltration YFP complementation was analyzed using a Leica TCS SP5 Confocal Laser Scanning Microscope (Leica). YFP was excited with a 488 nm laser and RFP with a 561 nm laser. YFP fluorescence was detected between 535 nm and 560 nm, RFP fluorescence between 600 nm and 625 nm. Quantification of the mean fluorescence of split-YFP was done by normalization against RFP. Data were calculated as means of at least 20 images selected at random. Relative fluorescence was determined using ImageJ estimating the mean grey value of the different pictures within an area of around 5 pixels. The maximum grey value per pixel of YFP fluorescence was set as 225.
Expression of BvBBX19a or BvBBX19h fused to nYFP or unfused nYFP (as negative control for rBiFC) was detected via the HA-tag positioned at the C-terminus of nYFP (Fig. S1). Proteins were extracted from infiltrated leaf tissues by TCA precipitation38. 40 µg of proteins per lane were separated by SDS-PAGE. After blotting, the nitrocellulose membrane was blocked with 7% milk powder in TBS and probed with rat anti-HA antibody (Roche, 11867423001, 1:1,500 in TBS-T) and secondary αRat-HRP antibody (Millipore, NMM1767593, 1:10,000 in TBS-T) using the ECL detection assay (Bio-Rad) according to supplier’s manual.
Electronic supplementary material
Acknowledgements
Financial support was given by the DFG (German Research Foundation) through Priority Program 1530 ‘Flowering time control – from natural variation to crop improvement’ to C.J. (JU205/24-1) and A.B. (BA985/14-1). We thank Jeanette Schermuly, Monika Bruisch and Kerstin Wulbrandt for technical assistance and Christian Renicke for support in rBiFC.
Author Contributions
N.D. and M.E. designed the experiments and analysed the data. N.D. performed phenotyping, genotyping and expression analysis. M.E. performed yeast-2-hybrid assays, ratiometric Bimolecular Fluorescence Complementation (rBiFC) and immunoblots. C.J., A.B. supervised the project and together with N.H. wrote the manuscript with input from all authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34328-4.
References
- 1.Biancardi, E., Campbell, L. G., Skaracis, G. N. & De Biaggi, M. Genetics and Breeding of Sugar Beet, 1–367 (Science Publishers Inc., Enfieldt, NH, USA, 2005).
- 2.Melzer, S., Müller, A. E. & Jung, C. Genetics and Genomics of Flowering Time Regulation in Sugar Beet. In Genomics of Plant Genetic Resources 3–26 (Springer Netherlands, 2014).
- 3.Tränkner C, et al. Deciphering the complex nature of bolting time regulation in Beta vulgaris. Theor. Appl. Genet. 2017;130:1649–1667. doi: 10.1007/s00122-017-2916-2. [DOI] [PubMed] [Google Scholar]
- 4.Tränkner, C. et al. A Detailed Analysis of the BR1 Locus Suggests a New Mechanism for Bolting after Winter in Sugar Beet (Beta vulgaris L.) Frontiers in Plant Science7 (2016). [DOI] [PMC free article] [PubMed]
- 5.Pin PA, et al. An Antagonistic Pair of FT Homologs Mediates the Control of Flowering Time in Sugar Beet. Science. 2010;330:1397–1400. doi: 10.1126/science.1197004. [DOI] [PubMed] [Google Scholar]
- 6.Pin PA, et al. The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr. Biol. 2012;22:1095–1101. doi: 10.1016/j.cub.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Omolade OO, Müller AE, Jung C, Melzer S. BvPRR7 is a cold responsive gene with a clock function in beets. Biologia Plantarum. 2016;60:95–104. doi: 10.1007/s10535-015-0568-0. [DOI] [Google Scholar]
- 8.Dally N, Xiao K, Holtgräwe D, Jung C. The B2 flowering time locus of beet encodes a zinc finger transcription factor. Proc. Natl. Acad. Sci. USA. 2014;111:10365–10370. doi: 10.1073/pnas.1404829111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Höft, N., Dally, N., Hasler, M. & Jung, C. Haplotype Variation of Flowering Time Genes of Sugar Beet and Its Wild Relatives and the Impact on Life Cycle Regimes. Frontiers in Plant Science8 (2018). [DOI] [PMC free article] [PubMed]
- 10.Höft N, Dally N, Jung C. Sequence variation in the bolting time regulator BTC1 changes the life cycle regime in sugar beet. Plant Breeding. 2018;137:412–422. doi: 10.1111/pbr.12579. [DOI] [Google Scholar]
- 11.Shim JS, Kubota A, Imaizumi T. Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration. Plant Physiol. 2017;173:5–15. doi: 10.1104/pp.16.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valverde F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 2011;62:2453–2463. doi: 10.1093/jxb/erq449. [DOI] [PubMed] [Google Scholar]
- 13.Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends in Plant Sci. 2013;18:575–83. doi: 10.1016/j.tplants.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song YH, et al. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA. 2014;111:17672–17677. doi: 10.1073/pnas.1415375111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tripathi P, Carvallo M, Hamilton EE, Preuss S, Kay SA. Arabidopsis B-BOX 32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc. Natl. Acad. Sci. USA. 2017;114:172–177. doi: 10.1073/pnas.1616459114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khanna R, et al. The Arabidopsis B-box zinc finger family. The Plant Cell. 2009;21:3416–20. doi: 10.1105/tpc.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C-Q, Guthrie C, Sarmast MK, Dehesh K. BBX19 Interacts with CONSTANS to Repress FLOWERING LOCUS T Transcription, Defining a Flowering Time Checkpoint in Arabidopsis. The Plant Cell. 2014;26:3589–3602. doi: 10.1105/tpc.114.130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chia TYP, Müller AE, Jung C, Mutasa-Goettgens ES. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Exp. Bot. 2008;59:2735–2748. doi: 10.1093/jxb/ern129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Dijk H. Evolutionary change in flowering phenology in the iteroparous herb Beta vulgaris ssp. maritima: a search for the underlying mechanisms. J. Exp. Bot. 2009;60:3143–55. doi: 10.1093/jxb/erp142. [DOI] [PubMed] [Google Scholar]
- 20.Hautekèete N-C, Piquot Y, Van Dijk H. Life span in Beta vulgaris ssp. maritima: the effects of age at first reproduction and disturbance. J. Ecol. 2002;90:508–516. doi: 10.1046/j.1365-2745.2002.00688.x. [DOI] [Google Scholar]
- 21.Hébrard C, et al. Epigenomics and bolting tolerance in sugar beet genotypes. J. Exp. Bot. 2016;67:207–225. doi: 10.1093/jxb/erv449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trap-Gentil M-V, et al. Time course and amplitude of DNA methylation in the shoot apical meristem are critical points for bolting induction in sugar beet and bolting tolerance between genotypes. J. Exp. Bot. 2011;62:2585–2597. doi: 10.1093/jxb/erq433. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez V, Takahashi Y, Le Gourrierec J, Coupland G. Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 2016;86:426–40. doi: 10.1111/tpj.13183. [DOI] [PubMed] [Google Scholar]
- 24.Gil K-E, et al. Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. The Plant Journal. 2017;89:128–140. doi: 10.1111/tpj.13351. [DOI] [PubMed] [Google Scholar]
- 25.Bai B, et al. OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice. Plant Science. 2016;247:25–34. doi: 10.1016/j.plantsci.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Tiwari SB, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytologist. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 27.Simon S, Ruhl M, de Montaigu A, Wotzel S, Coupland G. Evolution of CONSTANS Regulation and Function after Gene Duplication Produced a Photoperiodic Flowering Switch in the Brassicaceae. Mol Biol Evol. 2015;32:2284–301. doi: 10.1093/molbev/msv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayama R, et al. PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 2017;36:904–918. doi: 10.15252/embj.201693907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung C, Müller AE. Flowering time control and applications in plant breeding. Trends in Plant Sci. 2009;14:563–573. doi: 10.1016/j.tplants.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Pfeiffer N, et al. Genetic analysis of bolting after winter in sugar beet (Beta vulgaris L.) Theor. Appl. Genet. 2014;127:2479–2489. doi: 10.1007/s00122-014-2392-x. [DOI] [PubMed] [Google Scholar]
- 31.Meier, U. Growth stages of mono- and dicotyledonous plants. Phenological growth stages and BBCH-identification keys of beet. Federal Biological Research Centre for Agriculture and Forestry, Braunschweig, Germany (1993).
- 32.Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minervini G, et al. Isoform-specific interactions of the von Hippel-Lindau tumor suppressor protein. Scientific Reports. 2015;5:12605. doi: 10.1038/srep12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grefen C, Blatt MR. A 2 in 1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC) Biotechniques. 2012;53:311–14. doi: 10.2144/000113941. [DOI] [PubMed] [Google Scholar]
- 35.Koncz C, Schell J. The Promoter of Tl-DNA Gene 5 Controls the Tissue-Specific Expression of Chimeric Genes Carried by a Novel Type of Agrobacterium Binary Vector. Molecular & General Genetics. 1986;204:383–396. doi: 10.1007/BF00331014. [DOI] [Google Scholar]
- 36.Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr. Opin. in Biotechnol. 2006;17:147–154. doi: 10.1016/j.copbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal. 2003;33:949–56. doi: 10.1046/j.1365-313X.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 38.Shultz RW, Settlage SB, Hanley-Bowdoin L, Thompson WF. A trichloroacetic acid-acetone method greatly reduces infrared autofluorescence of protein extracts from plant tissue. Plant Molecular Biology Reporter. 2005;23:405–409. doi: 10.1007/BF02788888. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.