Abstract
The evolutionary history of African savannah tree species is crucial for the management of their genetic resources. In this study, we investigated the phylogeography of Parkia biglobosa and its modeled distribution under past and present climate conditions. This tree species is very valued and widespread in West Africa, providing edible and medicinal products. A large sample of 1610 individuals from 84 populations, distributed across 12 countries in Western and Central Africa, were genotyped using 8 nuclear microsatellites. Individual-based assignments clearly distinguished 3 genetic clusters, extreme West Africa (EWA), center of West Africa (CWA), and Central Africa (CA). Overall, estimates of genetic diversity were moderate to high, with lower values for populations in EWA (allelic richness after rarefaction [AR] = 6.4, expected heterozygosity [HE] = 0.78, and observed heterozygosity [HO] = 0.7) and CA (AR = 5.9, HE = 0.67, and HO = 0.61) compared with populations in CWA (AR = 7.3, HE = 0.79, and HO = 0.75). The overall population differentiation was found to be moderate (FST = 0.09). A highly significant isolation by distance pattern was detected, with a marked phylogeographic signature suggesting possible effects of past climate and geographic barriers to migration. Modeling the potential distribution of the species showed a contraction during the last glaciations followed by expansion events. The exploratory approximate Bayesian computation conducted suggests a best-supported scenario in which the cluster CWA traced back to the ancestral populations and a first split between EWA and CWA took place about 160000 years before present (BP), then a second split divided CA and CWA, about 100000 years BP. However, our genetic data do not enable us to conclusively distinguish among a few alternative possible scenarios.
Keywords: approximate Bayesian computation, genetic diversity, microsatellites, savannah, sub-Saharan Africa, tree species
Phylogeographic studies are crucial for a better understanding of the genetic patterns and the evolutionary history of tree species. Such studies are mainly available for temperate tree species, but recently, an increasing amount of research has focused on tropical tree species. The literature addressing phylogeographic studies of sub-Saharan Africa trees, using DNA markers, is limited to a few cases. These include savannah tree species, such as Acacia senegal (Odee et al. 2012), Adansonia digitata (Kyndt et al. 2009; Leong Pock Tsy et al. 2009), Khaya senegalensis (Sexton et al. 2015), Vitellaria paradoxa (Allal et al. 2011; Logossa et al. 2011), as well as rainforest and montane tree species, such as Milicia excelsa (Daïnou et al. 2010) and Prunus africana (Kadu et al. 2013; Mihretie et al. 2015). The genetic patterns identified often reflect the restrictions to gene flow that shaped the phylogeographical or spatial genetic structure of these species (e.g., Hardy et al. 2013). Abrupt changes in the pattern of genetic variation are usually interpreted as indication of the presence of isolating factors (i.e., cultural, geographical, biological). Major geographical barriers to gene flow in sub-Saharan Africa have been suggested. The paleo-lake Mega-Chad probably caused a very long isolation and led to substantial genetic differences between western populations of A. digitata and those of southern and eastern Africa (Leong Pock Tsy et al. 2009). Similarly the Rift Valley system is considered responsible for an influence on the different gene pools of P. africana (Kadu et al. 2013; Mihretie et al. 2015). Furthermore, the Dahomey Gap, a savannah corridor interrupting the zonal West African rain forest, has been proposed as a barrier to gene flow for rain forest species (Salzmann and Hoelzmann 2005; Duminil et al. 2013; Demenou et al. 2016).
The history of African savannahs over geological times is not well understood. It seems that savannah woodlands gradually replaced lowland rainforests in the regions of central and northern Sahara during the Miocene, 23–25 million years ago (Mya) and, at the end of it, the present configuration of African vegetation started to take shape. During the early Pliocene (5–3.5 Mya), the area covered by savannah woodlands decreased during periods of higher precipitation and expanded during periods of ice-age aridity (Coetzee 1993). This long-term trend was coupled with alternating wet–dry cycles paced by orbital precession driving the monsoon system (Trauth et al. 2009). Thus, from 2.5 Mya to 12000 years before present (BP), that is, during most of the Quaternary, the savannah flora in West Africa was influenced by an alternation of glacial and interglacial periods, especially since 800000 years ago onwards, with 9 major and 12 minor glacial cycles (Hamilton and Taylor 1991). The last 2 glacial maxima occurred during the Pleistocene, from 160000 to 130000 and from 24000 to 12000 years BP (Maley 1996).
Between 24000 and 12000 years BP, the boundary between the Sahara and the Sahel was positioned 300 km further south than currently. A progressive northward spread of woody plants from the south, along rivers and lakes, has been documented since 11500 years BP (Neumann et al. 2009). Very few sites contain sediments from the Late Pleistocene, but paleontological studies based on marine shore sediments of West Africa indicated that over at least the past 700000 years, the vegetation has expanded and retracted repeatedly in response to climatic changes (Dupont 1993). The boundary between the Sahara and the Sahel has been continuously moving on a north-south axis, due to the moving front of the West African monsoon (Larrasoaña et al. 2013), with a strong influence on vegetation dynamics in the region, including the peaks in savannah expansion that coincided with contractions of the Guineo-Congolian forests (Iloh et al. 2017).
During the mid-Holocene (ca. 6000 years BP) the Sahel was a mosaic of various vegetation types, depending on local site conditions, and was strongly influenced by fires (Salzmann 2000), with elements of the Sudanian flora expanding into the Sahelian grassland. The late Holocene was characterized by an increasing aridity and, since 4500 years BP, pastoralists started migrating southward, from the Sahara into the Sahel, pushed by drought. Since 3000 BP, the human impact on the vegetation in the Sahel has become evident (Ballouche and Neumann 1995). Today savannah parklands represent a traditional agroforestry system, common to the semi-arid zones of West Africa where landscapes are characterized by cultivated crops growing under a discontinuous cover of scattered trees, largely part of the original woodland and maintained after clearing for crop cultivation (Boffa 1999).
We focused our study on the phylogeography of the African locust bean (Parkia biglobosa [Jacq.] G. Don.), one of the most important tree species in the savannah parklands of West Africa, both from an economic and a biological perspective (Boffa 1999). The results could provide hints about the underlying effects of large past climate perturbations, which caused cycles of expansions and contractions in the distribution of less described African savannah tree species. Under current environmental conditions the African locust bean has a wide range extending from Senegal to Uganda, between latitudes 3°N and 15°N, with fragmented habitats in the eastern part of its distribution (Hall et al. 1997). The species is threatened by overexploitation, bush fires, and a progressive habitat degradation leading to fragmentation of tree populations (Gaisberger et al. 2017). Furthermore, overgrazing by domestic animals causes a lack of regeneration and an over-aging of tree individuals in savannah parklands; additional potential threats are also envisaged as a result of the absence or declining number of pollinators (Nikiema 1993; Ouedraogo 1995; Ræbild et al. 2012). Knowledge about the evolutionary history of the species and the present patterns of genetic variation is crucial for the development of genetic conservation strategies and breeding programs, in view of expected future climatic changes. Given the high socioeconomic importance of P. biglobosa, with this study we wished to address a critical knowledge gap on its phylogeography.
Parkia biglobosa is monoecious and highly outcrossing (>95%), pollinated by megabats over long distance and by insects over short distance. Polyads probably offer a selective advantage for this animal-mediated pollination. Each infructescence contains up to 25 pods, each with a full subfamily inside. Seeds are mainly disseminated by primates, rodents, and birds (Lompo et al. 2017). The first range-wide study on the genetic variation of this species was conducted by Sina (2006) using allozyme markers. High intrapopulation genetic diversity was observed, while a moderate genetic differentiation (FST = 0.13) was estimated among the 64 western and central African populations. A re-analysis of these data by Lompo et al. (2017) showed a high genetic diversity in the central part of West Africa (Benin, Burkina Faso, Ghana, Ivory Coast, Mali, Togo). However, when morphometric traits of leaves and seeds were considered, a clinal variation was detected (e.g., Millogo 2014; Ouedraogo 2015).
In savannah parklands, P. biglobosa is very closely associated with the shea tree (V. paradoxa). These 2 species have a similar distribution that covers a 6000 km wide belt across sub-Saharan Africa (Hall et al. 1996, 1997). Both reproduce sexually, are animal-pollinated, have fruits with zoochorous dispersal, and are particularly vulnerable to fire and overexploitation due to their high socioeconomic value. In addition, both are commonly part of agroforestry systems. Allal et al. (2011) have shown that V. paradoxa presents 2 clearly defined genetic clusters that distinguish western from eastern populations, while additional subclusters can be detected, especially in the eastern part of the distribution. The authors concluded that the phylogeography of V. paradoxa can be explained by the climate of the last glacial maximum (LGM). Thus, we assumed that the same driving factors shaped the phylogeography of P. biglobosa. Considering the diverse ecological conditions of savannahs over geological epochs, and the varying degree of genetic isolation that potentially affected some tree populations, we expected to find different levels of nonadaptive genetic diversity across its range. From the results of an earlier study based on allozymes, the expectation was to find a higher diversity in the central part of the range. In addition, given the long-distance gene flow, we envisaged detecting low genetic differentiation. These indications required a validation through a broader sampling effort and a more advanced methodological approach in the analyses.
We based our study on a representative sample of the species range. We used data obtained from 8 nuclear microsatellites and employed an exhaustive sampling of 1610 individuals across 84 populations in 12 western and central African countries. Recent methods were used for the inference of genetic structure of P. biglobosa, to investigate its demographic history and to examine likely changes in suitable habitat for the species, by comparing projected LGM and contemporary climate conditions. In particular, we addressed the following questions: 1) have climatic oscillations of the late Pleistocene affected the genetic structure of the species and, if so, 2) is it possible to elucidate what have been potential immigration paths and source populations? 3) Finally, is it possible on the basis of the information generated to suggest how priority locations for the conservation of the genetic resources of the species should be distributed across the species’ range?
Materials and Methods
Core Collection of Germplasms
Sample collections of P. biglobosa were conducted in 1989, 1990, and 1994 by the Centre National de Semences Forestières (CNSF), Burkina Faso, across 12 countries in West and Central Africa. Our study was based on a comprehensive sampling (less intensive in the south-eastern part of the species’ distribution) and included 84 sample sites (called “populations” in other sections of this article), distributed from Senegal to Chad, between the latitudes 6°N and 15°N (Supplementary Material 1, Table S1). In each population, about 30 maternal trees, distant at least 100 m from each other, were considered for seed collection. Collections of open-pollinated seeds were stored separately per maternal tree in the CNSF seed bank at 4°C. In 2014, seed samples were transferred to the Department of Forest Genetics, Austrian Research Centre for Forests, and stored at −21 °C until DNA extraction.
Data for Species Distribution Modeling
Data on the occurrence of P. biglobosa were compiled from the following sources: seed stands database (CNSF, Burkina Faso), data from the Herbarium Senckenbergianum (Germany), data from the herbarium of Aarhus University (Denmark), the West African Vegetation Database (Schmidt et al. 2012), georeferenced photo records of African plants (Dressler et al. 2014), observation records from a transect study in western Burkina Faso (Heubes 2012), georeferenced point locations from a detailed distribution map in Nigeria (Amusa et al. 2014) and point locations obtained from the Global Biodiversity Information Facility (GBIF 2017). In total, 2032 occurrence records from 14 African countries (Benin, Burkina Faso, Cameroon, Central African Republic, Chad, Ghana, Guinea, Guinea-Bissau, Ivory Coast, Mali, Niger, Nigeria, Senegal, and Togo) were collated.
DNA Isolation and Amplification
Genomic DNA was extracted from frozen seeds. An embryo of 1 seed per maternal tree was chosen for DNA extraction using the DNeasy 96 Plant Kit protocol from Qiagen (Germany) following the manufacturer’s instructions. Quantity and quality of the DNA were evaluated using a ND-1000 spectrophotometer (NanoDrop, Inc.). DNA samples were genotyped using 8 microsatellite loci developed by Lassen et al. (2014). DNA was amplified in 2 PCR multiplex reactions: multiplex assay 1 (loci PbL03, PbL15, PbL22) and multiplex assay 2 (loci PbL02, PbL09, PbL11, PbL12, PbL21). Primer pairs were labeled using the fluorochromes FAM (PbL03, PbL12), Atto565 (PbL09, PbL21), Atto550 (PbL02, PbL15), and Atto532 (PbL11, PbL22). Multiplex PCR amplifications were performed in a 10 μL reaction volume containing 2–10 ng of genomic DNA, 5 μL HotStarTaq Master Mix (Qiagen), double distilled water, and 0.3 μM of forward and reverse primers. PCR was conducted on a TC-412 Programmable Thermal Controller (Techne, Inc.) using the following cycling program: denaturation at 95 °C for 15 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 90 s, extension at 72 °C for 60 s, and a final extension at 72 °C for 30 min. Genotyping was outsourced to a commercial provider (Ecogenics, Balgach, Switzerland). PCR products were separated on an ABI3730 DNA sequencer (Applied Biosystems, Foster City, CA) and automatically sized using GeneScan 500 ROX size standard (Applied Biosystems) and analyzed with GeneMapper 3.7 (Applied Biosystems).
Data Analysis
Distribution Modeling
To discuss contemporary patterns of genetic variation, we spatially overlaid the modeled current potential distribution of P. biglobosa and the hind-cast LGM potential distribution, about 22000 years ago. The initial 2032 occurrence records were cleaned (according to Vinceti et al. 2013) and a “systematic sampling” method was applied (Fourcade et al. 2014). A grid of 30 arc-minutes (∼54 km at the equator) resolution was created and 1 occurrence per grid cell randomly sampled, resulting in a total of 223 occurrence records finally used for the model. A potential distribution model was calculated using the maximum entropy (MaxEnt) algorithm (Phillips et al. 2006) with a set of 20 initial environmental variables and species occurrence points as inputs (Elith et al. 2006; Vinceti et al. 2013). Altitude and 19 bioclimatic variables, representing contemporary baseline climatology (1950–2000) were obtained from the WorldClim 1.4 dataset (Hijmans et al. 2005) at 2.5 min (∼4.5 km at the equator) spatial resolution (http://www.worldclim.org/current). Highly cross-related variables (correlation coefficient >0.8 or <−0.8) were highlighted and ranked according to the number of correlated variables (Fourcade et al. 2014). The variable with the highest rank was retained, the redundant ones rejected. If 2 variables had the same rank, we used the sum of the absolute correlation values to select the most representative variable. Based on this method, the following 10 predictor variables were used for the final model: annual mean temperature, maximum temperature of the warmest month, range of the annual temperature, annual precipitation, precipitation of the wettest month, precipitation of the driest month, variation coefficient of annual precipitation, precipitation of the warmest quarter, precipitation of the coldest quarter, elevation above sea level. Ten thousand random points were used as background points for model training across sub-Saharan Africa and a 10-fold cross-validation option (10:1 ratio of training vs. test samples) was implemented; this was summarized subsequently into a single layer with average logistic output values across the replicates. We applied a 3-fold metric developed by Ramírez-Villegas et al. (2010) to test the model performance: 1) 10-fold average area under the Receiver Operating Characteristic (ROC) curve (AUC) of test data above 0.7, 2) standard deviation of the test AUC of the 10 different folds below 0.15, and 3) proportion of potential distribution area with standard deviation above 0.15 below 10%.
The past climate conditions were characterized by the median values of 3 downscaled General Circulation Models for the LGM (about 22000 years ago), obtained from the WorldClim website (http://www.worldclim.org/paleo-climate1) at the same spatial resolution as the contemporary baseline data. To assess the effects of past climate change on suitable habitat of P. biglobosa, the model results under contemporary climate conditions were projected to the LGM. The individual projections for each single LGM model and the differences with present-day projections can be found in Supplementary Material 1, Figures S4–S6.
The logistic output layers were restricted to 20 countries where P. biglobosa is native (Hall et al. 1997), and a binary raster of suitable versus unsuitable habitat (Scheldeman and van Zonneveld 2010) was created for the current and LGM climate scenario applying a commonly used thresholds (“maximum training sensitivity plus specificity”), based on the ROC curve (Liu et al. 2005). To visualize the predicted change of suitable habitat of P. biglobosa from LGM to present, we combined both binary rasters in one single map.
Genetic Diversity and Populations’ Differentiation
Mean number of alleles per locus (A), effective number of alleles (Ae), allelic richness after rarefaction (AR), expected heterozygosity (HE) expressing gene diversity (Nei 1978), observed heterozygosity (HO), and inbreeding coefficient (F) were estimated in SPAGeDi 1.5 (Hardy and Vekemans 2002). Significance of F was tested using 10000 permutations (2-sided test). A sample of 18 gene copies was used for AR. The number of private alleles (Ap) was calculated using GenAlEx 6.501 (Peakall and Smouse 2012). Pairwise FST and do (Gregorius 1974) were used to determine genetic distances. Allelic evenness (Gregorius 1990), a diversity independent measure of evenness which can vary from 0 (complete unevenness) to 1 (complete evenness), was obtained from GSED 3.0 (Gillet 2010).
The inverse distance weighted (IDW) interpolation function implemented in ArcGIS 10.1 (ESRI, Redlands, CA) was used to show the geographic patterns of HE and AR for all populations. As described by Murphy et al. (2008), we derived a continuous surface map using a linearly weighted combination of all the sample points such that each input point has a local influence that diminishes with distance to minimize the effect of irregular sampling.
We tested for the presence of null alleles using a population inbreeding model inferred in INEST 1.0 (Chybicki and Burczyk 2009).
The genetic structure of P. biglobosa was investigated using the Bayesian approach in STRUCTURE version 2.3.4 (Pritchard et al. 2000). We used the admixture model with allele frequencies all correlated, 10 independent repetitions per presumed subpopulation (K), which ranged from 1 to 10 with 800000 Markov Chain Monte Carlo repetitions and a 200000 burn-in period. The optimal number of clusters was inferred using the method of Evanno et al. (2005) implemented in STRUCTURE HARVESTER (Earl and von Holdt 2012). To further visualize the pattern of genetic relationship among populations, we conducted a principal coordinate analysis (PCoA) in GenAlEx 6.501 (Peakall and Smouse 2012) based on pairwise population location matrix and pairwise genetic distance matrix.
We used SPAGeDi 1.5 (Hardy and Vekemans 2002) to provide the global estimates of genetic differentiation whereby FST is based on allele identity and RST on allele size. Significance of FST > 0 and RST > FST was tested using 10000 permutations (1-sided test) (Hardy et al. 2003).
We further calculated the population differentiation (Dj) as proposed by Gregorius and Roberds (1986) to identify the most representative population for each cluster detected by STRUCTURE by using GSED 3.0 (Gillet 2010). As the real population sizes are unknown, we used the program option “equal population size.”
Genetic barriers between populations showing genetic discontinuity were highlighted using BARRIER v 2.2 (Manni et al. 2004). Monmonier’s maximum difference algorithm was employed (Monmonier 1973) to identify edges associated with high rates of change among studied populations. Barriers were computed using 1000 bootstrapped pairwise genetic distance matrices (chord-distance) from Microsatellite Analyser 4.05 (Dieringer and Schlötterer 2003). The robustness or the significance of computed barriers was assessed by the bootstrap score which represent the percentage of matrices contributing to strength of barrier over 1000 matrices.
To test for isolation by distance (IBD), we performed a paired Mantel test (Mantel 1967) in GenAlEx 6.501 (Peakall and Smouse 2012) estimating the correlation between a matrix of geographical distances among populations and a matrix of genetic distances given by pairwise FST. The statistical significance of the relationship between pairwise population genetic and geographic distances was assessed using 999 permutations.
Inference of Demographic History
The demographic history in West and Central Africa was investigated using the approximate Bayesian computation (ABC), DIYABC version 2.1.0. (Cornuet et al. 2014). The analysis was performed using 3 main clusters identified by STRUCTURE and BARRIER (Results section): extreme West Africa (EWA), center of West Africa (CWA), and Central Africa (CA). ABC simulations were run using only those microsatellite loci for which a complete dataset was available. We assumed a generation time of 30 years acknowledging that the definition of generation time is a controversial issue in forest tree species (Petit and Hampe 2006). As reported by Logossa et al. (2011), according to empirical observations by Okullo et al. (2004), the main tree species occurring in the savannah ecosystem examined in this study reach full reproductive maturity at ages that vary between 20 and 50 years. The generation length used in a study similar to ours for V. paradoxa was 30 years. Although P. biglobosa reaches maturity at 5–7 years of age, it reaches the canopy only at ca. 30 years and, at that stage, the tree also reaches its full reproductive potential, that is, its maximum height and expansion of the crown (Hall et al. 1997; Heuzé et al. 2018).
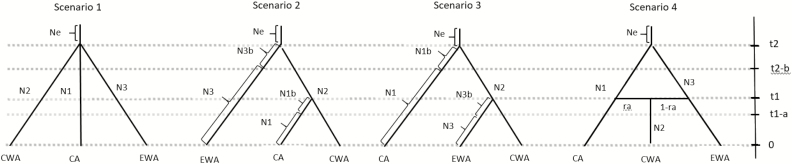
Three scenarios were compared with quantify the likelihood of a population expansion, decrease or constant size after the divergence of modern populations from the ancestral population (Supplementary Material 2—ABC, Figure S1) for each of the 3 populations, corresponding to the 3 main genetic clusters: CA, CWA, and EWA. These preliminary simulations provided also an indication of which of the 3 main genetic clusters diverged first from the ancestral population. The ABC analysis had an exploratory value. The scenario testing was limited to time spans of 10000 generations and focused on testing the likelihood of 4 simple scenarios (Figure 1), which were developed based on the results from STRUCTURE and BARRIER (see more explanations in Results section). Each scenario was replicated by choosing in turn a different population as the one that traced back to the ancestral population, disregarding the indications provided by the preliminary simulations about this aspect, to validate the results of the preliminary tests (see the full set of 12 scenarios compared in Supplementary Material 2—ABC, Figure S2). In the scenario with a simultaneous split of the 3 main genetic clusters, what varies is the selection of the cluster more closely associated to the ancestral population. All the details about competing scenarios, including summary statistics, are provided in Supplementary Material 2—ABC. In all scenarios, t# refers to time-scale (time priors scaled by generation time; t2 = timing of the first split of clusters, t1 = timing of the most recent split of clusters) and N# refers to effective population size of the corresponding populations (ancestral population “e,” and intermediate populations “2b” and “3b”) during the 2 main time periods (i.e., 0 to t1 and t1 to t2).
Figure 1.
The 4 scenarios compared in approximate Bayesian computation. EWA, Extreme West Africa group; CWA, Central West Africa group; CA, Central Africa group; t# = divergence time-scaled by generation time; N# = effective population size; ra = admixture rate. Scenario 1: the 3 clusters (EWA, CWA, CA) split at the same time from the ancestral population (see additional explanations in Supplementary Material 2—ABC); scenario 2: cluster EWA diverged earlier from CWA than CA; scenario 3: CA diverged earlier from CWA than EWA; scenario 4: EWA and CA split from the ancestral population and CWA resulted from their admixture. Posterior probabilities of each scenario are presented in Supplementary Material 2—ABC, Table S4, scenarios from 1 to 4).
Results
Genetic Diversity and Population Differentiation
The 8 microsatellite loci were highly polymorphic and we did not find significant evidence of null alleles or linkage disequilibrium. A total of 217 alleles were detected among the 1610 genotypes of the 84 populations. The number of alleles per locus ranged from 17 (PbL11) to 50 (PbL22), with an average of 27 alleles per locus (Supplementary Material 1, Table S2).
Private alleles were found in 32% of the studied populations (ranging from 1 up to 3 private alleles) with no obvious geographic pattern (Table 1). Genetic diversity was lower in populations originating from EWA and CA compared with populations from CWA (Tables 1 and 2). For instance, AR was estimated to be only 4.95 in Segueya (Guinea, EWA) and 4.79 in Bendah (Chad, CA), but 7.5 in Peperkou (Benin, CWA) (Table 1). A similar pattern was observed for the other diversity measures, such as HE, with a value of 0.82 in Logobou and Koupela (Burkina Faso, CWA) compared with only 0.61 in Bendah (Chad, CA) or 0.67 in Bokaria (Guinea, EWA). The FIS for all populations was not different from zero (FIS = 0.03, P > 0.05). In 10 populations, F values were relatively high and significantly different from zero (F = 0.10–0.16, P < 0.05) (Table 1). The IDW geographic pattern of genetic diversity showed the highest intrapopulation diversity in Burkina Faso, Northern Ivory Coast, and South-Western Mali (Figure 2; Supplementary Material 1, Figure S1). Estimates of evenness ranged from 0.40 to 0.64 and the average value per cluster was higher in CA and EWA compared with CWA (Table 1).
Table 1.
Genetic diversity indices, evenness, amount of genetic differentiation and rank of the 84 populations of Parkia biglobosa for genetic conservation in each cluster
| Cluster | Country | Population | ID | N | N A | A R | H E | H O | F | P | A P | e | D j |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EWA | Senegal | Nemanding | 1 | 15 | 5.63 | 5.01 | 0.72 | 0.68 | 0.06 | 0.25 | 0 | 0.52 | 0.34 |
| Senegal | Kolda | 2 | 28 | 8.50 | 6.04 | 0.77 | 0.75 | 0.02 | 0.62 | 0 | 0.52 | 0.24 | |
| Senegal | Touba Baria | 3 | 18 | 6.63 | 5.13 | 0.69 | 0.60 | 0.13 | 0.01 | 0 | 0.54 | 0.34 | |
| Senegal | Simbandya | 4 | 15 | 6.63 | 5.59 | 0.74 | 0.71 | 0.05 | 0.34 | 0 | 0.51 | 0.26 | |
| Senegal | Baghere | 5 | 25 | 7.38 | 5.72 | 0.76 | 0.71 | 0.07 | 0.08 | 1 | 0.52 | 0.27 | |
| Senegal | Sare Yoba | 6 | 20 | 7.00 | 5.71 | 0.78 | 0.76 | 0.02 | 0.60 | 0 | 0.54 | 0.30 | |
| Guinea | Segueya | 7 | 16 | 5.88 | 4.95 | 0.68 | 0.60 | 0.12 | 0.02 | 0 | 0.59 | 0.38 | |
| Guinea | Konkoure | 8 | 14 | 6.13 | 5.34 | 0.72 | 0.72 | 0.00 | 0.95 | 0 | 0.52 | 0.34 | |
| Guinea | Bokaria | 9 | 11 | 5.25 | 4.97 | 0.67 | 0.57 | 0.16 | 0.01 | 0 | 0.64 | 0.39 | |
| Guinea | Timbo | 10 | 18 | 8.00 | 6.27 | 0.77 | 0.80 | −0.04 | 0.37 | 2 | 0.52 | 0.43 | |
| CWA1 | Mali | M’besso | 11 | 28 | 9.75 | 6.96 | 0.79 | 0.72 | 0.08 | 0.01 | 0 | 0.40 | 0.23 |
| Mali | Fourou | 12 | 19 | 8.00 | 6.53 | 0.76 | 0.72 | 0.07 | 0.10 | 0 | 0.51 | 0.26 | |
| Mali | Zanga | 13 | 26 | 10.00 | 6.69 | 0.75 | 0.74 | 0.02 | 0.60 | 2 | 0.43 | 0.22 | |
| Mali | Lofini | 14 | 29 | 10.50 | 7.18 | 0.77 | 0.77 | 0.00 | 0.91 | 0 | 0.45 | 0.23 | |
| Mali | Katele | 15 | 18 | 8.50 | 6.86 | 0.79 | 0.79 | 0.01 | 0.77 | 0 | 0.52 | 0.24 | |
| Mali | Djeou | 16 | 24 | 10.38 | 7.15 | 0.78 | 0.75 | 0.04 | 0.27 | 0 | 0.46 | 0.20 | |
| Mali | Nafegue | 17 | 26 | 10.75 | 7.27 | 0.79 | 0.74 | 0.07 | 0.02 | 3 | 0.44 | 0.23 | |
| Mali | Dioumatenea | 18 | 31 | 10.75 | 6.87 | 0.78 | 0.79 | −0.01 | 0.74 | 2 | 0.42 | 0.19 | |
| Mali | Borioni | 19 | 32 | 10.38 | 6.77 | 0.73 | 0.74 | −0.01 | 0.77 | 0 | 0.40 | 0.21 | |
| Ivory Coast | Minignan | 20 | 19 | 8.88 | 6.66 | 0.74 | 0.74 | −0.01 | 0.86 | 0 | 0.47 | 0.36 | |
| CWA2 | Ivory Coast | Sogo-Boundiali | 21 | 21 | 8.63 | 6.42 | 0.76 | 0.78 | −0.02 | 0.57 | 0 | 0.50 | 0.29 |
| Ivory Coast | Karamogola | 22 | 30 | 9.50 | 6.40 | 0.76 | 0.75 | 0.00 | 0.89 | 1 | 0.46 | 0.23 | |
| Ivory Coast | Ngoloblasso | 23 | 19 | 9.13 | 7.18 | 0.78 | 0.78 | 0.00 | 0.92 | 0 | 0.44 | 0.25 | |
| Ivory Coast | Kafiofi | 24 | 26 | 10.13 | 7.14 | 0.79 | 0.79 | 0.00 | 0.97 | 1 | 0.49 | 0.25 | |
| Ivory Coast | Tenindieri | 25 | 16 | 7.50 | 6.08 | 0.70 | 0.62 | 0.12 | 0.01 | 0 | 0.53 | 0.29 | |
| Ivory Coast | Tiorgo | 26 | 20 | 9.50 | 7.04 | 0.75 | 0.74 | 0.02 | 0.48 | 0 | 0.51 | 0.28 | |
| Ivory Coast | Dolekahaa | 27 | 25 | 10.00 | 6.69 | 0.76 | 0.77 | −0.01 | 0.80 | 1 | 0.46 | 0.21 | |
| Ivory Coast | Broundougou | 28 | 10 | 5.75 | 5.68 | 0.78 | 0.79 | −0.01 | 0.94 | 0 | 0.61 | 0.35 | |
| Ivory Coast | Ouangofitini | 29 | 33 | 10.00 | 6.58 | 0.75 | 0.73 | 0.02 | 0.52 | 1 | 0.44 | 0.26 | |
| Burkina Faso | Niangoloko | 30 | 17 | 8.75 | 7.08 | 0.80 | 0.79 | 0.02 | 0.59 | 2 | 0.51 | 0.29 | |
| Burkina Faso | Bandougou | 31 | 17 | 8.75 | 6.87 | 0.80 | 0.76 | 0.06 | 0.12 | 0 | 0.48 | 0.26 | |
| Burkina Faso | Koutoura | 32 | 18 | 8.75 | 6.80 | 0.78 | 0.77 | 0.02 | 0.68 | 2 | 0.51 | 0.29 | |
| Burkina Faso | Toussiana | 33 | 22 | 9.38 | 6.70 | 0.78 | 0.80 | −0.02 | 0.54 | 0 | 0.48 | 0.24 | |
| Burkina Faso | Darsalamy | 34 | 19 | 8.50 | 6.37 | 0.74 | 0.69 | 0.07 | 0.08 | 1 | 0.45 | 0.31 | |
| CWA3 | Burkina Faso | Kampti | 35 | 23 | 8.38 | 6.32 | 0.75 | 0.77 | −0.03 | 0.47 | 0 | 0.42 | 0.27 |
| Burkina Faso | Kour-Poni | 36 | 19 | 8.25 | 6.41 | 0.75 | 0.74 | 0.01 | 0.75 | 0 | 0.49 | 0.25 | |
| Burkina Faso | Pa | 37 | 13 | 8.00 | 6.95 | 0.78 | 0.71 | 0.09 | 0.04 | 0 | 0.48 | 0.33 | |
| Burkina Faso | Gaoua | 38 | 16 | 8.00 | 6.42 | 0.73 | 0.70 | 0.04 | 0.30 | 1 | 0.54 | 0.27 | |
| Burkina Faso | Lere | 39 | 15 | 7.63 | 6.26 | 0.76 | 0.74 | 0.02 | 0.59 | 0 | 0.52 | 0.26 | |
| Burkina Faso | Dano | 40 | 19 | 7.88 | 6.11 | 0.75 | 0.74 | 0.02 | 0.67 | 1 | 0.55 | 0.26 | |
| Burkina Faso | Baupiel | 41 | 10 | 6.13 | 5.88 | 0.70 | 0.69 | 0.03 | 0.62 | 0 | 0.64 | 0.34 | |
| Burkina Faso | Lan | 42 | 15 | 8.38 | 6.98 | 0.79 | 0.72 | 0.1 | 0.02 | 0 | 0.54 | 0.33 | |
| Burkina Faso | Silmissin | 43 | 29 | 11.13 | 7.35 | 0.80 | 0.80 | 0.01 | 0.76 | 1 | 0.44 | 0.24 | |
| Burkina Faso | Toeghin | 44 | 15 | 8.00 | 6.58 | 0.78 | 0.74 | 0.05 | 0.23 | 0 | 0.53 | 0.26 | |
| Burkina Faso | Barssea | 45 | 30 | 10.50 | 6.93 | 0.78 | 0.74 | 0.05 | 0.09 | 0 | 0.47 | 0.18 | |
| Burkina Faso | Koupela | 46 | 29 | 12.00 | 7.53 | 0.82 | 0.80 | 0.02 | 0.40 | 1 | 0.44 | 0.24 | |
| Burkina Faso | Tiebele | 47 | 28 | 9.88 | 6.54 | 0.79 | 0.82 | −0.04 | 0.20 | 1 | 0.50 | 0.24 | |
| Burkina Faso | Tenkodogo | 48 | 20 | 9.75 | 7.38 | 0.81 | 0.77 | 0.06 | 0.10 | 0 | 0.47 | 0.26 | |
| Burkina Faso | Fada N’Gourma | 49 | 23 | 9.63 | 6.96 | 0.82 | 0.78 | 0.05 | 0.14 | 0 | 0.56 | 0.26 | |
| Burkina Faso | Pama | 50 | 19 | 8.88 | 7.00 | 0.80 | 0.80 | 0.01 | 0.88 | 0 | 0.53 | 0.28 | |
| CWA4 | Burkina Faso | Kantchari | 51 | 17 | 8.38 | 6.91 | 0.81 | 0.80 | 0.01 | 0.81 | 0 | 0.50 | 0.32 |
| Burkina Faso | Logobou | 52 | 22 | 10.00 | 7.36 | 0.79 | 0.82 | −0.04 | 0.24 | 0 | 0.49 | 0.27 | |
| Ghana | Tanga | 53 | 22 | 10.00 | 6.94 | 0.79 | 0.76 | 0.05 | 0.18 | 0 | 0.52 | 0.21 | |
| Ghana | Tansia | 54 | 16 | 8.00 | 6.23 | 0.75 | 0.72 | 0.04 | 0.37 | 1 | 0.46 | 0.31 | |
| Ghana | VRWS | 55 | 18 | 9.25 | 7.00 | 0.76 | 0.71 | 0.07 | 0.05 | 0 | 0.50 | 0.29 | |
| Togo | Timbou | 56 | 11 | 7.13 | 6.47 | 0.76 | 0.73 | 0.05 | 0.37 | 0 | 0.51 | 0.30 | |
| Togo | Nanergou | 57 | 19 | 8.13 | 6.23 | 0.78 | 0.70 | 0.11 | 0.01 | 1 | 0.56 | 0.27 | |
| Togo | Konkoate | 58 | 16 | 9.00 | 6.91 | 0.75 | 0.76 | −0.01 | 0.71 | 1 | 0.47 | 0.26 | |
| Togo | Twaga | 59 | 10 | 6.50 | 6.18 | 0.71 | 0.74 | −0.04 | 0.47 | 1 | 0.50 | 0.30 | |
| Togo | Magnan | 60 | 10 | 6.75 | 6.49 | 0.76 | 0.78 | −0.02 | 0.68 | 0 | 0.54 | 0.30 | |
| Togo | Bassar | 61 | 18 | 8.38 | 6.28 | 0.72 | 0.72 | 0.01 | 0.81 | 1 | 0.45 | 0.27 | |
| Togo | Binah | 62 | 22 | 9.75 | 7.02 | 0.75 | 0.74 | 0.01 | 0.67 | 0 | 0.45 | 0.27 | |
| Benin | Cotiakoua | 63 | 26 | 10.88 | 7.30 | 0.79 | 0.77 | 0.03 | 0.36 | 0 | 0.46 | 0.20 | |
| Benin | Toukountouna | 64 | 21 | 9.25 | 6.80 | 0.77 | 0.74 | 0.04 | 0.26 | 0 | 0.48 | 0.26 | |
| Benin | Natitingou | 65 | 22 | 8.75 | 6.35 | 0.75 | 0.80 | −0.06 | 0.11 | 0 | 0.55 | 0.28 | |
| Benin | Peperkou | 66 | 10 | 7.88 | 7.54 | 0.80 | 0.79 | 0.02 | 0.67 | 0 | 0.54 | 0.30 | |
| Benin | Onklou | 67 | 10 | 6.75 | 6.44 | 0.73 | 0.73 | 0.01 | 0.85 | 0 | 0.58 | 0.34 | |
| Niger | Kaka Sakara | 68 | 13 | 7.13 | 6.40 | 0.76 | 0.76 | 0.01 | 0.79 | 0 | 0.58 | 0.31 | |
| Niger | Bouma | 69 | 11 | 7.38 | 6.92 | 0.82 | 0.79 | 0.03 | 0.52 | 0 | 0.60 | 0.31 | |
| Niger | Saborijan | 70 | 12 | 6.25 | 5.71 | 0.72 | 0.72 | 0.01 | 0.88 | 0 | 0.50 | 0.37 | |
| Nigeria | Maidiguri | 71 | 25 | 8.38 | 6.21 | 0.78 | 0.73 | 0.07 | 0.03 | 1 | 0.60 | 0.34 | |
| Nigeria | Auchi | 72 | 10 | 7.63 | 7.28 | 0.82 | 0.71 | 0.14 | 0.01 | 1 | 0.64 | 0.39 | |
| Nigeria | Ilorin | 73 | 15 | 6.88 | 5.85 | 0.76 | 0.68 | 0.11 | 0.02 | 0 | 0.63 | 0.41 | |
| Nigeria | Ibadan | 74 | 19 | 7.63 | 5.80 | 0.72 | 0.67 | 0.08 | 0.08 | 0 | 0.46 | 0.47 | |
| CA | Cameroon | M’bor | 75 | 15 | 6.75 | 5.53 | 0.65 | 0.67 | −0.02 | 0.71 | 0 | 0.57 | 0.26 |
| Cameroon | Nakong | 76 | 15 | 6.88 | 5.77 | 0.68 | 0.59 | 0.14 | 0.00 | 1 | 0.64 | 0.28 | |
| Cameroon | N’gaïa | 77 | 19 | 7.13 | 5.50 | 0.65 | 0.61 | 0.07 | 0.11 | 0 | 0.58 | 0.20 | |
| Cameroon | Mbaiboum | 78 | 20 | 6.38 | 5.35 | 0.66 | 0.61 | 0.07 | 0.11 | 0 | 0.62 | 0.24 | |
| Cameroon | Wourouwate | 79 | 12 | 6.13 | 5.58 | 0.64 | 0.60 | 0.07 | 0.22 | 0 | 0.57 | 0.33 | |
| Cameroon | Bogdibo | 80 | 18 | 6.00 | 4.91 | 0.61 | 0.55 | 0.11 | 0.03 | 0 | 0.52 | 0.27 | |
| Chad | Bendah | 81 | 21 | 6.00 | 4.79 | 0.61 | 0.61 | 0.00 | 0.99 | 0 | 0.57 | 0.27 | |
| Chad | Koutoumire | 82 | 16 | 6.25 | 5.14 | 0.63 | 0.60 | 0.05 | 0.27 | 1 | 0.60 | 0.29 | |
| Chad | N’Dobenjton | 83 | 12 | 6.00 | 5.48 | 0.64 | 0.65 | −0.01 | 0.77 | 0 | 0.55 | 0.28 | |
| Chad | Talati | 84 | 19 | 5.75 | 4.84 | 0.62 | 0.60 | 0.02 | 0.58 | 1 | 0.64 | 0.27 | |
| All populations | 19.17 | 8.23 | 6.38 | 0.75 | 0.73 | 0.03 | 0.43 | 0.40 | 0.52 | — | |||
N, number of samples; NA, number of observed alleles; AR, allelic richness (expected number of alleles among 18 gene copies) (k = 18); HE, expected heterozygosity; HO, observed heterozygosity; F, inbreeding coefficient; Ap, number of private alleles; e, evenness; Dj, allelic differentiation; VRWS, Volta Region Woodland Savannah. P value for test F = 0.
aThe most representative populations.
Table 2.
Measures of genetic variation for each of the 6 Parkia biglobosa clusters based on 8 microsatellite loci
| Clusters | N | N A | A R (k = 18) | H E | H O | F |
|---|---|---|---|---|---|---|
| EWA | 180 | 13.88 | 6.44 | 0.781 | 0.700 | 0.103 |
| CWA 1 | 252 | 17.50 | 7.23 | 0.779 | 0.749 | 0.038 |
| CWA 2 | 293 | 19.63 | 7.22 | 0.782 | 0.755 | 0.035 |
| CWA 3 | 323 | 18.88 | 7.27 | 0.796 | 0.762 | 0.043 |
| CWA 4 | 395 | 20.75 | 7.39 | 0.792 | 0.743 | 0.062 |
| CA | 167 | 11.63 | 5.88 | 0.669 | 0.606 | 0.094 |
| All populations | 1610 | 27.13 | 8.03 | 0.830 | 0.730 | 0.114 |
N, number of samples; NA, number of observed alleles; NE, effective number of alleles; AR, allelic richness (expected number of alleles among 18 gene copies) (k = 18); HE, expected heterozygosity; HO, observed heterozygosity; F, inbreeding coefficient. All F values were significantly different from zero (P < 0.001).
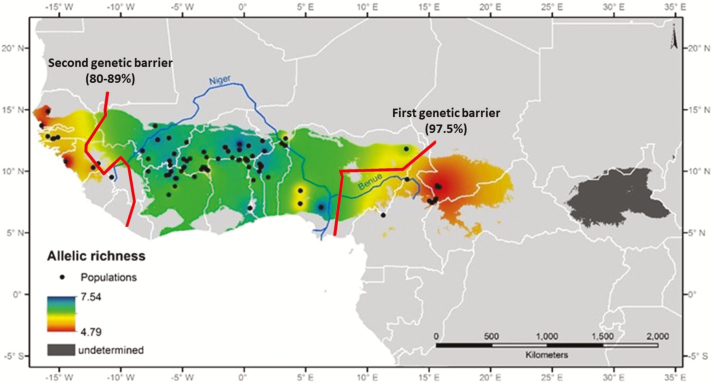
Figure 2.
IDW interpolation of allelic richness (AR) calculated for 84 Parkia biglobosa populations using 8 SSRs loci and restricted to the modeled distribution area of the species. The color gradient indicates the level of allelic richness across the species’ range. Red lines represent genetic boundaries detected by BARRIER. See online version for full colors.
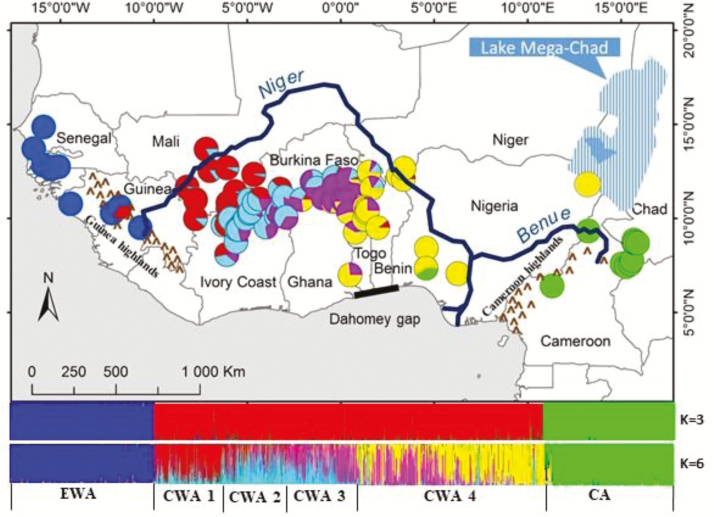
STRUCTURE revealed a strong differentiation resulting in 3–6 clusters (Figure 3). Six clusters (K = 6) were identified as most likely (Supplementary Material 1, Figures S2 and S3). However, at K = 3 clusters form very sharp boundaries, while at K = 6 one of the previous clusters (CWA) is further subdivided into 4 additional clusters showing considerable admixture.
Figure 3.
Spatial distribution of genetic clusters. Colors of the pie charts refer to the 6 clusters identified by STRUCTURE (EWA, CWA 1, CWA 2, CWA 3, CWA 4, CA) and the proportion of the pie charts sectors to the relative number of individuals assigned. The histograms of individual assignments to genetics clusters are given for K = 3 and K = 6 clusters. See online version for full colors.
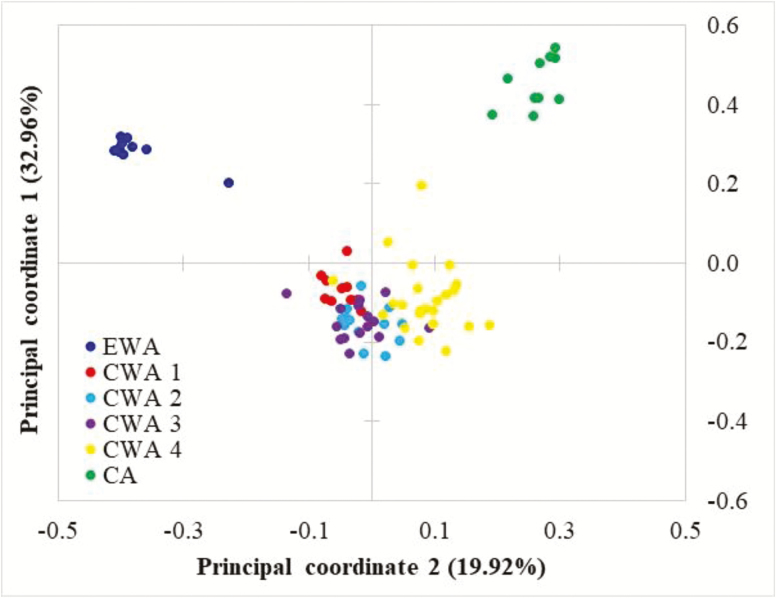
The identification of 3 clusters in STRUCTURE was also supported by the results from the PCoA (Figure 4), with the first 2 principal components explaining 52.9% of the total molecular variation, and by the results from BARRIER (Figure 2).
Figure 4.
PCoA showing the patterns of genetic structure of Parkia biglobosa grouping into 3 clusters corresponding to 3 geographic regions (EWA, CWA, CA). CWA is subdivided into 4 subclusters (CWA 1, CWA 2, CWA 3, and CWA 4). See online version for full colors.
By using Monmonier’s method, 2 main genetic barriers were detected which identified areas of abrupt changes in the spatial pattern of genetic variation. The first genetic barrier differentiated populations of CA from CWA with a bootstrap score of 97.5%, while the second barrier separated populations of EWA (Senegal and Guinea) from complementary populations with a bootstrap score varying from 80% to 89% (Figure 2).
The results derived from different analytical methods are aligned in indicating that the gene pool of P. biglobosa is split into 3 components (EWA, CWA, CA). About 12% of the nuclear variation was found among the 3 main genetic clusters (Tables 3–5). In all instances, that is, when the dataset was divided into 3 or 6 clusters, or when all 84 populations were treated separately, RST was always significantly larger than FST at a 95% confidence level, indicating a phylogeographic structure (Table 3).
Table 3.
Global FST and RST based on a different number of STRUCTURE clusters (K = 3 and 6) and based on all populations (sampling sites)
| Number of clusters/populations | Identified clusters/populations | F ST | R ST |
|---|---|---|---|
| 3 | EWA, CWA, CA | 0.117*** | 0.172* |
| 6 | EWA, CWA 1, CWA 2, CWA 3, CWA 4, CA | 0.074*** | 0.115** |
| 84 | Populations | 0.088*** | 0.122** |
For all comparisons, 8 microsatellite loci for Parkia biglobosa were used. Added numbers to CWA refer to subclusters. Asterisks indicate significance (*P < 0.05, **P < 0.01, ***P < 0.001) for FST > 0 and RST > FST.
Pairwise comparisons of FST and RST values for the 6 genetic clusters showed that RST values were significantly larger than the respective pairwise FST values for all clusters except CA (Table 4). Pairwise comparisons of FST and RST values for each main genetic cluster versus the respective complementary set of all other populations, RST was always significantly larger than FST only for CA (Table 5).
Table 4.
Pairwise RST (FST in italic) based on 6 STRUCTURE clusters
| Genetic clusters | EWA | CWA 1 | CWA 2 | CWA 3 | CWA 4 | CA |
|---|---|---|---|---|---|---|
| EWA | 0.10ns | 0.12ns | 0.12ns | 0.16ns | 0.43*** | |
| CWA 1 | 0.10 | 0.02* | 0.04* | 0.09*** | 0.22** | |
| CWA 2 | 0.12 | 0.01 | 0.01ns | 0.05* | 0.25** | |
| CWA 3 | 0.11 | 0.02 | 0.01 | 0.04ns | 0.28*** | |
| CWA 4 | 0.12 | 0.05 | 0.03 | 0.03 | 0.22** | |
| CA | 0.16 | 0.14 | 0.15 | 0.15 | 0.13 |
For all comparisons, 8 microsatellite loci for Parkia biglobosa were used. Added numbers to CWA refer to subclusters. Asterisks indicate significance (*P < 0.05, **P < 0.01, ***P < 0.001) for FST > 0 and RST > FST.
Table 5.
Pairwise FST (RST in italic) for 3 different groupings of populations, excluding 1 cluster in turn
| Clusters/populations | F ST (RST) |
|---|---|
| EWA vs. all other populations | 0.095 (0.103 ns) |
| CWA vs. all other populations | 0.079 (0.070 ns) |
| CA vs. all other populations | 0.118 (0.220**) |
For all comparisons, 8 microsatellite loci for Parkia biglobosa were used. Asterisks indicate significance (**P < 0.01) for FST > 0 and RST > FST.
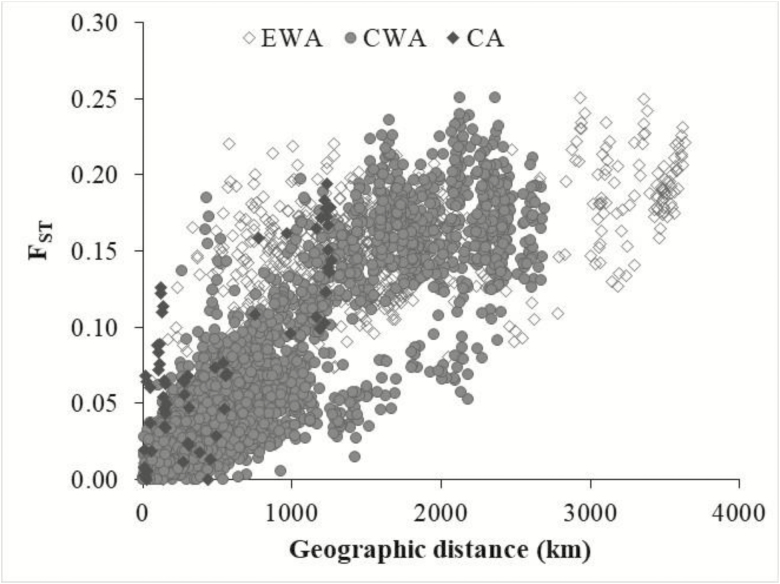
F ST values between 2 populations were used as genetic distance and were plotted against the respective pairwise geographic distance (Figure 5). The positive correlation between the matrix of genetic distances and geographical distances was significant (r = 0.82, P = 0.001).
Figure 5.
Relationship between pairwise FST and geographic distances among 84 populations of Parkia biglobosa. The symbols show population pairs from the 3 main genetic clusters.
Demographic History
The 4 simple scenarios tested (Figure 1) were based on results from the species distribution modeling and analyses of genetic diversity, which provided indications on how to cluster populations in the ABC simulations. The lower diversity of EWA and CA populations suggested that they were populations founded through colonization of new areas from CWA, located beyond barriers. These barriers were accounted for in the ABC as simulations of colonization bottlenecks at the splitting events between populations.
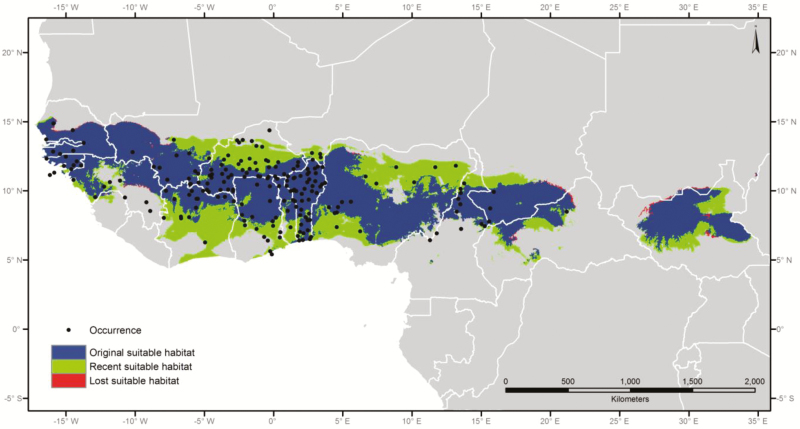
The MaxEnt distribution model that we developed for P. biglobosa under contemporary and projected LGM climate conditions can be considered accurate and stable as it fulfilled the 3 performance metrics (Ramírez-Villegas et al. 2010): 1) 0.893, 2) 0.021, and 3) 0.1%. The comparison of modeled potential distributions represented in Figure 6 clearly shows an expansion of about 47% of the original predicted suitable habitat from LGM to the present and only a very small area that had become unsuitable since the LGM.
Figure 6.
Predicted change of suitable habitat for Parkia biglobosa comparing projected LGM and contemporary climate conditions. Blue areas represent continued suitable habitat from LGM until present (original suitable habitat); light green indicates areas that were probably not suitable at the LGM, but are suitable at present (recent suitable habitat); and red areas represent potential suitable habitat during LGM but no longer at present (lost suitable habitat). See online version for full colors.
The first ABC pilot simulation, computed considering all populations as 1 large cluster, consistently with climate modeling, indicated that the best supported scenario was an expansion (Supplementary Material 2—ABC, Table S3). The second pilot simulation, computed grouping populations in 3 main clusters (EWA, CWA, and CA), indicated that the best supported scenario was an expansion after the divergence of modern populations from the ancestral population, for all 3 gene pools (Supplementary Material 2—ABC, Table S3). These preliminary results were incorporated in the following simulations. In the final competing ABC models, the highest posterior probability (0.304, 95% confidence intervals: 0.240–0.368) was found for scenario 2 (Figure 1; Supplementary Material 2—ABC, Table S4).
The posterior parameters showed that the effective population size of the ancestral population was lower than that of CWA suggesting an expansion event at time t2. In the best supported scenario (scenario 2), the hierarchical split assumed that the divergence of EWA occurred before the divergence of CWA and CA. CWA was set to trace back to the ancestral population and EWA diverged from CWA at t2; CWA and CA diverged more recently at t1.
The median values of effective population sizes for this scenario were 104000, 64200, and 83400 for N1 (CA), N2 (CWA), and N3 (EWA), respectively (Supplementary Material 2—ABC, Table S5). The median values of the divergence times, t1 and t2, were 3060 (95% CIs: 792–7140) and 5410 (95% CIs: 1150–9560) (Supplementary Material 2—ABC, Table S5). Considering a 30-year generation time, t1 scaled at 91800 years and t2 scaled at 162300 years. Results suggest a reasonable goodness of fit of observed data to scenario 2, given the results shown by PCA (Supplementary Material 2—ABC, Figure S3). All observed summary statistics were not statistically different from the simulated statistics (with a minor exception of mean Garza–Williamson’s M of the pairs CA–CWA and CA–EWA) computed from the parameter values drawn from the estimated posterior distribution (Supplementary Material 2—ABC, Table S7).
We further tested the 4 most likely scenarios (in which CWA diverged from the ancestral population) over a higher number of generations (t2 = 200000, t1 = 10000) and the likelihood of a second competing scenario seems to increase but the model fit results are worse than in the previous modeling attempts (results not shown).
Discussion
Hotspot of Intraspecific Diversity in CWA
The populations of EWA and CA showed lower genetic diversity than populations in CWA (Table 2). In particular, populations from Burkina Faso, Benin, Ghana, Togo, and western Nigeria displayed high estimates of genetic diversity. A previous study on 24 populations of West and Central Africa, using chloroplast markers, already indicated that populations in CWA are genetically diverse (Ouedraogo 2015). The haplotype distribution showed that the hypothesized most ancient haplotype was identified in Burkina Faso and Ivory Coast and was separated from the eastern and western groups by 5 and 2 mutations, respectively (Ouedraogo 2015). The high genetic diversity in this region may be explained by the presence of ancient refugial populations or by the coalescence of migration routes (Petit et al. 2003). The phylogeographic patterns of a substantial number of African rainforest species have been studied. We compared our results with those from other large-scale studies based on microsatellite markers focused on sub-Saharan African tree species that show overlapping ranges with P. biglobosa. In this study, the global estimates of genetic diversity in P. biglobosa were high. Although comparisons between different studies present constraints due to methodological differences, average values of expected heterozygosity found in P. biglobosa (0.83) were higher than in K. senegalensis (Sexton et al. 2015; HE = 0.64), M. excelsa (Daïnou et al. 2010; HE = 0.46–0.61) and V. paradoxa (HE = 0.27–0.65 by Allal et al. 2011; HE = 0.57–0.71 by Logossa et al. 2011). The visualization of genetic diversity using IDW interpolation on nuclear microsatellites matched nearly perfectly with the distribution of allozyme diversity (Lompo et al. 2017). Unfortunately, genetic data from the most eastern, isolated part of the range in South Sudan (where the species has been reported by Hall et al. 1997) were not available, either for the previous research based on allozymes or for our study.
Phylogeographic Structure Reveals Genetic Divides
Although P. biglobosa has a continuous distribution from east (Central African Republic) to west (Senegal) sub-Saharan Africa, our data indicated the presence of clear genetic divides, separating populations in the central part of West Africa from those in the eastern part of the range (Cameroon and Chad) and of the most western part of the range (Senegal and Guinea). Our data also revealed a strong phylogeographic signal. Regardless of how our dataset was organized, that is, considering all populations separately or subdividing them into different numbers of genetic clusters, RST was always significantly larger than FST. This suggests that not only genetic drift and constraints to gene flow, but also mutations, have contributed to genetic differentiation, at least in some of the populations or clusters. Thus, we compared values of FST and RST for different combinations of populations. Only for the genetic cluster CA was the value of RST significantly different from FST, indicating that in CA mutation have played a very important role and CA diverged putatively earlier than EWA and CWA (Tables 3–5). This is not aligned with the results from the ABC that points to a best supported scenario in which a split between EWA and CWA took place before the divergence of CA from CWA (scenario 2).
The observed east-west structure was detected also in the shea tree (Allal et al. 2011), which has a range distribution similar to that of P. biglobosa and likely shares a similar phylogeographic pattern. However, the degree of differentiation found in P. biglobosa is relatively high, despite the fact that the species is often pollinated by megabats (Eidolon helvum), able to fly over long stretches, and that seeds are also translocated over significant distances by primates (Lompo et al. 2017). Thus, we expected that this substantial long-distance gene flow would have blurred the genetic differentiation resulted from major changes in the species range that occurred before the last glaciation. As expected, nuclear genetic differentiation in P. biglobosa was approximately half of what was detected in V. paradoxa (Allal et al. 2011). In another sub-Saharan African tree species, the African wild cherry (P. africana) (Kadu et al. 2013), genetic differentiation was higher, while Sexton et al. (2015) and Daïnou et al. (2010) found lower genetic differentiation estimates in African mahogany (K. senegalensis) and African teak (M. excelsa), respectively, compared with our study.
The LGM climate model suggests that the Sudanian savannah, the main habitat of P. biglobosa, existed throughout glacial periods but the original area of the species during the LGM was restricted to suitable habitats along a narrow belt across West Africa and to potential refugia in CA (Figure 6). During the postglacial period, the different populations likely experienced an increase in their range due to an expansion of suitable habitat. This is in line with our ABC simulations showing that an expansion of the gene pool has the highest posterior probability (Supplemental Material 2—ABC). The species is adapted to savannah conditions and might have moved southward, toward the Equator, accompanying the retreat of Guineo-Congolian vegetation in drier periods, and then again moved northwards during wetter periods. Based on a generally accepted hypothesis regarding Pleistocene vegetation changes in Africa, we would expect a wider distribution of savannah species during glacial periods, but this case is not supported by our niche modeling approach, which suggests, on the contrary, a narrower distribution of P. biglobosa’s range during the glacial maxima, without a trace of latitudinal range shift (Figure 6). This discrepancy highlights gaps in current understanding of the dynamics in sub-Saharan tree species range shifts during the Pleistocene.
The available information supports the hypothesis that the range of the savannah biome extended as a continuous band from Western to Eastern Africa during the LGM and was characterized by features limiting migration. Lake Chad expanded to more than 350000 square kilometres during the Holocene, from 11000 to 5000 years ago (Schuster et al. 2005). Some authors hypothesized that it may have represented a large geographical barrier, isolating western and eastern African populations of baobab (A. digitata) (Leong Pock Tsy et al. 2009) and gum Arabic tree (A. senegal) (Odee et al. 2012). However, the expansion of Lake Chad occurred over a relatively short period and might not have been sufficient to cause strong genetic differentiation.
It is more likely that the Adamawa Plateau, with an average elevation of 1000 m above sea level (the highest peak is 2650 m), represented a significant genetic barrier for P. biglobosa. Allal et al. (2011) have already suggested that these highlands, which form a barrier in Cameroon between the forested south and the northern savannah, may have contributed to the speciation of the shea tree (V. paradoxa) into 2 subspecies, paradoxa in the west and nilotica in the east. On the other end of West Africa, the Guinea Highlands could have represented a significant barrier and may have affected the gene pool of P. biglobosa, whose tree density is known to decline at elevations above 1000 m (Hall et al. 1997). These highlands extend from the southern Fouta Djallon highlands through South-Eastern Guinea, Northern Sierra Leone, Liberia, and North-Western Ivory Coast, with an altitude between 300 and 500 m (the highest peak is 1945 m). Our results (Figure 2) point to the Guinea Highlands as a potentially more significant genetic barrier than the Adamawa Plateau.
The Guinea highlands are a source of some of West Africa’s largest rivers, including Niger, Gambia, and Senegal rivers. Evidence that rivers may function as barriers for gene dispersal in plant species is limited (Cazé et al. 2016; Nazareno et al. 2017) and we do not expect they acted as a genetic barrier for P. biglobosa. On the contrary, big African rivers represent an important transport system and, consequently, they may have contributed to human-induced gene flow in plants. Our data do not allow verification of this hypothesis.
In the West African region, the influence of the Dahomey Gap on plant gene flow has been well described for some rain forest tree species. The observed genetic clusters in Erythrophleum ssp. (Duminil et al. 2013) have been associated to the presence of this north to south savannah corridor, located in Togo and Benin, that appeared ca. 3000 years ago. The current abundant density of P. biglobosa within the Dahomey Gap can suggest that this corridor has been mainly a barrier for rain forest tree species rather than for tree species typical of drier forest ecosystems, although a potential influence on genetic structure has been hypothesized for V. paradoxa (Allal et al. 2011), which is also a savannah tree species. Based on our modeled past distribution of P. biglobosa, the range of the species already extended from Western to Eastern Africa during the LGM, and this counters the suggestion of a Dahomey Gap contribution to the genetic structure of P. biglobosa.
The complex of physical barriers of varying intensity described above, particularly the mountain plateaux, could have contributed to separating populations of P. biglobosa into the 3 genetic clusters that are observable today (Figures 3–5). Our exploratory ABC analysis produced a best supported scenario which predicts a divergence in P. biglobosa populations, leading to the current genetic structure, with a first split between EWA and CWA populations around 160000 years ago. A second split took place between CA and CWA approximately 60000 years later, shortly before, or just at the beginning of the penultimate glacial period.
These results are exploratory and should be considered with caution, due to the fact that ABC inferences are strongly influenced by the initial selection of priors, such as the estimation of generation time or mutation rate. In absence of a priori hypotheses on the timing of the potential populations’ changes, broad priors were selected in the pilot simulations, which included more than one glacial cycle. They support a picture in which an expansion of the species range has taken place starting from a smaller ancestral population (Ne) and in which the lower diversity of EWA and CA clusters can be explained by their splitting from the most diverse cluster (CWA) and their colonization of new areas separated from CWA by gene flow barriers. However, the available genetic data do not enable conclusively discriminating among 4 scenarios that emerge as possible alternatives. In fact, the best supported scenario (n. 2) shows a posterior probability exceeding the posterior probability of 3 other scenarios by a factor of 2–3, which is not sufficient to definitely single out a best supported scenario. In addition, pairwise FST–RST comparisons (Tables 3–5) do not support scenario 2 but rather scenario 3, which suggest an earliest split of CA. Finally, the posterior distribution of the parameter “ra” (admixture rate) (Supplementary Material 2—ABC, Table S6) does not support the case of a greater isolation of EWA due to early divergence of this cluster (scenario 2) or its low genetic contribution to CWA (scenario 4).
Historic Settlements and Current Genetic Differentiation
Our findings indicate that the split of the main genetic clusters in P. biglobosa predates the impact of human populations. However, it is interesting to note that the current spatial genetic structure of P. biglobosa reflects closely the distribution of historic settlements of indigenous people in western Africa around 1300 to 1100 years ago. The regions formerly occupied by the main ethnolinguistic groups have boundaries that coincide with the delimitation of different genetic clusters found in P. biglobosa. The major ethnolinguistic groups were Senegambian, Mande, Gur, and Kur (Murdock 1959). The Senegambian settled in the basin of the Senegal River in the most western part of West Africa (including Senegal, Gambia, Guinea-Bissau, and West of Guinea; EWA, Figure 3). The river Niger was home to the other 3 communities, Mande in the northern part of West Africa (Mali; CWA1, Figure 3), Gur in the central part of West Africa (Burkina Faso and northern parts of Ivory Coast, Ghana, Togo, Benin, and Nigeria; CWA2, CWA3, CWA4, Figure 3) and Kur in the southern part of coastal West African countries from Liberia to Nigeria. The genetic cluster CWA embeds sub-clusters that are less differentiated among them compared with CA and EWA. Evidence shows that past migratory movements in West Africa have taken place largely along a north-south axis (Pourtier 1995). Thus, Mande and Gur from landlocked dry areas, such as Mali and Burkina Faso, moved to costal countries in the south. We presume that the genetic differentiation in P. biglobosa that took place before the establishment of historical settlements in the region has probably not been significantly affected by humans, at least in the western part of the range of the species. In the central part of West Africa a more recent, human-induced movement, primarily but not exclusively along a north-south axis, cannot be excluded as a driving factor that has shaped the current distribution of genetic diversity.
Implications for Genetic Conservation
Dryland forests are characterized by higher population densities, in and around forests, than moist tropical forests and patterns of extraction of nontimber forest products are also typically higher (Murphy and Lugo 1986). However, forests in dryland biomes are exposed to extended droughts and regional warming, which, combined with a growing human population, can lead to an increased risk of land degradation. Therefore, savannah ecosystems should be targeted with priority by conservation efforts (Solbrig et al. 1996). Given this general threat and considering that part of the range of P. biglobosa is highly affected by overexploitation and fire (Gaisberger et al. 2017), it is crucial to take measures to avoid a significant loss of genetic diversity in this species and the consequent reduction in its adaptive potential in the long-term. The findings from most analyses presented (i.e., STRUCTURE; Barrier, PCoA, AMOVA) concur to indicate the presence of 3 main clusters in the genepool of P. biglobosa. In addition, our findings clearly show that CWA is a hotspot of genetic diversity, thus efforts to conserve genetic resources of P. biglobosa should focus on this region. However, considering that populations in EWA and CA may present important adaptive characteristics to different environmental conditions, selected populations in these 2 clusters should be identified as well, to optimally conserve a representative sample of the genetic diversity of P. biglobosa. For this purpose, we ranked all populations in each genetic cluster and subcluster, based on a decreasing genetic representativeness, to guide priority setting in the selection of what sites should host genetic conservation units (see Dj results in Table 1). To improve the current study, other indicators could be examined to identify priority populations for conservation of genetic resources (e.g., see Vinceti et al. 2013, for the definition of conservation priorities for P. africana across the range of the species). In addition, sampling efforts should be further expanded to cover less investigated regions of occurrence of P. biglobosa, in particular the southern part of Benin, the southern part of Ivory Coast, where the species seems to be present, and South Sudan. Given the potential adaptive significance of a strong north-south climate gradient in the distribution of the species, the adaptive genetic diversity of P. biglobosa in these countries still needs to be assessed for conservation purposes. Moreover, the results of the ABC analysis could be improved in future by using a larger number of microsatellites. Nevertheless, our recommendations are an important first step in the design of a genetic conservation strategy for P. biglobosa.
Supplementary Material
Supplementary data are available at Journal of Heredity online.
Funding
This work was supported by the Austrian Development Agency and the CGIAR Research Program on Forests, Trees and Agroforestry (FTA).
Acknowledgments
The present study is part of Djingdia Lompo’s PhD thesis at the University of Natural Resources and Life Sciences, Vienna, Austria. We are grateful to Bioversity International, the European Union, the Food Agriculture Organisation (FAO) and national institutions from Benin, Burkina Faso, Cameroon, Chad, Ghana, Guinea, Ivory Coast, Mali, Niger, Nigeria, Senegal, and Togo that supported and facilitated the collection of seed germplasms of Parkia biglobosa. We thank Dr Anders Ræbild and Prof. Erik D. Kjær at the Department of Geosciences and Natural Resource Management, University of Copenhagen (Denmark), for their valuable suggestions and comments on the manuscript and Mattia Manica for statistical advice. Thanks also to 2 anonymous reviewers whose constructive comments helped to greatly improve the manuscript.
Data Availability
The primary data underlying these analyses, sampling locations, and microsatellite genotypes have been deposited in Dryad (doi: 10.5061/dryad.15pg4g4/1).
References
- Allal F, Sanou H, Millet L, Vaillant A, Camus-Kulandaivelu L, Logossa ZA, Lefèvre F, Bouvet JM. 2011. Past climate changes explain the phylogeography of Vitellaria paradoxa over Africa. Heredity (Edinb). 107:174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amusa O, Adesoye A, Ogunkanmi A, Omoche O, Olowe O, Akinyosoye S, Omodele T. 2014. Genetic diversity of Parkia biglobosa from different agroecological zones of Nigeria using RAPD markers. Int J Biodivers. 2014:1–6. [Google Scholar]
- Ballouche A, Neumann K. 1995. A new contribution to the Holocene vegetation history of the West African Sahel: pollen from Oursi, Burkina Faso and charcoal from three sites in the northeast Nigeria. Veg Hist Archaeobot. 4:31–39. [Google Scholar]
- Boffa JM. 1999. Agroforestry parklands in sub-Saharan Africa. FAO conservation guide 34. Rome: Food and Agriculture Organization of the United Nations (FAO) p. 230. [Google Scholar]
- Cazé ALR, Mäder G, Nunes TS, Queiroz LP, de Oliveira G, Diniz-Filho JAF, Bonatto SL, Freitas LB. 2016. Could refuge theory and rivers acting as barriers explain the genetic variability distribution in the Atlantic Forest?Mol Phylogenet Evol. 101:242–251. [DOI] [PubMed] [Google Scholar]
- Chybicki IJ, Burczyk J. 2009. Simultaneous estimation of null alleles and inbreeding coefficients. J Hered. 100:106–113. [DOI] [PubMed] [Google Scholar]
- Coetzee JA. 1993. African flora since the terminal Jurassic. In: P., Goldblatt, editor. Biological relationships between Africa and South America. New Haven (CT): Yale University Press; p. 37–61. [Google Scholar]
- Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics. 30:1187–1189. [DOI] [PubMed] [Google Scholar]
- Daïnou K, Bizoux JP, Doucet JL, Mahy G, Hardy OJ, Heuertz M. 2010. Forest refugia revisited: nSSRs and cpDNA sequences support historical isolation in a wide-spread African tree with high colonization capacity, Milicia excelsa (Moraceae). Mol Ecol. 19:4462–4477. [DOI] [PubMed] [Google Scholar]
- Demenou BB, Piñeiro R, Hardy OJ. 2016. Origin and history of the Dahomey Gap separating West and Central African rain forests: insights from the phylogeography of the legume tree Distemonanthus benthamianus. J Biogeogr. 43:1020–1031. [Google Scholar]
- Dieringer D, Schlötterer C. 2003. Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes. 3:167–169. [Google Scholar]
- Dressler S, Schmidt M, Zizka G. 2014. Introducing African plants—a photo guide—an interactive identification tool for continental Africa. Taxon. 63:1159–1161. [Google Scholar]
- Duminil J, Brown RP, Ewédjè EE, Mardulyn P, Doucet JL, Hardy OJ. 2013. Large-scale pattern of genetic differentiation within African rainforest trees: insights on the roles of ecological gradients and past climate changes on the evolution of Erythrophleum spp (Fabaceae). BMC Evol Biol. 13:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont LM. 1993. Vegetation zones in NW Africa during the Brunhes chron reconstructed from marine palynological data. Quat Sci Rev. 12:189–202. [Google Scholar]
- Earl DA, von Holdt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 4:359–361. [Google Scholar]
- Elith J, Graham CH, Anderson RP, Dudík M, Ferrier S, Guisan A, Hijmans RJ, Huettmann F, Leathwick JR, Lehmann A, et al. . 2006. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 29:129–151. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- Fourcade Y, Engler JO, Rödder D, Secondi J. 2014. Mapping species distributions with MAXENT using a geographically biased sample of presence data: a performance assessment of methods for correcting sampling bias. PLoS One. 9:e97122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaisberger H, Kindt R, Loo J, Schmidt M, Bognounou F, Da SS, Diallo OB, Ganaba S, Gnoumou A, Lompo D, et al. . 2017. Spatially explicit multi-threat assessment of food tree species in Burkina Faso: a fine-scale approach. PLoS One. 12:e0184457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet EM. 2010. GSED—genetic structure from electrophoresis data, Version 3.0. Abteilung Forstgenetik und Forstpflanzenzüchtung: Universität Göttingen. [Google Scholar]
- Global Biodiversity Information Facility 2017. GBIF occurrence. [cited 2017 Sep 14]. Available from: www.GBIF.org [Google Scholar]
- Gregorius HR. 1974. Genetischer abstand zwischen populationen. I. Zur konzeption der genetischen abstandsmessung. Silvae Genet. 23:22–27. [Google Scholar]
- Gregorius HR. 1990. A diversity-independent measure of evenness. Am Nat. 136: 701–711. [Google Scholar]
- Gregorius HR, Roberds JH. 1986. Measurement of genetical differentiation among subpopulations. Theor Appl Genet. 71:826–834. [DOI] [PubMed] [Google Scholar]
- Hall JB, Aebischer DP, Tomlinson HF, Osei-Amaning E, Hindle JR. 1996. Vitellaria paradoxa. A monograph.Bangor: School of Agricultural Sciences Publication. Number 8. University of Wales. [Google Scholar]
- Hall JB, Thomlinson HF, Oni PI, Buchy M, Aebischer DP. 1997. A monograph of Parkia biglobosa. Publication number 9. Bangor (UK): School of Agricultural and Forest Sciences, University of Wales. [Google Scholar]
- Hamilton AC, Taylor D. 1991. History of climate and forests in tropical Africa during the last 8 million years. Clim Change. 19:65–78. [Google Scholar]
- Hardy OJ, Born C, Budde K, Daïnou K, Dauby G, Duminil J, Ewédjé E-EBK, Gomez C, Heuertz M, Koffi GK, et al. . 2013. Comparative phylogeography of African rain forest trees: a review of genetic signatures of vegetation history in the Guineo-Congolian region. C R Geosci. 345:284–296. [Google Scholar]
- Hardy OJ, Charbonnel N, Fréville H, Heuertz M. 2003. Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics. 163:1467–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2:618–620. [Google Scholar]
- Heubes J. 2012. Modelling the impact of future climate and land use change on vegetation patterns, plant diversity and provisioning ecosystem services in West Africa. Frankfurt (Germany): Goethe University. [Google Scholar]
- Heuzé V, Thiollet H, Tran G, Edouard N, Lebas F. 2018. African locust bean (Parkia biglobosa & Parkia filicoidea). Feedipedia, a programme by INRA, CIRAD, AFZ and FAO. 13:49. [cited 2018 Jan 25]. Available from: https://feedipedia.org/node/268
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 25:1965–1978. [Google Scholar]
- Iloh AC, Schmidt M, Muellner-Riehl AN, Ogundipe OT, Paule J. 2017. Pleistocene refugia and genetic diversity patterns in West Africa: insights from the liana Chasmanthera dependens (Menispermaceae). PLoS One. 12:e0170511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadu CA, Konrad H, Schueler S, Muluvi GM, Eyog-Matig O, Muchugi A, Williams VL, Ramamonjisoa L, Kapinga C, Foahom B, et al. . 2013. Divergent pattern of nuclear genetic diversity across the range of the Afromontane Prunus africana mirrors variable climate of African highlands. Ann Bot. 111:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndt T, Assogbadjo AE, Hardy OJ, Glele Kakaï R, Sinsin B, Van Damme P, Gheysen G. 2009. Spatial genetic structuring of baobab (Adansonia digitata, Malvaceae) in the traditional agroforestry systems of West Africa. Am J Bot. 96:950–957. [DOI] [PubMed] [Google Scholar]
- Larrasoaña JC, Roberts AP, Rohling EJ. 2013. Dynamics of green Sahara periods and their role in hominin evolution. PLoS One. 8:e76514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KM, Kjær ED, Ouédraogo M, Nielsen LR. 2014. Microsatellite primers for Parkia biglobosa (Fabaceae: Mimosoideae) reveal that a single plant sires all seeds per pod. Appl Plant Sci. 2. doi:10.3732/apps.1400024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong Pock Tsy JM, Lumaret R, Mayne D, Vall AO, Abutaba YI, Sagna M, Rakotondralambo Raoseta SO, Danthu P. 2009. Chloroplast DNA phylogeography suggests a West African centre of origin for the baobab, Adansonia digitata L. (Bombacoideae, Malvaceae). Mol Ecol. 18:1707–1715. [DOI] [PubMed] [Google Scholar]
- Liu C, Berry PM, Dawson TP, Pearson RG. 2005. Selecting thresholds of occurrence in the prediction of species distributions. Ecography. 28:385–393. [Google Scholar]
- Logossa ZA, Camus-Kulandaivelu L, Allal F, Vaillant A, Sanou H, Kokou K, Bouvet JM. 2011. Molecular data reveal isolation by distance and past population expansion for the shea tree (Vitellaria paradoxa C.F. Gaertn) in West Africa. Mol Ecol. 20:4009–4027. [DOI] [PubMed] [Google Scholar]
- Lompo D, Vinceti B, Gaisberger H, Konrad H, Duminil J, Ouedraogo M, Sina S, Geburek T. 2017. Genetic conservation in Parkia biglobosa (Fabaceae: Mimosoideae)—what do we know?Silvae Genet. 66:1–8. [Google Scholar]
- Maley J. 1996. The African rain forest: main characteristics of changes in vegetation and climate from the upper cretaceous to the quaternary. Proc R Soc Edinb. 104B:31–73. [Google Scholar]
- Manni F, Guérard E, Heyer E. 2004. Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier’s algorithm. Hum Biol. 76:173–190. [DOI] [PubMed] [Google Scholar]
- Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209–220. [PubMed] [Google Scholar]
- Mihretie Z, Schueler S, Konrad H, Bekele E, Geburek T. 2015. Patterns of genetic diversity of Prunus Africana in Ethiopia: hot spot but not point of origin for range-wide diversity. Tree Genet Genomes. 11:1–13. [Google Scholar]
- Millogo AMD. 2014. Etude des caractéristiques morphologiques et de la viabilité des semences de Parkia biglobosa (Jacq.) R. Br. ex G. Don.—germoplasme de conservation à long terme à 4°C. Mémoire de Master, Institut de Développement Rural—Université Polytechnique de Bobo-Dioulasso. [Google Scholar]
- Monmonier M. 1973. Maximum-difference barriers: an alternative numerical regionalization method. Geogr Anal. 3:245–261. [Google Scholar]
- Murdock GP. 1959. Africa, its peoples and their culture history. New York: McGraw- Hill Book Co. [Google Scholar]
- Murphy MA, Evans JS, Cushman SA, Storfer A. 2008. Representing genetic variation as continuous surfaces: an approach for identifying spatial dependency in landscape genetic studies. Ecography. 31:685–697. [Google Scholar]
- Murphy PG, Lugo AE. 1986. Ecology of dry forests. Annu Rev Ecol Syst. 17:67–88. [Google Scholar]
- Nazareno AG, Dick CW, Lohmann LG. 2017. Wide but not impermeable: testing the riverine barrier hypothesis for an Amazonian plant species. Mol Ecol. 26:3636–3648. [DOI] [PubMed] [Google Scholar]
- Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 89:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann K, Fahmy A, Lespez L, Ballouche A, Huysecom E. 2009. The early holocene palaeoenvironment of Ounjougou (Mali): phytoliths in a multiproxy context. Palaeogeogr Palaeoclimatol Palaeoecol. 276: 87–106. [Google Scholar]
- Nikiema A. 1993. Regeneration of Parkia biglobosa (Jacq.) R. Br; ex G. Don in an agroforestry system. A pilot study in Burkina Faso [MSc thesis]. Wageningen Agricultural University, Wageningen, the Netherlands. [Google Scholar]
- Odee DW, Telford A, Wilson J, Gaye A, Cavers S. 2012. Plio-Pleistocene history and phylogeography of Acacia senegal in dry woodlands and savannahs of sub-Saharan tropical Africa: evidence of early colonisation and recent range expansion. Heredity (Edinb). 109:372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okullo JBL, Hall JB, Obual J. 2004. Leafing, flowering and fruiting of Vitellaria paradoxa subsp. nilotica in savanna parklands in Uganda. Agroforest Syst.. 60: 77–91. [Google Scholar]
- Ouedraogo AS. 1995. Parkia biglobosa (Leguminosae) en Afrique de l’Ouest: Biosystématique et Amélioration [Dissertation; ]. Wageningen Agricultural University, Wageningen, the Netherlands. [Google Scholar]
- Ouedraogo M. 2015. Improving and conserving sahelian fruit trees: a case study of Parkia biglobosa (Jacq.) Benth [IGN PhD]. Frederiksberg: Department of Geosciences and Natural Resource Management, University of Copenhagen. [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research–an update. Bioinformatics. 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit R, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, et al. . 2003. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 300:1563–1565. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Hampe A. 2006. Some evolutionary consequences of being a tree. Annu Rev Ecol Evol Syst. 37: 187–214. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modeling of species geographic distributions. Ecol Model. 190:231–259. [Google Scholar]
- Pourtier R. 1995. Atlas de la Zone Franc en Afrique Subsaharienne. Paris: La documentation française. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ræbild A, Hansen UB, Kambou S. 2012. Regeneration of Vitellaria paradoxa and Parkia biglobosa in a parkland in Southern Burkina Faso. Agroforest Syst. 85:443–453. [Google Scholar]
- Ramírez-Villegas J, Khoury C, Jarvis A, Debouck DG, Guarino L. 2010. A gap analysis methodology for collecting crop genepools: a case study with phaseolus beans. PLoS One. 5:e13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann U. 2000. Are savannas degraded forests? A holocene pollen record from the Sudanian zone of NE Nigeria. Veg Hist Archaeobot. 9:1–15. [Google Scholar]
- Salzmann U, Hoelzmann P. 2005. The Dahomey gap: an abrupt climatically induced rain forest fragmentation in West Africa during the late holocene. Holocene. 15:190–199. [Google Scholar]
- Scheldeman X, van Zonneveld M. 2010. Training manual on spatial analysis of plant diversity and distribution. Rome: Bioversity International. [Google Scholar]
- Schmidt M, Janßen T, Dressler S, Hahn K, Hien M, Konaté S, Lykke AM, Mahamane A, Sambou B, Sinsin B, et al. . 2012. The West African vegetation database. Biodiver Ecol. 4:105–110. [Google Scholar]
- Schuster M, Roquin C, Duringer PH, Brunet M, Fontugne M, Mackaye HT, Vignaud P, Ghienne J-F. 2005. Highlighting Holocene Lake Mega-Chad paleoshorelines from space. Quat Sci Rev. 24:1821–1827. [Google Scholar]
- Sexton GJ, Frere CH, Kalinganire A, Uwamariya A, Lowe AJ, Godwin ID, Prentis PJ, Dieters MJ. 2015. Influence of putative forest refugia and biogeographic barriers on the level and distribution of genetic variation in an African savannah tree, Khaya senegalensis (Desr.) A. Juss. Tree Genet Genomes. 11:103. [Google Scholar]
- Sina S. 2006. Reproduction et Diversité Génétique chez Parkia biglobosa (Jacq.) G. Don [PhD thesis]. Wageningen Agricultural University, Wageningen, the Netherlands. [Google Scholar]
- Solbrig OT, Medina E, Silva JF, editors. 1996. Biodiversity and Savanna ecosystem processes. Berlin, Heidelberg, Germany: Springer. [Google Scholar]
- Trauth MH, Larrasoaña JC, Mudelsee M. 2009. Trends, rhythms and events in Plio-Pleistocene African climate. Quat Sci Rev. 28:399–411. [Google Scholar]
- Vinceti B, Loo J, Gaisberger H, van Zonneveld MJ, Schueler S, Konrad H, Kadu CA, Geburek T. 2013. Conservation priorities for Prunus africana defined with the aid of spatial analysis of genetic data and climatic variables. PLoS One. 8:e59987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The primary data underlying these analyses, sampling locations, and microsatellite genotypes have been deposited in Dryad (doi: 10.5061/dryad.15pg4g4/1).