Abstract
The polysomnogram (PSG) analysis is considered the golden standard for sleep staging under the clinical environment. The electroencephalogram (EEG) signal is the most important signal for classification of sleep stages. However, in-vivo signal recording and analysis of EEG signal presents us with a few technical challenges. Electrocardiogram signals on the other hand, are easier to record, and can provide an attractive alternative for home sleep monitoring. In this paper we describe a method based on deep neural network (DNN), which can be used for the classification of the sleep stages into Wake (W), rapid-eye-movement (REM) and non-rapid-eye-movement (NREM) sleep stage. We apply the sleep stage stacked autoencoder to constitute a 4-layer DNN model. In order to test the accuracy of our method, eighteen PSGs from the MIT-BIH Polysomnographic Database were used. A total of 11 features were extracted from each electrocardiogram recording The experimental design employs cross-validation across subjects, ensuring the independence of the training and the test data. We obtained an accuracy of 77% and a Cohen’s kappa coefficient of about 0.56 for the classification of Wake, REM and NREM.
Keywords: Sleep stage, Electrocardiogram (ECG), Deep neural network (DNN), Stacked autoencoder (SAE)
Introduction
Sleep is important for the recovery, integration and memory consolidation of the body. Sleep can roughly be divided into three main stages: (1) wakefulness (W), (2) rapid-eye-movement (REM) stage and (3) non-rapid-eye-movement (NREM) (N1-N2-N3-N4). These stages alternate in cycles of about 90 min. However, the rapid pace of life in modern society and harmful habits can lead to sleep disorders which can often induce or aggravate cardiovascular and cerebrovascular diseases. Therefore, monitoring and assessment of the quality of sleep is an important task for the overall well-being of a person [1–3].
The conventional method of automatic sleep staging essentially involves spectral analysis of EEG signal collected using polysomnography (PSG). Since EEG devices are bulky, collecting and recording EEG signals during sleep can be cumbersome. Therefore, many researchers have studied alternative signals such as heart rate, respiratory rate, and electrocardiogram (ECG) for sleep analysis. In 2006, Redmond and Heneghan [4] presented a successful machine learning approach using cardio-respiratory monitoring and analysis for sleep stages classification. In 2010, Mendez et al. [5] proposed a method for sleep staging with time-varying spectral features and Hidden Markov Models from heart rate variability. In 2013, Ebrahimi et al. [6] used fourteen parameters of heart rate variability including time domain features, entry and normalized energy from empirical mode decomposition (EMD) to distinguish between REM and NREM. Several studies [7–10] have reported the effectiveness of various signals and methods for the analysis of sleep stages. Most of the researchers have used a combination of features extracted from one or more of respiratory, ECG and actigraphy signals [11]. While the use of multiple signal streams has shown to be effective, recording them requires the user to wear multiple sensor-based wearable devices. For the analysis of these signals and for training machine learning models to perform sleep staging, some studies have shown that neural networks [12, 13] can be effective learning algorithms. However, most of the papers use shallow neural networks due to their ease of fitting. However, one drawback of shallow neural networks is that their power to represent complex functions is limited. To address these problem, in this paper, we propose a new sleep staging method based on the ECG signal as the only source signal stream, and use deep neural networks (DNN) to develop an algorithm to produce high quality sleep stage identification. We show that our technique is stable and is performs well for the sleep stage classification problem. We train a 4-layer DNN based on stacked autoencoders (SAE) to establish the nonlinear mapping between the ECG signal and the sleep stage.
The rest of the paper is structured as follows. In section two, we describe the database used and the steps used for processing the RR interval time series. We also describe the feature extraction from the RR interval series and evaluate the performance of our method. In section three, the SAE is combined with a softmax classifier to generate the entire DNN model of sleep staging. At this stage the 11-dimensional RR interval feature vector is used as the input and a 3-dimensional sleep stage vector is generated as the output. In section four, we describe the result of the classifier designed in section three. In section five we discuss the implication of our results and provide our conclusions.
Materials and methods
Data collation
The proposed approach presented in this paper was tested on the MIT/BIH Polysomnographic Database (MITBPD). This database is freely available on the PhysioNet web page [14].There are 18 recordings in total. The subjects were aged between 32 and 56 years (mean age 43 years); and their weights varied between 89 and 152 kg (mean weight 119 kg). Records slp01a and slp01b are segments of the same subject and slp02a and slp02b are segments of another subject. All the remaining 14 records are from different subjects. An ECG signal, an invasive blood pressure signal, an EEG signal and a respiration signal were included in all recordings. Other signals such as respiratory effort signal, an electro-oculogram (EOG) signal, a stroke volume signal and an earlobe oximeter signal were included in some records. All physiological signals were sampled at a rate of 250 Hz.
Preprocessing
The RR intervals were derived directly from the QRS annotations. We observed some instances of missing ECG signal. Therefore we removed the QRS annotations from the record slp03 in the range 176–163 and part of the record of slp60 in the range 540–549. If the RR interval time series contains abnormal points, they will adversely affect the results. Therefore, to remove signal noise, the raw RR data is processed in the following manner:
Step 1 Remove the segments marked with apnea and body movement time.
Step 2 Use the lower-upper-threshold method to remove the data <0.7 times the average RR interval and more than 1.3 times the average RR interval.
Step 3 Use the 3 σ criterion to remove the gross error.
Step 4 Interpolate the RR interval using a cubic spline and resample at 5 Hz to rebuild the RR interval.
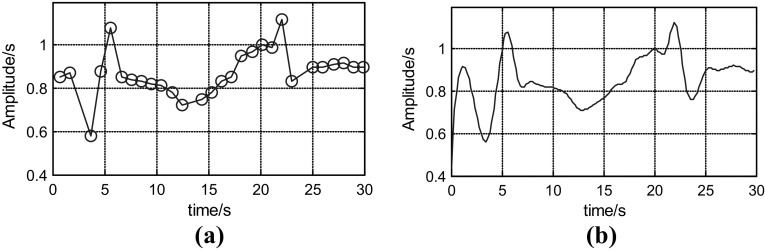
The RR interval after removal of outliers and RR interval after resampling are showed in Fig. 1.
Fig. 1.
a The RR interval after removal of outliers. b The RR interval after resampling
Feature extraction
Feature extraction is a critical step and a prerequisite to perform classification. Previous studies have shown [6, 15] that time-domain features, spectral features and non-linear features are important for the study of sleep staging. Therefore, in our study we extract different features from each of these categories.
Time-domain features
Time domain features analyze the variation of RR intervals using statistical methods. They are the simplest features to extract and are the most intuitive measures used to characterize HRV. We extract three time domain features: the Mean, the SD (standard deviation), and the RMSSD (the root mean square of successive differences between adjacent normal cycles) of the RR intervals. The values of Mean, SD and RMSSD were calculated as follows:
| 1 |
| 2 |
| 3 |
Spectral features
We used the fast Fourier transform (FFT) algorithm to estimate the power spectral density of the processed RR intervals. In research literature, the frequency band of the RR interval series is usually divided into three parts: very low frequency (VLF) band which ranges from 0 to 0.04 Hz; low frequency (LF) band which ranges from 0.04 to 0.15 Hz; and the high frequency (HF) band which ranges from 0.15 to 0.4 Hz. The LF, HF, TF(the sum of LF and HF), the LF-HF ratio represented as LF/HF, and the and were selected as the spectral features. The values of and were calculated as follows:
| 4 |
| 5 |
Non-linear features
Although the information from the time and frequency domains can be used to represent the HRV intuitively, they cannot fully reflect the changes in the HRV. In recent years, non-linear features have been introduced for the analysis of HRV. These non-linear features provide a new method for the study of sleep staging.
Detrended fluctuation analysis (DFA) can detect long range power-law correlations and identify intrinsic fluctuations in the investigated nonlinear data by removing polynomial noise [16].
The root mean square (RMS) of the signal fluctuation from this trend line is calculated as:
| 6 |
where N stands for the total length of the RR interval after resampling, represents the cumulative profile of the signal obtained by summing up the distances of the RR interval from the mean and represents these regression lines. In this study, DFA was used to extract a single feature: derived from every 0.5-min segment. This feature represents the impact of the autonomic nervous system (ANS) activity on the HRV signals. This method was used to remove the external and random fluctuations from the HRV signals.
Sample entropy is usually used to measure the regularity and the complexity of physiological and clinical time series [17]. Sample entropy quantifies the regularity of a time series by matching a pattern of length m with any other pattern of the same length with a tolerance of r; this comparison is repeated by extending the range of comparison to length m + 1. The value of the sample entropy is calculated as:
| 7 |
where m is the dimension of the embedding and
| 8 |
| 9 |
| 10 |
In this study, the values of m = 5 and r = 0.2 × SD were used.
Performance evaluation
To test the reliability and accuracy of our designed classifiers, Cohen’s kappa statistics [18, 19] was used. Accuracy is a basic index to evaluate a classifier. k is regarded as a more effective evaluation due to the fact that it takes into account the prior probability of the specific class. Cohen’s Kappa is calculated as
| 11 |
where is the relative observed agreement among raters, and is the proportion of agreement expected by chance. For every-stage, and stand for the corresponding proportions for that specific stage. If ≤ 0, then the observed agreement is even worse than that expected by chance. = 1 means that all the samples were classified into their expected classes. A higher value of k indicates a stronger agreement between the results of our designed classifier and the expected results.
The deep neural network model of sleep staging
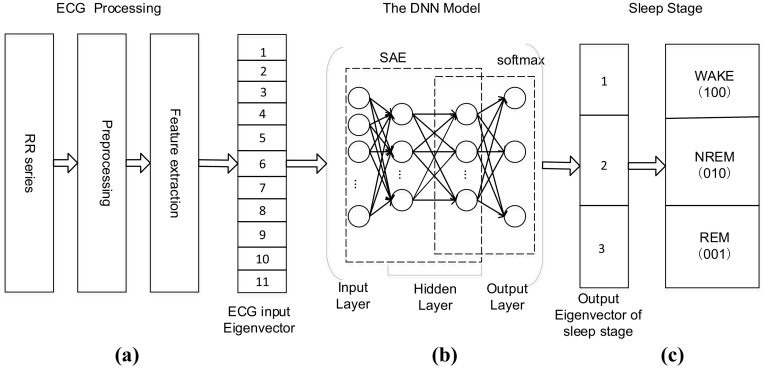
In this section, the entire DNN model consisting of the SAE combined with a softmax classifier attached to the top is described. As illustrated in Fig. 2, our DNN-based sleep stage classifier contains two steps: (1) Use the training set to train the DNN model; (2) Use the testing data through the trained DNN model to obtain the test result for determining the sleep stage.
Fig. 2.
The working principle of the DNN-based sleep staging classifier. a The ECG signal is processed to obtain a data set. b The DNN model is based on SAE. c The test result is used to estimate the sleep stage
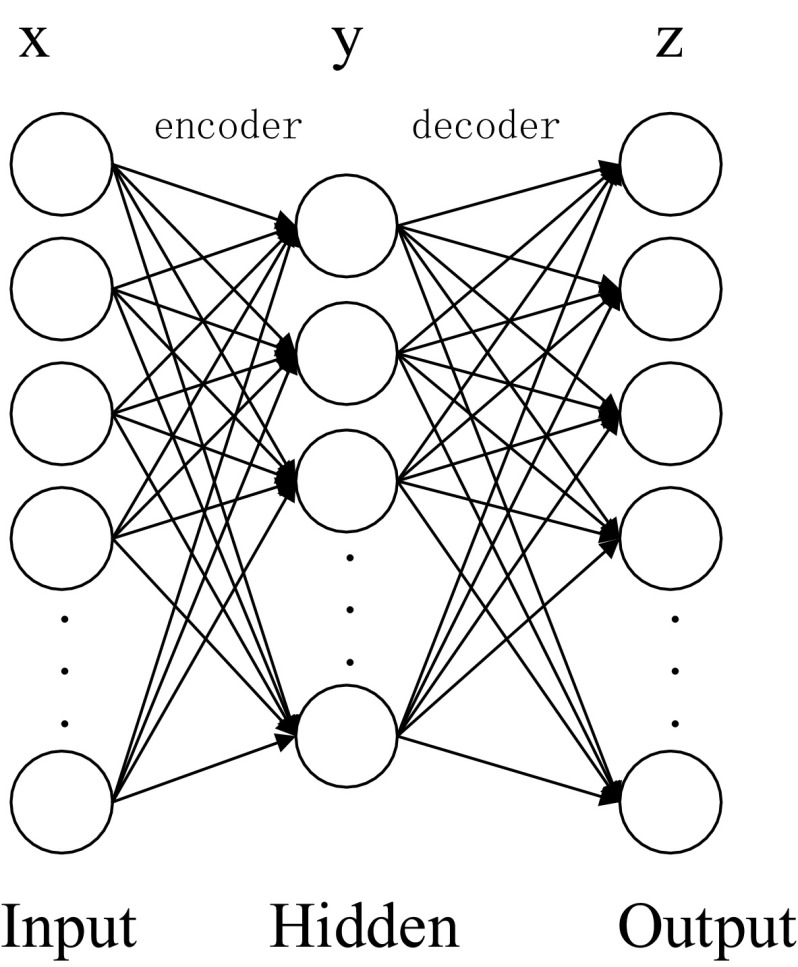
Autoencoder
The stacked autoencoder, referred to as the SAE in this paper, consists of a series of autoencoders which are stacked on top of each other. A typical autoencoder is a neural network consisting of three fully connected layers. As shown in Fig. 3, these layers are: (a) the input layer, (b) the hidden layer, and (c) the output layer. Let be a set of input training samples, where ( is the number of training samples). The input value can be represented by ,and the components of the weight matrix can be expressed as where and , where is the number of neurons in the input layer and is the number of neurons in the hidden layer. The autoencoder transforms the input vector to the hidden vector via the encoder . In this study, we use the logistic sigmoid function which can be written as follows:
| 12 |
The output of the neurons of the hidden layer, called encoding, is obtained by the following formula:
| 13 |
where is the bias of the hidden layer neuron . The autoencoder attempts to reconstruct the input vector via the decoder to produce the reconstructed vector . The values of the output layer, also called the decoding, is given by
| 14 |
here is the bias of the neuron of the output layer. The parameters of this neural network are optimized to minimize the average reconstruction error as follows:
| 15 |
here is a loss function. The Gradient Descent method is used to update the weight matrices and the bias vectors according to formulas (16), (17) and (18).
| 16 |
| 17 |
| 18 |
where represents the learning rate.
Fig. 3.
A basic schema of an autoencoder
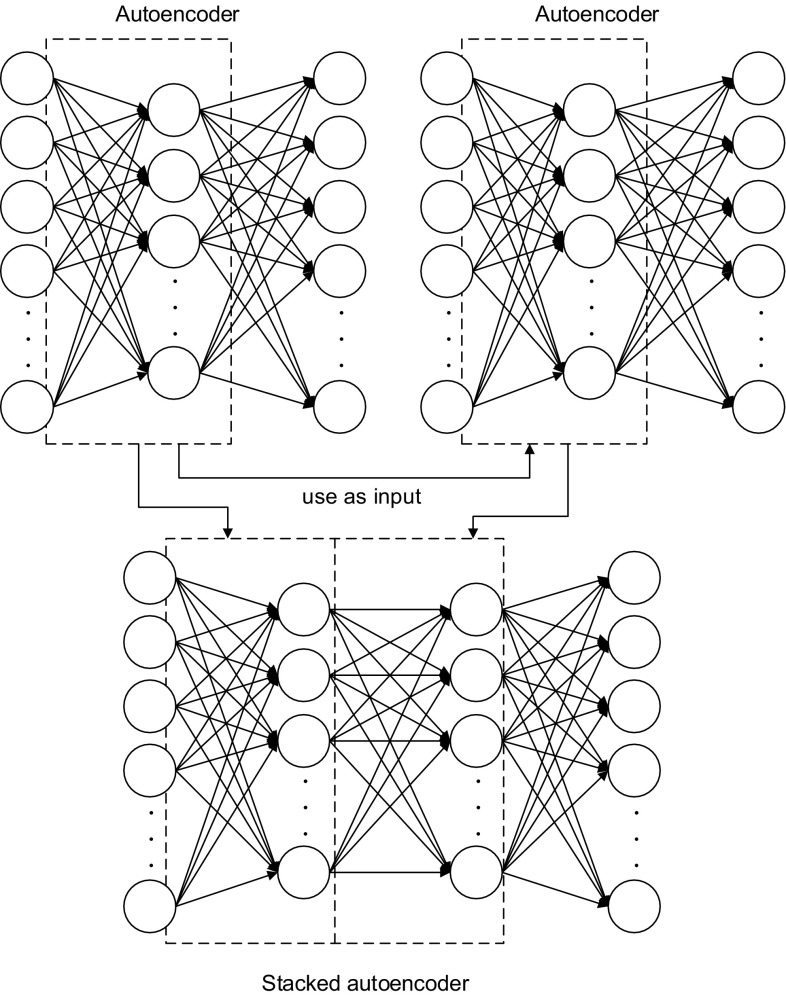
SAE
The DNN model based on SAE used in our study is constructed using several autoencoder layers. The working principle of the SAE is illustrated in Fig. 4. A SAE is a network that combines the hidden layers of the subsequent autoencoders. That is, the output layer of the previous autoencoder is discarded after the training phase, and the output of the hidden layer is used as the input of the next autoencoder. Here, we can say that each hidden layer is a higher-level abstraction of the previous layer [20] with the last hidden layer containing the high-level structures and representative information of the sleep stage. Therefore, the information from the output layer is an effective for estimation of the sleep stage. To employ the DNN model based on SAE for estimating the sleep stage, the real sleep stage must be added to the top layer.
Fig. 4.
The structure of the SAE used in this study. It contains an input layer, two hidden layers, and an output layer
Training the DNN model of sleep staging
In this section, we describe the standard technique of training stacked autoencoders. The training procedure of our sleep staging algorithm contains the SAE unsupervised pre-training and supervised fine-tuning steps. The input samples for training the model are a set of features, denoted by , (where m is the number of training samples), is a feature vector containing 11-dimensional features extracted from the RR interval series of the subjects. The output samples for training the model are a set of sleep staging vectors, expressed as , where is a 3-dimensional sleep staging marker matrix vector. The same process was performed for the test data to obtain the relationship between the ECG signal and the sleep stage. The number of hidden layers is preset to , and the number of nodes is preset in each hidden layer. Then we initialize the network parameters including the training epochs, the learning rate, the batch-size and the number of iterations. In the pre-training stage, we randomly initialize the weight matrices and the bias vectors. Then is used as the input to train the first hidden layer, and the successive hidden layers are trained in a greedy layer-wise manner which uses the output of the previous hidden layer as the input to the next hidden layer. In the fine-tuning stage, we use the output of the last hidden layer as the input to the softmax layer and randomly initialize the values . The backpropagation (BP) algorithm with the gradient-based optimization technique is used to update the parameters of the entire network in a top-down manner.
Testing out DNN based sleep stage model
To evaluate the generalizability of our method, we obtained our results using 10-fold cross-validation. After processing, we obtained 9040 samples, and the number of WAKE, NREM and REM segments were 2748, 5650 and 642 respectively. We divided all the sleep staging samples into 10 random copies. The quality of every copy is the same. In each experiment, nine of them were selected as training samples and one was used as the test sample. In all experiments, each test sample was used only once. With this experimental design, we were able to assess the overall performance of our method.
The test set includes the untrained samples of the ECG eigenvector, and can be expressed as (where is the number of test samples), is a feature vector containing the 11-dimensional features extracted from RR interval series of the subjects and is the input for testing the trained DNN model. This result can be expressed as . As shown in Fig. 2b, c, is used to estimate the sleep stage.
Results
In this paper, we used our proposed framework to estimate the WAKE-NREM-REM sleep staging cycle.
The features used for training our algorithm consisted of time domain features, spectral features and non-linear features. The DNN model was trained with 4 layers, and the number of nodes in each layer was 11, 30, 20, and 3 respectively. In the pre-training stage, the learning rate was set to 1 for each layer, the training epoch was 20 and the batch-size was set to 100. In the fine-tuning stage, the learning rate was set to 2 for each layer, the batch-size was set to 80 and the number of iterations was set to 100. The test sets were used to obtain the estimations of the sleep staging through the trained network. 10-fold cross-validation was used to evaluate the generalizability of our method.
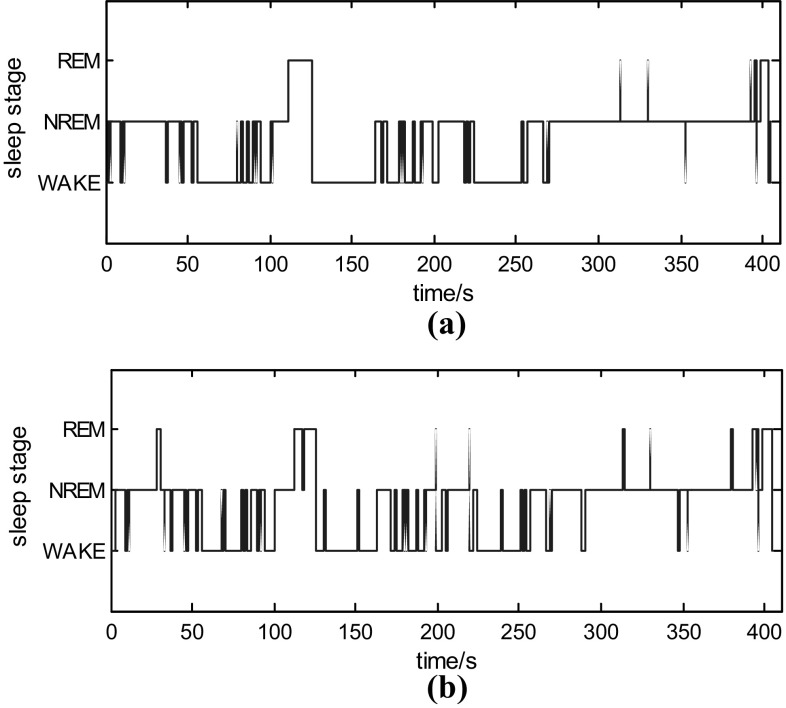
One of the test results of the original manually scored hypnogram and the estimated sleep stage is shown in Fig. 5. The confusion matrices and the performance of our method are presented at Tables 1 and 2. In Table 1, the numbers refer to the number of corresponding epochs. From Table 2 we can see that the NREM discrimination rate is the highest, and the REM discrimination rate is the lowest (Table 3). In table 3, we can see that the performance of R-N classification in related works for reference and our work.
Fig. 5.
Comparison of the original manually scored hypnogram and the estimated hypnogram in one experiment. a Original manually scored hypnogram. b Estimated hypnogram
Table 1.
Confusion matrix from the 10-fold cross-validation
| WAKE | NREM | REM | |
|---|---|---|---|
| WAKE | 1958 | 781 | 55 |
| NREM | 600 | 4527 | 173 |
| REM | 172 | 352 | 422 |
Table 2.
Classification performance
| WAKE | NREM | REM | Overall | |
|---|---|---|---|---|
| Accuracy | 0.71 | 0.80 | 0.65 | 0.77 |
Table 3.
Performance of R-N classification in related works for reference
| References | Modalities | No of features | N | Average k | Average accuracy |
|---|---|---|---|---|---|
| Redmond [4] | RIP, ECG | 27 | 22 | 0.42 | 0.72 |
| Kurihara [21] | HR, movement in bed | – | 45 | 0.46 | 0.73 |
| Our work | ECG | 11 | 16 | 0.56 | 0.77 |
| Fonseca [7] | RIP, ECG | 70 | 48 | 0.56 | 0.80 |
| Willemen [10] | RRI, RIP movement | 375 | 85 | 0.62 | 0.81 |
N number of subjects for studied, RIP respiratory inductance plethysmography, HR heart rate
Because the accuracy depends both on the characteristics of the datasets and the specific type of classifier employed, it is difficult to compare the results. However, the accuracy and the Cohen’s kappa coefficient of the Wake, NREM and REM classification obtained in this study are 77% and 0.56, which is an improvement over the results reported in Kurihara et al. and Redmond [4, 21]. Although, Willemen et al. reported a higher classification accuracy of 81% and higher kappa of 0.62 for W-N-R, they had used a set of 375 features, collected not only from ECG but also from the respiratory signals [10].
Conclusion
In this study, we used 11 features derived from the ECG signal for the classification of Wake-REM-NREM sleep staging. Considering that the inter-annotator agreement among experts annotating sleep staging had a Cohen’s kappa coefficient of about 0.8, the kappa of 0.56 achieved with this effort is acceptable. The experimental results show that the values of the RR interval can be used to estimate the different sleep stages accurately, and can be used as a supplement to traditional sleep staging methods. Compared with the traditional PSG sleep monitoring technique, this method greatly reduces the physiological parameters required for testing the sleep monitoring process. Thus, by reducing the number of devices a user must wear for sleep tracking, it also greatly reduces the adverse factors that interfere with sleep. This helps to better simulate realistic scenarios of natural sleep. But, in order to achieve a higher accuracy of the sleep staging, the algorithm for the processing of the RR interval and the parameter adjustment of the DNN model need to be further improved. We believe that a continuous improvement of this method, can enable continuous and unobtrusive sleep monitoring at home. This can open up great opportunities for research and take sleep research out of the sleep labs to provide important parameters for the natural screening of conditions such as obstructive sleep apnea syndrome. We also believe that this research has great prospects in the development of monitoring for sleep and respiratory diseases.
Acknowledgements
This work was supported by the National Natural Science Foundation for Young Scholars of China (Grant No. 61403276), Tianjin Research Program of Application Foundation and Advanced Technology (14JCYBJC42400).
Conflict of interest
The authors (Ran Wei, Xinghua Zhang, Jinhai Wang, Xin Dang) declare that they have no conflict of interests in relation to the work in this article.
Human and animal rights
Approval was obtained from the CSULB Institutional Review Board for experiments involving human subjects.
References
- 1.Carley DW, Farabi SS. Physiology of sleep. Diabetes Spectr. 2016;29(1):5–9. doi: 10.2337/diaspect.29.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolpert EA. A manual of standardized terminology. Techniques and scoring system for sleep stages of human subjects. Arch Gen Psychiatry. 1969;20(2):246–247. doi: 10.1001/archpsyc.1969.01740140118016. [DOI] [Google Scholar]
- 3.Nascimento AP, Passos VMM, Pedrosa RP, Brasileiro-Santos MDS, Barros IMLD, Costa LOBF, Lima AMJD. Sleep quality and stress tolerance in patients with obstructive sleep apnea. Rev Bras Med Esp. 2014;20(2):115–118. doi: 10.1590/1517-86922014200201357. [DOI] [Google Scholar]
- 4.Redmond SJ, Heneghan C. Cardiorespiratory-based sleep staging in subjects with obstructive sleep apnea. IEEE Trans Biomed Eng. 2006;53(3):485–496. doi: 10.1109/TBME.2005.869773. [DOI] [PubMed] [Google Scholar]
- 5.Mendez MO, Matteucci M, Castronovo V, Ferini-Strambi L. Sleep staging from heart rate variability: time-varying spectral features and hidden Markov models. Biomed Eng Technol. 2010;3(3–4):246–263. doi: 10.1504/IJBET.2010.032695. [DOI] [Google Scholar]
- 6.Ebrahimi F, Setarehdan SK, Ayala-Moyeda K, Nazeran H. Automatic sleep staging using empirical mode decomposition, discrete wavelet transform, timedomain, and nonlinear dynamics features of heart rate variability signals. Comput Methods Programs Biomed. 2013;112(1):47–57. doi: 10.1016/j.cmpb.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Fonseca P, Long X, Radha M, Haakma R. Sleep stage classification with ECG and respiratory effort. Physiol Meas. 2015;36(10):2027–2040. doi: 10.1088/0967-3334/36/10/2027. [DOI] [PubMed] [Google Scholar]
- 8.Domingues A, Paiva T, Sanches JM. Hypnogram and sleep parameter computation from activity and cardiovascular data. IEEE Trans Biomed Eng. 2014;61(6):1711–1719. doi: 10.1109/TBME.2014.2301462. [DOI] [PubMed] [Google Scholar]
- 9.Kurihara Y, Watanabe K. Sleep-stage decision algorithm by using heartbeat and body-movement signals. IEEE Trans Syst Man Cybern Part A Syst Hum. 2012;42(6):1450–1459. doi: 10.1109/TSMCA.2012.2192264. [DOI] [Google Scholar]
- 10.Willemen T, Van Deun D, Verhaert V, Vandekerckhove M, Exadaktylos V, Verbraecken J, Van Huffel S, Haex B, Vander Sloten J. An evaluation of cardiorespiratory and movement features with respect to sleep-stage classification. IEEE J Biomed Health Infor m. 2014;18(2):661–669. doi: 10.1109/JBHI.2013.2276083. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto K, Kuriyama H, Tajima S. Actigraphic detection of REM sleep based on respiratory rate estimation. J Med Bioeng. 2013;2(1):20–25. [Google Scholar]
- 12.Ebrahimi F, Mikaeili M, Estrada E, Nazeran H. Automatic sleep stage classification based on EEG signals by using neural networks and wavelet packet coefficients. Conf Proc IEEE Eng Med Biol Soc 2008;2008:1151–4. [DOI] [PubMed]
- 13.Park H, Park KS, Jeong DU. Hybrid neural-network and rule-based expert system for automatic sleep stage scoring. Eng Med Biol Soc. 2000;2:1316–1319. [Google Scholar]
- 14.Goldberger AL. Components of a new research resource for complex physiologic signals, physiobank, physiotoolkit, and physionet. Am Heart Assoc J Circ. 2000;101(23):1–9. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 15.Singh J, Sharma RK, Gupta AK. A method of REM-NREM sleep distinction using ECG signal for unobtrusive personal monitoring. Comput Biol Med. 2016;78:138–143. doi: 10.1016/j.compbiomed.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Penzel T, Kantelhardt JW, Grote L, Peter JH, Bunde A. Comparison of detrended fluctuation analysis and spectral analysis for heart rate variability in sleep and sleep apnea. IEEE Trans Biomed Eng. 2003;50(10):1143–1151. doi: 10.1109/TBME.2003.817636. [DOI] [PubMed] [Google Scholar]
- 17.Richman JS, Moorman JR. Physiological time-series analysis using approximate entropy and sample entropy. Am J Physiol Heart Circ Physiol. 2000;278(6):H2039–H2049. doi: 10.1152/ajpheart.2000.278.6.H2039. [DOI] [PubMed] [Google Scholar]
- 18.Rosenfield GH, Fitzpatrick-Lins K. A coefficient of agreement as a measure of the matic classification accuracy. Photogramm Eng Remote Sens. 1986;52(2):223–227. [Google Scholar]
- 19.Xu S, Lorber MF. Interrater agreement statistics with skewed data: evaluation of alternatives to Cohen’s kappa. J Consult Clin Psychol. 2014;82(6):1219. doi: 10.1037/a0037489. [DOI] [PubMed] [Google Scholar]
- 20.Li Xiang, Peng Ling, Yuan Hu, Shao Jing, Chi Tianhe. Deep learning architecture for air quality predictions. Environ Sci Pollut Res. 2016;23(22):22408–22417. doi: 10.1007/s11356-016-7812-9. [DOI] [PubMed] [Google Scholar]
- 21.Kurihara Y, Watanabe K. Sleep-stage decision algorithm by using heart-beat and body-movement signals. IEEE Trans Syst Man Cyber. 2012;42(6):1450–1459. doi: 10.1109/TSMCA.2012.2192264. [DOI] [Google Scholar]