Abstract
This paper presents an impedance-based biosensor for rapid and simultaneous detection of Salmonella serotypes B, D, and E with very low concentration. The biosensor consists of a focusing region, and three detection regions. The cells focusing was achieved using a ramp down electroplated vertical electrode pair along with tilted thin film finger pairs that generate p-DEP forces to focus and concentrate the bacterial cells into the center of the microchannel, and direct them toward the detection region. The detection regions consist of three interdigitated electrode arrays (IDEA), each with 20 pairs of finger coated with a mixture of anti-Salmonella antibody and crosslinker to enhance the adhesion to IDEA. The impedance changes as the target Salmonella binds to the antibody. The biosensor has showed excellent performance as proven by the detection of a single Salmonella serotype B, and simultaneous detection of two Salmonella serotypes B and D with a limit of detection (LOD) of 8 Cells/ml in ready-to-eat turkey samples, the addition of focusing capability improved the measured signal by a factor of between 4–4.5, the total detection time of 45 minutes, selectivity of the sensor on different types of bacterial cells, and the ability to distinguish between dead and live cells.
Introduction
The Center for Disease Control and Prevention (CDC) estimates that around 48 million people in America get sick, 128,000 are hospitalized, and 3,000 die of foodborne diseases for each year1. Salmonella is ranked as the number one in the five pathogens that contribute to domestic foodborne illnesses resulting in hospitalization and death2. In 2013, the Economic Research Service (ERS) from USDA has reported that the annual cost due to infection of Salmonella in food source is estimably 3.6 billion in US dollar, while the aggregate cost due to food recall is 77 billion annually3. Among all the foodborne pathogens, Salmonella typhimurium is the second most common serotype of Salmonella found in humans4. So a method that can provide rapid, selective and accurate detection of Salmonella in food products is in urgent need for a better food safety.
Currently, microbiological culture and colony counting as a traditional method remains to be the most commonly used technique for food industry5. This method relies on enrichment and selective cultures and subsequent colony counting6,7. It is the official food screening procedure established by FDA for pathogens detection of clinical and food products8. In addition, it requires 2−5 days by trained professionals for a definitive result, which makes this method time consuming, labor intensive and costly9. Nucleic acid based assays such as PCR, qPCR10, mPCR11 are well established techniques for rapid pathogens detection in food products with high specificity, sensitivity and low detection limit12. It is popular among the food industry because it reduced the testing time to 24 hours13. If the processing plant/company does not have its own lab, additional time is needed to transport samples to a lab that can perform PCR14. PCR cannot distinguish between live and dead bacteria and false positives may occur15,16. BioRad has implemented a DNA cleaning step to get rid of residual dead cells DNA which may reduce the rate of false positive results17. Immunological methods such like enzyme-linked immunosorbent assay (ELISA) is based on specific binding of antibody to antigen18. ELISA and its relevant techniques including IMS-ELISA, ELISA-PCR are based on antibody-antigens binding process. The detection step is rapid but is done after an enrichment culture, e.g., the commercially available Solus Scientific Testing Solutions can detect Salmonella in 36 hrs, respectively19. The long test turnaround time not only cuts into a product’s short shelf-life but also increases the product cost due to the need for storage space and labor needed to transport the products in and out of storage. Alternatively, if food products are released before testing is completed, the company risks releasing product that may cause a foodborne illness or outbreaks of foodborne diseases, economic losses from medical costs associated with foodborne illnesses and product recalls, and risks damage to the company’s brand or even survival. Therefore, there is an urgent need for testing device/methods that could offer a rapid, accurate detection of foodborne pathogens.
Recent development in biosensor has focused on key issues such as rapid detection, limit of detection, feasibility of operation and low cost. The biosensors are mainly based on the immunoassay principle and thus grouped into three major categories: (1) Electrochemical sensors including: (a) Amperometric biosensor. With combination of Immunomagnetic beads and amperometric biosensor, Salmonella were detected using screen – printed carbon electrode with the detection limit of 89 CFU/ml20. (b) Potentiometric biosensor. Shaibani et al. has described a portable nanofiber-light addressable potentiometric sensor than can detect E. coli low to 100 CFU/ml with only 1 hour21. (c) Impedimetric sensors. For example, a glassy carbon electrode modified with graphene oxide and carbon nanotubes has demonstrated a LOD of 25 CFU/ml22. And Wan et al., describe a signal-off impedimetric immune-biosensor using AuNP to detect E. coli with LOD of 100 CFU/ml23. (2) Optical based biosensor. This includes: (a) surface plasmon resonance (SPR). A SPR biosensor based on ultra-low fouling and functionalizable poly brushes has successfully detected E. coli and Salmonella in cucumber and hamburger for 57 CFU/ml and 17 CFU/ml, and 7.4 × 103 CFU/ml and 11.7 × 103 CFU/ml, respectively24. (b) Surface enhanced plasmon resonance (SERS). For example, with the use of magnetic gold nanoparticles achieved LOD for S.aureus and Salmonella of 35 CFU/ml and 15 CFU/ml, respectively25. (c) Colorimetric. For example, Suaifan et al. has designed an optical colorimetric biosensor for detection of Staphylococcus aureus and achieved 7 CFU/ml in pure culture and 40 CFU/ml in food culture as LOD26. (3) Mass based biosensor. This includes (a) acoustic wave-based biosensors. For example, a micro-nano-bio acoustic system for Salmonella at 2 cells/µl27. (b) quartz crystal microbalance (QCM) method. For example, a QCM-based platform using ssDNA aptamers has achieved detection of 103 CFU/ml of Salmonella within only 1 hour28. (c) Cantilever and based biosensor. For example, a dynamic-mode cantilever biosensor without surface functionalization has detected E. coli at 100 Cells/ml29. Although many of these methods have provided promising research results, none of them have been commercialized yet. This might be due to the limit of detection, the use of microbead/nanoparticles coated antibody may have complicated the detection process. The USDA/FDA set a zero-tolerance requirement for ready to eat (RTE) products. For example, in RTE poultry products, the testing method must meet the AOAC standard for certification, i.e.,1 cell/325 gr of product.
Impedimetric based biosensing technique, as one kind of the electrochemical biosensor, has proven to be a promising technique for detection of foodborne pathogens in terms of its detection speed, accuracy, and sensitivity30. The impedance of the electrodes will be changed by capturing bacterial cells on electrodes.
In this study, an impedance-based biosensor for simultaneous detection of multiple Salmonella serotypes has been designed, fabricated, characterized, and validated using ready to eat Turkey (RTE) samples. Detection of S. Typhimurium with various concentrations, with and without cross-linker, were utilized using RTE Turkey samples and pure culture samples. The device’s selectivity, differentiation between dead and live cells were also validated. Initially, the surface of the detection electrodes were immobilized with bioreceptors (in our case antibodies), thus, the labeling procedure was eliminated. Impedance change were achieved after direct capture of target bacteria by specific antibody on surface of the electrode31. An impedance analyzer was used to monitor the impedance changes.
Dielectrophoresis (DEP) is a particle manipulation technique defined as particles movement in medium in presence of electrical fields (E-field). Particles that suspended in a medium can be manipulated and polarized by non-uniform E-field. The particle – medium interface will attract and accumulate all the charges inside the particles and the medium to their interface edges. An electric double layer will be formed by the accumulated electric charges. The relative quantity of these accumulated charges on the edges of the particle and the medium will be determined by the polarization of both medium and particle. Under E-field, positive and negative charges attract and bind to each other, and thus a dipole will be created. And the relative difference of positive and negative charges will determine the orientation of the dipoles created32. The net dipoles from the E-field results in a force that attract the opposite charges and repulse the identical charges. In the case of the uniform E-field when that generated by same size and shape of the electrodes with AC/DC source applied, particles under that E-field will be stationary because forces exerted on the particles from positive and negative charge will cancel each other, so the net force will be zero. However, the E-field will be non-uniform and inhomogeneous when created by different size, shape or non-parallel electrodes with AC/DC source applied. The net force on the particles in that field will not be zero. The movement of the spherical particle is the result of DEP force, and the force can be expressed as33:
where εm is the permittivity of the medium, r is the radius of the particle, is the gradient of the external E-field. FCM is the Clausius-Mossotti factor, which has information about the dielectric properties of the particle and the medium, also the frequency dependence of DEP force. FCM is given by34:
εp* and εm* are the complex permittivity of the particle and medium, respectively. From the equation above, complex permittivity of the particle and medium are frequency dependent. The relation of and can be changed by adjusting frequency of the E-field. When , the particle will be pulled to the region where the E-field is stronger, which is observed as positive DEP (pDEP) force. When , the particle will be pushed away to weaker E-field region, which is observed as negative DEP (nDEP) force. DEP force will accelerate the particle inside the medium, and the acceleration is contributed by DEP force and the drag force. The drag force increases with the velocity of the particle moving inside the medium. When the DEP force equals to drag force, the particle reaches its equilibrium force. Usually, the particle reaches to its terminal velocity inside the medium after only a very short time35.
Results
Biosensor design
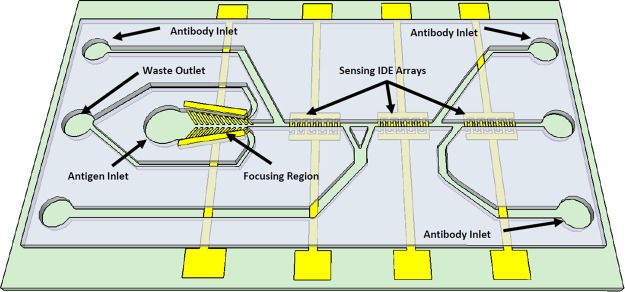
The biosensor consists of two regions fabricated in SU8 microchannels as shown in Fig. 1. The focusing region is used for concentrating the Salmonella sample by getting rid of over 67% volume of the testing material. This has improved the device’s ability to detect bacteria at a low concentration. The focusing has the length of 3 mm and it consists of a vertical electrode pair along with 45° tilted thin film finger pairs with a ramp down channel with a start and end width of from 300 µm, and 75 µm, respectively. This electrode generates p-DEP forces that pushes the Salmonella cells toward the centerline of the channel and direct them toward the sensing microchannel. This has resulted in a concentrated sample. The bulk fluid will flow toward the outer channel into the waste outlets. The sensing region consists of three set of interdigitated electrode (IDE) arrays fabricated in the same microchannel that incorporate impedance measurement principles to detect the presence/ absence of Salmonella. Each electrode array has 20 finger pairs with finger length, width, and spacing between the fingers of 30 µm, 10 µm, and 10 µm, respectively. The bonding pads are stretched away to the sides of the device and used for impedance measurements. The sensing microchannel has a width and height of 25 µm, 25 µm, respectively. The three sensing electrodes were pre-functionalized by delivery specific anti-Salmonella antibody serotypes B, D and E, each for one electrode through independent antibody inlets without causing cross-contamination. All the antibodies were pre-linked with crosslinker. After the channel is filled with antibody solutions, the flow was stopped for 1 hour to ensure efficient adhesion of the antibodies to IDE arrays. The Salmonella samples were tested by flowing them through the sample inlet toward the focusing microchannel and continue flowing toward the sensing IDE arrays. After the sensing channel was filled with the testing sample, the flow was stopped for 30 minutes to facilitate the contact and binding between Salmonella antigens and the corresponding Salmonella antibody. It is noted that each device was used for one test to eliminate the possibility of sample contamination.
Figure 1.
3D schematic of the impedance biosensor showing the focusing IDE array and three sets of the IDE arrays for three sensing regions. Each of the region could be used for sensing different serotypes of Salmonella without causing any cross-contamination.
Focusing demonstration using microbeads
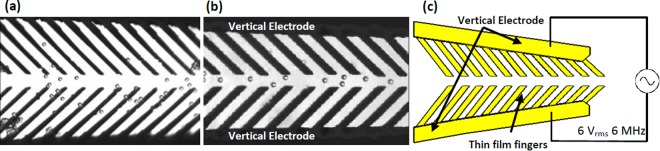
We first used polystyrene microbeads to test the focusing capability of the device. The microbeads were introduced into the channel via the sample inlet. When a AC signal (6 V peak-to-peak at 6 MHz from an AC power supply) was applied across the bonding pad, the focusing electrodes generates a non-uniform E-field and p-DEP forces the microbeads to move toward the center of the channel toward the region of high E-field. Figure 2 shows microbeads position before and after focusing turning on.
Figure 2.
Optical Image of 4 µm microbeads (a) microbeads were dispersive at the beginning position of focusing region. (b) microbeads were concentrated to center of the channel at then middle position of the focusing region when focusing function is on. (c) schematics of the focusing electrode along with the applied voltage and frequency.
Dose response
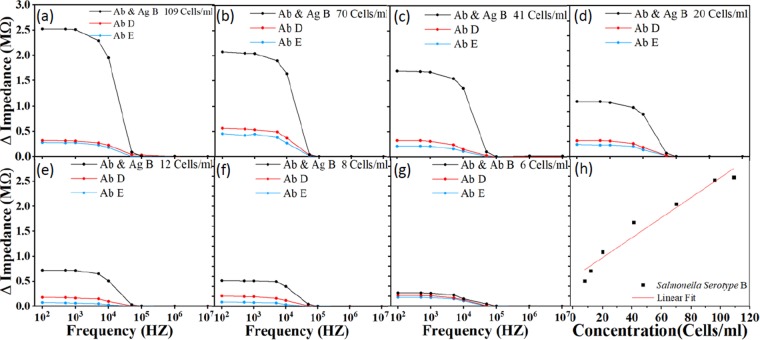
The variations of the amount of substance on the surface of the electrodes cause the impedance to change. Impedance increases after antibody is immobilized on the IDE arrays compared to the IDE array immersed with just the antibody. After the Salmonella were selectively bound to the Salmonella antibody, the impedance was changed. The variation in impedance response is based on the concentration of the Salmonella as shown in Fig. 3, the sensor was tested with antibody serotypes B, D and E for each of the concentration. The impedance increases with higher concentration. The detection limit of the sensor was found to be 8 Cells/ml. For all concentrations, impedance decreases as function of frequency. All the sample concentration is pre-confirmed by culture method, so the number of bacteria colony is known before being delivered to the sensor for testing. The device cannot quantify the number of detected bacterial cells but it informs the presence or absence of bacterial cells. Experimentally, the impedance across the IDE array were measured and recorded after immobilizing the antibodies, and after the exposure of the target molecules (e.g., Salmonella B). Then, the antibody impedance was subtracted from the total impedance (after the exposure of the target molecules) and plotted the impedance difference, i.e., ΔImpedance (MΩ). To demonstrate the limit of detection, the device was tested using Salmonella B with a concentration of 6 Cells/ml. The impedance value is close to that of the antibody B, D and E. For example, the impedance reference reading after antibody coating was 5.22 MΩ, while the impedance after exposure to Salmonella B was 5.48 MΩ. The difference, 0.26 MΩ, is very small and hard to distinguish. Thus, 8 Cells/ml is the limit of detection. The Salmonella concentration was plotted as a function of impedance change from 8 Cells/ml to 109 Cells/ml in Fig. 3(h). The figure demonstrates that the variation of impedance change is directly proportional to bacterial cell concentration. In addition, the figure shows a linear correlation, ΔImpedance = Slope ∗ Concentration + Δo, where Δo is the intersection of the linear fit and the Y-axis. The correlation coefficient is 0.955 and the sensitivity (Slope) is 0.02 MΩ per bacteria concentration decade. The limit of detection is 8 Cells/ml.
Figure 3.
Impedance response for different concentrations of Salmonella. The sensor was first functionalized with crosslinked Salmonella antibody type B, D and E. Salmonella serotype B with concentrations of (a) 109, (b) 70, (c) 41, (d) 20, (e) 12, (f) 8 Cells/ml, spiked with ready-to-eat turkey sample was then delivered to the sensor for detection. The limit of detection was found to be 8 Cell/ml. (g) shows the variation of impedance change is directly proportional (linear) to bacterial cell concentration. The linear correlation coefficient is 0.955 and the sensitivity (Slope) is 0.02 MΩ per bacteria concentration decade. Ab stands for antibody, Ag stands for antigen.
Simultaneous detection
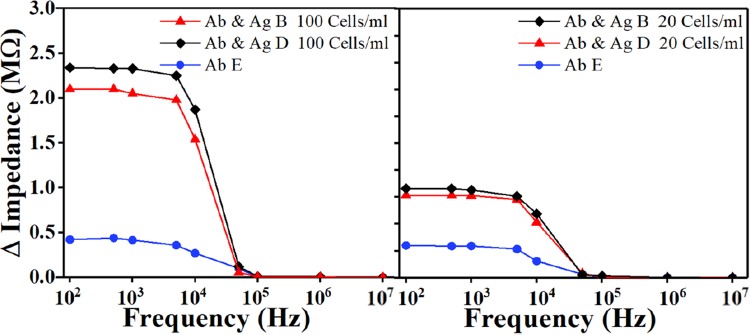
Initially, the three mixtures of antibodies (B, D and E) were immobilized on the detection electrodes independently without causing any cross contamination such that Salmonella type B was placed on the first sensing electrode, D on the second sensing electrode, and E on the third sensing electrode. The impedance of each sensing electrode was recorded. After the Salmonella were injected to the device, the impedance was measured again. The experiments were performed using RTE turkey spiked with Salmonella type B and D with several concentrations. The impedance differences between antibodies and antigens were then calculated for each sensing region. The impedance change that demonstrates the device ability to simultaneously detect two Salmonella serotype B and D were plotted in Fig. 4. The results demonstrated a high impedance change using the electrode with Salmonella antibody B and D, while the other one electrode with antibody E had a weak signal.
Figure 4.
Sensor’s ability to simultaneously detect Salmonella serotypes B and D were tested by flowing two equal quantities mixture of the two different Salmonella serogroups for (a) 100 Cells/ml, and (b) 20 Cells/ml concentration. The figure shows the impedance change response as a function of frequency. The impedance of Salmonella antibodies B and D were subtracted from the measured impedance after the exposure of the target molecules and plotted the impedance difference. Ab stands for antibody, Ag stands for antigen.
Salmonella detection with and without focusing function
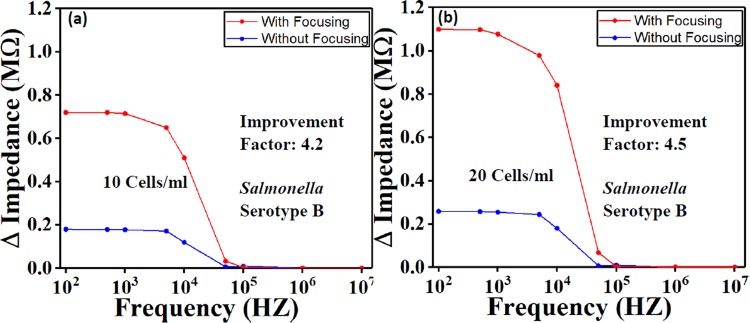
Similar electrical field (amplitude and frequency) was applied to the biological sample to concentrate cells towards the sensing region. Experiments were conducted on Salmonella detection with and without the use of the focusing effect. The results demonstrated that the signal was improved by a factor range between 4–4.5 (Fig. 5).
Figure 5.
The comparison with and without focusing function on Salmonella with 2 concentrations (a) 10 Cells/ml and (b) 20 Cells/ml. The result has showed that the focusing function has improved the signal response by a factor up to 4.5.
Selectivity on Salmonella and E. coli
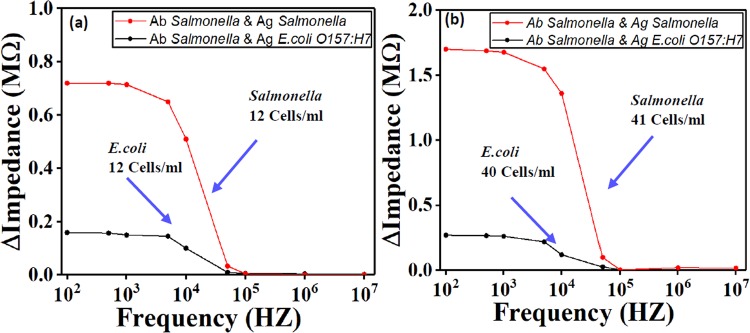
In order to test the sensor’s selectivity on different types of antigen, Salmonella and E. coli O157:H7 were used. The electrodes were coated with Salmonella antibody type B. the tested samples were spiked with Salmonella type B and E. coli. They were then introduced into the device separately in two different experiments. The result in Fig. 6 demonstrated that the device with E. coli antigens had a weak signal response while the other device with Salmonella type B antigens has a high signal response.
Figure 6.
Test of sensor’s specificity to Salmonella serotype B for two concentrations (a) around 12 Cells/ml and (b) around 40 Cells/ml. The result show that E. coli O157:H7 wound not bind to Salmonella antibody due to specificity. Ab stands for antibody, Ag stands for antigen.
Live and dead Salmonella cells differentiation
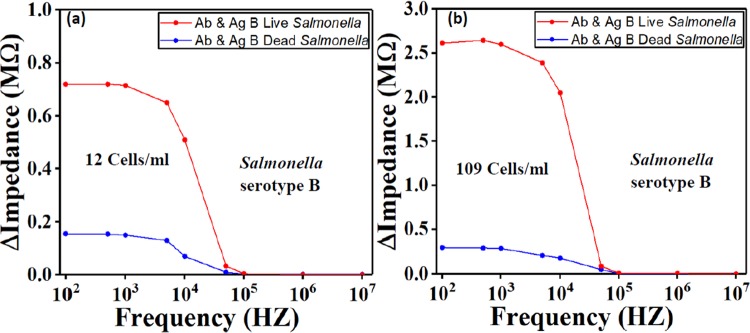
The biosensor was tested with live and dead Salmonella cells to demonstrate its ability to differentiate between them. The Salmonella bacteria were killed by brief exposure to high temperature which resembles real-life event. Two devices were coated with the anti-Salmonella serotype B, while dead and live bacteria were pumped into the two devices separately, each into one device. Figure 7 indicates that the impedance value obtained from the dead bacteria was close to baseline impedance, which was expected because the membrane of Salmonella antigen was damaged and thus lost the function to bonding with Salmonella antibody.
Figure 7.
Comparison of live and dead Salmonella serotype B for two concentrations (a) 12 Cells/ml and (b) 109 Cells/ml. The results show the sensor’s capability to differentiate between live and dead Salmonella. Ab stands for antibody, Ag stands for antigen.
Discussion
This paper presents design and fabrication of an impedance-based MEMS biosensor for different Salmonella strains detection. Rapid and quantitative detection of Salmonella in food source has been achieved. The design has single channel with focusing electrode and three sensing electrodes that make the sensor suitable for detections of three Salmonella serotypes simultaneously. The results demonstrate that the impedance increases when concentration of target bacteria increases, with the limit of detection found to be as low as 8 Cells/ml. Microbeads were used as an alternative for Salmonella cells in order to demonstrate the device focusing capability. The optimum parameters including frequency and AC voltage (Vp-p) obtained with microbeads, were used for focusing Salmonella. The relative permittivity of the microbeads, and Salmonella are 436, and 4.5–6.537,38, respectively. The small difference may affect the optimum value of the operation frequency. However, it is noted that the frequency of operation is the dominant parameter that affect the focusing of microbeads and cells, especially the solution is the same for both experiments. By changing the frequency, we can switch between n-DEP, and p-DEP. The testing results demonstrated that the addition of focusing region have improved the response sensitivity by a factor between 4–4.5, while the total detection time was 45 minutes. The sensor can also differentiate dead from live cells and has a good selectivity on different Salmonella serotypes.
The results demonstrate the device advantages in terms of detection limit, required detection time and ease of operation. The impedance-based sensor in this study is suitable for detecting multiple pathogens simultaneously, which hasn’t been achieved by any other research groups. The ability to detect very low concentration (8 Cells/ml) in 45 minutes has granted the sensor a great potential to be used in the food industry. The current detection techniques which include bacterial culture, PCR, and ELISA do not allow timely assessment of safety of food products39,40 because they require 2–5 days9,14, and 1–2 days41,42, and 2–3 days5–7, respectively. In addition, PCR cannot distinguish between live and dead bacteria and false positives may occur43,44. Multiple biosensing techniques demonstrated high sensitivity including electrochemical biosensors, optical biosensors, and mass-based biosensors. However, none of these techniques met the USDA/FDA requirements. In addition, those devices can only do detection of one type of pathogens at a time.
The USDA/FDA set a zero-tolerance requirement for RTE poultry products. The testing method must meet the AOAC standard for certification, i.e.,1 cell/325 gr of product. Therefore, use of our device for RTE poultry products will require a short enrichment step. Due to the high sensitivity of our existing device, 8 Cells/ml, the length of the enrichment culture will be much shorter than with other technologies, such as ELISA and PCR. The duration of the enrichment step with our device is expect it to be around 6 hrs. This means the total detection time for our device will be approximately 7 hrs, which would mean that the results could be available within a typical production plant shift. In experiment, approximately 32.5 Salmonella cells were inoculated on 325 gr RTE Turkey and enriched in 2,925 ml BPW at 1:10 dilution and at 37 °C. After 4 hrs, the Salmonella concentration reached 7 Cells/ml. In addition, by applying the new USDA’s Laboratory Guidebook which suggests that the enrichment culture is done by inoculating 325 gr RTE poultry into 975 ml BPW at 1:4 dilution, a less time will be needed to reach the same pathogen concentration. For example, when the same number of Salmonella cells (32.5 cells) inoculated on 325 gr RTE Turkey sample, it took only 2 hours to reach 8 Cells/ml, which can be detected with our biosensing device. The low limit of detection (8 Cells/ml) indicates that our device will be able to approach the 1 cell/325 gr requirement within a production plant shift.
Methods
Biosensor fabrication
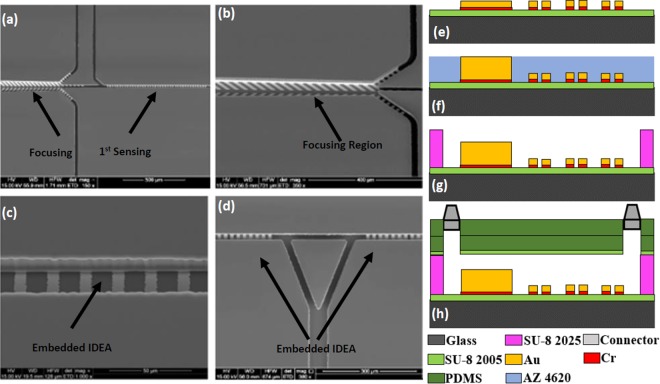
The biosensor was fabricated on a 2 × 1.5 inch2 glass substrate using surface micromachining in following steps: (1) The glass slide was first cleaned using Piranha solution (hydrogen peroxide and sulfuric acid with a ratio of 1:3) for 4 minutes. (2) A layer of SU-8 (Microchem) with a thickness of 4 μm was spin coated on the surface of the glass slide, prebaked, exposed to UV light and hard bake at 150 °C for 30 minutes. (3) Chromium (Cr) and gold (Au) thin layers were deposited using DC sputtering with thickness of 50 nm and 150 nm respectively. (4) The Au thin film was patterned to form the IDE arrays, bonding pads, focusing fingers and seed layer for electroplating the focusing side walls. (5) Positive photoresist (AZ4620) with 15 µm thickness was patterned as a mold for electroplated Au to create the sidewalls. The Au was then electroplated for 15 μm. This is followed by Cr layer etching. (6) The microchannel was formed using SU-8 2025 negative photoresist with a thickness of 25 μm. (7) Two thick layers of polydimethylsiloxane (PDMS) were prepared and cured. The first one is used to cover the microchannel, while the second layer serves as top cover with fluidic connectors. Oxygen plasma treatment was used on the first layer to change its surface to be more hydrophilic before it was aligned and bonded to the glass substrate. Then it was baked on a hotplate at 65 °C for 5 minutes to ensure the bonding between the PDMS and the glass substrate is secured. The second PDMS slab with inlet and outlet fluidic connectors were then bonded to the first layer of PDMS using oxygen plasma. Epoxy glue was used to seal the fluidic connectors further to improve the device reliability. Scanning electron micrographs (SEMs) of the fabricated devices are shown in Fig. 8(a,d). Figure 8(e,h) is the fabrication of the sensor step by step. A completely packaged sensor in PCB board in Fig. 9 has shown the fluidic connectors, tubes and electrical wires.
Figure 8.
SEM of the sensor: (a) Focusing and 1st Sensing region, (b) Magnified view of the focusing region. The excess waste will leave through the channels on two sides while the bacteria stay in the center and enter the sensing region, (c) Magnified view of the IDE arrays for sensing region embedded in the microchannel, (d) IDE arrays for 2nd and 3rd Sensing regions. The triangle part is used to split the fluid during antibodies collection for 2nd and 3rd sensing regions to reduce cross-contamination. Cross-sectional view demonstrating multiple layers of the sensor: (e) Electrode traces on top of the SU8 and glass slide, (f) Electroplating to increase the height of the focusing wall, (g) SU8 microchannel, (h) PDMS and fluidic connectors as seal.
Figure 9.
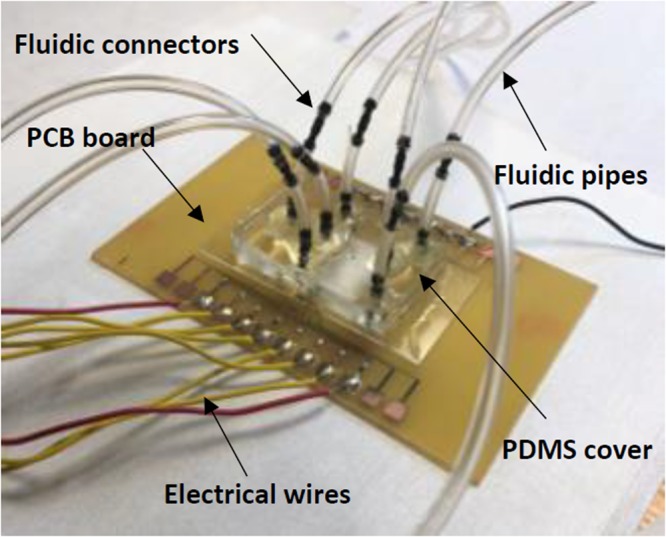
Completely fabricated sensor on a PCB board showing the fluidic connectors, tubes and wires.
Microbeads for focusing function demonstration
The 4 μm diameter DUKE STANDARDSTM Microbead with 1% coefficient of variation was purchased directly from ThermoScientific. The microbeads were made from polystyrene and suspended in water-based solution before use. The relative permittivity of the microbeads to free space is 3.8 (the permittivity of free space is 8.854187817 × 10−12 C2/(Nm2)).
Culture and cell preparation Salmonella culture preparation
Ready-to-eat turkey breast (RTE) turkey breast was purchased from a grocery store and stored at 4 °C until use. Each 325 g RTE turkey breast was weighted and placed in a sterile bag, 2925 ml buffered peptone water was poured into the bag, and the bag was shaken for one minute. The supernatant was then filtered through a 100 μm and then a 20 μm cell strainer (pluriSelect Life Science, Leipzig, Germany) to remove big debris which may block the biosensor microchannels. The filtered rinse was used freshly to dilute Salmonella cells or aliquoted and frozen at −20 °C until used.
Salmonella typhimurium preparation
An avirulent Salmonella enterica Typhimurium strain (ΔsipB, Cmr) was used to spike RTE turkey breast. An overnight culture (37 °C, 200 rpm, in LB broth) of S. enterica Typhimurium was harvested by centrifugation at 4000 rpm 10 minutes and washing with sterile distilled water 3 times and then suspended in 25% sterile glycerol. The cell suspension was aliquoted and frozen at −80 °C until used. At the same time, one aliquot was serially diluted and plated on LB agar plates to determine the cell concentration. The cell concentration was determined to be 2 × 109 Cells/ml. Before the test, one aliquot Salmonella suspension and several aliquots of RTE turkey rinse were thawed on ice. The Salmonella suspension was then diluted with the of filtered RTE turkey rinse to desired concentrations.
Antibody preparation
Rabbit anti-Salmonella O antiserum poly B, D, and E (Becton, Dickinson and Company, Franklin Lakes, NJ) were used as capture antibodies. The crosslinker, sulfosuccinimidyl 6-[3-(2-pyridyldithio) propionamido] hexanoate (sulfo-LC-SPDP) (Fisher Scientific, Hampton, NH), was used for antibody immobilization. Briefly, for each test, 8 μl of each antiserum was diluted with 292 μl filtered chicken rinse, mixed with 300 µl sulfo-SPDP (20 mM water solution), and then incubated at room temperature for 1 hour. To reduce the disulfide bond of the thiolated antibody, 200 μl DTT (0.1 M sodium acetate buffer, 0.1 M NaCl, pH 4.5) (Fisher Scientific, Hampton, NH) was then added into the tube to react for 30 min at room temperature before the antiserum mixture was loaded into the biosensor. The 1:100 diluted antiserum-crosslinker mixtures were delivered into the device using three syringe pumps through the antibodies’ inlets, respectively. The electrode surfaces were then functionalized with the three antiserum-crosslinker mixtures, one for each channel and without causing any cross contamination. The flow was stopped to all the three channels and allowed 1 hour for the antibodies to get immobilized on the gold electrodes to achieve good bonding to the electrodes thus providing high specificity. A washing step was performed after 1 hour to remove any unbounded antibodies by pumping distilled water inside the three channels.
Antibody crosslinker
A new immobilization method using cross-linker was used to exploit natural and direct covalent bond formation between gold electrode and a thiolated antibody. The cross-linker we used is Sulfo-LC-SPDP, it is a thiolation reagent that introduce available sulfhydryl groups to an antibody. A monolayer coating of the thiolated antibody onto the gold surface can be easily achieved because the reaction potential between the gold and sulfur is very strong, which achieved 50% in signal response45,46. In our experiments, all the antibodies were prepared with crosslinker.
Antibody immobilization
The mixtures of antibody with cross-linker were delivered to the device through inlets using syringe pumps at a constant flow rate. Subsequently, the electrode surface was then functionalized with the antibodies, one for each sensing region without causing any cross contamination. The flow was stopped for 1 hour to allow the antibodies to get immobilized on the gold electrodes firmly to achieve a good and specific bonding between the antibodies and the gold electrodes. A washing step by pumping DI water into the channel is then performed after 1 hour of the immobilization to remove excess antibodies and other waste materials. The flow for all fluids was set to be 2 µL/min to prevent fluids from flowing in random fashion.
Antigen capturing
Ready to eat turkey (RTE) breast meat was stored at 3 °C before use. An avirlent S. enterica Typhimurium strain (ΔsipB, cat+) (serotype B) was used for spiking turkey meat. The RTE samples spiked with Salmonella strain were then introduced into the channel through the sample inlet by a syringe pump at a constant flow rate. After the channel was filled with sample, the flow was stopped for 30 minutes to allow the biological interaction between the antibodies and the antigens to form antibodies-antigens binding. The same washing step and flow rate were used again to remove unbounded cells and excess reactants. Impedance was then measured by Agilent 4294A impedance analyzer. The device will be discarded after being used for one time.
Acknowledgements
This project is supported in part by USDA-Capacity Building Grant, and Coulter Foundation/MU partnership.
Author Contributions
J.L. conducted the fabrication, testing and data analysis. I.J., A.A., and L.Z. helped with fabrication and lab equipment. M.A. conceived the experiments and design. Z.S., S.Z., M.E.-D. prepared biological sample. All authors reviewed the manuscript.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.CDC. Estimates of Foodborne Illness in the United States | Estimates of Foodborne Illness | CDC. Center for disease control and prevention 3 (2016). Available at: https://www.cdc.gov/foodborneburden/index.html. (Accessed: 31st May 2018).
- 2.Scallan E, et al. Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.UDSA. USDA ERS - Cost Estimates of Foodborne Illnesses. (2014). Available at: http://www.ers.usda.gov/data-products/cost-estimates-of-foodborne-illnesses/cost-estimates-of-foodborne-illnesses/#Salmonella (non-typhoidal). (Accessed: 31st May 2018).
- 4.Solutions, S. Regulatory Compliance. Available at: https://www.fsis.usda.gov/wps/portal/fsis/topics/regulatory-compliance/. (Accessed: 31st May 2018).
- 5.Bitton Gabriel. Microbiology of Drinking Water. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2014. [Google Scholar]
- 6.Jasim, I. et al. An impedance biosensor for simultaneous detection of low concentration of Salmonella serotypes in poultry samples. In TRANSDUCERS 2017 - 19th International Conference on Solid-State Sensors, Actuators and Microsystems 726–729, 10.1109/TRANSDUCERS.2017.7994151 (IEEE, 2017).
- 7.Magnitude & Burden of Waterborne Disease in the U.S. | Healthy Water | CDC. Available at: https://www.cdc.gov/healthywater/burden/index.html. (Accessed: 31st May 2018).
- 8.WHO | Publications on water, sanitation and health: 2017. Who (2018).
- 9.Krauss S, Webster RG. Avian Influenza Virus Surveillance and Wild Birds: Past and Present. Avian Dis. 2010;54:394–398. doi: 10.1637/8703-031609-Review.1. [DOI] [PubMed] [Google Scholar]
- 10.Abdullah, A., Jasim, I., Alalem, M., Dweik, M. & Almasri, M. MEMS based impedance biosensor for rapid detection of low concentrations of foodborne pathogens. In 2017 IEEE 30th International Conference on Micro Electro Mechanical Systems (MEMS) 381–385, 10.1109/MEMSYS.2017.7863421 (IEEE, 2017).
- 11.Li F, et al. Sextuplex PCR combined with immunomagnetic separation and PMA treatment for rapid detection and specific identification of viable Salmonella spp., Salmonella enterica serovars Paratyphi B, Salmonella Typhimurium, and Salmonella Enteritidis in raw meat. Food Control. 2017;73:587–594. doi: 10.1016/j.foodcont.2016.09.009. [DOI] [Google Scholar]
- 12.Chin WH, et al. Direct PCR – A rapid method for multiplexed detection of different serotypes of Salmonella in enriched pork meat samples. Mol. Cell. Probes. 2017;32:24–32. doi: 10.1016/j.mcp.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Bashir R. Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotechnol Adv. 2008;26:135–150. doi: 10.1016/j.biotechadv.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Bonetta S, et al. Detection of pathogenic Campylobacter, E. coli O157:H7 and Salmonella spp. in wastewater by PCR assay. Environ. Sci. Pollut. Res. 2016;23:15302–15309. doi: 10.1007/s11356-016-6682-5. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Mustapha A. EMA-Real-Time PCR as a Reliable Method for Detection of Viable Salmonella in Chicken and Eggs. J. Food Sci. 2010;75:M134–M139. doi: 10.1111/j.1750-3841.2010.01525.x. [DOI] [PubMed] [Google Scholar]
- 16.He Y, Chen C-Y. Quantitative analysis of viable, stressed and dead cells of Campylobacter jejuni strain 81-176. Food Microbiol. 2010;27:439–446. doi: 10.1016/j.fm.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, et al. Biosensors based on modularly designed synthetic peptides for recognition,detectionand live/dead differentiation of pathogenic bacteria. Biosens. Bioelectron. 2016;80:9–16. doi: 10.1016/j.bios.2016.01.041. [DOI] [PubMed] [Google Scholar]
- 18.Mansfield LP, Forsythe SJ. The detection of Salmonella using a combined immunomagnetic separation and ELISA end-detection procedure. Lett. Appl. Microbiol. 2000;31:279–283. doi: 10.1046/j.1472-765x.2000.00811.x. [DOI] [PubMed] [Google Scholar]
- 19.Pathogen Testing Solutions Salmonella E. coli Listeria. Available at: http://www.solusscientific.com/pathogen-testing-solutions/. (Accessed: 4th September 2018).
- 20.Fei J, Dou W, Zhao G. Amperometric immunoassay for the detection of Salmonella pullorum using a screen - printed carbon electrode modified with gold nanoparticle-coated reduced graphene oxide and immunomagnetic beads. Microchim. Acta. 2016;183:757–764. doi: 10.1007/s00604-015-1721-3. [DOI] [Google Scholar]
- 21.Shaibani PM, et al. Portable Nanofiber-Light Addressable Potentiometric Sensor for Rapid Escherichia coli Detection in Orange Juice. ACS Sensors. 2018;3:815–822. doi: 10.1021/acssensors.8b00063. [DOI] [PubMed] [Google Scholar]
- 22.Jia F, et al. Impedimetric Salmonella aptasensor using a glassy carbon electrode modified with an electrodeposited composite consisting of reduced graphene oxide and carbon nanotubes. Microchim. Acta. 2016;183:337–344. doi: 10.1007/s00604-015-1649-7. [DOI] [Google Scholar]
- 23.Wan J, et al. Signal-off impedimetric immunosensor for the detection of Escherichia coli O157:H7. Sci. Rep. 2016;6:19806. doi: 10.1038/srep19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaisocherová-Lísalová H, et al. Low-fouling surface plasmon resonance biosensor for multi-step detection of foodborne bacterial pathogens in complex food samples. Biosens. Bioelectron. 2016;80:84–90. doi: 10.1016/j.bios.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, et al. Gold nanoparticles enhanced SERS aptasensor for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Biosens. Bioelectron. 2015;74:872–877. doi: 10.1016/j.bios.2015.07.033. [DOI] [PubMed] [Google Scholar]
- 26.Suaifan GARY, Alhogail S, Zourob M. Rapid and low-cost biosensor for the detection of Staphylococcus aureus. Biosens. Bioelectron. 2017;90:230–237. doi: 10.1016/j.bios.2016.11.047. [DOI] [PubMed] [Google Scholar]
- 27.Papadakis G, et al. Micro-nano-bio acoustic system for the detection of foodborne pathogens in real samples. Biosens. Bioelectron. 2018;111:52–58. doi: 10.1016/j.bios.2018.03.056. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, et al. QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chem. 2017;221:776–782. doi: 10.1016/j.foodchem.2016.11.104. [DOI] [PubMed] [Google Scholar]
- 29.Leahy S, Lai Y. A cantilever biosensor based on a gap method for detecting E. coli in real time. Sensors Actuators, B Chem. 2017;246:1011–1016. doi: 10.1016/j.snb.2017.02.144. [DOI] [Google Scholar]
- 30.Ivnitski D, Abdel-Hamid I, Atanasov P, Wilkins E, Stricker S. Application of electrochemical biosensors for detection of food pathogenic bacteria. Electroanalysis. 2000;12:317–325. doi: 10.1002/(SICI)1521-4109(20000301)12:5<317::AID-ELAN317>3.0.CO;2-A. [DOI] [Google Scholar]
- 31.Mantzila Aikaterini G., Maipa Vassiliki, Prodromidis Mamas I. Development of a Faradic Impedimetric Immunosensor for the Detection ofSalmonellatyphimuriumin Milk. Analytical Chemistry. 2008;80(4):1169–1175. doi: 10.1021/ac071570l. [DOI] [PubMed] [Google Scholar]
- 32.Kang Y, Li D, Kalams SA, Eid JE. DC-Dielectrophoretic separation of biological cells by size. Biomed. Microdevices. 2008;10:243–249. doi: 10.1007/s10544-007-9130-y. [DOI] [PubMed] [Google Scholar]
- 33.Pohl HA, Pollock K, Crane JS. Dielectrophoretic force: A comparison of theory and experiment. J. Biol. Phys. 1978;6:133–160. doi: 10.1007/BF02328936. [DOI] [Google Scholar]
- 34.Morgan, H. & Green, N. G. AC electrokinetics: colloids and nanoparticles. (Research Studies Press, 2003).
- 35.Hyoung Kang K, Xuan X, Kang Y, Li D. Effects of dc-dielectrophoretic force on particle trajectories in microchannels. J. Appl. Phys. 2006;99:064702. doi: 10.1063/1.2180430. [DOI] [Google Scholar]
- 36.Relative Permittivity - the Dielectric Constant. Available at: https://www.engineeringtoolbox.com/relative-permittivity-d_1660.html. (Accessed: 4th September 2018).
- 37.Weaver JC, Schoenbach KH. Biodielectrics. IEEE Trans. Dielectr. Electr. Insul. 2003;10:715–716. doi: 10.1109/TDEI.2003.1237322. [DOI] [Google Scholar]
- 38.Esteban Ferrer, D. Electric polarization properties of single bacteria measured with electrostatic force microscopy Theoretical and practical studies of Dielectric constant of single bacteria and smaller elements (2014). [DOI] [PubMed]
- 39.Pathogen Testing Solutions Salmonella E. coli Listeria. Available at: http://www.solusscientific.com/pathogen-testing-solutions/. (Accessed: 4th September 2018).
- 40.Lee KM, et al. Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control. 2015;47:264–276. doi: 10.1016/j.foodcont.2014.07.011. [DOI] [Google Scholar]
- 41.Deisingh AK, Thompson M. Strategies for the detection of Escherichia coli O157:H7 in foods. J Appl Microbiol. 2004;96:419–429. doi: 10.1111/j.1365-2672.2003.02170.x. [DOI] [PubMed] [Google Scholar]
- 42.Lazcka O, et al. Pathogen detection: a perspective of traditional methods and biosensors. Biosens.bioelectron. 2007;22:1205–1217. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 43.Hagens S, Loessner MJ. Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol. 2007;76:513–519. doi: 10.1007/s00253-007-1031-8. [DOI] [PubMed] [Google Scholar]
- 44.Velusamy V, et al. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol Adv. 2010;28:232–254. doi: 10.1016/j.biotechadv.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Finklea HO, Avery S, Lynch M, Furtsch T. Blocking Oriented Monolayers of Alkyl Mercaptans on Gold Electrodes. Langmuir. 1987;3:409–413. doi: 10.1021/la00075a024. [DOI] [Google Scholar]
- 46.Bain CD, Evall J, Whitesides GM. Formation of monolayers by the coadsorption of thiols on gold: variation in the head group, tail group, and solvent Formation of Monolayers by the Coadsorption of Thiols on Gold: Variation in the Head Group, Tail Group, and Solvent’. Advances. 1989;111:7155–7164. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.