Abstract
This letter presents an automated obstructive sleep apnoea (OSA) detection method with high accuracy, based on a deep learning framework employing convolutional neural network. The proposed work develops a system that takes single lead electrocardiography signals from patients for analysis and detects the OSA condition of the patient. The results show that the proposed method has some advantages in solving such problems and it outperforms the existing methods significantly. The present scheme eliminates the requirement of separate feature extraction and classification algorithms for the detection of OSA. The proposed network performs both feature learning and classifies the features in a supervised manner. The scheme is computation-intensive, but can achieve very high degree of accuracy—on an average a margin of more than 9% compared to other published literature till date. The method also has a good immunity to the contamination of the signals by noise. Even with pessimistic signal to noise ratio values considered here, the methods already reported are not able to outshine the present method. The software for the algorithm reported here can be a good contender to constitute a module that can be integrated with a portable medical diagnostic system.
Keywords: Artificial neural network (ANN), Convolutional neural network (CNN), Electrocardiography, Obstructive sleep apnoea (OSA), Polysomnography (PSG)
Introduction
Health problems due to poor sleep and sleep disorders are very common in urban as well as rural population in recent times. Among the different sleep disorders, sleep apnoea syndrome (SAS) or obstructive sleep apnoea (OSA) is one of the common varieties, characterized by the recurrent cessation of breathing during sleep. However, such problems emanating from sleep disorders are often remain undiagnosed and untreated in their earlier stages. One of the reasons for this could be the lack of easy diagnostic procedures to detect sleep disorders like OSA [1–4].
Polysomnography (PSG) is the gold standard method for sleep apnoea diagnosis. PSG consists of an overnight recording of different physiological signals such as electroencephalogram (EEG), electrooculogram (EOG), electromyogram (EMG), electrocardiogram (ECG), airflow, oxygen saturation in arterial blood, respiratory efforts, snoring, and body position etc. PSGs are carried out in sleep laboratories with attending systems and specialized staffs. The medical domain-experts have the skill to correlate these recordings, to detect the possible occurrence of sleep apnoea in a patient. It is understandable that PSG is expensive, time consuming, and labour intensive procedure. Hence, there exists a need for developing reliable diagnostic alternatives in sleep study using fewer biological signals that can provide effective diagnosis and treatment of patients with sleep related complaints [5–8].The effect of sleep apnoea on the single chanel ECG recordings is one of the new approaches towards OSA study. It has been observed that patients with OSA have an escalated possibility of fatal and non-fatal cardiovascular events and an enhanced risk of sudden death from cardiac causes during the sleeping hours [5, 9, 10].
The aim of the present paper is to maximize the accuracy of the information obtained from single chanel ECG recordings only regarding the occurrence of sleep apnoea, and thus to help in the journey towards much simplified diagnostic technique with minimal number of physiological signals for the detection of OSA.
In this context, the present work contributes in developing an automated obstructive sleep apnoea (OSA) detection method, based on a deep learning framework employing convolutional neural network (CNN) using single lead electrocardiography (ECG) signals [1, 6]. A CNN consists of a single or more convolutional layers, with a subsampling layers in some cases, and after that there are fully connected layer(s) like those in conventional artificial neural network (ANN). One of the motivations for using the CNNs is that their training is easier and they have much smaller number of parameters compared to those in fully connected neural networks with identical number of hidden units. Additionally, the architecture of the CNN exploits the structure of the 2D input signals, e.g. images, multichannel speech signals etc. This is attained by means of local connections and attached weights, with a subsequent process known as pooling that yields features which are invariant to translation. The proposed scheme is computation-intensive, but can achieve very high degree of accuracy- on an average a margin of more than 9% compared to results given in other published literature till date.
Detailed scheme of work
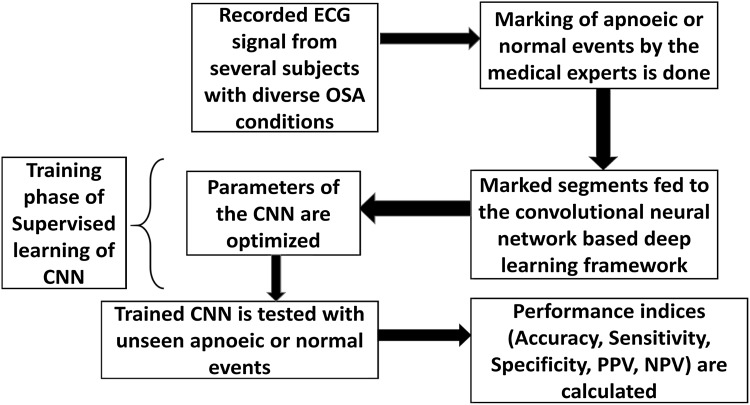
The proposed scheme consists of a layered structure akin to conventional neural network, where each layer has its distinct functions for supervised learning. The training phase of the proposed network uses a deep learning framework. The important merit of deep learning compared to earlier ANNs and other machine-learning algorithms is its ability to find newer features which are well connected to the restricted number of features that can already be obtained from the data in a training data set. Recordings of single lead ECG waveforms from several subjects with diverse OSA conditions are used. Every recorded wave is considered to consist of several sequential segments in tandem, each of 1 min duration. Each segment is marked and assessed by medical experts to indicate whether the segment portrays any apnoeic event or not. These waveform segments from the subjects form the input dataset, and are fed to the Convolutional Neural Network based deep learning framework. In different initial layers of the network, the various features are learnt from the training data and the final layers of the network classify those features into proper classes, like apnoeic/normal episodes. A flowchart of the methodology used in the paper is shown in Fig. 1. In the testing phase, the trained network is used to process input data from segments other than those considered for training, and the ability of the network to correctly identify the OSA condition is examined. Thus the methodology reported in this paper, does away with the necessity of using separate feature extraction and classification algorithms for the detection of OSA.
Fig. 1.
Flowchart of the proposed methodology
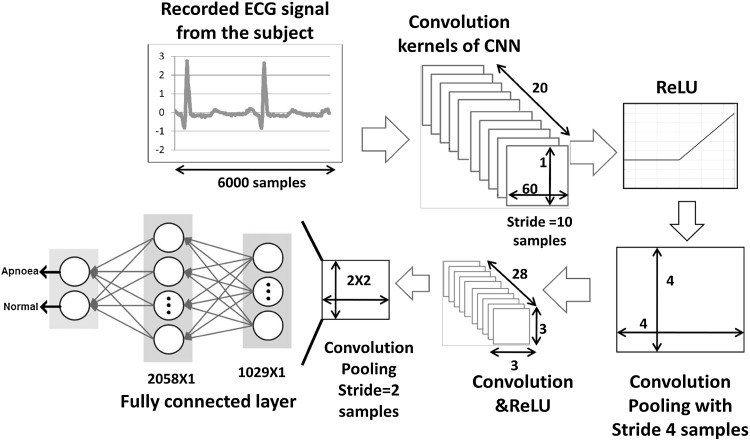
Details of the CNN structures can be found in different sources [11, 12]. For majority of the applications, these variants of networks have been used for image analysis. In the work presented here, it is used to process time series data. The proposed network is discussed in the following paragraphs and the complete scheme is shown in Fig. 2. It is clearly seen in Fig. 2, that the CNN structure consists of a number of layers. The descriptions of the layers are as follows:
Input Layer This layer receives the recorded single lead ECG signals from different subjects with sleep disorders. The dimensions of different layers are depicted in Fig. 2. Every input data has 6000 samples for each one minute segment.
Convolution layer This layer accomplishes the ‘convolution filtering’ of the input data to extract the important features from the data and the information propagates through the subsequent layers. The convolution operation for 1D signal (sequence) can be mathematically expressed as , where s[n] is the signal and h[n] is the convolution kernel. In the CNN, each convolution kernel represents a genre of features, e.g., nature of temporal variation, sharp edges in the data, subtle variations of amplitude etc. In the network considered here, twenty convolution kernels have been used and the weights of the kernels have been randomly initialized. Subsequently, the weights are updated via back propagation learning. In the first convolution layer, the dimension of the filter structure can be represented as [1 × 60 × 20]. This structure is obtained by trial and error method, in the same way as the number of neurons are selected in a layer of conventional neural network.
- ReLU layer It is the abbreviation of Rectified Linear Units. This layer performs a thresholding operation based on a nonlinear decision mapping function. In the present work, the function is
Pooling layer The task of this layer is dimension reduction. Max pooling is a very popular technique [9] for such an operation. Average pooling and L2-norm pooling are also in use. These pooling methods perform very well, when the inputs are images. To suit the 1D input signals in the time domain, a modification is incorporated by replacing the simple pooling operation with convolutional pooling. It is done simply by introducing another convolution layer, but with a different slide along time axis during kernel shifting. This can be considered as another novelty of the present scheme.
Fully connected layer This layer is similar to those in conventional Neural Networks and as revealed by its name, each neuron in this layer is connected to all activations in the preceding layer. It is noted that this layer actually performs the classification task, whereas the earlier layers perform feature learning. Gradient based back propagation algorithm is used for network updating. However, one can choose any other updating algorithm as dictated by the specific problem in which it is applied.
These layers can be repeated n number of times, depending upon the level of accuracy required.
Fig. 2.
Schematic of the proposed method using CNN
In the present work, the layers are repeated twice to achieve desired level of accuracy, which is shown in Fig. 2. The CNN is trained using Intel Core i5-4460T; RAM: 16 GB with NVIDIA GTX 4GB graphics card.
Results and discussion
The performance of the present scheme has been tested on apnoea-ECG dataset, available in Physionet [1]. Here, single lead ECG recordings of 35 subjects with OSA have been used. Table 1 shows the physiological parameters of the population under study. The time duration of each recording is 7–10 h. Each data in the dataset is labeled and scored by experts on minute-by-minute basis, as stated earlier. These annotations indicate the presence or absence of sleep apnoea during that interval. Each 1 min record can be assumed as one ‘episode’ or an ‘event’. Further details of the database can be found in [1, 2]. The performance of proposed scheme is evaluated in terms of accuracy, sensitivity and specificity [3, 6]. As these parameters are well known, their expressions are not given here.
Table 1.
Physiological parameters of the population
| Sub. no. | Length in minutes | Non-apnea minutes | Apnea minutes | AHI | Age | Sex |
|---|---|---|---|---|---|---|
| 01 | 524 | 149 | 375 | 63 | 44 | M |
| 02 | 470 | 261 | 209 | 37.7 | 46 | M |
| 03 | 466 | 454 | 12 | 0.13 | 44 | F |
| 04 | 483 | 483 | 0 | 0 | 39 | M |
| 05 | 506 | 190 | 316 | 34 | 55 | M |
| 06 | 451 | 451 | 0 | 0 | 31 | M |
| 07 | 510 | 270 | 240 | 21 | 58 | M |
| 08 | 518 | 194 | 324 | 48 | 55 | M |
| 09 | 509 | 342 | 167 | 18.5 | 43 | M |
| 10 | 511 | 415 | 96 | 10 | 39 | 11 |
| 11 | 458 | 445 | 13 | 5 | 52 | M |
| 12 | 528 | 471 | 57 | 33 | 40 | M |
| 13 | 507 | 215 | 292 | 18.7 | 57 | M |
| 14 | 491 | 52 | 439 | 79.5 | 38 | M |
| 15 | 499 | 299 | 200 | 15.9 | 63 | M |
| 16 | 516 | 451 | 65 | 24 | 53 | M |
| 17 | 401 | 400 | 1 | 0 | 27 | F |
| 18 | 460 | 458 | 2 | 0 | 27 | M |
| 19 | 488 | 81 | 407 | 56.2 | 54 | M |
| 20 | 514 | 250 | 264 | 43 | 51 | M |
| 21 | 511 | 391 | 120 | 19 | 53 | M |
| 22 | 483 | 481 | 2 | 0 | 27 | F |
| 21 | 511 | 391 | 120 | 19 | 53 | M |
| 22 | 483 | 481 | 2 | 0 | 27 | F |
| 23 | 528 | 409 | 119 | 14.3 | 43 | M |
| 24 | 430 | 429 | 1 | 0 | 31 | M |
| 25 | 511 | 220 | 291 | 48 | 55 | M |
| 26 | 521 | 177 | 344 | 15.1 | 57 | M |
| 27 | 499 | 11 | 488 | 75 | 60 | M |
| 28 | 496 | 62 | 434 | 75 | 60 | M |
| 29 | 471 | 471 | 0 | 0 | 41 | F |
| 31 | 558 | 42 | 516 | 93.5 | 29 | F |
| 32 | 539 | 114 | 425 | 71.8 | 29 | F |
| 33 | 474 | 471 | 3 | 0.13 | 28 | F |
| 34 | 476 | 472 | 4 | 0.38 | 30 | F |
| 35 | 484 | 484 | 0 | 0 | 31 | M |
| Mean | 494.02 | 317.7 | 176.3 | 26.1 | 43.6 |
50% of the available data is used for training the network and the rest is used for testing. The output of the proposed network gives a scoring between 0 and 1 to indicate whether the ECG episode is an apnoeic event or normal. During training phase, the target or output was set as for an apnoeic event and for a normal breathing event. Hence, under ideal network performance after training, the output of the network should be for an apnoeic episode and for a normal breathing event. During practical testing, the outputs are not ideal. For example, in an apnoeic event the output sometimes comes as . Hence, a thresholding of the output is used in testing phase, which makes the larger of the two elements of the output matrix equal to 1 and the other equal to 0.
The results as well as a comparison of performance of the proposed scheme with existing methodologies, is shown in Table 2. It is evident that all performance parameters are quite high for the proposed scheme compared to those of existing methods when tested on the same dataset, which establishes the positive contribution of the proposed methodology. A detailed view of the performance of the proposed scheme can be obtained in Table 3.
Table 2.
Comparison of performance of the proposed scheme vis-à-vis existing methodologies (50% training and 50% testing data)
Table 3.
Confusion matrix and performance indices of the proposed scheme
| Truth | ||
|---|---|---|
| Apnea | Non apnea | |
| Results | ||
| Apnea | True positive (TP) | False positive (FP) |
| 4878 | 46 | |
| Non apnea | False negative (FN) | True negative (TN) |
| 109 | 5754 | |
| Total | 4987 | 5800 |
Moreover, the robustness of the feature learning ability of the deep network is verified by decreasing the amount of training data and correspondingly increasing the testing data.
The results are shown in Table 4. It can be seen that even with lesser amount of training data, the performance of the scheme is acceptable, which indicates the versatility of learning the inherent features of a signal by a deep learning framework.
Table 4.
Effect of number of training data on the performance of the proposed scheme
| Percentage of data (training: testing) | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| 50:50 | 98.91 | 97.82 | 99.20 |
| 40:60 | 98.80 | 97.33 | 98.67 |
| 30:70 | 96.11 | 96.03 | 97.11 |
| 20:80 | 94.33 | 93.88 | 95.67 |
The initial layers of the network employ convolution operation which has a filtering property. The convolution kernels get updated during the training phase. In the process, one or more of the kernels assume filtering property that expunge(s) any contaminating random noise.
Thus, the proposed scheme is also immune to ambient noise up to certain extent. Hence, the developed scheme is capable of handling noisy ECG data without sacrificing much of its performance. It is an added advantage of the proposed scheme. The effect of noise on the performance has been examined by adding synthetic Gaussian zero mean white noise to the data, and the results are given in Table 5.
Table 5.
Effect of noise on the performance of the proposed scheme with 50% training and 50% testing data
| SNR in dB | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| 60 | 98.52 | 97.11 | 98.86 |
| 40 | 96.33 | 95.27 | 96.16 |
| 30 | 93.12 | 92.28 | 94.76 |
| 20 | 89.09 | 89.01 | 89.10 |
| 10 | 79.28 | 79.10 | 80.01 |
Conclusions
This letter reports a deep learning framework using CNN for automated detection of obstructive sleep apnoea (OSA) using single lead ECG signals. The main contribution of the work is enhancement in the OSA detection accuracy compared to the existing methods. The present scheme eliminates the requirement of separate feature extraction and classification algorithms. A part of the proposed network performs feature learning and the other part classifies the features in a supervised manner. The performance of the present scheme is compared with the existing methods and it has been observed that the present method increases the classification accuracy on an average by a margin of more than 9%. The proposed scheme is capable of learning inherent features with less training data and can also eliminate the effect of random noise in the data up to a certain extent.
The drawback of the scheme is its computational burden, which is high as it employs a deep learning methodology. As signal records for OSA are long (7–10 h) and the expert observation is mostly an off-line process, this limitation does not pose any hindrance in its real life application. However, the computational burden is not to the extent that would debar the implementation of the algorithm by a microcontroller, since deep learning based systems have already been realized using ARM microcontrollers. Thus the software module for implementing the complete algorithm reported in this work, may be a good candidate for integration with a portable health monitoring system, and since it works on ECG signals, the necessity of a dedicated polysomnographic system is dispensed with. The resilience of the system to the corrupting effect of noise is also excellent. Even with a pessimistic figure for the corruption (e.g. a signal-to-noise ratio of 30 dB), the performance indices are not inferior to those obtained by the existing methods. Furthermore, the scheme is a generic one that can be used for other physiological signals, like EEG, EOG, EMG etc. apart from ECG for the same purpose.
Acknowledgements
This work is supported by the grant received by Jadavpur University under UPE-II scheme of UGC, Government of India.
Conflict of interest
All the authors declare that they have no conflict of interest in relation to the work in this article.
Ethical approval
This study meets ethical standards for engineering studies at the Jadavpur University. No humans or animals were involved in this study, thus no review by ethical committee is required.
References
- 1. Penzel T, Moody GB, Mark RG et al. The apnoea-ECG database. In: Computers in cardiology 2000. Cambridge: IEEE Engineering in Medicine and Biology Society; 2000. p. 255–58.
- 2.Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng C-K, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101(23):e215–e220. doi: 10.1161/01.CIR.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 3.Hassan AR. Computer-aided obstructive sleep apnoea detection using normal inverse Gaussian parameters and adaptive boosting. Biomed Signal Proc Control. 2016;29:22–30. doi: 10.1016/j.bspc.2016.05.009. [DOI] [Google Scholar]
- 4.Varon C, Caicedo A, Testelmans D, et al. A novel algorithm for the automatic detection of sleep apnea from single-lead ECG. IEEE Trans Biomed Eng. 2015;62(9):2269–2278. doi: 10.1109/TBME.2015.2422378. [DOI] [PubMed] [Google Scholar]
- 5.Hassan AR, Haque MA. Computer-aided obstructive sleep apnea screening from single-lead electrocardiogram using statistical and spectral features and bootstrap aggregating. Biocybern Biomed Eng. 2016;36(1):256–266. doi: 10.1016/j.bbe.2015.11.003. [DOI] [Google Scholar]
- 6.Kumar TS, Kanhangad V. Automated obstructive sleep apnoea detection using symmetrically weighted local binary patterns. IET Electron Lett. 2017;53(4):212–214. doi: 10.1049/el.2016.3664. [DOI] [Google Scholar]
- 7.Koley B, Dey D. On-line detection of apnea/hypopnea events using SpO2 signal: a rule-based approach employing binary classifier models. IEEE J Biomed Health Inform. 2014;18(1):231–239. doi: 10.1109/JBHI.2013.2266279. [DOI] [PubMed] [Google Scholar]
- 8.Koley B, Dey D. Real-time adaptive apnea and hypopnea event detection methodology for portable sleep apnea monitoring devices. IEEE Trans Biomed Eng. 2013;60(12):3354–3363. doi: 10.1109/TBME.2013.2282337. [DOI] [PubMed] [Google Scholar]
- 9.Khalil MM, Rifaie OA. Electrocardiographic changes in obstructive sleep apnoea syndrome. Respir Med. 1998;92:25–27. doi: 10.1016/S0954-6111(98)90027-0. [DOI] [PubMed] [Google Scholar]
- 10.Pépin JL, Defaye P, Vincent E, Christophle-Boulard S, Tamisier R, Lévy P. Sleep apnea diagnosis using an ECG Holter device including a nasal pressure (NP) recording: validation of visual and automatic analysis of nasal pressure versus full polysomnography. Sleep Med. 2009;10:651–656. doi: 10.1016/j.sleep.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Ravı D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, Yang GZ. Deep learning for health informatics. IEEE J Biomed Health Inform. 2017;21(1):4–21. doi: 10.1109/JBHI.2016.2636665. [DOI] [PubMed] [Google Scholar]
- 12.Lee KB, Cheon S, Kim CO. A convolutional neural network for fault classification and diagnosis in semiconductor manufacturing processes. IEEE Trans Semicond Manuf. 2017;30(2):135–142. doi: 10.1109/TSM.2017.2676245. [DOI] [Google Scholar]