Abstract
Aquaculture is the fastest growing industry worldwide. Aquatic diseases have had enormous economic and environmental impacts in the recent past and the emergence of new aquatic pathogens, particularly viruses, poses a continuous threat. Nevertheless, little is known about the diversity, abundance and evolution of fish viruses. We used a meta-transcriptomic approach to help determine the virome of seemingly healthy fish sold at a market in Sydney, Australia. Specifically, by identifying and quantifying virus transcripts we aimed to determine (i) the abundance of viruses in market fish, (ii) test a key component of epidemiological theory that large and dense host populations harbour a greater number of viruses compared to their more solitary counterparts and (iii) reveal the relative roles of virus–host co-divergence and cross-species transmission in the evolution of fish viruses. The species studied comprised both shoaling fish—eastern sea garfish (Hyporhamphus australis) and Australasian snapper (Chrysophrys auratus)—and more solitary fish—eastern red scorpionfish (Scorpaena jacksoniensis) and largetooth flounder (Pseudorhombus arsius). Our analysis identified twelve potentially novel viruses, eight of which were likely vertebrate-associated across four viral families and that exhibited frequent cross-species transmission. Notably, the most solitary of the fish species studied, the largetooth flounder, harboured the least number of viruses while eastern sea garfish, a densely shoaling fish, had the highest number of viruses. These results support the emerging view that fish harbour a large and largely uncharacterised virome.
Keywords: fish, virome, meta-transcriptomics, virus evolution, phylogenetics
1. Introduction
Fish are an important source of food, nutrition and income for millions of people globally (Pulvenis 2016). It is estimated that fish provide approximately 16 per cent of the animal protein consumed by the world’s population (Tidwell and Allan 2001) and aquaculture production is the fastest growing industry worldwide (Broitman et al. 2017). Aquatic diseases are a major factor limiting aquaculture production. Indeed, the growing global demand for seafood and the subsequent expansion of aquaculture provide opportunities for the transmission of novel viruses. Globally, both farmed and wild aquatic animal populations are affected by a number of emerging diseases and the substantial economic losses from pathogens, which are often viral in origin, remain a significant threat (Zhang and Gui 2015). Nevertheless, until recently (Shi et al. 2018), detailed knowledge of fish viruses and their evolution, including how frequently they are able to jump species boundaries, was scarce. There is therefore an urgent need to dive deeper into this unexplored aquatic virosphere, which may be of considerable practical importance to aquaculture and provide important general insights into virus ecology and evolution.
Much of our current understanding of the viruses that infect fish is based on the study of pathogenic viruses in symptomatic hosts (Crane and Hyatt 2011). A meta-transcriptomics (i.e. bulk RNA-sequencing) approach offers a powerful alternative, potentially revealing the entire virus composition (both DNA and RNA viruses) associated with an individual animal—that is, its virome—and that can be performed in the absence of overt disease (Li et al. 2015; Shi et al. 2016, 2018; Geoghegan et al. 2018). In addition, this approach can provide important insights into key aspects of virus evolution, including the factors that may mediate differences in virus composition between species, and the frequency with which viruses jump species boundaries over evolutionary time, which in turn may help reveal key determinants of the process of disease emergence (Geoghegan et al. 2016, 2017). More broadly, understanding the ecological, evolutionary, anthropogenic and immunological factors that influence virus composition may assist surveillance efforts for emerging viruses before they become established in diverse host populations and in a variety of environments (Geoghegan and Holmes 2017; Delwart 2007; Lopes et al. 2014; Avarre 2017). Indeed, recent metagenomic studies have greatly accelerated the pace of virus discovery, transforming our understanding of virus diversity and providing information on their likely evolutionary origins (Delwart 2007; Li et al. 2015; Shi et al. 2016, 2018; Simmonds et al. 2017; Zhang et al. 2018).
Recent work suggests that fish harbour a greater number of viruses than any other class of vertebrate, and it is striking that most families of RNA viruses previously thought to only infect mammals have recently been described in bony fish (Shi et al. 2018). This in turn suggests that the evolutionary history of these viruses spans the entire history of the vertebrates and perhaps longer. In particular, as fish have ancient evolutionary origins, with the earliest organisms classified as fish first appearing during the Cambrian period approximately 540 million years ago (Shu et al. 1999), it is possible that some fish viruses will occupy more basal phylogenetic positions than those observed in other vertebrate hosts, including mammals, amphibians and birds (Lauber et al. 2017). As a notable example, the recent identification of hepadnaviruses in fish has revealed both that these viruses have ancient vertebrate origins (Lauber et al. 2017) and that there have been more instances of host jumping than previously realized, including a potential jump from aquatic to terrestrial vertebrates (Dill et al. 2016). Importantly, there is no evidence of fish viruses causing human disease or establishing a productive infection, which largely reflects the phylogenetic distance between fish and humans, along with major differences in cell types and cell receptors. However, the frequency with which plant viruses are found in human faecal samples provides compelling evidence for the passive transmission of viruses through food (Chau et al. 2017) and the consumption of raw fish has been associated with bacterial (group B Streptococcus) disease in humans (Zhang et al. 2006; Tan et al. 2016).
The transmission of viruses among fish predominantly occurs horizontally via faeces, via contaminated water or by the use of unpasteurized wild fish products in aquaculture (Kurath and Winton 2011). Fish also exhibit diverse population ecologies that are likely to be important for viral diversification and transmission. In particular, fish can be characterized by very low population density (i.e. solitary fish) to very high population density (i.e. shoaling fish). For example, it is estimated that some fish schools can occupy nearly 5 km3, with population densities between 0.5 and 1 fish per cubic meter, totalling approximately three billion fish in a single school (Radakov and Mills 1974). Contact between donor and recipient hosts is an obvious necessity for virus transmission, such that host ecology, behaviour and geographical separation all likely impact the probability of virus emergence (Parrish et al. 2008; Engering et al. 2013; Dennehy 2017). Although theory suggests that host population density is central to viral spread and epidemic potential (Anderson and May 1982), few studies have determined the effect of density-dependent transmission in natural host systems. In fish, for example, it is possible that close contact while shoaling facilitates virus transmission between hosts (Johnson et al. 2011), such that the high fish stock densities in aquaculture will greatly assist viral emergence, although this hypothesis is yet to be tested.
To address key questions in the ecology and evolution of fish viruses we performed a meta-transcriptomic survey of healthy fish that were purchased from a fish market in Sydney, Australia. In particular, by measuring the number of viral transcripts in pooled samples, we aimed to determine how virus composition and abundance varies between species, determine the relative frequencies of cross-species transmission and virus–host co-divergence, and test the idea that large and dense host populations harbour a greater number of viruses compared to their solitary counterparts, which will in turn make them a more important source of emerging viruses. To this end, we studied both shoaling fish species—the eastern sea garfish (Hyporhamphus australis) and Australasian snapper (Chrysophrys auratus)—as well as more solitary fish—the eastern red scorpionfish (Scorpaena jacksoniensis) and the largetooth flounder (Pseudorhombus arsius).
2. Materials and methods
2.1 Fish sample collection
Dead fish were purchased from a fish market in Sydney, Australia on the day of catch and initially stored on ice. Fish had been caught by commercial fisheries in coastal waters in New South Wales, Australia, from similar overall habitats. The species studied were: eastern sea garfish (H. australis), Australasian snapper (C. auratus), eastern red scorpionfish (S. jacksoniensis) and largetooth flounder (P. arsius). To increase the likelihood of virus discovery, twelve individuals from each species were purchased and analysed. Liver and gill tissues were immediately dissected and stored separately in RNALater before being transferred to a −80 °C freezer.
2.2 Transcriptome sequencing
Deep transcriptome sequencing was performed on fish liver and gill samples from the four fish species (ninety-six samples total). Frozen tissue was partially thawed and submerged in lysis buffer containing 1 per cent ß-mercaptoethanol and 0.5 per cent Reagent DX before homogenisation with TissueRupture (Qiagen). The homogenate was centrifuged to remove any potential tissue residues and RNA from the clear supernatant was extracted using the Qiagen RNeasy Plus Mini Kit. RNA was quantified using NanoDrop (ThermoFisher) and tissues from each tissue type and species were pooled resulting in a total of eight samples (four gill and four liver) to 3 μg per pool (250 ng per tissue sample). For library construction, the TruSeq Total RNA Library Preparation Protocol was used. To facilitate virus discovery, host ribosomal RNA (rRNA) was depleted using the Ribo-Zero-Gold Epidemiology Kit. Paired-end (100 bp) sequencing of the RNA library was performed on the HiSeq 2500 platform (Illumina). All library preparation and sequencing was carried out by the Australian Genome Research Facility (AGRF).
2.3 Virus discovery in fish
Sequencing reads were first quality trimmed then assembled de novo using Trinity RNA-Seq (Haas et al. 2013). The assembled contigs were annotated based on similarity searches against the NCBI nucleotide (nt) and non-redundant protein (nr) databases using BLASTn and Diamond (BLASTX) (Buchfink et al. 2015), and an e-value threshold of 1 × 10−5 was used to maximize sensitivity, meaning that we would not expect to observe a sequence match by chance alone. We removed non-viral hits, such as host contigs with similarity to viral sequences (e.g. endogenous viral elements), as well as any contigs with high similarity to plant viruses, which were more likely to be derived from food sources. Transcript abundance was estimated using RSEM (Li and Dewey 2011) implemented within Trinity. To identify very low abundance hits, an additional Diamond search was performed on the non-assembled raw RNA sequence reads against a custom database of RNA viruses.
2.4 Inferring the evolutionary history of fish viruses
To infer the evolutionary (phylogenetic) relationships of the viruses contained in the fish samples, the translated viral contigs were combined with protein sequences obtained from GenBank using the top search results from BLAST (see Table 1 for more details of the sequences analysed). The sequences retrieved were then aligned with those generated here using MAFFT v.3.4, employing the E-INS-I algorithm. Ambiguously aligned regions were removed using trimAl v.1.2 (Capella-Gutiérrez et al. 2009). To estimate phylogenetic trees, we selected the optimal model of amino acid substitution identified using the Bayesian Information Criterion as implemented in Modelgenerator v0.85 (Keane et al. 2006) and analysed the data using the maximum likelihood approach available in PhyML v3.1 (Guindon et al. 2010) with 1000 bootstrap replicates. Phylogenetic trees were annotated with FigTree v.1.4.2.
Table 1.
Amino acid identity, contig length and relative frequency of the viruses identified in this study.
| Host | Virus species | Contig length (nt) | % Relative abundance in library | Closest match (GenBank accession number) | % Amino acid identify |
|---|---|---|---|---|---|
| Eastern sea garfish (Hyporhamphus australis) | Eastern sea garfish astrovirus | 483 | 0.0000809% | Wenling plagiopsetta astrovirus ORF1ab | 54% |
| (AVM87176.1) | |||||
| Eastern sea garfish bunya-like virus | 888 | 0.0000546% | Xingshan nematode virus-3 RdRp | 48% | |
| (APG79357.1) | |||||
| Eastern sea garfish hepatitis B virus | 2, 304 | 0.0002588% | Bluegill hapadnavirus polymerase | 65% | |
| (YP_009259541.1) | |||||
| Eastern sea garfish picornavirus | 378 | 0.0000150% | Eel picornavirus 1 polyprotein | 61% | |
| (YP_008531322.1) | |||||
| Eastern sea garfish rhabdovirus | 2, 829 | 0.0010890% | Beihai rhabdo-like virus-2 RdRp | 54% | |
| (YP_009333449.1) | |||||
| Australasian snapper (Pagrus auratus) | Australasian snapper hepatitis B virus | 2, 400 | 0.0033941% | White sucker hepatitis B virus polymerase | 46% |
| (YP_009165599.1) | |||||
| Australasian snapper noda-like virus | 843 | 0.0000831% | Feline fesavirus-4 hypothetical protein | 36% | |
| (AII82234.1) | |||||
| Eastern red scorpionfish (Scorpaena jacksoniensis) | Eastern red scorpionfish astrovirus | 906 | 0.0003463% | Wenling righteye flounder astrovirus ORF1ab | 67% |
| (AVM87607.1) | |||||
| Eastern red scorpionfish picornavirus | 2, 013 | 0.0015272% | Guangdong spotted longbarbel catfish picornavirus polyprotein | 29% | |
| (AVM87450.1) | |||||
| Eastern red scorpionfish flavivirus | 519 | 0.0000234% | Wenzhou shark flavivirus polyprotein | 54% | |
| (AVM87250.1) | |||||
| Largetooth flounder (Pseudorhombus arsius) | Largetooth flounder astrovirus | 201 | 0.0000043% | Wenling rattails astrovirus-5 ORF1ab | 67% |
| (AVM87155.1) | |||||
| Largetooth flounder picorna-like virus | 315 | 0.0000801% | Octopus Beihai picorna-like virus-21 hypothetical protein 2 | 67% | |
| (YP_009333574.1) |
To determine the relative frequencies of cross-species transmission versus virus–host co-divergence, we reconciled the co-phylogenetic relationship between viruses isolated from fish and their hosts using the Jane v.4 co-phylogenetic software package (Conow et al. 2010). This method uses a polynomial time dynamic programming algorithm in conjunction with a genetic algorithm to find optimal solutions to reconcile co-phylogenies. Virus phylogenies were first inferred using PhyML v3.1 (Guindon et al. 2010) as described above, excluding all non-fish viruses. Host trees at fish order-level were constructed using topologies from the literature (Betancur et al. 2017). We used ‘event costs’ associated with phylogenetic incongruences between trees that were conservative towards co-divergence and defined here as: 0 for co-divergence, 1 for duplication, 1 for host-jumping and 1 for extinction. Finally, to assist visualisation of these data, tanglegrams for each virus family were constructed using TreeMap v3.0 (Charleston 2011). Lines between the trees connect the fish host (left) with its virus (right). We utilised the ‘untangle’ function, which rotates the branches of one tree to minimise the number of crossed lines.
3. Results
3.1 Abundance of viruses in market fish
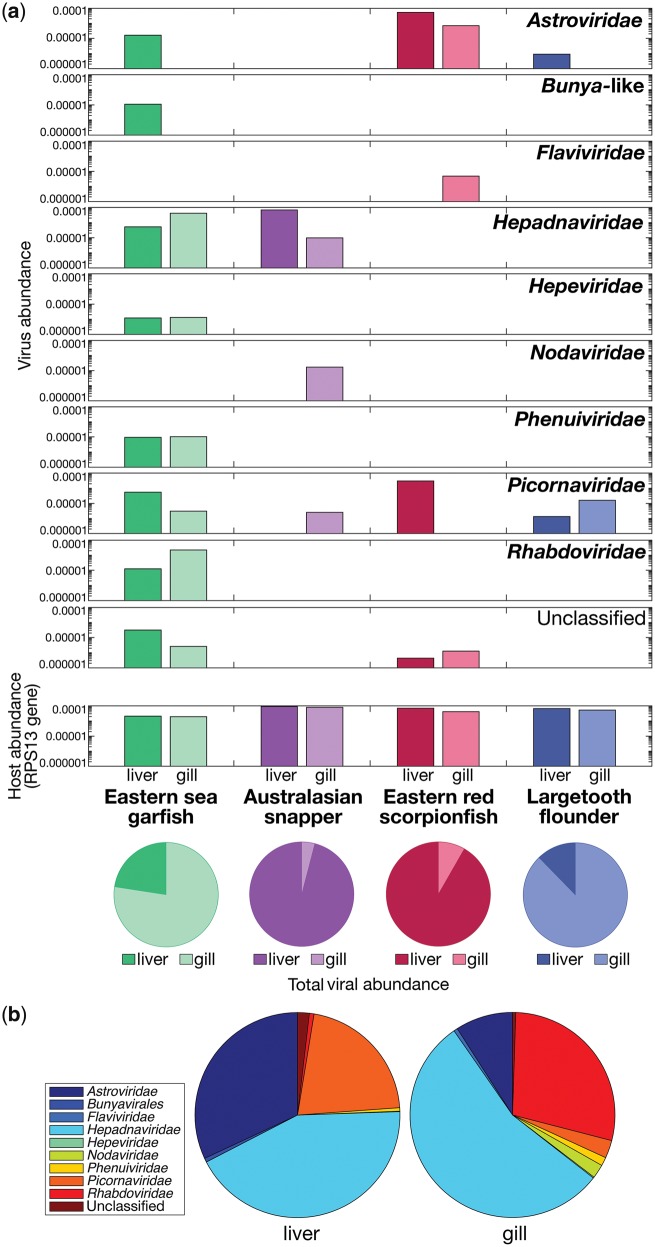
We used a meta-transcriptomics approach to characterise the viral transcripts of four fish species sold at a fish market in Sydney, Australia—eastern sea garfish, Australasian snapper, eastern red scorpionfish and largetooth flounder—and from this make insights into virus evolution. These species belong to different taxonomic orders of fish, although are all members of the superorder Acanthopterygii. We extracted total RNA from the liver and gill tissue of these animals, which were then organised into eight libraries for high-throughput RNA sequencing. Ribosomal RNA-depleted libraries resulted in a median of 47,034,084 (range 44,459,814–49,455,480) reads per pool. Reads were assembled de novo into a median of 220,203 contigs (range 60,251–448,659). An assessment of host reference gene ribosomal protein S13 (RPS13) revealed similar abundances (0.002–0.009% of reads), implying similar sequencing depth across libraries (Fig. 1a).
Figure 1.
(a) Relative abundance of viral contigs within each RNA sequencing library for each fish species and tissue type, falling across nine viral families. The relative abundance of the host reference gene, ribosomal protein S13, is also included for each library. The proportion of viral contigs (including those that are unclassified) in each tissue type is shown below in pie charts. (b) Total relative abundance of viral contigs within each tissue type falling into each viral family.
Our analysis revealed the relative abundance of viral families present in the non-rRNA transcriptome data (Fig. 1a). Among the four fish species sampled, we found virus transcripts that could be assigned to eight different viral families: the Astroviridae, Flaviviridae, Hepadnaviridae, Hepeviridae, Nodaviridae, Phenuiviridae, Picornaviridae and Rhabdoviridae as well as from the order Bunyavirales. With the exception of the Hepadnaviridae, all are RNA viruses. Although the approach used is able to identify DNA microbes (Eden et al. 2017), no other DNA viruses were identified in these data, such that any present in the samples tested are generating transcripts at very low frequency. The Hepadnaviridae, that possess reverse-transcribed DNA genomes, were the most abundant comprising ∼43 per cent and ∼55 per cent of the total viral reads in the liver and gill tissues, respectively (Fig. 1b). In liver tissues, the Astroviridae comprised ∼32 per cent and the Picornaviridae comprised ∼21 per cent of the total virome (Fig. 1b). In gill tissues, Astroviridae comprised ∼9 per cent and Rhabdoviridae comprised ∼28 per cent of the total virome (Fig. 1b).
It is notable that the eastern sea garfish, a highly densely shoaling fish, had the highest number of distinct virus species compared to the other fish sampled (P < 0.01), containing potentially novel viruses belonging to seven and five viral families in the liver and gill tissue, respectively, and a total viral abundance (i.e. relative to that of all transcripts) of 0.002 per cent (Fig. 1a). In contrast, largetooth flounder, the most solitary fish species studied here, possessed viruses that fell into only two viral families with a total relative viral abundance of only 0.00004 per cent. Interestingly, viruses were more abundant in the gill tissues of eastern sea garfish and largetooth flounder, yet more abundant in liver tissues in the case of the Australasian snapper and eastern red scorpionfish (P < 0.01 in all cases). Taken together, however, there was no significant difference in viral abundance between liver and gill tissues (P > 0.05).
A comparison of viral family abundance between fish species revealed marked differences (Fig. 2). Picornaviruses (or picorna-like viruses) and astroviruses were the most wide-spread viruses across species and were particularly abundant in eastern red scorpionfish. Both eastern sea garfish and Australasian snapper possessed novel viruses that fell within the Hepadnaviridae. These viruses were present in liver and gill tissues from both species and have been identified in an increasingly large number of fish species (Lauber et al. 2017; their phylogenetic relationships are discussed in more detail below). In addition, we discovered transcripts from a virus belonging to the Flaviviridae—a family comprised single-stranded, positive-sense RNA viruses that is commonly associated with vector-borne transmission in terrestrial vertebrates—in the eastern red scorpionfish. Other single-stranded RNA viruses fell into families including the Nodaviridae, discovered in Australasian snapper, and the Phenuiviridae and Hepeviridae, within eastern sea garfish.
Figure 2.
Relative abundance of viral contigs within each RNA sequencing library for each fish species and tissue type, normalized for each viral family.
3.2 Phylogenetic relationships of the viral sequences determined here
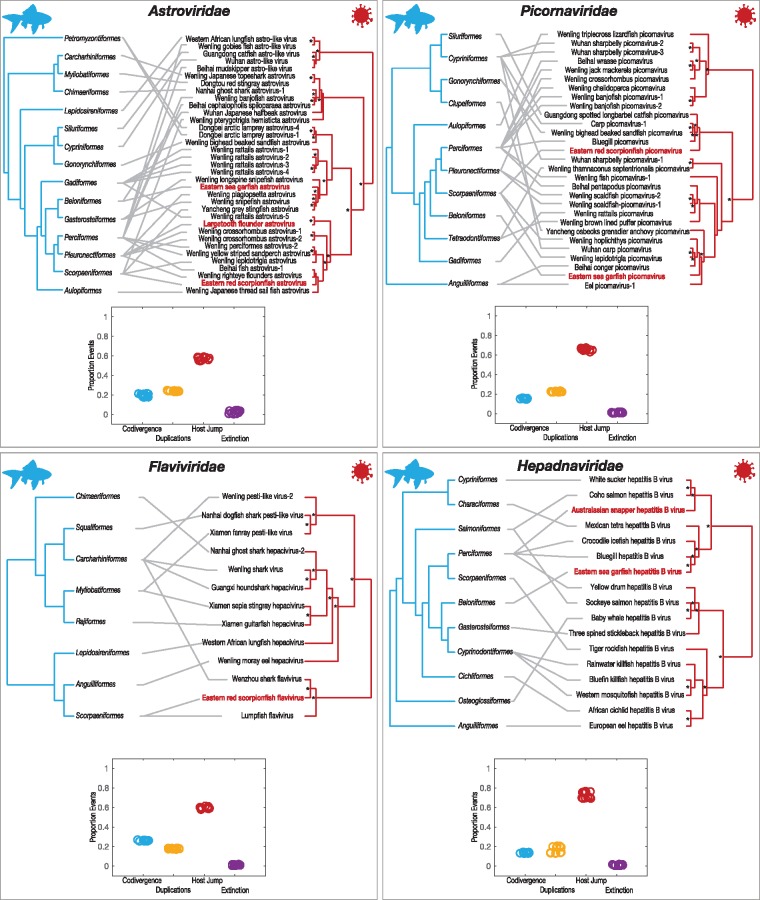
Although multiple contigs covering various genomic regions were present for all viruses, because our main motivation was to reveal phylogenetic patterns (rather than genomic characterisation), we necessarily focused on the most conserved viral regions that comprise the RNA-dependent RNA polymerase (RdRp), or the polymerase (P) ORF in the case of the hepadnaviruses, and that are routinely used for virus species demarcation. For the most abundant viral contigs (Table 1), we inferred phylogenetic trees to reveal their evolutionary histories in the context of their closest relatives that were obtainable from GenBank. In total, we identified 12 distinct and potentially novel virus species. Among the viruses identified were those from families that have only recently been found in fish (Shi et al. 2018); this suggests that these viruses were directly infecting the fish sampled rather than being associated with the aquatic environment or a co-infecting organism (see below). As a case in point, we identified astrovirus sequences in three fish species, all of which shared amino acid similarity to other fish astroviruses (Fig. 3). Interestingly, all three viruses were most phylogenetically similar to astroviruses previously isolated from hosts within the fish order Pleuronectiformes (Fig. 3). Specifically, eastern sea garfish astrovirus shared 54 per cent amino acid similarity to Wenling plagiopsetta astrovirus ORF1ab isolated from crested flounders, eastern red scorpionfish astrovirus shared 67 per cent similarity to Wenling righteye flounder astrovirus ORF1ab, and largetooth flounder astrovirus shared 67 per cent identity to Wenling rattails astrovirus-5 ORF1ab (Table 1). Although the latter comprised a relatively short viral contig (Table 1), any phylogenetic uncertainty is reflected in the bootstrap values across the tree.
Figure 3.
Phylogenetic relationships of likely vertebrate-associated viruses discovered from assembled contigs: (a) Astroviridae, (b) Picornaviridae, (c) Flaviviridae and (d) Hepadnaviridae. The maximum likelihood phylogenetic trees show the topological position of the newly discovered potential viruses (bold text), in the context of their closest relatives (major genera are labelled). Fish viruses are coloured to correspond to host order, as indicated in the fish order phylogeny. All branches are scaled to the number of amino acid substitutions per site and trees were mid-point rooted for clarity only. An asterisk indicates node support of >70% bootstrap support.
A potentially novel flavivirus in the eastern red scorpionfish was related (with strong bootstrap support) to Wenzhou shark flavivirus (Shi et al. 2018), sharing 54 per cent amino acid similarity in the polyprotein. This phylogenetic pattern suggests a viral species jump between distantly related Carcharhiniformes and Scorpaenidae (see Fig. 3), although a greater sampling effort is obviously needed to determine whether intermediate hosts are involved. Both viruses fell basal to the invertebrate-specific and vector-borne members of the genus Flavivirus (Fig. 3). The only other fish virus that has been identified in this genus is lumpfish flavivirus (Skoge et al. 2018). Other fish viruses within the Flaviviridae group with the Hepaciviruses and Pestiviruses, although it is highly likely that more fish flaviviruses will be identified with additional sampling.
Interestingly, potentially novel hepadnaviruses were identified in the eastern sea garfish and Australasian snapper (Fig. 3). Eastern sea garfish hepatitis B virus was most closely related to bluegill hepatitis B virus, with 65 per cent amino acid similarity (Dill et al. 2016), and as a pair these viruses were more closely related to mammalian hepatitis B viruses (i.e. the genus Orthohepadnavirus) than to other fish hepatitis B viruses. Conversely, the more divergent Australasian snapper hepatitis B virus shared 46 per cent amino acid similarity to white sucker hepatitis B virus (Hahn et al. 2015) and Coho salmon hepatitis B virus (Lauber et al. 2017). These viruses fell into clades that form an in-group to the recently described and divergent hepadna-like viruses, Nackednavirus, found in a number of fish species (Lauber et al. 2017).
Novel picornavirus sequences were detected in eastern sea garfish and eastern red scorpionfish that are related to other fish picornaviruses (Fig. 3). Specifically, Eastern sea garfish picornavirus shared 61 per cent amino acid identity to eel picornavirus-1 isolated from a diseased European eel of the distantly related order Anguilliformes (Fichtner et al. 2013). Similarly, eastern red scorpionfish picornavirus shared only 33 per cent amino acid identity to its closest known relative, the Guangdong spotted longbarbel catfish picornavirus that infect hosts from order Siluriformes. While many picornaviruses have been associated with mortality in fish hosts, little is known about their epidemiology and disease potential including those identified here (Mor and Phelps 2016).
To examine the frequency of cross-species transmission among fish viruses, we first inferred tanglegrams depicting pairs of rooted phylogenetic trees that display the evolutionary relationship between each virus family and their fish hosts (Fig. 4). Despite our limited sample of fish viruses it was obvious from this analysis that cross-species transmission has been common and occurred among all hosts and viruses. For example, the Perciformes harboured viruses in the Astroviridae, Picornaviridae and Hepadnaviridae that appeared to regularly jump species boundaries. To examine the frequency of cross-species transmission in a more quantitative manner, we performed a reconciliation analysis that determined the range of optimal co-phylogenetic solutions for each virus family (Fig. 3). This revealed that cross-species transmission was the most common evolutionary event of those studied, with virus–host co-divergence consistently less frequent, and lineage duplication and extinction playing a much more minor role. Importantly, however, these results are likely to change as the number of fish viruses identified increases with future metagenomic studies.
Figure 4.
Tanglegrams of rooted phylogenetic trees for each virus family of the vertebrate-associated fish viruses described here, constructed using TreeMap3 v3.0 (48). Viruses identified in this study are indicated in red, while all other viruses have previously been identified in fish. The ‘untangle’ function was used to maximise the congruence between the host (left) and virus (right) phylogenies. Below each tanglegram, reconciliation analysis of each virus family using Jane (47) illustrates the range of the proportion of possible events. The ‘event costs’ associated with incongruences between trees were conservative towards co-divergence and defined here as: 0 for co-divergence, 1 for duplication, 1 for host-jumping and 1 for extinction. An asterisk on the virus trees indicates node support of >70% bootstrap support.
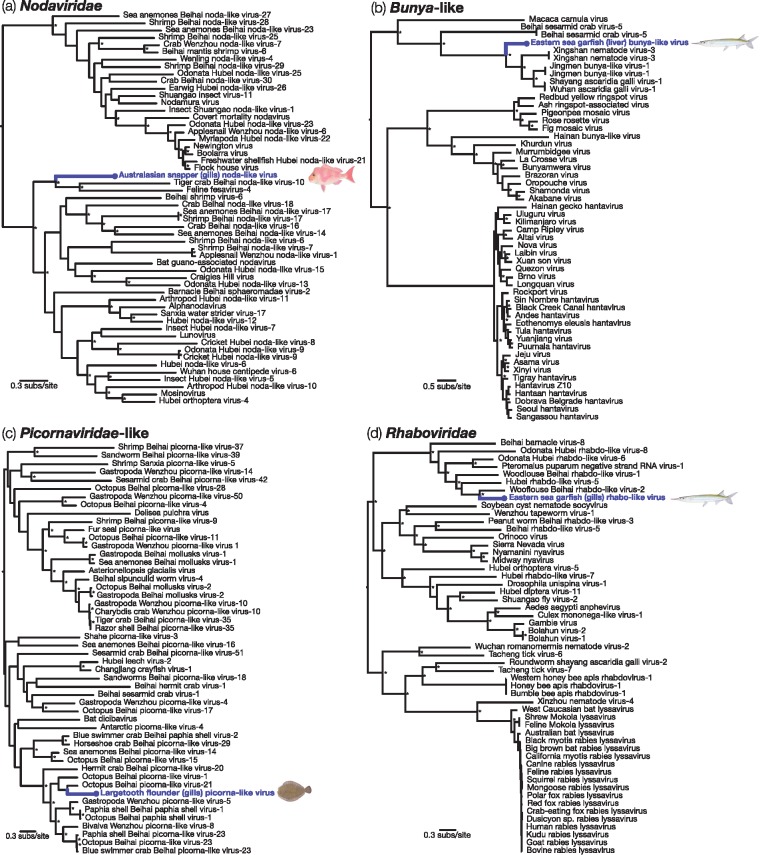
Finally, we also identified a number of viral transcripts in these fish liver and gills samples that were more closely related to invertebrate-associated viruses: this implies that they more likely originated from invertebrates within the fish, rather than from the fish themselves. The majority of these viruses belonged to RNA viruses from the families Nodaviridae and Rhabdoviridae, that were particularly abundant in gill tissue, as well as from the order Bunyavirales, and from various unclassified invertebrate-associated viruses. Similarly, we found a picorna-like virus sampled from the largemouth flounder that harboured genetic similarity to picorna-like viruses from marine invertebrates, specifically the octopus and hermit crab Beihai picorna-like viruses (Table 1; Fig. 4). Phylogenetic analysis of all these viruses revealed that their closest genetic relatives were indeed from invertebrate hosts (Table 1; Fig. 5).
Figure 5.
Phylogenetic relationships of likely invertebrate-associated viruses discovered from assembled contigs: (a) Nodaviridae, (b) Bunyaviridae-like, (c) Picornaviridae-like and (d) Rhabdoviridae. The maximum likelihood phylogenetic trees show the topological position of the newly discovered potential viruses (blue), in the context of their closest relatives. All branches are scaled to the number of amino acid substitutions per site and trees were mid-point rooted for clarity only. An asterisk indicates node support of >70% bootstrap support.
4. Discussion
To help determine the abundance and evolution of viruses in fish, we identified viral transcripts from a meta-transcriptomic analysis of four fish species purchased from a fish market in Sydney, Australia. As these species included two shoaling fish and two solitary fish, these data also provided an opportunity to address how host population density might affect virus composition. Overall, our analysis identified twelve potentially novel virus species, eight of which were distinctly vertebrate-associated.
Two relatively highly abundant viral transcripts were assigned to the Hepadnaviridae and discovered in Australasian snapper and eastern sea garfish. Until recently, exogenous hepadnaviruses had only been identified in mammals (genus Orthohepadnavirus) and birds (Avihepadnavirus), although endogenous versions of these viruses were found in a wider array of host genomes (Cui and Holmes 2012; Gilbert et al. 2014; Dill et al. 2016; ). However, several hepadnaviruses have since been discovered in fish and amphibians (Hahn et al. 2015; Dill et al. 2016; Lauber et al. 2017). These observations, along with the discovery of more divergent hepadna-like viruses in fish (i.e. Nackednavirus) (Lauber et al. 2017), strongly suggests that hepadnaviruses have existed for the entire evolutionary history of the vertebrates (Gilbert et al. 2014; Suh et al. 2014), which may also be true of many families of RNA viruses (Shi et al. 2018). Notably, the hepadnaviruses in Australasian snapper and eastern sea garfish were separated by a large phylogenetic distance, with eastern sea garfish hepatitis B virus more closely related to mammalian hepatitis B viruses than to other fish hepadnaviruses. This supports the growing view that, in the case of the hepadnaviruses as well as many other viruses, cross-species transmission occurs frequently on a backbone of virus-host co-divergence (Geoghegan et al. 2017; Shi et al. 2018), and that these species jumps can occasionally cover very large phylogenetic distances. Indeed, cophylogenetic reconciliation analysis revealed a high frequency of host jumps within the Hepadnaviridae compared to other virus families studied here, although these patterns are highly dependent on taxa number and composition and are likely to change as the number of sequenced fish viruses inevitably increases.
Picornaviruses and astroviruses were the most widespread among the fish species studied here, and all shared strong sequence similarity to other fish viruses. Interestingly, both picornaviruses and astroviruses are single-stranded positive-sense RNA viruses with small icosahedral capsids and no external envelope, and it is possible that these phenotypic features aid their preservation in harsh marine environments. While astroviruses have only recently been discovered in fish (Shi et al. 2018), they are known to persist in aquatic environments and have been detected in seabirds (Chu et al. 2012) and marine mammals (Rivera et al. 2010). The astroviruses identified here fell across a large group of fish astroviruses that formed a sister group to mammalian (Mamastrovirus) and avian (Aviastrovirus) viruses. Picornaviruses have previously been associated with morbidity and mortality in fish (Barbknecht et al. 2014; Hahn et al. 2015), and we identified two potentially novel picorna viruses and one picorna-like virus in the fish studied here. Picornaviruses were recently shown to have one of the highest frequencies of cross-species transmission in a comparison of nineteen virus families (Geoghegan et al. 2017). Indeed, our phylogenetic analyses shows that the evolutionary history of picornaviruses exhibits major incongruences with that of their hosts, indicative of multiple cross-species transmission events, again on a backbone of likely long-term virus–host co-divergence (Shi et al. 2018). In accord with this general macroevolutionary pattern, the picornaviruses sampled in fish do not form a monophyletic group.
A large-scale analysis of virus diversity across the vertebrate phylogeny also suggested that viruses in fish often fall basally to the viruses found in birds and mammals (Shi et al. 2018), indicative of ancient virus–host associations (although, again, our sample of other vertebrate taxa is small). In addition, viruses that are more commonly associated with vector-borne transmission, for example Alphavirus, Dimarhabdovirus, and Flavivirus, are also found in fish (Shi et al. 2018). We identified a potentially novel flavivirus in the eastern red scorpionfish, making it the third member of the genus Flavivirus sampled from ray-finned fish. In the phylogeny, eastern red scorpionfish flavivirus is most closely related to a flavivirus identified in cartilaginous fish, Wenzhou shark flavivirus (Shi et al. 2018), strongly suggesting that fish flaviviruses may be even more wide spread than currently sampled. These viruses fall basal to the vector-borne and invertebrate-associated flaviviruses and shared common ancestry with Tamana bat virus that has no known vector (Shi et al. 2018).
It has previously been shown that species forming large conspecific groups that are characterised by high contact rates exhibit greater viral richness (that is, diversity and abundance) compared to species with small group sizes (Ezenwa et al. 2006; Lindenfors et al. 2007; Gay et al. 2014; Webber et al. 2017). This finding supports classic epidemiological theory that larger populations that have higher contact rates have an increased likelihood of acquiring and transmitting viruses (May and Anderson 1979). Here, we hypothesised that close contact while shoaling may facilitate virus transmission between fish (Dennehy 2017). It was therefore notable that the most solitary of the fishes we studied here, the largetooth flounder, harboured the smallest number of viral transcripts. Conversely, eastern sea garfish, a highly densely shoaling fish, harboured the greatest number of viral transcripts. As such, these data tentatively support the notion that more frequent intra-host contacts increase the potential for viral diversification and spread. Clearly, a broader comparison of many more fish species is required to truly understand how fish host ecology, especially population density, might facilitate virus diversity, abundance and ultimately, evolution.
Although this initial study focused on only four fish species commonly sold in markets, we identified twelve potentially unknown viruses, and found examples of likely novel viruses in every species sampled. Since the species analysed were market-bought rather than being directly sampled during fishing trips, it is possible that we missed those viruses with short intra-host durations of infection or those at very low abundance. However, that we found viruses in all species sampled provides further support for the proposition that fish harbour a very large number of diverse viruses (Lauber et al. 2017; Shi et al. 2018) that will gradually be revealed with more extensive sampling. Finally, these data show that fish species commonly sold as food may contain a wide range of fish viruses (many unknown), although the major differences in virus biology mean that they likely pose no risk for cross-species transmission to humans.
Ethics
Biosafety approved by Macquarie University (5201700856).
Acknowledgements
We thank efishalbum.com for fish images in Figs 2, 3 and 5, which were used with permission. ECH is funded by an ARC Australian Laureate Fellowship (FL170100022).
Data availability
Data are available through Dryad.
Conflict of interest: None declared.
References
- Anderson R. M., May R. M. (1982) ‘Coevolution of Hosts and Parasites’, Parasitology, 85: 411–26. [DOI] [PubMed] [Google Scholar]
- Avarre J.-C. (2017) ‘Molecular Tracing of Aquatic Viruses: Where Epidemiology Needs to Meet Genomics’, Frontiers in Microbiology, 8: 1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbknecht M. et al. (2014) ‘Characterization of a New Picornavirus Isolated from the Freshwater Fish Lepomis Macrochirus’, Journal of General Virology, 95: 601–13. [DOI] [PubMed] [Google Scholar]
- Betancur R. et al. (2017) ‘Phylogenetic Classification of Bony Fishes’, BMC Evolutionary Biology, 17: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broitman B. R. et al. (2017) ‘Dynamic Interactions among Boundaries and the Expansion of Sustainable Aquaculture’, Frontiers in Marine Science, 4: 1–15. [Google Scholar]
- Buchfink B., Xie C., Huson D. H. (2015) ‘Fast and Sensitive Protein Alignment Using DIAMOND’, Nature Methods, 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009) ‘trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses’, Bioinformatics, 25: 1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charleston M. A. (2011) ‘TreeMap’ <https://sites.google.com/site/cophylogeny/home>.
- Chau M. L. et al. (2017) ‘Group B Streptococcus Infections Caused by Improper Sourcing and Handling of Fish for Raw Consumption, Singapore, 2015-2016’, Emerging Infectious Disease, 23: 1982–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D. K. et al. (2012) ‘A Novel Group of Avian Astroviruses in Wild Aquatic Birds’, Journal of Virology, 86: 13772–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conow C. et al. (2010) ‘Jane: A New Tool for the Cophylogeny Reconstruction Problem’, Algorithms for Molecular Biology, 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane M., Hyatt A. (2011) ‘Viruses of Fish: An Overview of Significant Pathogens’, Viruses, 3: 2025–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Holmes E. C. (2012) ‘Endogenous Hepadnaviruses in the Genome of the Budgerigar (Melopsittacus Undulatus) and the Evolution of Avian Hepadnaviruses’, Journal of Virology, 86: 7688–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart E. L. (2007) ‘Viral Metagenomics’, Reviews in Medical Virology, 17: 115–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy J. J. (2017) ‘Evolutionary Ecology of Virus Emergence’, Annals of the New York Academy of Sciences, 1389: 124–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill J. A. et al. (2016) ‘Distinct Viral Lineages from Fish and Amphibians Reveal the Complex Evolutionary History of Hepadnaviruses’, Journal of Virology, 90: 7920–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden J.-S. et al. (2017) ‘Francisella tularensis Ssp. holarctica in Ringtail Possums, Australia’, Emerging Infectious Diseases, 23: 1198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engering A., Hogerwerf L., Slingenbergh J. (2013) ‘Pathogen–Host–Environment Interplay and Disease Emergence’, Emerging Microbes & Infections, 2: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezenwa V. O. et al. (2006) ‘Host Traits and Parasite Species Richness in Even and Odd‐Toed Hoofed Mammals, Artiodactyla and Perissodactyla’, Oikos, 115: 526. [Google Scholar]
- Fichtner D. et al. (2013) ‘Characterization of a Novel Picornavirus Isolate from a Diseased European Eel (Anguilla anguilla)’, Journal of Virology, 87: 10895–9., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay N. et al. (2014) ‘Parasite and Viral Species Richness of Southeast Asian Bats: Fragmentation of Area Distribution Matters’, International Journal of Parasitology: Parasites and Wildlife, 3: 161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. L., Duchêne S., Holmes E. C. (2017) ‘Comparative Analysis Estimates the Relative Frequencies of co-Divergence and Cross-Species Transmission within Viral Families’, PLoS Pathogens, 13: e1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. L. et al. (2018) ‘Virological sampling of inaccessible wildlife with drones’, Viruses, 10: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. L., Holmes E. C. (2017) ‘Predicting Virus Emergence amid Evolutionary Noise’, Open Biology, 7: 170189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. L., Senior A. M., Holmes E. C. (2016) ‘Pathogen Population Bottlenecks and Adaptive Landscapes: Overcoming the Barriers to Disease Emergence’, Proceedings of the Royal Society B: Biological Sciences, 283: 20160727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C. et al. (2014) ‘Endogenous Hepadnaviruses, Bornaviruses and Circoviruses in Snakes’, Proceedings of the Royal Society B, 281: 20141122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Haas B. J. et al. (2013) ‘De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis’, Nature Protocols, 8: 1494–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C. M. et al. (2015) ‘Characterization of a Novel Hepadnavirus in the White Sucker (Catostomus Commersonii) from the Great Lakes Region of the United States’, Journal of Virology, 89: 11801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. B. et al. (2011) ‘Parasite Transmission in Social Interacting Hosts: Monogenean Epidemics in Guppies’, PLoS One, 6: e22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M. et al. (2006) ‘Assessment of Methods for Amino Acid Matrix Selection and Their Use on Empirical Data Shows That Ad Hoc Assumptions for Choice of Matrix Are Not Justified’, BMC Evolutionary Biology, 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurath G., Winton J. (2011) ‘Complex Dynamics at the Interface between Wild and Domestic Viruses of Finfish’, Current Opinion in Virology, 1: 73–80. [DOI] [PubMed] [Google Scholar]
- Lauber C. et al. (2017) ‘Deciphering the Origin and evolution of Hepatitis B Viruses by Means of a Family of Non-Enveloped Fish Viruses’, Cell Host & Microbe, 22: 387–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011) ‘RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome’, BMC Bioinformatics, 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-X. et al. (2015) ‘Unprecedented Genomic Diversity of RNA Viruses in Arthropods Reveals the Ancestry of Negative-Sense RNA Viruses’, eLife, 4: e05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenfors P. et al. (2007) ‘Parasite Species Richness in Carnivores: Effects of Host Body Mass, Latitude, Geographical Range and Population Density’, Global Ecology and Biogeography, 16: 496–509. [Google Scholar]
- Lopes A. M. et al. (2014) ‘Molecular Evolution and Antigenic Variation of European Brown Hare Syndrome Virus (EBHSV)’, Virology, 468-470: 104–12. [DOI] [PubMed] [Google Scholar]
- May R. M., Anderson R. M. (1979) ‘Population Biology of Infectious Diseases: Part II’, Nature, 280: 455–61. [DOI] [PubMed] [Google Scholar]
- Mor S. K., Phelps N. B. D., (2016) ‘Picornaviruses of Fish’, in Godoy MG. (ed.) Aquaculture Virology, pp. 337–348. San Diego: Academic Press. [Google Scholar]
- Parrish C. R. et al. (2008) ‘Cross-Species Virus Transmission and the Emergence of New Epidemic Diseases’, Microbiology and Molecular Biology Reviews, 72: 457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvenis J. F. (2016). ‘Fisheries and Aquaculture Topics’. The State of World Fisheries and Aquaculture (SOFIA). Topics Fact Sheets. FAO Fisheries and Aquaculture Department.
- Rivera R. et al. (2010) ‘Characterization of Phylogenetically Diverse Astroviruses of Marine Mammals’, Journal of General Virology, 91: 166–73. [DOI] [PubMed] [Google Scholar]
- Shi M. et al. (2018) ‘The Evolutionary History of Vertebrate RNA Viruses’, Nature, 556: 197–202. [DOI] [PubMed] [Google Scholar]
- Shi M. et al. (2016) ‘Redefining the Invertebrate RNA Virosphere’, Nature, 540: 539–43. [DOI] [PubMed] [Google Scholar]
- Simmonds P. et al. (2017) ‘Virus Taxonomy in the Age of Metagenomics’, Nature Reviews. Microbiology, 15: 161–8. [DOI] [PubMed] [Google Scholar]
- Shu D. G. et al. (1999) ‘Lower Cambrian Vertebrates from South China’, Nature, 402: 42–6. [Google Scholar]
- Skoge R. H. et al. (2018) ‘New Virus of the Family Flaviviridae Detected in Lumpfish (Cyclopterus Lumpus)’, Archives of Virology, 163: 679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A. et al. (2014) ‘Early Mesozoic Coexistence of Amniotes and Hepadnaviridae’, PLoS Genetics, 10: e1004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. et al. (2016) ‘Group B Streptococcus Serotype III Sequence Type 283 Bacteremia Associated with Consumption of Raw Fish, Singapore’, Emerging Infectious Diseases, 22: 1970–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidwell J. H., Allan G. L. (2001) ‘Fish as Food: Aquaculture’s Contribution: Ecological and Economic Impacts and Contributions of Fish Farming and Capture Fisheries’, EMBO Reports, 2: 958–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber Q. M. R., Fletcher Q. E., Willis C. K. R. (2017) ‘Viral Richness Is Positively Related to Group Size, but Not Mating System, in Bats’, EcoHealth, 14: 652–61. [DOI] [PubMed] [Google Scholar]
- Radakov D. V., Mills H. (1974) ‘Schooling in the Ecology of Fish’, Quarterly Reviews of Biology, 49: 373. [Google Scholar]
- Zhang T. et al. (2006) ‘RNA Viral Community in Human Feces: Prevalence of Plant Pathogenic Viruses’, PLoS Biology, 4: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Gui J.-F. (2015) ‘Virus Genomes and Virus–Host Interactions in Aquaculture Animals’, Science China. Life Sciences, 58: 156–69. [DOI] [PubMed] [Google Scholar]
- Zhang T., Gui J.-F., Shi M., Holmes E. C. (2018) ‘Using Metagenomics to Characterize an Expanding Virosphere’, Cell, 172: 1168–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available through Dryad.
Conflict of interest: None declared.