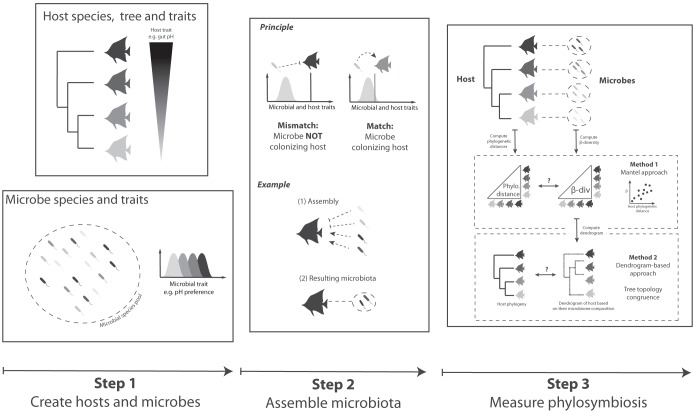
FIG 1.
Overview of the simulation workflow. Each simulation was run in three steps. First, host and microbial species and traits were independently simulated. Second, microbiotas were assembled for each host independently by following a set of ecological rules. Assembly was either neutral, a pure ecological filtering process (as depicted in the figure) based on the match between host trait (e.g., gut pH) and microbial trait (e.g., pH preference), or a mix of ecological filtering and competition. Third, phylosymbiosis was measured using two alternative methods: one using the Mantel test (Mantel approach), the other using a topological distance metric between the host species tree and the clustering tree of microbiota compositional dissimilarities (dendrogram-based approach).