Abstract
Medical imaging plays an integral role in the identification, segmentation, and classification of brain tumors. The invention of MRI has opened new horizons for brain-related research. Recently, researchers have shifted their focus towards applying digital image processing techniques to extract, analyze and categorize brain tumors from MRI. Categorization of brain tumors is defined in a hierarchical way moving from major to minor ones. A plethora of work could be seen in literature related to the classification of brain tumors in categories such as benign and malignant. However, there are only a few works reported on the multiclass classification of brain images where each part of the image containing tumor is tagged with major and minor categories. The precise classification is difficult to achieve due to ambiguities in images and overlapping characteristics of different type of tumors. In the current study, a comprehensive review of recent research on brain tumors multiclass classification using MRI is provided. These multiclass classification studies are categorized into two major groups: XX and YY and each group are further divided into three sub-groups. A set of common parameters from the reviewed works is extracted and compared to highlight the merits and demerits of individual works. Based on our analysis, we provide a set of recommendations for researchers and professionals working in the area of brain tumors classification.
Keywords: Human brain cancer diagnosis and analysis, Magnetic resonance imaging, Human brain tumor multi-classification
Introduction
A brain tumor is one of the major reason of an increase in mortality among adults and children in the world causing a high burden for families and health care systems. A brain tumor is a collection or mass of abnormal cells produced in the brain from parenchyma or adjacent brain parts. The brain cancer may cause severe disabilities which significantly constrain patients’ daily activities and reduce their quality of life [1]. Comprehensive statistical studies of the brain cancer epidemiology of the world have been published [2–6].
In the United States, the registered brain cancer cases increased significantly; 1.5 million cases in 2013; 1.6 million in 2014, and 1.658 million in 2015 [7].Its fatality rate also is around 33% and this rate is rising [8]. Another study tells that it has risen to 300% over past three decades [9].
The brain tumors which are mainly divided into two categories: benign and malignant. The malignant tumors are cancerous while the benign tumors do not. Though various classification schemes have been used for brain tumor categorization, however, the scheme suggested by the World Health Organization (WHO) is considered as a standard. It categorized tumor types into four grades. As the grade of tumor increases, it becomes more malignant i.e. grade 1 and 2 are considered as benign whereas grade 3 and 4 are malignant. Detailed categorization is presented by WHO in Rousseau, Audrey [10].
Different types of tumors are treated differently. The treatment methods include surgery, radiotherapy, chemotherapy, biological therapy, steroids therapy and other therapies using nanotechnology [11, 12]. Before using any treatment, it is required to understand the present status of the brain tumor such as its location, its growth rate, and all related factors. There are two main methods of obtaining this information: Surgery and Imaging. Considering the cost, risk and time factors, imaging methods are preferable for disease diagnostic either pre-treatment, inter-treatment or post-treatment [13–16].
Researchers have employed various imaging modalities and techniques for binary to the multiclass classification of brain tumors by digitally processing of brain images. The process of analyzing and understanding the brain image containing tumor is normally divided among various steps i.e. preprocessing, image enhancement, segmentation, feature selection, and classification [17]. Various mathematical and statistical approaches have been used to carry out different image processing steps. These techniques may include wavelets, machine learning methods, discrete cosine transform or hybrid methods [18–21]. Here are a few of the practical applications of medical image analysis:
Locating abnormal region and other pathologies
Measuring tissue sizes
Computer-guided surgery
Computer aided diagnosis
Radiotherapy
Treatment planning
Study of anatomical structure
Identification of malignant parts within tumor area in order to minimize the risk of sampling errors in biopsy
Classification results are promising for binary classification, however, for multi-classification, a lot of work is required to bring it to an acceptable level [22, 23]. In this study, we review research work on brain tumor multiclass classification only. We have tried to keep scope limited to multiclass classification approaches based on conventional MRI, however, works including multi-parametric classification and brain disease classification have also been included due to two main reasons: (1) researchers have presented single parameter and multi-parameter based classification as comparative study and (2) conventional and advanced MRI are used collectively to produce better results [24].
Tumor classification phases using brain images
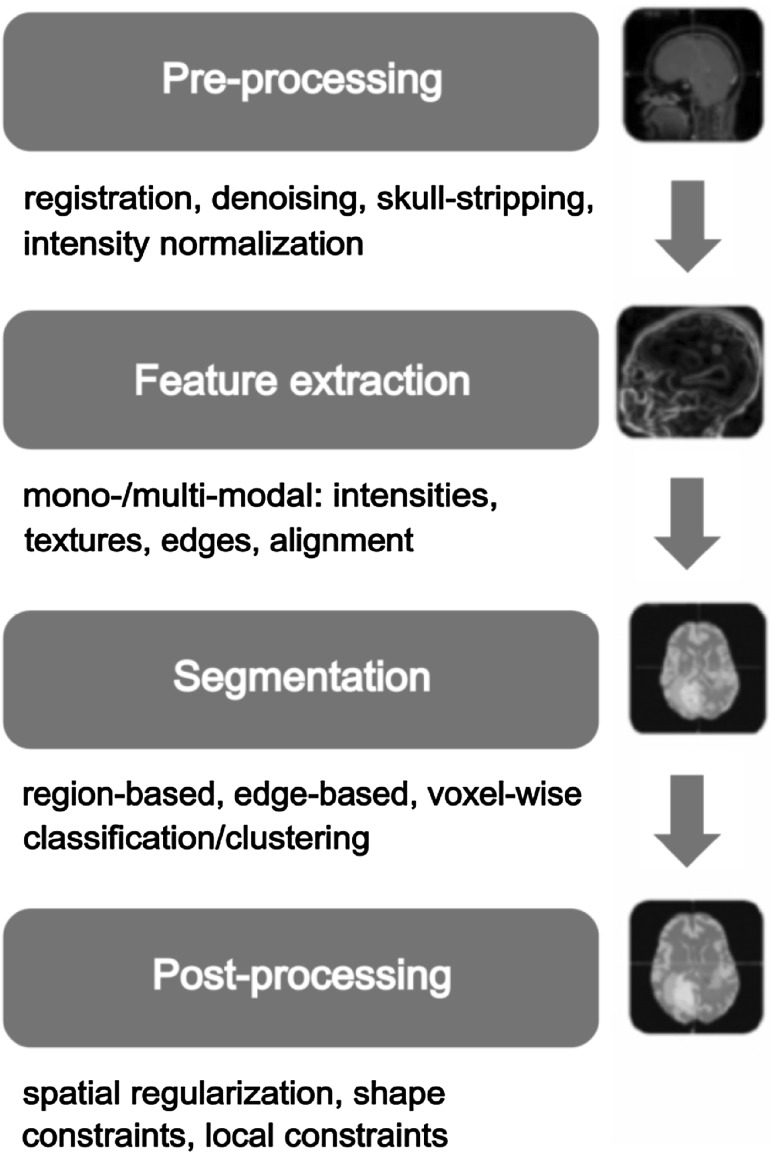
There is little work done on tumor grading [25]. Tumor identification and classification is a most challenging task due to various imaging features that create ambiguity. This ambiguity is due to different factors like brain tumor characteristics, characteristics of tumor surrounding regions and imaging modality characteristics. Low contrast images, unknown shape, and size of the tumor, unknown noise, tumor characteristic variation from patient to patient and partial volume effect are important to note. A single region of the tumor (Gliomas) may contain heterogeneous sub-regions of higher and lower grade [26, 27]. Brain tumor characteristics include iso-intensity (different brain tumor tissues may have same signals as that of tumor), hypointensity (image may have darker shades than actual tumor tissues), hyperintensity (high-intensity areas), tumor heterogeneity and tumor homogeneity [28]. Ambiguities due to surrounding regions may include different type of edema (swelling) like perilesional edema [29]. Tumor intensities and shapes vary from patient to patient and same gray scales may be found in different tumor types. Multiclass classification or grading of brain tumors using MRI is considered more difficult and challenging task [30]. For example, conventional anatomical MRI is considered insufficient in determining Glioma [31] and grades of Glioma [32]. Different sequences of MRI could help in tumor grading using texture analysis [25]. The following figure presents an overview of grading process:
In Fig. 1, filled blocks represent the essential phases whereas unfilled boxes show optional steps. Rest of the paper is organized as follows: Section three presents the study methodology. Section 4 discusses the brain tumor and its different categorizations. Section 5 discusses the pipeline of brain tumor classification. Findings and recommendations are given in Sect. 6 and future work is discussed in Sect. 7.
Fig. 1.
Brain tumor classification process
Methodology
As the first step of the study, we identified information sources like conferences, journals, research groups and other published material. Instead of citing specific platforms, we used search engines like Google Scholar, Pubmed, Pubmed Central, IEEE Xplore, WebMD and Medscape to search papers published on different platforms. We used following search queries in order to retrieve relevant information: “Brain MRI classification”, “Brain tumor”, “classification”, “CNS tumor classification”, “Medical imaging for brain tumor”, “Brain tumor multi-classification”, “Brain tumor MRI classification”, “Brain tumor grading in MRI”, “Multiclass segmentation”. We have reviewed papers published from 2010 to 2016 however important references therein if found useful, are also included. We have collected 161 papers and reviewed 31 papers.
We have focused on anatomical MRI-based multiclass classification work only. Research works with following issues are excluded: work on binary classification (benign and malignant), based on other imaging modalities, with missing required details or ambiguous details and effect of time on tumor progression [18, 33–36].
Brain tumor and its categorization
WHO have graded brain tumors into four grades: I, II, III and IV. A tumor does not evolve directly in high grade. It starts from low grade and gradually moves to a higher level. In other words, it starts as benign and with time it becomes malignant. With the rise of tumor grade, its treatment becomes difficult to impossible [37]. Moreover, surgical methods of brain tumor detection and cure are very painful, costly, highly risky and time-consuming. It is therefore very required to develop methods and procedures to detect such lesions in the brain at an early stage for a proper cure without facing any risk. There are two major categories of brain tumors based on their origin: primary brain tumor that originate within the brain whereas secondary (metastatic tumors) brain tumors originate at different locations of the body and move to the brain. Primary brain tumors are found at different locations in children and adults. In adults, it is normally found in anterior two-thirds of cerebral hemispheres and in children, it is commonly found in posterior cranial fossa [38]. Like a metastatic tumor, primary tumor can expand and affect other parts of the brain and central nervous system. Different researchers have classified tumors differently based on tumor characterizations that could be formulated on:
the location of tumor origin (medulla, midbrain, and pons) [39]
the degree of ‘locality’; (whether diffuse of focal) [40]
the path and degree of tumor growth [41]
the degree of brainstem enlargement [42]
the degree of exophytic growth [43]
the presence of necrosis, cysts, or hemorrhage [44]
the presence of hydrocephalus [44]
However, WHO classification is considered as standard in the medical world. WHO has classified the tumors into 120 types with around 20 major categories and sub-categories. Tumors are named based on their stem cells i.e. Glioma tumor is caused by Glial cells, the area of the brain from which tumor is evolved like pituitary adenoma is named as it originates in pituitary tissues. WHO has assigned grades to each tumor type based on tumor growth rate [45]. There are four grades as listed below: Grade 1-very slow growing; Grade 2-fast growing, Grade 3-faster growing, Grade 4-fastest growing. Current research shows that WHO classification system based on histology and morphology will be extended by a system based on molecular markers of tumor differentiation and progression [46].
Brain tumor imaging and dataset acquisition
To ease the process of brain tumor detection and analysis, a good number of imaging modalities have been developed. A short list of highly used medical imaging modalities in brain study includes single photon emission computed tomography (SPECT), computed tomography (CT), positron emission tomography(PET), medical resonance imaging (MRI) and its different applications and functional magnetic resonance imaging (fMRI). There is enough literature to provide a detailed overview of different medical imaging modalities [15, 47]. These imaging modalities could be divided into two major groups: invasive and non-invasive. Invasive techniques can cause damage to human body or the area under scan however non-invasive techniques are harmless, hence preferred. Most of the imaging techniques use contrast enhancement agents such as gadolinium.
Different imaging sequences have varying capabilities to separate different brain tissues [29]. Tumor grade could be obtained using contrast enhancement on MRI and is commonly used in clinical systems [8, 48–51]. Researchers have shown that MRI-based tumor grading is potentially capable and viable method and have mostly used MRI and its applications for detection and classification of brain tumors. MRI applications include T1-weighted imaging with contrast enhancement and T2-weighted images. There are three major variants of MRI: (1) no contrast enhancement (2) contrast enhancement (3) perfusion and dynamic contrast enhancement. Conventional MRI sequences like T1W and T2W contrast enhanced MRI are found useful in grading gliomas and other types of tumors into a low and high grade [52].
In order to rank tumors among different grades defined by WHO, researchers find variants of MRI useful like diffusion imaging: diffusion tensor imaging-DTIand diffusion weighted imaging-DWI, vascular imaging (dynamic contrast enhancement MRI-DCE, dynamically susceptibility weighted imaging-DSC), metabolic imaging (MR spectroscopy-MRSI), molecular imaging (positron emission tomography) functional MRI and other multi-parametric imaging [25, 26, 53, 54].
In this work, we only review the grading and classification approaches that are based on the anatomical structure of the brain and use machine learning approaches. Quality data set is basic ingredient of tumor classification process. There are three main ways to prepare good quality data set: (1) custom collection (2) from open online repositories (3) synthetic imaging. In the custom collection, the researcher collects original patient images from radiology department of some hospital or health unit. This is a time taking process but is most useful as a geo-local brain tumor real time data is available that can help in the production of other metadata like area wise, gender wise tumor intensities. There are various online data repositories that provide MRI of brain tumors. Such repositories are either maintained by hospitals or university research centers. Real tumor images may not show a deep level of variation or other required parameters may be missing. To overcome this deficiency, imaging software is used to produce fake tumor images (that may have their own issues). "Appendix A" presents resources list used in current review research.
Brain tumor classification process
Preprocessing
Preprocessing in medical image analysis is an optional step. It is due to the availability of good quality image data sets acquired through careful imaging process. There are multiple preprocessing steps applied either all or few by researchers in their studies [55, 56]. These steps include skull stripping [22, 57], noise removal [6, 58, 59], eddy current effect removal [60, 61], band reduction i.e. converting color image into grey image [58], image enhancement, image restoration and image standardization [12]. Image morphology is also considered whenever functional or time series data is being analyzed.
An image is represented as a matrix of color or intensity values of pixels. A small set of values (normally a small matrix) is used to manipulate and modify the image pixel values for a specific purpose [62]. Such small matrices are known as filters. Application of filters to an image is either performed in the spatial domain or in the frequency domain. Filters are normally divided among two groups: linear and non-linear. All preprocessing phases apply different filters to image in order to highlight or extract important information. An image could be improved using preprocessing techniques however if not applied appropriately, there may be adverse effects [63–65].
Segmentation
Brain tumor classification schemes using images may or may not perform segmentation [66]. In this study, we review both approaches. Although segmentation is an open research area (https://www.miccai2015.org/) however, segmentation based schemes fail if abnormalities in the brain are not possible to be segmented spatially. Different type of variation in brain images, like statistical and geometrical, also limit the performance of classification approaches [67–69].
There are three types of image segmentation methods: manual, semi-automatic and fully automatic [70]. Manual segmentation is performed by Radiologists whereas fully automatic approach requires computer aided design (CAD) software to perform this task with high computational cost. In order to reduce computation, some CAD systems provide a semi-automatic approach in which part of the task is done by human expert and rest is performed by the machine [71, 72]. A lot of work on segmentation can be found in the literature. We discuss here the segmentation techniques used in our reviewed multiclass classification work only.
Approaches using unsupervised learning need to initialize the clustering algorithm. Based on some similarity measure, these clusters are merged based on predefined stopping criteria.Juan-Albarracín et al. [87], used K-means++ algorithm with 100 different initializations that are further reduced to 10. Two other unsupervised segmentation approaches i.e. Gaussian Mixture Model (GMM) and Gaussian Hidden Markova Random Field (GHMRF), are used and compared in this work.
Support Vector Machine classifier could effectively partition the image into two classes using maximum-margin. Images in spatial domain cannot be partitioned using linear segregator. However if a transform could convert non-linear data into high dimensional space, linear separation becomes possible. Liu et al. [73], have used linear kernel method for segmentation in by applying Gabor Wavelet transform on images.
Chaddad [63], have used multilevel thresholding Otsu method in their study. This method defines a set of thresholds and each threshold is tried in order to obtain a minimal interclass variance.
Gradient vector flow (GVF) are active contour-based methods used in image segmentation. Contours or curves are defined within image domain and that can move under the influence of internal and external factors. Internal factors are defined within curves whereas external factors are present in image data. These methods are used for object boundary detection [74].
Multilevel thresholding Single level thresholding based segmentation produces low-quality results whereas multi-level thresholding can improve the classification of pixels into normal and abnormal regions. In this method calculation of threshold is based on variance maximization between classes. In order to make thresholding process more robust, a median filter is used that enhances the variance.
Super-pixel based segmentation methods group multiple pixels into meaningful atomic regions that can replace the firm structure of pixel grid. There are multiple super-pixel based approaches that have been used by researchers in computer vision and image processing. Such approaches include graph-based methods (NC05, GS04, SL08, GCa10, GCb10), gradient based algorithms (MS02, QS08, WS91, TP09) and SLIC superpixels.
Feature extraction
In order to classify pictorial data, it is required to identify important features present in images that lead to categorization. Such features could be grouped into basic and complex features [75, 76]. Basic quantitative features may include average, variance, correlation, contrast, entropy, sum entropy, difference entropy, energy, homogeneity, kurtosis, inverse difference moment, kth moment and inertia. Using basic features, a variety of derived features could be extracted. Multiple algorithms have been developed to extract high-value complex features [35, 77–80]. In this study, we have grouped complex features into three major groups. There may be more candidate features for a particular domain but we have listed only those which are used in research under review:
Spatial domain features
We have grouped different types of features which could be obtained directly from an image and are considered to lie in the spatial domain. These include spatial features like points and lines; textural features like coarseness, contrast, directionality and regularity [81].
Topological Features,
Gray Level Co-occurrence Matrix (GLCM),
Neighborhood Gray Tone Difference Matrix (NGTDM),
Gray level co-occurrence linked list (GLCLL),
Dominant Grey level run length matrix (DGLRLM),
Spatial gray level dependency matrix (SGLDM),
Motif co-occurrence matrix (MCM),
Color co-occurrence histograms (CCH),
Laplacian of Gaussian (LoG),
Rotation Invariant Local Binary Patterns (RILBP),
Intensity based features (IBF),
Directional Gabor Texture Features (DGTF),
Rotation Invariant Circular Gabor Features (RICGF),
Counting Label Occurrence Matrix (CLOM),
Apparent Diffusion Coefficients (ADC).
Wavelets and frequency domain features
These are the features extracted from images after converting them into frequency or any other domain [82]. In this category, we have placed the frequency domain features and other transformation based features as listed below:
Gabor filters (GF),
Rotation invariant circular Gabor filter(RICGF),
Discrete wavelet transform (DWT,)
Discrete Cosine Transform (DCT),
Haar wavelets,
Daubechies 4 wavelets,
Contextual features
The features which are not directly related to the image are referred here as contextual features. Usually, medical image processing works at pixel or voxel level that is fine-grained image processing however local and global contextual features may lead to better results. Contextual features are either coarse-grained information or attached textual information. Template matching or atlas based approaches in medical imaging are the prominent examples.
This category includes different type of features like patient meta-data i.e. age, tumor history etc. Pathological features like tumor area, enhancement, necrosis, edema, neovascularization, and bleeding. Sometimes, such features are also referred as clinical features [83]. Pathological features too are grouped in this category [84].
Feature reduction
As the current research exhibits, there are a huge number of features used for image segmentation and classification. In order to enhance the accuracy and processing speed, it is essential to eliminate low-value features and retain only high-value features [85, 86]. Feature selection or reduction phases are normally divided into two sub-phases: feature ranking and feature subset selection [58]. In this review, we found various ranking and subset selection methods to extract high-value features. Popular feature reduction methods include Principal Component Analysis, Linear Discriminant Analysis, Independent Component Analyses and student’s t test. The complete list of methods can be found in next sections. However, feature reduction, sometimes, could adversely affect the system performance [63].
Multiclass classification
In literature, an ample research work is reported in tumor classification. However, most of the research is based on binary classification i.e. benign and malignant. A few research could be found on multiclass classification. In this section, we intend to review multiclass classification work only. Classification work is either based on tumor area segmentation based approaches or non-segmentation based approaches. To formulate review criteria, we have grouped the multiclass classification work into two main categories: Segmentation based and non-segmentation based. These two categories are further divided into sub-categories by looking at the type of features used for classification.
Following list shows the grouping of review work:
- Segmentation based multi-classification
- Spatial domain features based classification
- Wavelet and frequency features based classification
- Contextual and Hybrid features based classification
- Non-segmentation based multi-classification
- Spatial domain features based classification
- Wavelet and frequency features based classification
- Contextual and Hybrid features based classification
Our review shows that there may be little work that lies in some of our coined category and on the other hand there may be enough work that can be placed under a different category (Table 1).
Table 1.
Segmentation oriented spatial feature based multi-classification
| Study | Imaging modalities | Pre-processing | Segmentation method | Dataset and tumor types | Feature set | Feature reduction | Classification | Evaluation |
|---|---|---|---|---|---|---|---|---|
| Kumar et al. [88] | T1 T2 Post cont. T1 FLAIR |
– | Gradient vector flow (GVF) | Patients = 55 Images = 428 AS = 118 GBM = 59 MED = 97 MEN = 88 MET = 66 Image size = 256 × 256 |
GLCM = 16 LoG = 16 DGTF = 100 RICGF = 40 RILBP = 36 IBF = 10 |
PCA | ANN with GDBPM weight estimation | Accuracy = 92% |
| Singhal et al. [8] | C-MET PET FDG PET |
– | Manual segmentation | Patients = 102 OG = 15 PAS = 01 AS Grade-II = 24 AS Grade-III = 14 GBM = 43 |
Tumor/normal (T/N) ratio | Student t test and Mann–Whitney test | p value | T/N ratio |
| Sachdevaet al. [28] | T1 post contrast T2, FLAIR |
– | Manual segmentation | Images = 428 Patients = 55 AS = 118 GBM = 59 MED = 97 MEN = 88 MET = 66 NR = 428 Image size = 256 × 256 |
Size = 218 LoG = 16 GLCM = 16 RILBP = 36 IBF = 10 DGTF = 100 RICGF = 40 |
PCA | New ANN based classifier ANN |
Accuracy = 86% |
| Soltaninejad et al. [60] | FLAIR T2 |
– | Super-pixel based method with nearly similar intensity features, manual segmentation | Patients = 21 GLM MEN PADN NrSh Image size = 256 × 256 |
Features = 38 GLCM = 16 GTSDM = 22 |
– | SVM | Accuracy = 80% |
| Ryu et al. [25] | T1-weighted T2-weighted DWI FLAIR |
– | Manual ROI segmentation | Patients = 40 AS = 8 AAS = 10 GBM = 21 Image size = 448 × 256 |
GLCM = 10 SD = 6 Entropy and kurtosis |
Student t test | ADC maps and histogram analysis, 5th percentile ADC values, ROC Analysis | Sensitivity = 82% Specificity = 90% Accuracy = 84.4% |
| Juan-Albarracín et al. [87] | T1, T1c, T2, FLAIR and T1d | Denoising, skull stripping, biase field correction, super resolution | Unsupervised segmentation | Patients = 21 C1:non-brain non-tumor necrosis C2: Surrounding edema C3: non-enhancing tumor C4: Enhancing tumor C0: every other thing |
Texture features = 20 | PCA | K-means Fuzzy K-means, GMM clustering, GHMRF technique |
Dice = 74% PPV = 71% Sensitivity = 81% Kappa = 99% |
Segmentation based multiclass classification
A significant number of segmentation techniques have been developed by researchers to mark and extract the region of interest. In this section, we review the multiclass classification techniques that employ the segmentation process before classification.
Spatial domain features based classification
In this section, we review tumor classification work based on spatial domain features. A comparative list of parameters is exhibited in Table 2.
Table 2.
Segmentation oriented wavelets and frequency feature multi-classification
| Study | Imaging | Preprocessing | Segmentation | Dataset | Feature extraction | Feature reduction | Classification | Evaluation |
|---|---|---|---|---|---|---|---|---|
| Liu et al. [73] | T1 | Denoising | SVM based segmentation | Patients = 18 PCNSL = 08 GBM = 10 Image size = 512 × 256 |
Gabor wavelet features | – | LDA | Accuracy = 99% Sensitivity = 100% Specificity = 98% |
| Hussain et al. [22] | T1, T2 | Skull stripping | Gradient method, region growing method | Images = 10 Normal = 5 Abnormal = 5 |
Haar wavelets, Textural features | – | FFBNN, dynamic neuro fuzzy Technique |
(Accuracy, sensitivity, specificity) > 98% |
Kumar et al. [86] attempted to group brain tumors among different classes. These classes include Glioblastoma Multiforme (GBM), Astrocytoma (AS), Meningioma (MEN), child tumor-Medulloblastoma (MED) and secondary tumor-Metastatic (MET) and normal regions (NR). The used approach is simple that employed Gradient Vector Flow (GVF) for segmentation without any preprocessing and features are extracted from GVF based ROIs. High-value features are filtered using PCA and ANN with GDBPM weight estimation method is used to label the ROIs. The authors have used only one parameter for evaluation i.e. accuracy.
Rehman and Azim [33] used ANNs for tumor classification among different classes. The images used in classification contain Anaplastic grade III, Glioblastoma grade IV, Astrocytoma grade I & II and Pilocytic. No proper information about dataset is provided in this work. They obtain segmented MRI through their custom segmentation algorithm without performing any preprocessing. Features are extracted in the form of ANNs weights and then ANNs are used for classification purpose. However, results reported in [33] are not clear.
Singhal et al. [8], presented a comparative study of MRI and PET for classification of glioma. They used manual segmentation process after acquiring images of both sequences. They proved that PET can better grade gliomas than MRI. Contrast enhancement can improve high-grade gliomas to a limited extent and even zero improvements in some cases. However, the use of C-Methionine has been found a useful tracer (contrast enhancement agent) in PET. They used tumor/normal ratio as classification parameter. We included this study to only show that other modalities can perform the same task.
Sachdeva et al. [19] have presented semiautomatic segmentation and classification method using post contrast T1 weighted MRI to find the multiclass classification of cancer tumors. They considered six classes (primary tumors-AS, childhood tumor-MED, GBM, secondary tumor-MET, MEN and normal) of brain tumors in classification. The proposed system consists of four main modules. To mark tumor regions, content based active contour model is used. This model allows the user to mark the ROIs manually that are saved as segmented ROIs (SROIs). Second module deals with feature extraction using SROI and third module performs features reduction using principal component analysis (PCA). The last module classifies the SROIs based on extracted features using artificial neural networks (ANN). A good survey of different classification algorithms used in MRI classification is elaborated in [86].
Soltaninejad et al. [60] attempted to classify four types of brain tumors Gliomas, Meningiomas, Pituitary adenoma and Nerve sheath tumors using MRI obtained through FLAIR and T2 protocols. MRI is 3D volumetric data acquired through different protocols. As preprocessing step, eddy current effect removal using realignment is used and for image segmentation, two methods have been used: manual and super-pixel based segmentation. Statistical textural features are extracted for classification. Features are extracted for individual and combined protocols. Finally, SVM is used to classify data using different combinations of extracted features. The obtained accuracy is 80%.
Texture analysis is used by Ryu et al. [25], to categorize gliomas tumor into its different grades. There is no precise definition of image texture however it is a rich source of information and can easily be perceived by humans’ visual system. Features found in texture analysis are classified into two main categories: local features and global features. Textural parameters representing tumor heterogeneity can be associated with different biological features of tumors like metabolism, stage, and prognosis. T1 and T2 weighted images are used in this study. Apparent diffusion coefficients and histogram parameters are used to grade the gliomas. Two statistical parameters: entropy, skewness and 5th percentile values are found differentiating parameters. Both skewness and entropy are found higher for high-grade tumors and lower for low-grade tumors whereas the behavior of 5th percentile is opposite. Authors have discussed a list of distinguishing features in gliomas grading.
Juan-Albarracín et al. [87] devised an automatic and unsupervised approach to segment glioblastoma in MRI. Although the work could be categorized as segmentation task, however, we include it here as it identifies the tumor and further, it tags with glioblastoma: a type of classification and secondly it shows the viability of unsupervised approaches. They use post contrast T1W, T2W and FLAIR images provided by MICCAI 2013 competition. They perform denoising using a Manjón filter, skull stripping using Brain Suite Software, bias field correction using an N4 algorithm and super resolution using the Manjon algorithm as preprocessing steps. They extract four type of spatial domain features and reduce them using PCA. For unsupervised classification, they use different algorithms i.e. k-means, fuzzy k-means, GMM and GHMRF (Lung et al. [17]). Evaluation of designed algorithm is performed using four measures with average values: Dice = 70%, PPV = 68%, sensitivity = 77% and kappa = 98%. The obtained results are not superior but comparable to supervised methods.
Discussion
Kumar et al. [88], identify few shortcoming in previous research like small amount of studies in segmenting and multiclass classification of brain tumors, application of multiclass classification on few tumor types i.e. Meningiomas, Gliomas and Metastatic tumors, use of small data sets, absence of classification study in child tumors and absence of effort to differentiate Astrocytoma and Gliobalstoma Multiforme. They use a large number of features that are further reduced using PCA that leads to computing intensive solution. The strength of work is large dataset consisting of real patient images but evaluation is reported with one parameter only. Rehman and Azim [33] used custom unpublished and unavailable segmentation method for Magnetic Resonance Imaging classification. Manual segmentation makes this solution human expert dependent and non-scalable [8]. Sachdeva et al. [19]., have used a large data set to devise their own classifier. The approach could better perform if more levels of the network are considered or deep learning based approaches are used. Work of Soltaninejad et al. [60], and Ryu et al. [25], use small datasets. ADC components of DWI can produce better results if used with T1 and T2 [25]. However, the use of manual segmentation restricts the scalability of proposed technique. Juan-Albarracín [87] proved the suitability of unsupervised approaches for multi-classification, however, more work needs to be done with such approaches especially when working with only tumor types and sub-types.
Wavelet and frequency domain features based classification
Liu et al. [73], used Gabor wavelets to differentiate between primary central nervous system lymphoma (PCNSL) and glioblastoma multiform (GBM). They used T1W images and using Gabor wavelets, texture features are extracted. Detailed differences between different types of tumors can be obtained using Gabor wavelets with varying directions and frequencies. For preprocessing phase, wavelet decomposition is performed to remove noise. To segment the tumor shape, SVM is used and in the final phase and to evaluate Gabor texture features, LDA is used.
Hussain et al. [22], published their approach to identify and extract Cerebrospinal fluid (CSF), white matter (WM), gray matter (GM), edema and tumor. Although this work does not lie in the scope of this paper however it shows multiclass classification of brain tissues. They perform preprocessing to remove non-cortical tissues in normal brain images. Segmentation process on healthy brain images and abnormal brain images are applied separately. Pathological parts of abnormal brain image are separated into edema and tumor. After applying preprocessing step, spatial features are extracted for segmentation and classification. They define new segmentation method that helps in better classification. No separate classification approach has been used instead the segmentation method plays the role of classified too. The claimed accuracy is more than 98%.
Table 3 lists the parameters extracted from work reviewed in this section.
Table 3.
Datasets and their sources
| S. No | Paper | Acquisition method | Source |
|---|---|---|---|
| 1 | Kumar et al. [88] | Custom developed Jan 2010–May 2011 | Department of Radio-diagnosis, Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh, India |
| 2 | Singhal et al. [8] | Custom developed 1998–2006 | Charles F. Kettering Memorial Hospital, Wright State University, Dayton, Ohio |
| 3 | Sachdeva et al. [19]. | Online repository | BRATS dataset, 2012, 2013 http://martinos.org/qtim/miccai2013/ |
| 4 | Soltaninejad et al. [60] | Custom developed | Didn’t mention |
| 5 | Ryu et al. [25] | Custom developed | Didn’t mention |
| 6 | Juan-Albarracín et al. [87] | Online repository | BRATS 2013 |
| 7 | Liu et al. [73] | Custom development | Didn’t mention |
| 8 | Hussain et al. [22] | Not given | Didn’t mention |
| 9 | Watanabe et al. [89] | Custom developed Aug 2006–Nov 2011 | Department of Neurosurgery, Graduate School of Biomedical and Health Sciences, Hiroshima University, Hiroshima, Japan |
| 10 | Naeini et al. [84] | Custom developed Apr 2000–Dec 2011 | David Geffen School of Medicine, University of California–Los Angeles |
| 11 | Martínez-Cortés et al. [83] | Didn’t mention | Didn’t mention |
| 12 | Cheng et al. [90] | Custom developed 2005–2010 | Nanfang Hospital and General Hospital, Tianjin Medical University |
| 13 | Chaddad [63] | Online repository | Cancer Imaging Archive (http://www.cancerimagingarchive.net/) |
| 14 | Zulpe et al. [78] | Custom developed | Didn’t mention |
| 15 | Rajini et al. [91] | Custom developed | Department of Radiology, Rajah Muthiah Medical College Hospital (RMMCH), Tamil Nadu, India |
| 16 | Javed et al. [66] | Online repository | Harvard medical brain database http://www.med.harvard.edu/AANLIB/home.html |
| 17 | Al-Shaikhli et al. [30] | Online repository Custom Developed Online repository |
1-Brain web for simulated brain database (http://brainweb.bic.mni.mcgill.ca/brainweb/) 2-Brain tumor segmentation database 3-Whole brain atlas (http://www.med.harvard.edu/aanlib/home.html) |
| 18 | Nasir et al. [92] | Not given | Didn’t mention |
| 19 | Lahmiri et al. [94] | Online repository | Harvard Medical School (http://www.med.harvard.edu/aanlib/home.html) |
| 20 | Saritha [95] | Online repository | Harvard Medical School (http://www.med.harvard.edu/aanlib/home.html) |
| 21 | Kalbkhani et al. [96] | Online repository | Harvard Medical School (http://www.med.harvard.edu/aanlib/home.html) |
| 22 | Zöllner et al. [93] | Custom developed | Heidelberg University, Mannheim, Germany |
| 23 | Schi et al. [97] | Custom developed Apr 2005–Aug 2011 | Department of Radiology, Charité—Universitätsmedizin Berlin, Campus Virchow Klinikum, Berlin, Germany |
| 24 | Bentley et al. [98] | Custom developed Jul 2007–Oct 2010 | Purdue University, University of Minnesota |
| 25 | Caulo et al. [99] | Custom developed Jan 2008–Sep 2012 | University G. d’Annunzio of Chieti-Pescara, Chieti, Italy |
| 26 | Guzmán-De-Villoria et al. [100] | Custom developed Feb 2004–Apr 2009 | Servicio de Radiodiagnóstico. Hospital General Universitario Gregorio Marañón, Madrid, Spain |
| 27 | Lin et al. [101] | Custom developed Jan 2006–Dec 2012 | National Defense Medical Center, Taipei, Taiwan, Republic of China |
| 28 | Vidyarthi et al. [102] | Custom developed Oct 2013–Apr 2014 | Didn’t mention |
| 29 | Wang et al. [103] | Custom developed May 2004–Nov 2011 | Hospital of Xi’an Jiaotong University |
| 30 | Pan et al. [104] | Online repository | BRATS 2014 https://sites.google.com/site/miccaibrats2014/ |
Discussion
A few work based on frequency domain features is found in the literature. It shows that spatial and hybrid domain features are most useful. Although frequency domain and wavelets based features are very useful in other imaging modalities like DWI, fMRI, their use in anatomical images is limited. Hussain et al. [22]. have performed binary classification but if considered in detail they perform three type of tumor classification CSF, WM, and GM. The work shows the usability of wavelet features, however, the results are weak and the dataset is very small that lead to less reliable output.
Hybrid features based classification
Watanabe et al. [89] opted diffusion-weighted MRI to grade the Meningioma. They label grade 1, grade 2 and grade 3 gliomas. The work is unique in a sense that it only grades one type of tumor and its subtypes. They manually segment the images and to improve the efficiency, using T2W and FLAIR images are used in which cystic parts are divided into two groups: hyper-intense and hypointense area. Contrast-enhanced T1W images are used to differentiate necrotic components and to differentiate hemorrhagic lesions non-enhanced T1W images. Apparent diffusion components are calculated and used for further processing. They use contextual data like edema size, shape of tumor, bone destruction, contrast enhancement status and cyst status to group the patients. ADC coefficients could be considered as spatial features. High-grade gliomas show low ADC values whereas low-grade gliomas show high ADC values and similar behavior is observed in the case of Meningioma. Hence ADC is inversely correlated with histological grade of Meningioma.
Naeini et al. [84] worked on the hypothesis that subtypes of GBM can cause varying features in Magnetic Resonance Imaging. To test the hypothesis, they collected the data of 46 patients with histologically confirmed GBM. They used two MRI sequences: contrast-enhanced T1W, T2W, and FLAIR. Mesenchymal cells are found associated with GBM and may have different signatures i.e. from subtypes. This relationship is studied under the title of Radiogenomics. No preprocessing is performed however using custom scripts three type of ROIs is marked. These ROIs are, first, contrast enhancement (hyperintensity) second, central necrosis (hypointensity) on post contrast T1 W and third T2 hyperintensity on either T2W or T2W/FLAIR images. There are three subtypes of GBM named proneural (PN), proliferative (PROLIF) and Mesenchyme (MES). Five biomarkers are used to classify the tumor among GBM subtypes. Volume ratio can be used as a biomarker to differentiate MES GBM subtypes.
Martínez-Cortés et al. [83], applied the Bayesian model for brain tumor classification using statistical, histological and clinical features. Magnetic Resonance Imaging is obtained through T1, T2, FLAIR, and contrast enhanced T1 3D are used in this study. They manually label ROIs without any preprocessing. Values of different parameters are generated manually and automatically. Complete details of different features used in classification are missing however good information about used data set is present in this work.
Cheng et al. [90] attempted to classify three types of tumors: glioma, pituitary tumor, and Meningioma. They use contrast-enhanced T1W images. Instead of using original image ROIs, they use augmented ROIs with manual segmentation. Before segmentation, intensity normalization is performed. After this, they extract three type of features i.e. GLCM, BoW and intensity histogram, various spatial and contextual features are used for classification and LDA is applied for feature reduction. Finally, they use three statistical classifiers to label the segments. These classifiers are SVM, sparse representation based classification (SRC) and KNN. Among all three feature sets, BoW is found to be more informative and discriminative, producing the best results. Authors suggest that better classification results could be achieved by applying better-preprocessing techniques i.e. noise removal and tuning the parameters of three feature models.
Chaddad [63] studied the classification of GBM from normal brain. He found Gaussian Mixture Model (GMM) based features sufficient to differentiate between GBM and normal brain tissue. The system operates in six phases. In the first phase, images are acquired using T1-weighted, T2-weighted and FLAIR sequences and preprocessing is performed on them. In preprocessing, filtering to remove noise and normalization of gray scales is performed. Automatic segmentation is performed using a multi-thresholding technique which resulted in normal and tumors areas. GMM based features are extracted and classification is performed using three classifiers i.e. Naive Bayes, Support Vector Machine and Probabilistic Neural Networks. To filter high-value features, PCA is used. It is observed that use of PCA has reduced the computational time however it also reduced the efficiency of the classifiers.
Table 4 lists the parameters extracted from work reviewed.
Table 4.
Segmentation oriented hybrid and contextual feature classification
| Study | Imaging modalities | Pre-processing | Segmentation method | Dataset | Feature set | Feature reduction | Classification | Evaluation |
|---|---|---|---|---|---|---|---|---|
| Watanabe et al. [89] | T1, T2, FLAIR | – | Manual segmentation | Patients = 77 GLM Grade I = 42 (Fibrous = 19, Meningiothelial = 10, Transitional = 6, Others = 7) GLM Grade-II = 31 (Atypical = 31) GLM Grade-III = 04 (Anaplastic = 4) |
ADC maps Contextual features |
– | Kruskal–Wallis test, logistic regression | Log likelihood = −56 p value = 0.05 |
| Naeini et al. [84]. | T1, T2, T1c, FLAIR | – | Automatic Segmentation through custom script | Patients = 46 GBM-MES = 22 GBM-PN = 17 GBM-PROLIF = 07 |
5 = Biomarkers | – | AUC | Sensitivity = 70% Specificity = 74% |
| Martínez-Cortés et al. [83] | T1, T2, FLAIR, T13D | – | Manual segmentation | Patients = 97 GLM = 85 MET = 12 |
LoG, Gabor Filters, GLCM, LBP/VAR histogram, fractal dimensions and lacunarity, clinical features | – | SVM | Average AUC = 90% |
| Chaddad [63] | T1–WI T2–WI FLAIR |
Noise removal Normalization |
Otsu multi-thresholding | Patients = 17 GBM Normal Size = 512 × 512 |
GMM = 9 Wavelet = 3 (db1, coif1) |
PCA | NB, SVM, PNN | *False Alarm = 15.68% Missed = 15.68% Accuracy = 68.62% |
| Cheng et al. [90] | T1c | Intensity normalization | Manual delineation | Patients = 233 Images = 3064 MEN = 708 GLM = 1426 PT = 930 Image size = 512 × 512 |
GLCM BoW, Intensity Histogram, SIFT descriptors, raw patch |
LDS | SVM, KNN, SRC | GLCM Accuracy = 84.75% Sensitivity = 74% Specificity = 89% BoW – Accuracy = 88.19% Sensitivity = 81% Specificity = 92% Intensity hist. – Accuracy = 82.31% Sensitivity = 65% Specificity = 85% |
Discussion
Features used by Naeini et al. [84], are pathological features. To work with such features, a detailed knowledge of histopathology and 3D images are required. Moreover, the proposed method uses multiple tests that make the approach compute intensive. The method is non-invasive and can alternate microarray analysis (an invasive method).
Martínez-Cortés et al. [83], has incorporated clinical features in the classification of brain tumors. The work shows the usability of clinical features however the work is based on manually marked ROIs. This restricts the scalability of proposed scheme. Dataset size is considerable with a lot of spatial features and a few clinical features. Description of clinical features is provided however spatial features are not described in detail. Further, no comparison of results based on spatial features and spatial + clinical features is provided in order to find out the result differences.
Chaddad [63], used three classifiers to classify tumors. Highest performance is obtained using Naïve Bayes classifier before and after feature reduction through PCA. However, the use of PCA has reduced the performance. This study shows that spatial features are more useful as compared to wavelet based features. Dataset used is small in quantity.
A large size data set is used by Cheng et al. [90]. For ROI delineation, they have used a manual approach that is time-consuming and incorporates human knowledge instead of automatic processing. Parameters used in feature extraction are tuned manually that limits the scalability of proposed method. Authors proposed few performance enhancement techniques that need to be tested for the use of discriminative visual dictionary learning, sparse coding-based feature coding and use of more complicated preprocessing steps.
Non-segmentation based multi-classification
There are approaches of multiclass classification that do not employ the explicit segmentation process. In this section, such approaches have been reviewed. Tables 5, 6 and 7 list the analysis parameters for spatial domain feature based classification, wavelets, and frequency domain feature based classification and hybrid features based classification respectively.
Table 5.
Segmentation-free spatial feature based multi-classification
| Study | Imaging | Preprocessing | Dataset | Feature set | Feature reduction | Classification | Evaluation |
|---|---|---|---|---|---|---|---|
| Zulpe et al. [78] | T2 weighted PD |
Noise suppression, contract enhancement, intensity equalization, outlier elimination | Patients = 4 Images = 80 AS = 20 MEN = 20 Sarcoma = 20 Metastatic Bronchogenic Carcinoma = 20 Image size = 256 × 256 |
GLCM = 44 Textural features = 16 |
NA | FFNN | Accuracy = 97.5% |
| Rajini et al. [91] | T1 weighted | Noise removal | Images = 110 Astrocytoma = Not Reported Glioblastoma = Not Reported Glioma = Not Reported Metastatic = Not Reported Pituitary macro = Not Reported |
GLCM based Features = 6 |
– | Decision tree | Average: Sensitivity = 96 Specificity = 100 Accuracy = 99 |
| Javed et al. [66] | T2 weighted | – | Patients = 48 Normal = 25 Glioma = 25 Sarcoma = 25 MEN = 25 Image size = 256 × 256 |
NGTDM | Fuzzy weights | SVM | Accuracy = 80% |
| Al-Shaikhli et al. [30] | T1, T2, PD | – | Patients = 200 Normal = 50 ODG = 50 MET Carcinoma = 50 GBM = 50 Image size = 256 × 256 |
GLCM = 4 Topological = 3 |
– | Sparse coding, linear SVM | Recall = 92.5% Precision = 94.87% Accuracy = 93.75% |
| Nasir et al. [92] | T2 weighted | – | Images = 104 GLM G-I = 12 (AS) GLM G-III = 12 (AAS) GLM G-IV = 12 (GBM) MEN = 12, MET Adenocarcinoma = 12 MET Bronchogenic Carcinoma = 12 Sarcoma = 12 Normal = 20 Image size = 256 × 256 |
Moments | – | Normalized cross correlation (NCC) | Accuracy = 98% Sensitivity = 97% Specificity = 99% FPR = 0.44% FNR = 3.13% PPV = 95% |
Table 6.
Segmentation-free Wavelet and frequency features based multi-classification
| Study | Imaging modalities | Preprocessing | Dataset | Feature set | Feature reduction | Classification | Evaluation |
|---|---|---|---|---|---|---|---|
| Lahmiri et al. [94] | T2-weighted | – | Images = 56 Normal = 5 Alzheimer = 9 Glioma = 13 Herps = 07 MB Carcinoma = 08 MS = 14 Image size = 256 × 256 |
DWT features LL1 (LL2, HL2, LH2, HH2), HL1, LH1, HH1 |
PCA | SVM, ROC characteristics | Sensitivity = 95% Specificity = 70% Accuracy = 92% |
| Saritha [95] | T2-weighted | – | Images = 75 Normal = 15 Stroke = 15 Infectious disease = 15 Degenerative disease = 15 Brain tumor = 15 Size = 256 × 256 |
Wavelet entropy based Spider web plots 8 level wavelet entropy, 3-features for each 8-part image |
– | PNN | Sensitivity = 100% Specificity = 100% Accuracy = 100% |
| Kalbkhani et al. [96] | T2-weighted | Image denoising using GARCH model | 80 total image 10 = Alzheimer 10 = Alzheimer P 10 = Glioma 10 = Huntington 10 = Meningioma 10 = Pick 10 = Sarcoma Size = 256 × 256 |
4 features (H, value, GarchStat, critical value) of each class GARCH = 24 NCSE features Wavelet Coeff. = 61,440 |
PCA, LDA | kNN and SVM | Average % Precision = 97.73% Sensitivity = 97.62% Specificity = 99.66% |
Table 7.
Segmentation-free hybrid features based multi-classification
| Study | Imaging | Preprocessing | Dataset | Feature set | Feature reduction | Classification | Evaluation |
|---|---|---|---|---|---|---|---|
| Zöllner et al. [93] | – | Un-supervised C-means fuzzy clustering technique | 101 glioma patients 06 = Grade I 32 = Grade II 14 = Grade III 49 = Grade IV |
Parametric features of Relative cerebral blood volume (rCBV) + patient ages = 8 + 1 | PCC, PCA, and ICA | SVM | Accuracy = 87% Sensitivity = 83% Specificity = 91% |
| Schi et al. [97] | T1W, T2W, FLAIR, T2* | Contrast enhancement | 108 patients 29 = AS (II) 5 = oligo AS(II), 12 = ODG(II) 45 = AS(III) 9 = oligo AS(III) 8 = ODG(III) |
Morphological/contextual Features | – | Logistic regression | Accuracy = 75% PPV = 83% Sensitivity = 71% |
| Bentley et al. [98] | T1, T2 | – | 31 patients (dogs) 17 = AS 14 = ODG |
Contextual/MRI features = 18 | – | Multi parametric Logistic Regression | Sensitivity = 91% Specificity = 94% Variable with different parameters |
| Caulo et al. [99] | T1, T1c, T2, MRS, DWI, DTI, PWI | Manual | 110 Patients 21 = Diffuse AS-II 4 = Oligo AS-II 8 = ODG-II 13 = Anaplastic AS-III 1 = Analplastic oligo AS-III 3 = Analplastic ODG-III 59 = GBM-IV 1 = Gliosarcoma-IV |
Histological Features, Glioma Grading Index | – | ROC Analysis, AUC | Sensitivity = 83.9% Specificity = 96.2% Accuracy = 95.5% |
| Guzmán-De-Villoria et al. [100] | T1, T2, FLAIR, T2* | 129 patients 99 = Low grade (I, II) 30 = High grade (III, IV) |
Histological features | – | Multivariate logistic regression | AUC = 94% Sensitivity = 97.8% Specificity = 77% PPV = 93.7% NPV = 91% |
|
| Lin et al. [101] | T1, T2, FLAIR, DWI | – | 120 patients 90 = MEN G1 30 = MEN GII&GIII Image size = 256 × 256 |
Radiological features | – | Logistic regression | Score > 2 Sensitivity = 80% Specificity = 45.6% Score > 4 Sensitivity = 40% Specificity = 85.6% |
| Vidyarthi et al. [102] | NG | Noise removal-median filter | Patients = 150 CNC = 30 GBM = 30 Gliomas = 30 IVMM = 30 MET = 30 Image size = 256 × 256 |
DWT (HAAR), Gabor Filter 13 = Shape 56 = texture For each tumor type (hybrid feature set: texture and shape based) |
t test 99% significance |
K-NN | Average: Accuracy = 94% Precision = 92% Sensitivity = 92% FPR = 20% FNR = 8% |
| Wang et al. [103] | T1W, T2W | – | 154 patients 77 = LGA 48 = HGA 29 = SCM |
Histological features | – | p value, Chi Square | SVZ-contact rate = 83% |
| Pan et al. [104] | T1, T2, FLAIR | – | 195 patients 170 = high grade 25 = low grade Image size = 60 × 60 |
Auto-learning features | – | BPNN, CNN | Sensitivity = 55% Specificity = 60% |
Spatial domain features based classification
GLCM textural features are spatial features and are popular in brain tumor classification. Zulpeet. al. [78], used GLCM features to categorize four type of tumors: Astrocytoma, Meningioma, Metastatic bronchogenic carcinoma, and Sarcoma. T1 and T2 sequences are used to capture brain images. As preprocessing, noise removal is applied using a Gaussian filter. 16 GLCM features are extracted and used in classification.
Rajini et al. [91], analyzed Magnetic Resonance Imaging to classify brain tumor among five groups: Astrocytoma, Glioblastoma Multiforme, Glioma, Pituitary macro and metastatic. They preprocessed the image data set to remove noise using Wiener filter. At next stage, they extracted spatial features i.e. GLCM from images and used them to classify the tumor. With small data set, a low number of features and simple classification method results obtained are quite impressive. However few details are missing in this work like the complete specification of the data set, the justification for the use of only T1W images and justification for selecting a few features that produced high results.
Javed et al. [66] used textural features and invariant moments to classify brain tumors among four classes normal, glioma, meningioma and sarcoma using MRI T2W images. They did not perform any preprocessing and segmentation task. The images provide by Harvard Medical Brain database are already in good quality and most of the repositories are synthetic. Neighborhood gray tone difference matrix is used for feature representation and fuzzy weights are assigned to rank the features. They have designed rules to assign fuzzy weights (a use of rule-based approach). It is observed that such weight assignment can better counter the overlapping features. They use SVM machine learning approach for classification purpose. They showed that change of orientation and scale can reduce the accuracy. SVM classifier generates better results even if there are features with overlapping boundaries of different classes. The obtained results are around 80%.
Al-Shaikhli et al. [30], applied modified sparse coding and dictionary learning based approach for grading of brain tumors. They classified three type of tumors with normal: Normal, Glioma, Glioblastoma, and Carcinoma. They used topological and texture features to learn dictionary instead of using direct pixel values. They build topological matrix for an input image as a set of clusters depending upon the dissimilarity among clusters. The topological relationship is based on intersection probability of clusters. K-SVD method is used for learning and updating of the dictionary. Sparse coding and linear SVM are used as classification algorithms. Average accuracy obtained is around 94%.
Nasir et al. [92], attempted to classify and grade Glioma, Meningioma, Sarcoma, Metastatic Adenocarcinoma and Metastatic Bronchogenic Carcinoma using normalized cross-correlation (NCC). NCC is non-learning/training-based classifier that uses templates and matches the query images. Using image moments features, classification is performed. Although image moments are highly unstable, however by mapping them onto polynomial domain this deficiency could be eliminated. No preprocessing step is performed and image moments are calculated directly. Second order moment is found more useful than higher order moments. They used only one imaging sequence T2 W for tumor detection and classification. The results are appealing and technique used is computationally efficient. However larger the template set, better the performance is obtained.
Discussion
Zöllner et al. [93] has used feed forward neural networks for classification. The results may be biased due to images obtained from only four patients. The Larger size of data set with more variability is expected to produce better results. Rajini et al. [91] have used the good size of the dataset for classification but the class distribution details of images are missing which leads to a question on the explanation of results. Javed et al. [66], have used medium size dataset with a good mix of variability. Use of other anatomical modalities and larger size of the dataset can produce better results. They attempted to perform a comparison of their approach with existing methods however this comparison seems irrelevant. Proposed method attempt to classify tumor among four classes whereas the comparison is between binary classification (benign and malignant) of only two existing approaches. Further, there is a difference of classification approaches and datasets of compared studies. Al-Shaikhli et al. [30] have used datasets of three different types: simulated, original and atlases. The selection process of images from these datasets is not clear however the dataset size is large. Nasir et al. [92], have used a greater number of classes brain tumors. A large size data set is used in this study. As the main deficiency, they have used images obtained through T2 sequence only. As indicated by the author, if results obtained through other sequences are combined, proposed algorithm may perform better. Further if suitable preprocessing is done on ground truth, the result may improve more.
Wavelet and frequency feature based classification
Lahmiri et al. [94] used two-dimensional discrete wavelets transform to classify Magnetic Resonance Imaging into six classes. These classes include normal, Alzheimer disease, glioma, herpes encephalitis, metastatic bronchogenic carcinoma and multiple sclerosis. They use only T2W images from Harvard medical school. No preprocessing is performed and for feature extraction 2D DWT and its spatial/frequency domain components are used. Small wavelets can be used to extract fine-grained details whereas large wavelets can be used to find coarse-grained details. Features are reduced using PCA and classification is performed through SVM classification. Detailed features are extracted from LL band. Other bands may be explored for further information extraction and efficient classification.
Saritha [95], used discrete wavelet transform to extract features from digital images in order to classify them among different classes of a brain tumor. DWT is a good tool when images from single modality are under analysis or being processed at multi-resolutions. Wavelet transform decomposes the image into a range of scales from fine to coarsest. Hence it provides the image representation at various resolutions and as a result a good tool for feature extraction from images.
Generalized autoregressive conditional heteroscedasticity (GARCH) model is used by Kalbkhani et al. [96]. to classify seven brain diseases with normal. They are Alzheimer, Alzheimer plus Visual Agnosia, Glioma, Huntington, Meningioma, Pick, Sarcoma, and Normal. This approach works in six phases. In the first phase, 2D DWT is calculated. DWT captures both frequency and location information hence it could be considered as a hybrid approach. No image preprocessing is applied however as the second phase, extracted features are normalized in order to remove the outliers. To minimize the computation, high-value features are selected using PCA and LDA. The data set is obtained from Harvard Medical School. The results obtained show the average accuracy around 98%, however, the computational complexity is high.
Discussion
Wavelet transforms are considered a better way of extracting detailed features as compared to frequency transform due to multi-level feature extraction. Lahmiri et al. [94] used 50% images for training and remaining for testing. As seen in above studies, wavelet features produce inferior results than spatial features is evident in this study. They compare their results with a conventional 2D-DWT approach that uses PCA on HL and LH features and then classifies the images. We have observed that use of PCA, although reduces features and makes classifier computationally fast, yet reduction of features deteriorates classifier performance. Lahmiri et al. [94] has removed the PCA processing which ultimately resulted in improved performance. The results could be improved if images from other modalities are included in the experiment. Kalbkhani et al. [96] have used a regressive way of system testing with few training images. Results shown in this work are promising that show the use of small sample size to obtain acceptable results. Lahmiri et al. [94], has shown the use of LL DWT band for more useful features. The usability of this band in Kalbkhani et al. [96] research may further be explored that may result in precise results. Classification approach used in Saritha [95], is computationally efficient and results produced are 100%. Retraining of system with the addition of new data in the data set is mentioned as system limitation by Saritha [95]. Use of a large dataset is a direction to test the validity of results and need to be explored.
Hybrid feature based classification
Zöllner et al. [93] explored spatial and contextual features in order to grade the gliomas tumor in patients. They used standard anatomical pre/post contrast T1 weighted and T2 weighted and dynamic susceptibility contrast (DSC) images. As preprocessing step, they generate rCVB histograms which are later used for feature extraction. They use spatial domain features and contextual features in order to obtain better classification. Three different feature reduction techniques i.e. PCA, ICA, and PCC are applied and results are calculated using SVM classifier. Achieved classification accuracy is above 90% that is quite encouraging for grading gliomas tumor among 4 WHO grades.
Schi et al. [97], have used histopathology features to identify low-grade gliomas from high-grade gliomas. Such features include nuclear atypia, cellularity, pleomorphism, mitotic activity, vascular hyperplasia, the presence of necrosis and different proliferation markers. Conventional MRI analysis can produce good results however advanced MRI techniques like MRS and PET can produce better results. In this study, they use T1, T2, T2*, FLAIR, DWI and post-contrast signal characteristics offer-tumoral areas and brain lesions for differentiating grade II and grade III gliomas. Different morphological criteria are used to analyze the images. Retrospective design, heterogeneous histology and lower spatial and temporal resolutions of images are the limitations of this study.
Bentley et al. [98] presented an interesting research work. They worked on brain tumors of pet dogs and attempt to grade the tumors. They use MRI features and contextual features for their classification task. Two main observations by other researchers are considered in this work: contrast enhancement is associated with tumor grade and cysts. Areas of necrosis are not associated with tumor however they are common in higher grade tumors. This study uses few other contextual features that include intratumoral accumulation of fluid (ITF) and tumor surface contact with oligodendroglioma which has been stated for ventricular distortion and meningioma differentiation. Further ventricular distortion is more common with oligodendrogliomas than astrocytomas. Pre-contrast T1W, post contrast T1W and T2W images are used in this study. Reported accuracy and other measures are different for different classification parameters. However, MRI parameters and contextual information need to be investigated in human brain tumor identification and grading.
Caulo et al. [99] used conventional and advanced MR imaging to grade gliomas. In conventional MR imaging, they use pre and post contrast T1W, T2W, and FLAIR. Here we will present method and result associated with conventional imaging only. They made three evaluations: semi-quantitative analysis that uses conventional and advanced MRI sequences, a qualitative analysis that uses only conventional MRI sequences and quantitative analysis that use both conventional and advanced MRI imaging. The three studies show that use of advanced sequences provides additional information useful in classification. They use different histologic and MRI parameters to find glioma grade. This is the only study in our analysis that intends to grade gliomas among four categories.
Guzmán-De-Villoria et al. [100], studied the added value of advanced MRI sequences in brain tumor detection and classification. Feasibility of advanced MRI sequences is accepted however their additional value over MRI has yet not been quantified. They studied 129 patients with low-grade (grade I and II) and high-grade (grade III and IV). Different MRI sequences used include conventional MRI (T1W and T2W), dynamic contrast enhanced PWI, DWI, and MRS. They use histological features and MR variables to classify the images. They prove that advanced sequences have little added value to conventional MRI. The obtained results using conventional MRI are comparable to advanced sequences.
Lin et al. [101], investigated the MRI features of patients with intracranial meningiomas that could help in determining the pathological grade of the tumor. Meningiomas are found in three grades: grade-I are slow growing and non-cancerous, grade-II are cancerous and non-cancerous, grade-III are cancerous and can grow in a fast way. Researchers have identified different MRI features that can help in grading the meningioma. They use conventional MRI sequences for their study i.e. contrast-enhanced T1W, T2W, and FLAIR. In addition, DWI is also used to extract advanced features. No preprocessing and explicit segmentation are performed. From available images, they extract contextual and radiographic features. Multiple logistic regression is applied to classify the images. All the parameters are fed into a formula in order to find out a score that leads to the labeling of lesions in images. This study shows that we can formulate rules that can predict to a good level of certainty. However, this approach requires large data set to ensure its validity.
A good comparative study on brain tumor classification is presented by Vidyarthi et al. [102]. However, a major part of the study discusses the work on binary classification. In another work Vidyarthi et al. [102], attempt to classify high-grade brain tumors. In this work, 5 types of high-grade tumors are selected that include Gliomas, Central Neuro Cytoma, Intra Ventricular Malignant Mass Glioblastoma Multiforme, and Metastasis. They used all five stages of machine learning: data collection, preprocessing, feature extraction, feature reduction and classification. K-NN based classification approach is used to achieve around 94% accuracy.
Lasocki et al. [27] studied the correlation between histopathology grading and Magnetic Resonance Imaging analysis base grading. Both histopathologic and MRI-based tumor grading have pros and cons. For example, grading through histopathologic methods may suffer from error due to sampling error. In this work, they attempt to compare the grading process of two methods: histopathological and MRI-based. They prove that MRI-based methods of tumor grading outperform than histopathologic methods.
Wang et al. [103] worked on the classification of Astrocytoma, Solitary Cerebral Metastasis (SCM). The hypothesis is that whether the involvement of subventricular zone (SVZ) could help in differentiating astrocytoma and SCM. In this study, they use three sequences of MRI: T1W, T2W, and FLAIR. Astrocytoma is further classified into low-grade gliomas and high-grade gliomas. The distinction of astrocytoma and SCM is challenging due to overlapping features.
Pan et al. [104], used deep learning models for brain tumor grading. Deep learning is state of the art models used in machine learning based approaches. Usually, these are unsupervised learning models [105]. Dataset used is original, however, to create balance among two classes (high labeled and low labeled), they enhance the data set using synthetic techniques to balance both classes but that takes the results away from actual distribution. It is to note that this system does not extract explicit features. However, features are extracted as part of classification implicitly and images are labeled.
Discussion
Schi et al. [97], is more detailed work that deals with a bigger group of tumors and has enough detail of data set and tumor detection criteria. Three limitations of this work are mentioned by authors. The scheme is computationally efficient and obtained results are at acceptable level, however, require an expert second opinion. Although Bentley et al. [98], have worked on dog brain tumor, however, tumor classification is our interest, therefore, it is included in this study. The study is limited to two types of tumors only. Lin et al. [101], work focuses only on meningioma grading. They compare the single parameter and multi-parameter based grading. Caulo et al. [99], have worked on large types of tumors. However, authors have mentioned two major limitations of their work. In addition, they have also used conventional and advanced imaging sequences. Guzmán-De-Villoria et al. [100], have used a bigger size data set to differentiate among high grade and low-grade tumor by using mixed (conventional and advanced perfusion based) sequences. It is interesting to know in this work that results obtained using only conventional sequences are higher than combined results of conventional and advanced sequences. Authors have discussed multiple limitations of this study and by removing those, better results may be produced. Vidyarthi et al. [102], have used large dataset and achieved better results. They use their custom developed dataset, however, details about imaging modality are missing. Wang et al. [103], attempted to classify tumor among few categories i.e. four. The suggested classification measure is good but not strong enough to produce clinical strength results. Pan et al. [104] used deep learning methods for classification however they classified tumors among three classes only: normal, high grade and low grade. Generally saying their work shows the usability of deep learning in tumor grading, however, more work is required for multi-classification. Moreover, results obtained are low.
Summary and precautions
Different brain tumor classification schemes have been presented by the research community, however, classification scheme published by WHO is considered more comprehensive and standard. It is essential to analyze brain tumor before surgical resection and very required to obtain maximum information using invasive techniques to avoid time, money and pain. A variety of imaging techniques has been invented to capture fine details of the brain. MRI is considered a most successful way of imaging. MRI is divided into two categories: conventional or anatomical and advanced MRI. For tumor grading mostly used imaging is anatomical MRI i.e. T1, T2, T2* and FLAIR. However advanced techniques can produce better results. Researchers are using anatomical and advanced techniques in combination to obtain better results. Most of the data sets (20/30) are prepared by researchers under fine-tuned imaging parameters like TR, TE, and contrast. Few of the researchers have preprocessed their datasets. The reason is the availability of high-quality imaging machines that can be tuned to produce images according to requirements. We conclude that digital preprocessing have been implemented to some extent in some way in medical imaging cameras. Digital processing of the medical image is based on number and type of features extracted from the input. A variety of parameters is extracted however spatial domain features and hybrid features are more successful. Hybrid feature produces better results but they require the production of metadata with images. Classification process either involves segmentation or not. We have observed in this review that classification results obtained without segmentation are equally comparable. However, brain tumor segmentation is still an open area of research. Following list presents further findings:
Machine learning approaches are prevalent. In this context, supervised classification has more applications as compared to unsupervised methods (Kalbkhani et al. [96]). Machine learning is mainly applied on structural images however the application of ML on other imaging sequences are yet needed to be explored.
Studies that perform multiclass classification consider only a few classes whereas there are lot more types of brain tumor. Normally researchers have worked on the classification of glioma, meningioma and metastatic tumors are classified (Sachdeva et al. [19]).
Most of the studies use supervised learning approaches in which training and test objects are already labeled. The performance of supervised learning approaches is determined by size and quality of corpus (training data). Although unsupervised approaches do not face such limitations their results are far less than supervised methods. More research in unsupervised methods can produce good algorithms resulting in better performance (Juan-Albarracín et al. [87]).
Although advanced MRI sequences and techniques may produce better results to some extent however they are time-consuming and costly. There is the requirement to find out better computational models to be used with anatomical MRI scans for efficient and cost effective solutions.
Use of contextual information like age, family history, sex, tumor location, tumor shape is used little in research. The use of such information may lead to computationally efficient and more accurate methods of tumor detection and grading [106].
Texture analysis is affected by tumor shape. More research is required to link tumor shape with medical interpretation [73].
Change is data set requires fresh training of applied methods [95].
Some of the work did not show the proper grading of tumors used for classification purposes [60].
Most of the researchers have used small datasets to check the accuracy of their classification approaches. However, it is seen that with the increase in the size of the dataset, the classification accuracy declines [102].
Almost every researcher has used a different dataset that limits the comparative analysis of different classification approaches [29].
At present medical image processing is being done using Machine Learning approaches. Other less compute intensive approaches like rule based or case based reasoning may be tested.
More advanced machine learning approaches like deep learning etc. need to be tested [97].
Different survey and review work published focus only on binary classification work [29].
MRI does not provide sufficient information for characterization of Glioma and its different grades [26, 31].
Good thing is that most of the researchers have used actual brain images instead of synthetic images. However synthetic images may provide greater variety.
Application of feature reduction and feature selection techniques can reduce the computation time however also reduce the efficiency of the algorithm applied in general [93].
Conclusion and future work
This research has attempted to provide a ready reference for people interested in brain tumor multi-classification. We searched thoroughly and tried to include every multiclass classification of brain tumor published from currently. It is observed that a lot of work on binary classification has been published however work on multiclass classification is still in infancy stage. More research in this area is required for each phase. New image modalities are being invented, better algorithms for classification are being published, and a variety of features extracted from images are analyzed and yet more need to be tested. A big number of tumors are classified by WHO, however, in research, only a few (6–8 at max) are classified. The next phase of this work is the development of industry standard computer aided diagnosis systems that are still missing.
As our own future work, we intend to extend multiclass classification on brain tumor by including more tumor classes and subclasses. Deep learning and histopathological features are candidate tools for exploration.
Appendix A
See Table 3.
Conflict of interest
Authors declare that they have no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Lacy J, Saadati H, Yu J. Complications of brain tumors and their treatment. Hematol Oncol Clin N Am. 2012;26(4):779–796. doi: 10.1016/j.hoc.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Nayak L, Lee EQ, Wen PY. Epidemiology of brain metastases. Curr Oncol Rep. 2012;14(1):48–54. doi: 10.1007/s11912-011-0203-y. [DOI] [PubMed] [Google Scholar]
- 3.Crocetti E, et al. Epidemiology of glial and non-glial brain tumours in Europe. Eur J Cancer. 2012;48(10):1532–1542. doi: 10.1016/j.ejca.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 4.de Robles P, et al. The worldwide incidence and prevalence of primary brain tumors: a systematic review and meta-analysis. Neuro-oncology. 2015;17(6):776–783. doi: 10.1093/neuonc/nou283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamimi AF, et al. Epidemiology of malignant and non-malignant primary brain tumors in Jordan. Neuroepidemiology. 2015;45(2):100–108. doi: 10.1159/000438926. [DOI] [PubMed] [Google Scholar]
- 6.Saba T, Rehman A, Altameem A, Uddin M. Annotated comparisons of proposed preprocessing techniques for script recognition. Neural Comput Appl. 2014;25(6):1337–1347. doi: 10.1007/s00521-014-1618-9. [DOI] [Google Scholar]
- 7.Situ, Breast Carcinoma. In “Cancer Facts.” (2015). www.cancer.org.
- 8.Singhal T, et al. 11C-methionine PET for grading and prognostication in gliomas: a comparison study with 18F-FDG PET and contrast enhancement on MRI. J Nucl Med. 2012;53(11):1709–1715. doi: 10.2967/jnumed.111.102533. [DOI] [PubMed] [Google Scholar]
- 9.Rehman A, Saba T. Performance analysis of character segmentation approach for cursive script recognition on benchmark database. Digit Signal Process. 2011;21(3):486–490. doi: 10.1016/j.dsp.2011.01.016. [DOI] [Google Scholar]
- 10.Rousseau A, Mokhtari K, Duyckaerts C. The 2007 WHO classification of tumors of the central nervous system—what has changed? Curr Opin Neurol. 2008;21(6):720–727. doi: 10.1097/WCO.0b013e328312c3a7. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder A, et al. Treating metastatic cancer with nanotechnology. Nat Rev Cancer. 2012;12(1):39–50. doi: 10.1038/nrc3180. [DOI] [PubMed] [Google Scholar]
- 12.Rehman A, Saba T. An intelligent model for visual scene analysis and compression. Int Arab J Inf Technol. 2013;10(13):126–136. [Google Scholar]
- 13.Younus ZS, Mohamad D, Saba T, Alkawaz MH, Rehman A, Al-Rodhaan M, Al-Dhelaan A. Content-based image retrieval using PSO and k-means clustering algorithm. Arabian J Geosci. 2015;8(8):6211–6224. doi: 10.1007/s12517-014-1584-7. [DOI] [Google Scholar]
- 14.Roy S, Bandyopadhyay SK. Detection and quantification of brain tumor from MRI of brain and it’s symmetric analysis. Int J Inf Commun Technol Res. 2012;2(6):477–83.
- 15.Bauer S, et al. A survey of MRI-based medical image analysis for brain tumor studies. Phys Med Biol. 2013;58(13):R97. doi: 10.1088/0031-9155/58/13/R97. [DOI] [PubMed] [Google Scholar]
- 16.Al-Ameen Z, Sulong G, Rehman A, Al-Dhelaan A, Saba T, Al-Rodhaan M. An innovative technique for contrast enhancement of computed tomography images using normalized gamma-corrected contrast-limited adaptive histogram equalization. EURASIP J Adv Signal Process. 2015 [Google Scholar]
- 17.Lung JWJ, Salam MSH, Rehman A, Rahim MSM, Saba T. Fuzzy phoneme classification using multi-speaker vocal tract length normalization. IETE Tech Rev. 2014;31(2):128–136. doi: 10.1080/02564602.2014.892669. [DOI] [Google Scholar]
- 18.Sridhar D, Krishna IVM. Brain tumor classification using discrete cosine transform and probabilistic neural network. In: Signal processing image processing and pattern recognition (ICSIPR), 2013 international conference on. IEEE; 2013.
- 19.Sachdeva J, et al. A dual neural network ensemble approach for multiclass brain tumor classification. Int J Numer Methods Biomed Eng. 2012;28(11):1107–1120. doi: 10.1002/cnm.2481. [DOI] [PubMed] [Google Scholar]
- 20.Artzi M, et al. Classification of tumor area using combined DCE and DSC MRI in patients with glioblastoma. J Neuro-Oncol. 2015;121(2):349–357. doi: 10.1007/s11060-014-1639-3. [DOI] [PubMed] [Google Scholar]
- 21.Patwa N, Kavuri SP, Grandhi B, Hongrak K. Study on effectiveness of location-based advertising on food service industry in Sydney. J Bus Technovation. 2016;4(3):112–124. [Google Scholar]
- 22.Hussain SJ, Savithri TS, Sree Devi PV. Segmentation of tissues in brain MRI images using dynamic neuro-fuzzy technique. Int J Soft Comput Eng (IJSCE). 2012;1(6):416–23.
- 23.Rehman A, Saba T. Neural network for document image preprocessing. Artif Intell Rev. 2014;42(2):253–273. doi: 10.1007/s10462-012-9337-z. [DOI] [Google Scholar]
- 24.Joudaki S, Mohamad D, Saba T, Rehman A, Al-Rodhaan M, Al-Dhelaan A. Vision-based sign language classification: a directional review. IETE Tech Rev. 2014;31(5):383–391. doi: 10.1080/02564602.2014.961576. [DOI] [Google Scholar]
- 25.Ryu YJ, et al. Glioma: application of whole-tumor texture analysis of diffusion-weighted imaging for the evaluation of tumor heterogeneity. PLoS ONE. 2014;9(9):e108335. doi: 10.1371/journal.pone.0108335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung C, Metser U, Ménard C. Advances in magnetic resonance imaging and positron emission tomography imaging for grading and molecular characterization of glioma. In: Seminars in radiation oncology, vol. 25(3). WB Saunders; 2015. [DOI] [PubMed]
- 27.Lasocki A, et al. MRI grading versus histology: predicting survival of World Health Organization Grade II–IV astrocytomas. Am J Neuroradiol. 2015;36(1):77–83. doi: 10.3174/ajnr.A4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sachdeva J, et al. Segmentation, feature extraction, and multiclass brain tumor classification. J Digit Imaging. 2013;26(6):1141–1150. doi: 10.1007/s10278-013-9600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah SA, Chauhan NC. Techniques for detection and analysis of tumours from brain MRI images: a review. J Biomed Eng Med Imaging. 2016;3(1):09. doi: 10.14738/jbemi.31.1696. [DOI] [Google Scholar]
- 30.Al-Shaikhli SDS, Yang MY, Rosenhahn B. Brain tumor classification using sparse coding and dictionary learning. In: IEEE international conference on image processing, 2014.
- 31.Upadhyay N, Waldman AD. Conventional MRI evaluation of gliomas. Brit J Radiol. 2011;84(Spec Iss 2):107–11. doi:10.1259/bjr/65711810 [DOI] [PMC free article] [PubMed]
- 32.Van Cauter S, et al. Integrating diffusion kurtosis imaging, dynamic susceptibility-weighted contrast-enhanced MRI, and short echo time chemical shift imaging for grading gliomas. Neuro-oncology. 2014;16(7):1010–1021. doi: 10.1093/neuonc/not304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehman Y, Azim F. Comparison of different artificial neural networks for brain tumour classification via magnetic resonance images. In: Proceedings of 14th IEEE international conference on computer modelling and simulation; 2012.
- 34.Bobek-Billewicz B, et al. Anaplastic transformation of low-grade gliomas (WHO II) on magnetic resonance imaging. Folia Neuropathol. 2014;52(2):128–140. doi: 10.5114/fn.2014.43784. [DOI] [PubMed] [Google Scholar]
- 35.Nagpal J, Vidyarthi A, Mittal N. CLOM: counting label occurrence matrix for feature extraction in MR images. In: Signal processing and communication (ICSC), 2015 international conference on. IEEE; 2015.
- 36.Arnaud A, et al. Tumor classification and prediction using robust multivariate clustering of multiparametric MRI. Int Soc Magn Resonance Med. Toronto, Canada. 2015. http://www.ismrm.org/15/. https://hal.archives-ouvertes.fr/hal-01253584.
- 37.Hoegler D. Radiotherapy for palliation of symptoms in incurable cancer. Curr Probl Cancer. 1997;21(3):129–183. doi: 10.1016/S0147-0272(97)80004-9. [DOI] [PubMed] [Google Scholar]
- 38.Ricard D, et al. Primary brain tumours in adults. The Lancet. 2012;379(9830):1984–1996. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan AM, et al. Brainstem gliomas in children. Pediatric Neurosurg. 1996;24(4):185–192. doi: 10.1159/000121036. [DOI] [PubMed] [Google Scholar]
- 40.Stewart S, et al. Revision of the 1996 working formulation for the standardization of nomenclature in the diagnosis of lung rejection. J Heart Lung Transplant. 2007;26(12):1229–1242. doi: 10.1016/j.healun.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Figarella-Branger D, et al. Prognostic factors in intracranial ependymomas in children. J Neurosurg. 2000;93(4):605–613. doi: 10.3171/jns.2000.93.4.0605. [DOI] [PubMed] [Google Scholar]
- 42.Schubert LK, et al. A comprehensive assessment by tumor site of patient setup using daily MVCT imaging from more than 3800 helical tomotherapy treatments. Int J Radiat Oncol Biol Phys. 2009;73(4):1260–1269. doi: 10.1016/j.ijrobp.2008.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barkovich AJ, et al. Brain stem gliomas: a classification system based on magnetic resonance imaging. Pediatr Neurosurg. 1990;16(2):73–83. doi: 10.1159/000120511. [DOI] [PubMed] [Google Scholar]
- 45.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO Classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. https://www.miccai2015.org/. [DOI] [PMC free article] [PubMed]
- 46.Doolittle ND. State of the science in brain tumor classification. In: Seminars in oncology nursing, vol. 20(4). WB Saunders; 2004. [PubMed]
- 47.Biersack HJ, et al. Imaging of brain tumors with L-3-[123I] iodo-alpha-methyl tyrosine and SPECT. J Nucl Med. 1989;30(1):110–112. [PubMed] [Google Scholar]
- 48.Chen W, et al. 18F-FDOPA PET imaging of brain tumors: comparison study with 18F-FDG PET and evaluation of diagnostic accuracy. J Nucl Med. 2006;47(6):904–911. [PubMed] [Google Scholar]
- 49.Jadooki S, Mohamad D, Saba T, Almazyad AS, Rehman A. Fused features mining for depth-based hand gesture recognition to classify blind human communication. Neural Comput Appl. 2016 [Google Scholar]
- 50.Saba T, Al-Zahrani S, Rehman A. Expert system for offline clinical guidelines and treatment. Life Sci J. 2012;9(4):2639–2658. [Google Scholar]
- 51.Elarbi-Boudihir M, Rehman A, Saba T. Video motion perception using optimized Gabor filter. Int J Phys Sci. 2011;6(12):2799–2806. [Google Scholar]
- 52.Puttick S, et al. PET, MRI, and simultaneous PET/MRI in the development of diagnostic and therapeutic strategies for glioma. Drug Discov Today. 2015;20(3):306–317. doi: 10.1016/j.drudis.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 53.Saba T, Rehman A, Elarbi-Boudihir M. Methods and strategies on off-line cursive touched characters segmentation: a directional review. Artif Intell Rev. 2014;42(4):1047–1066. doi: 10.1007/s10462-011-9271-5. [DOI] [Google Scholar]
- 54.Norouzi A, Rahim MSM, Altameem A, Saba T, Rada AE, Rehman A, Uddin M. Medical image segmentation methods, algorithms, and applications. IETE Tech Rev. 2014;31(3):199–213. doi: 10.1080/02564602.2014.906861. [DOI] [Google Scholar]
- 55.Rad AE, Rahim MSM, Rehman A, Altameem A, Saba T. Evaluation of current dental radiographs segmentation approaches in computer-aided applications. IETE Tech Rev. 2013;30(3):210–222. doi: 10.4103/0256-4602.113498. [DOI] [Google Scholar]
- 56.Rad AE, Rahim MSM, Rehman A, Saba T. Digital dental X-ray database for caries screening. 3D Res. 2016;7(2):1–5. doi: 10.1007/s13319-016-0096-5. [DOI] [Google Scholar]
- 57.Husham A, Hazim Alkawaz M, Saba T, Rehman A, Saleh Alghamdi J. Automated nuclei segmentation of malignant using level sets. Microsc Res Tech. 2016;79(10):993–997. doi: 10.1002/jemt.22733. [DOI] [PubMed] [Google Scholar]
- 58.Vidyarthi A, Mittal N. Comparative study for brain tumor classification on MR/CT images. In: Proceedings of the third international conference on soft computing for problem solving. Springer; 2014.
- 59.Saba T, Rehman A, Sulong G. An intelligent approach to image denoising. J Theor Appl Inf Technol. 2010;17(2):32–36. [Google Scholar]
- 60.Soltaninejad M, et al. Brain tumour grading in different MRI protocols using SVM on statistical features. 2014;1–6.
- 61.Soleimanizadeh S, Mohamad D, Saba T, Rehman A. Recognition of partially occluded objects based on the three different color spaces (RGB, YCbCr, HSV) 3D Research. 2015;6(3):1–10. doi: 10.1007/s13319-015-0052-9. [DOI] [Google Scholar]
- 62.Saba T. Pixel intensity based cumulative features for moving object tracking (MOT) in darkness. 3D Res. 2016;7(10):1–6. [Google Scholar]
- 63.Chaddad A. Automated feature extraction in brain tumor by magnetic resonance imaging using gaussian mixture models. J Biomed Imaging. 2015;2015:8. doi: 10.1155/2015/868031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rehman A, Saba T. Document skew estimation and correction: analysis of techniques, common problems and possible solutions. Appl Artif Intell. 2011;25(9):769–787. doi: 10.1080/08839514.2011.607009. [DOI] [Google Scholar]
- 65.Rehman A,, Mohammad D, Sulong G, Saba T (2009). Simple and effective techniques for core-region detection and slant correction in offline script recognition. In: Proceedings of IEEE international conference on signal and image processing applications (ICSIPA’09), p. 15–20.
- 66.Javed U, et al. MRI brain classification using texture features, fuzzy weighting and support vector machine. Prog Electromagn Res B. 2013;53:73–88. doi: 10.2528/PIERB13052805. [DOI] [Google Scholar]
- 67.Abbas N, Saba T, Mohamad D, Rehman A, Almazyad AS, Al-Ghamdi JS. Machine aided malaria parasitemia detection in Giemsa-stained thin blood smears. Neural Comput Appl. 2016 [Google Scholar]
- 68.Jamal A, Hazim Alkawaz M, Rehman A, Saba T. Retinal imaging analysis based on vessel detection. Microsc Res Technol. 2017;00:1–13. doi: 10.1002/jemt.22867. [DOI] [PubMed] [Google Scholar]
- 69.Saba T, Rehman A, Al-Dhelaan A, Al-Rodhaan M. Evaluation of current documents image denoising techniques: a comparative study. Appl Artif Intell. 2014;28(9):879–887. doi: 10.1080/08839514.2014.954344. [DOI] [Google Scholar]
- 70.Saba T, Rehman A, Sulong G. Non-linear segmentation of touched roman characters based on genetic algorithm. Int J Comput Sci Eng. 2010;2(6):2167–2172. [Google Scholar]
- 71.Kharat KD, Kulkarni PP, Nagori MB. Brain tumor classification using neural network based methods. Int J Comput Sci Inform. 2012;1(4):2231–5292.
- 72.Saba T, Rehman A, Sulong G. Cursive script segmentation with neural confidence. Int J Innov Comput Inf Control (IJICIC) 2011;7(7):1–10. [Google Scholar]
- 73.Liu Y-h, et al. Classification of MR tumor images based on Gabor wavelet analysis. J Med Biol Eng. 2012;32(1):22–28. doi: 10.5405/jmbe.813. [DOI] [Google Scholar]
- 74.Iftikhar S, Fatima K, Rehman A, Almazyad AS, Saba T. An evolution based hybrid approach for heart diseases classification and associated risk factors identification. Biomed Res. 2017;28(8):3451–3455. [Google Scholar]
- 75.Rahim MSM, Norouzi A, Rehman A, Saba T. 3D bones segmentation based on CT images visualization. Biomed Res. 2017;28(8):3641–3644. [Google Scholar]
- 76.Rahim MSM, Rehman A, Kurniawan F, Saba T. Ear biometrics for human classification based on region features mining. Biomed Res. 2017;28(10):4660–4664. [Google Scholar]
- 77.Fu B, Ren Z. Frequency and space domain features for image classification using Gaussian mixture models. In: Embedded software and systems symposia, 2008. ICESS Symposia’08. International conference on. IEEE; 2008.
- 78.Zulpe N, Pawar V. GLCM textural features for brain tumor classification. IJCSI International Journal of Computer Science Issues. 2012;9(3):354–359. [Google Scholar]
- 79.Saba T, Rehman A, Sulong G. Improved statistical features for cursive character recognition. Int J Innov Comput Inf Control (IJICIC) 2011;7(9):5211–5224. [Google Scholar]
- 80.Saba T, Rehman A. Machine learning and script recognition. Lambert Academic Publisher; 2012. p. 56–68.
- 81.Haralick RM, Shanmugam K, Dinstein H. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;6:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 82.Mughal B, Muhammad N, Sharif M, Saba T, Rehman A. Extraction of breast border and removal of pectoral muscle in wavelet, domain. Biomed Res. 2017;28(11):5041–5043. [Google Scholar]
- 83.Martínez-Cortés T, et al. A Bayesian model for brain tumor classification using clinical-based features. In: IEEE International conference on image processing; 2014.
- 84.Naeini KM et al. Identifying the mesenchymal molecular subtype of glioblastoma using quantitative volumetric analysis of anatomic magnetic resonance images. Neuro-oncology. 2013;15(5):626–34. doi:10.1093/neuonc/not008. [DOI] [PMC free article] [PubMed]
- 85.Fern BM, Rahim MSM, Saba T, Almazyad AS, Rehman A. Stratified classification of plant species based on venation state. Biomed Res. 2017;28(13):5660–5663. [Google Scholar]
- 86.Kumar RS, Karnan M. Review of MRI image classification techniques. Int J Res Stud Comput Sci Eng. 2014;1(1):21–28. [Google Scholar]
- 87.Juan-Albarracín J, et al. Automated glioblastoma segmentation based on a multiparametric structured unsupervised classification. PLoS ONE. 2015;10(5):e0125143. doi: 10.1371/journal.pone.0125143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumar V, et al. Classification of brain tumors using PCA-ANN. In: IEEE World Congress on information and communication technologies (WICT); 2011.
- 89.Watanabe Y, et al. Preoperative histological grading of meningiomas using apparent diffusion coefficient at 3T MRI. Eur J Radiol. 2013;82(4):658–663. doi: 10.1016/j.ejrad.2012.11.037. [DOI] [PubMed] [Google Scholar]
- 90.Cheng J, et al. Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS ONE. 2015;10(10):e0140381. doi: 10.1371/journal.pone.0140381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rajini NH, Narmatha T, Bhavani R. Automatic classification of MR brain tumor images using decision tree. In: Proceedings of international conference on electronics, vol. 31; 2012.
- 92.Nasir M, Khanum A, Baig A. Classification of brain tumor types in MRI scans using normalized cross-correlation in polynomial domain. In: Frontiers of information technology (FIT), 2014 12th international conference on. IEEE; 2014.
- 93.Zöllner FG, Emblem KE, Schad LR. SVM-based glioma grading: optimization by feature reduction analysis. Zeitschrift für medizinische Physik. 2012;22(3):205–214. doi: 10.1016/j.zemedi.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 94.Lahmiri S, Boukadoum M. Classification of brain MRI using the LH and HL wavelet transform sub-bands. In: Circuits and systems (ISCAS), 2011 IEEE international symposium on. IEEE; 2011.
- 95.Saritha M, Joseph KP, Mathew AT. Classification of MRI brain images using combined wavelet entropy based spider web plots and probabilistic neural network. Pattern Recognit Lett. 2013;34(16):2151–2156. doi: 10.1016/j.patrec.2013.08.017. [DOI] [Google Scholar]
- 96.Kalbkhani H, Shayesteh MG, Zali-Vargahan B. Robust algorithm for brain magnetic resonance image (MRI) classification based on GARCH variances series. Biomed Signal Process Control. 2013;8(6):909–919. doi: 10.1016/j.bspc.2013.09.001. [DOI] [Google Scholar]
- 97.Shi J, et al. Stacked deep polynomial network based representation learning for tumor classification with small ultrasound image dataset. Neurocomputing. 2016;94:87–94.
- 98.Bentley RT, et al. Canine intracranial gliomas: relationship between magnetic resonance imaging criteria and tumor type and grade. Vet J. 2013;198(2):463–471. doi: 10.1016/j.tvjl.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Caulo M, et al. Data-driven grading of brain gliomas: a multiparametric MR imaging study. Radiology. 2014;272(2):494–503. doi: 10.1148/radiol.14132040. [DOI] [PubMed] [Google Scholar]
- 100.Guzmán-De-Villoria JA, et al. Added value of advanced over conventional magnetic resonance imaging in grading gliomas and other primary brain tumors. Cancer Imaging. 2014;14(1):1–10. doi: 10.1186/s40644-014-0035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin B-J, et al. Correlation between magnetic resonance imaging grading and pathological grading in meningioma: clinical article. J Neurosurg. 2014;121(5):1201–1208. doi: 10.3171/2014.7.JNS132359. [DOI] [PubMed] [Google Scholar]
- 102.Vidyarthi A, Agarwal P, Mittal N. “Machine learning based classification of high grade malignant brain tumors using diverse feature set. In: 2nd international conference on advances in computing and information technology (ICACIT); 2014.
- 103.Wang R, et al. Differentiation between solitary cerebral metastasis and astrocytoma on the basis of subventricular zone involvement on magnetic resonance imaging. PLoS ONE. 2015;10(7):e0133480. doi: 10.1371/journal.pone.0133480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pan Y, et al.: Brain tumor grading based on neural networks and convolutional neural networks. In: 37th IEEE annual international conference of engineering in medicine and biology society (EMBC); 2015. [DOI] [PubMed]
- 105.Mesnil G, et al.: Unsupervised and transfer learning challenge: a deep learning approach. In: ICML unsupervised and transfer learning, vol. 27; 2012. p. 97–110.
- 106.Weizman L, et al. Automatic segmentation of optic pathway gliomas in MRI. In: Biomedical imaging: from nano to macro, 2010 IEEE international symposium on. IEEE; 2010.