Abstract
Microbes are capable of producing alcohols, making them an important source of alternative energy that can replace fossil fuels. However, these alcohols can be toxic to the microbes themselves, retaring or inhibiting cell growth and decreasing the production yield. One solution is improving the alcohol tolerance of such alcohol-producing organisms. Advances in omics technologies, including transcriptomic, proteomic, metabolomic, and genomic technologies, have helped us understand the complex mechanisms underlying alcohol toxicity, and such advances could assist in devising strategies for engineering alcohol-tolerant strains. This review highlights these advances and discusses strategies for improving alcohol tolerance using omics analyses.
Keywords: Adaptive laboratory evolution, Alcohol tolerance, Bacteria, Omics technology
Introduction
Engineering microorganisms to produce biochemicals has attracted attention as a strategy for reducing the dependency on fossil fuels and developing alternate renewable energy sources. Microbial production of alcohols such as ethanol, propanol, butanol, and other short-chain alcohols has been refined by many researchers (Hanai et al. 2007; Keasling and Chou 2008; Stephanopoulos 2008; Gronenberg et al. 2013). The selection of a microbial production host for an industrial biotechnology process is primarily determined by its potential to efficiently produce the product of interest. Yeast and bacteria are frequently selected as host organisms for bioalcohol production (Lau et al. 2010; Yamamoto et al. 2013). The yeast Saccharomyces cerevisiae is a natural producer of ethanol that has been widely applied for the production of bioethanol. Conversely, bacteria have advantages such as rapid growth, the utilization of various carbon sources, and the availability of genetic and molecular tools (Rumbold et al. 2009; Koppolu and Vasigala 2016).
In the bioproduction of these alcohols, a major problem is that the toxicity of the alcoholic compounds slows or inhibits cell growth, decreasing the production yield. The alcohol tolerance of bacteria is generally inferior to that of yeast; thus, alcohol toxicity is a more serious problem for bioproduction using bacteria. One strategy to overcome this problem is to develop strains that have tolerance to the target compounds. Thus, it is important to understand the mechanisms underlying alcohol tolerance.
Bacterial alcohol stress response has been studied for more than 40 years, and the physiological effect of alcohol stress has been well described. These studies primarily investigated the mechanisms by which alcoholic compounds affect the bacterial membrane. For example, alcohols interact directly with the lipid bilayer because of their amphiphilicity, and membrane fluidity is altered by the insertion of alcohols into cellular membranes (Ingram 1976). These changes in fluidity increase membrane permeability and induce conformational changes in membrane proteins, and ethanol-induced membrane changes induce the expression of heat-shock and phage-shock proteins (Neidhardt et al. 1984; Brissette et al. 1990). Alcoholic compounds also cause the partial breakdown of membrane function. This membrane damage causes various perturbations to cells such as ion leakage or loss of energy. In this context, many studies have focused on the relationship between alcohol tolerance and membrane composition. For example, changes of fatty acid composition (increase in the amount of unsaturated fatty acids) are observed during adaptation to ethanol in Escherichia coli (Ingram 1976; Berger et al. 1980). As another example, modifications of the unsaturated/saturated fatty acid ratio are found in Clostridium acetobutylicum cell membranes during acetone-butanol fermentation (Lepage et al. 1987). Modification of membrane composition via genetic manipulation also confers alcohol tolerance (Grandvalet et al. 2008; Luo et al. 2009). These studies illustrate that modifying the membrane composition can partially mitigate the toxicity of alcohols.
Alcoholic compounds activate various stress response networks (Bury-Moné et al. 2009). For example, the regulatory mechanisms of envelope stress (Ades 2004), oxidative stress (Belkin et al. 1996), and the respiratory cycle (Garbe and Yukawa 2001) are affected by alcohols. These responses are induced by membrane damage and physiological changes of the cellular state (e.g., changes of membrane fluidity, protein misfolding, ion leakage). To establish a rational strategy for improving bacterial alcohol tolerance, it is necessary to understand the cellular activities related to alcohol toxicity. We believe that advances in omics technologies, including transcriptomic, proteomic, metabolomic, and genomic technologies, can help us understand the impact of bacterial alcohol stress.
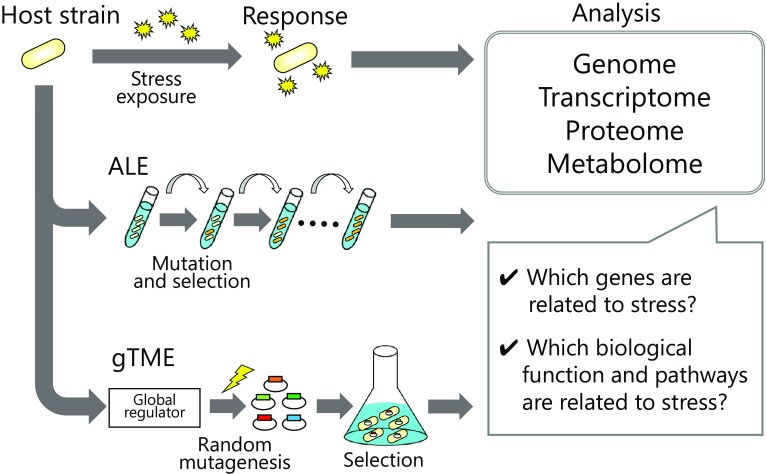
This review highlights advances in the use of omics technologies to understand alcohol tolerance in bacteria. First, we describe the comprehensive effect of alcohol toxicity using omics technologies. Further, we focus on several approaches for improving alcohol tolerance. We also focus on the alcohol stress response and tolerance of several bacterial species. E. coli has been used for biofuel production by engineering production pathways (Clomburg and Gonzalez 2010; Peralta-Yahya and Keasling 2010), and its well-characterized genetic background and well-developed genetic tools allow for flexible and economical process design for large-scale alcohol production. Likewise, C. acetobutylicum has been used for decades to produce butanol (Jones et al. 1982; Hermann et al. 1985). Most recently, cyanobacteria have attracted attention as promising industrial microorganisms for bioproduction because the cells can directly fix atmospheric carbon dioxide and convert it to a target compound using energy from photosynthesis (Nozzi et al. 2013; Lau et al. 2015). In this study, we review studies on alcohol tolerance in these species with an emphasis on improving alcohol production. Furthermore, to combine omics approaches with recent engineering approaches for strain improvement, it is possible to expand our search for phenotypes of alcohol tolerance (Fig. 1). We describe these promising approaches toward understanding and improving microbial tolerance to alcohol.
Fig. 1.
Strategy for the understanding alcohol-tolerance using omics technologies and recent engineering approaches for strain improvement. Adaptive laboratory evolution (ALE) is an approach for generating cells with improved growth and stress tolerance by mutations and natural selection. Global transcription machinery engineering (gTME) is an approach for obtaining various cellular phenotypes by reprogramming gene transcription using error-prone PCR. To combine omics analyses with these approaches, it is possible to expand our research for phenotypes of alcohol tolerance
Understanding alcohol tolerance using omics technologies
Alcoholic compounds activate various stress response networks by causing membrane damage. Comprehensive measurements made using omics technologies help us analyze the effect of alcohol stress on cellular states. Several studies provided lists of genes, proteins, and intracellular compounds that participate the alcohol stress response. For example, to screen genes related to this stress response, investigators analyzed gene expression in the presence and absence of alcohols in the culture medium. These findings should be a starting point for understanding the molecular mechanisms involved in ethanol stress tolerance, and thus, they represent fundamental knowledge for designing ethanol-tolerant cells. Recent studies on the bacterial alcohol stress response are summarized in Table 1.
Table 1.
Omics experiments with bacteria
| Species | Strain | Stress | Analysis | References |
|---|---|---|---|---|
| E. coli | Ethanol-tolerant strain LY01 | Ethanol | Transcriptome | Gonzalez et al. (2003) |
| E. coli | Transposon library, overexpression library | Ethanol | Enrichment | Goodarzi et al. (2010) |
| E. coli | Ethanol-evolved strains A–F | Ethanol | Transcriptome | Horinouchi et al. (2010) |
| E. coli | Mutated IrrE from Deinococcus radiodurans | Ethanol | Transcriptome, proteome | Chen et al. (2012) |
| E. coli | Fosmid library | Ethanol | Enrichment | Nicolaou et al. (2012) |
| E. coli | trans10 | Ethanol, n-butanol, Isobutanol | Metabolome | Wang et al. (2013) |
| E. coli | Genomic library | Ethanol | Enrichment analysis, transcriptome, proteome | Woodruff et al. (2013) |
| E. coli | Tolerant strain MTA156, MTA157, and MTA160 | Ethanol | DNAseq, RNAseq, ribosome profiling | Haft et al. (2014) |
| E. coli | Genomic library of solvent-tolerant Lactobacillus plantarum | Ethanol | Enrichment | Zingaro et al. (2014) |
| E. coli | Metagenomic and heterologous genomic libraries of sigma factor | Ethanol | Enrichment, transcriptome | Gaida et al. (2015) |
| E. coli | Ethanol-evolved strains A–F | Ethanol | Genome, transcriptome, metabolome | Horinouchi et al. (2015) |
| E. coli | High tolerance populations HT1-16 | Ethanol | Genome | Swings et al. (2017) |
| E. coli | Ethanol-tolerant mutant EM | Ethanol | Genome | Chen et al. (2018) |
| E. coli | Ethanol-tolerant ethanologenic strains | Ethanol | Genome | Lupino et al. (2018) |
| E. coli | IPA-tolerant strains A–F | Isopropanol | Genome, transcriptome | Horinouchi et al. (2017a) |
| E. coli | DH1 | n-Butanol | Transcriptome, proteome | Rutherford et al. (2010) |
| E. coli | Efflux pump library | n-Butanol | Enrichment | Dunlop et al. (2011) |
| E. coli | CRP mutation library | n-Butanol | Transcriptome | Lee et al. (2011) |
| E. coli | Genomic library, overexpression library | n-Butanol | Enrichment | Reyes et al. (2011) |
| E. coli | Adaptive mutants MG1–6 and MY1–4 | n-Butanol | Genome, transcriptome | Reyes et al. (2012) |
| E. coli | Adapted strains B500, G500, O500, H500, and P500 | n-Butanol and 3 abiotic stress | Genome, transcriptome, cross-stress tolerance | Dragosits et al. (2013) |
| E. coli | Transposon library | n-Butanol and 12 chemicals | Enrichment, transcriptome | Rau et al. (2016) |
| E. coli | Sigma70 mutant | n-Butanol | Transcriptome | Si et al. (2016) |
| E. coli | Fifty-five evolved strains | n-Butanol and ten chemicals | Genome, transcriptome, cross-stress tolerance | Horinouchi et al. (2017b) |
| E. coli | Butanol-tolerant evolved strain PKH5000 | n-Butanol | Transcriptome, phenotype microarray | Jeong et al. (2017) |
| E. coli | BW25113 and knockout strains | Isobutanol | Transcriptome | Brynildsen and Liao (2009) |
| E. coli | Isobutanol-tolerant mutant SA481 | Isobutanol | Genome | Atsumi et al. (2010) |
| E. coli | Isobutanol-tolerant clone G3.2, X3.5 | Isobutanol | Genome, transcriptome | Minty et al. (2011) |
| E. coli | CRP mutation library | Isobutanol | Transcriptome | Chong et al. (2014) |
| C. acetobutylicum | groESL overexpression strain | n-Butanol | Transcriptome | Tomas et al. (2004) |
| C. acetobutylicum | spo0A disrupt mutant | n-Butanol | Transcriptome | Tomas et al. (2003a) |
| C. acetobutylicum | groESL overexpression strain | n-Butanol | Transcriptome, metabolic flux | Tomas et al. (2003b) |
| C. acetobutylicum | Genomic library | n-Butanol | Enrichment | Borden and Papoutsakis (2007) |
| C. acetobutylicum | ATCC824 | n-Butanol, butyrate, acetate | Transcriptome | Alsaker et al. (2010) |
| C. acetobutylicum | Butanol-tolerant mutant Rh8 | n-Butanol | Proteome | Mao et al. (2010) |
| C. acetobutylicum | DSM1731 | n-Butanol | Proteome | Jia et al. (2012) |
| C. acetobutylicum | Butanol-tolerant mutant Rh8 | n-Butanol | Genome, proteome | Bao et al. (2014) |
| C. acetobutylicum | ATCC824 | n-Butanol | Metabolome | Wang et al. (2016) |
| C. acetobutylicum | Butanol-tolerant asporogenic strain JB200 | n-Butanol | Genome | Xu et al. (2017) |
| Synechocystis | sp. PCC 6803 | Ethanol | Proteome | Qiao et al. (2012) |
| Synechocystis | Δsll0794 | Ethanol | Proteome | Song et al. (2014) |
| Synechocystis | sp. PCC 6803 | Ethanol | Metabolome | Zhu et al. (2015) |
| Synechocystis | sp. PCC 6803 | Ethanol, n-butanol, hexane, salt | sRNA-seq | Pei et al. (2017) |
| Synechocystis | Recombinant strains UL 004 and UL 030 | Ethanol | Flow cytometry | Lopes da Silva et al. (2018) |
| Synechocystis | Tolerant mutant SY1043 | Isopropanol | Genome, single-cell screening | Hirokawa et al. (2018) |
| Synechocystis | sp. PCC 6803 | n-Butanol | Transcriptome | Anfelt et al. (2013) |
| Synechocystis | Evolved strain S1, S3, S4 | n-Butanol | Metabolome | Wang et al. (2014) |
| Synechocystis | ∆slr1037 mutant | n-Butanol | Metabolome | Niu et al. (2015) |
| Synechocystis | Evolved strain T(1)–T(4) | Isobutanol | Genome | Matsusako et al. (2017) |
Transcriptome analyses of E. coli under isobutanol stress revealed that cellular functions related to respiration, phosphate metabolism, and iron metabolism were perturbed (Brynildsen and Liao 2009). Further investigation via network component and knockout analyses illustrated that several transcription factors play important roles in the isobutanol response network. This study proposed that the isobutanol stress response is triggered by a malfunction of quinone (Brynildsen and Liao 2009). As another example, transcriptome and proteome analyses of E. coli exposed to n-butanol demonstrated that this stress activates several stress response machineries simultaneously, including cell envelope stress, oxidative stress, and acid stress responses. This stress also induces protein misfolding and activates efflux systems (Rutherford et al. 2010). These analyses allowed us to identify key genes involved in alleviating oxidative stress, protein misfolding, and other causes of growth defects. The genes and biological activities identified in these studies could be important assets for engineering alcohol-tolerant bacteria.
In E. coli, metabolomic analyses of its responses to ethanol, n-butanol, and isobutanol stresses revealed that several amino acids and osmoprotectants, such as isoleucine, valine, glycine, glutamate, and trehalose, are key metabolites that protect against these stresses (Wang et al. 2013). Several transcriptomic studies also demonstrated the relationship between alcohol stress and these low-molecular-weight compounds (Gonzalez et al. 2003; Horinouchi et al. 2010, 2017a; Swings et al. 2017). Although the roles of these compounds in the alcohol stress response remain unclear, they may mitigate the growth inhibition caused by alcohol stress. Further, alcohol can inhibit translation, and this inhibition can be mitigated by the addition of exogenous amino acids or deletion of repressors (Haft et al. 2014). Specifically, Haft et al. (2014) found that ethanol has detrimental effects on translational misreading, ribosome stalling, and the aberrant termination of polypeptide synthesis. They demonstrated that such effects on translation are caused by methionine depletion.
Transcriptomic analyses of C. acetobutylicum under n-butanol stress identified genes related to the n-butanol stress response (Tomas et al. 2003a, b, 2004). These studies identified important roles of the groESL chaperone system and the global regulator of sporulation spo0A. Further analyses suggested that the expression level of genes related to the functional categories of “glycolysis”, “amino acid biosynthesis and transport”, and “oxidative stress response” are changed by exposure to n-butanol stress (Alsaker et al. 2010). Although significant physiological differences exist between E. coli and C. acetobutylicum (such as cellular membrane structure, composition, and the ability to form spores), similar functional categories were noted (Alsaker et al. 2010). Although differences of cellular membrane structure strongly affect n-butanol tolerance, these similarities of functions response to n-butanol suggest that a common mechanism of alcohol toxicity exists between these species.
The alcohol stress responses of Synechocystis sp. have been analyzed using omics technologies since 2010 (summarized in Table 1). Several studies revealed that oxidative stress response is activated by alcohols. For example, transcriptomic analysis of Synechocystis spp. exposed to n-butanol demonstrated that oxidative stress response-related genes were upregulated (Anfelt et al. 2013). As another example, proteomic analysis of Synechocystis revealed that oxidative stress response is induced by ethanol (Qiao et al. 2012). Photosynthetic organisms may be more susceptible to the effects of alcohols because of the sensitivity to the redox state of key molecules such as plastoquinone and the intricate organization of the membrane-bound photosynthetic apparatus. Alleviating oxidative stress is a target for improving the alcohol tolerance of Synechocystis.
Adaptive laboratory evolution (ALE) of alcohol tolerance
Adaptive laboratory evolution is a powerful tool for analyzing phenotypic and genotypic changes during bacterial evolution (Dragosits and Mattanovich 2013; Winkler and Kao 2014). In this approach, cells are cultured under a selective environment for many generations, leading to adaptive evolution. Then, using omics technologies, we can obtain genome-wide information about adaptive phenotypic and genotypic changes resulting from the selective pressure. Advances in omics technologies, especially decreases in the cost of genome re-sequencing, have made ALE a standard approach for investigating and engineering desired phenotypes and analyzing alcohol tolerance in various microorganisms (summarized in Table 1).
In some cases, different studies identified different genes as contributing to alcohol tolerance even though the studies used the same stressor for ALE experiments. For example, Minty et al. (2011) identified mutations in mdh (malate dehydrogenase) and rph (defective ribonuclease PH) in E. coli evolved under isobutanol stress (Minty et al. 2011). However, neither change was found in a study of isobutanol-tolerant E. coli by Atsumi et al. (2010). Alternatively, mutations in tnaA (tryptophanase) and yhbJ (renamed as rapZ, RNase adaptor protein) identified by Atsumi et al. (2010) were not identified by Minty et al. (2011). These discordances may be caused by differences in experimental conditions, such as the parental strain, culture conditions, and selective pressure. Another possible cause is the diversity of the obtained alcohol-tolerant strains. Many different mutations are often acquired among the evolved strains obtained via multiple ALEs under the same experimental conditions. It is important to verify whether the identified mutations contribute to alcohol tolerance.
When bacterial cells become tolerant to one stress via ALE, they sometimes also become more tolerant to other stresses, a phenomenon called cross protection. However, they sometimes become more sensitive to other stresses, a phenomenon called collateral sensitivity. These scenarios provide valuable information regarding the mechanisms of stress tolerance. Notably, the existence of cross protection and collateral sensitivity among n-butanol and other abiotic stresses, such as acid stress, hyperosmotic stress, oxidative stress, and alkali stress, has been identified (Dragosits et al. 2013; Reyes et al. 2013; Horinouchi et al. 2017b). For example, an n-butanol-tolerant strain obtained by Dragosits et al. (2013) also exhibited tolerance to NaCl and low pH. This study found that mutations in iron-related genes represent a common genetic factor that drives bacterial tolerance across multiple stresses. Such cross-stress observations are important because they can provide insight into the pleiotropic effects of alcohol on cellular functions.
In addition to cross-stress behavior, the acquisition of stress tolerance in bacterial cells is sometimes accompanied by “fitness cost”, a reduction of fitness (growth or viability in this case) in the absence of stress. This phenomenon is well studied in the field of antibiotic resistance in bacteria (Lenski 1998; Andersson and Hughes 2010). Fitness in the absence of stress is important for biofuel production and alcohol tolerance. One possible approach to overcoming this problem is improving fitness in the culture condition (e.g., medium, temperature, scale of culture), which is not dependent on the presence or absence of alcohol stress. Several studies demonstrated the improvement the fitness in the absence of alcohol stress via ALE (Conrad et al. 2010; Jiang et al. 2012; Zhao et al. 2013).
Engineering alcohol tolerance
Based on information about target genes and metabolites obtained via omics analyses, it is possible to rationally engineer alcohol-tolerant bacterial strains. As described previously, several reports noted a relationship between alcohol tolerance and specific amino acids (Gonzalez et al. 2003; Horinouchi et al. 2010, 2017a; Wang et al. 2013; Haft et al. 2014). Based on these results, several groups investigated the biosynthesis or supplementation of these amino acids for improving alcohol tolerance (Gonzalez et al. 2003; Horinouchi et al. 2010, 2017a; Haft et al. 2014). In addition, it is possible to identify mutations that contribute to alcohol tolerance via genome re-sequencing analyses of tolerant strains obtained using ALE, and it is possible to evaluate the effects of specific mutations on alcohol tolerance using genome-editing technology (Pósfai et al. 1999; Wang et al. 2009; Jiang et al. 2013). Indeed, there are many examples in which alcohol tolerance has been improved by introducing mutations identified by ALE and genome re-sequencing into the genome (Atsumi et al. 2010; Minty et al. 2011; Reyes et al. 2012; Dragosits et al. 2013; Horinouchi et al. 2015, 2017a, b).
Another approach for improving alcohol tolerance is the heterologous expression of beneficial genes in a host strain. For example, overexpressing the GroESL chaperone from C. acetobutylicum (Abdelaal et al. 2015) in E. coli enhances alcohol tolerance, most likely by stabilizing or refolding proteins that are crucial for cell metabolism and survival. Overexpressing the phasin polyhydroxyalkanoate granule-associated protein (PhaP) from Azotobacter sp. strain FA8 (Mezzina et al. 2017) in E. coli also enhances alcohol tolerance PhaP from Azotobacter has chaperone activity and exerts a stress-alleviating effect in recombinant E. coli cells (de Almeida et al. 2011; Mezzina et al. 2015). In these approaches, beneficial genes associated with alcohol tolerance (e.g., GroESL or PhaP) were found in known alcohol-tolerant species such as C. acetobutylicum or Azotobacter spp. The advancement of genome sequencing technology will provide useful information about the genetic resources of various microbial species.
A tool called global transcription machinery engineering (gTME) was developed for improving cellular phenotypes (Alper and Stephanopoulos 2007). In this approach, random mutagenesis libraries of global transcription factors are generated by error-prone PCR to reprogram transcription and obtain various phenotypes. The mixture of libraries is cultured in the presence of a stressor to enrich for stress-tolerant mutants. Via this approach, alcohol tolerance was increased in E. coli by modifying the RNA polymerase sigma factor σ70 (Alper and Stephanopoulos 2007) and cAMP receptor protein (CRP) (Chong et al. 2013). Chong et al. (2014) performed transcriptomic analyses of CRP-engineered mutants and single-gene knockout experiments to characterize the functions of genes related to alcohol tolerance. They demonstrated that GadX (regulator of acid resistance system), HdeB (periplasmic acid stress chaperone), and several other genes were associated with isobutanol resistance (Chong et al. 2014). They also observed that the intracellular reactive oxygen species level was lower in the engineered mutants than in control strain when facing stress. These results indicated that the engineered mutants can withstand toxic isobutanol much better than the control strain.
Concluding remarks
The development of omics technologies has undoubtedly facilitated new insights into the mechanisms by which microorganisms tolerate alcohol. For example, recent studies using omics technologies demonstrated that biological functions and response networks related to intracellular redox states are involved in alcohol stress in several microorganisms. These biological functions could be important assets for engineering alcohol-tolerant bacteria. Although alcohol-tolerant strains have been engineered, the impact of alcohol stress at the whole-cell level is not fully understood. Such an understanding is complicated by the fact that various cellular functions are undoubtedly related to the toxicity of alcohols; likewise, controlling the biological functions related to alcohol tolerance will likely be complicated. We reason that valuable information will continue to be generated using omics technology. Although omics technologies provide new biological functions that may be related to alcohol tolerance, it is necessary to verify whether these biological functions contribute to alcohol tolerance.
Further, by combining omics approaches with ALE and gTME, we can expand our search for the phenotypes of alcohol tolerance. This will accelerate the research process by identifying novel genes, proteins, or biological functions related to alcohol tolerance. We expect that further developments in the omics space and novel methodologies will allow us to further characterize the mechanisms of alcohol tolerance.
Acknowledgements
This work was supported in part by Japan Society for the Promotion of Science KAKENHI Grants number JP17K15052 and JP18H04807.
Contributor Information
Takaaki Horinouchi, Phone: +81-6-6155-0456, Email: takaaki_horinouchi@riken.jp.
Chikara Furusawa, Phone: +81-6-6155-0489, Email: chikara.furusawa@riken.jp.
References
- Abdelaal AS, Ageez AM, Abd El-Hadi AEHA, Abdallah NA. Genetic improvement of n-butanol tolerance in Escherichia coli by heterologous overexpression of groESL operon from Clostridium acetobutylicum. 3 Biotech. 2015;5:401–410. doi: 10.1007/s13205-014-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ades SE. Control of the alternative sigma factor σE in Escherichia coli. Curr Opin Microbiol. 2004;7:157–162. doi: 10.1016/j.mib.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Alper H, Stephanopoulos G. Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Alsaker KV, Paredes C, Papoutsakis ET. Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol Bioeng. 2010;105:1131–1147. doi: 10.1002/bit.22628. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reversein resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- Anfelt J, Hallström B, Nielsen J, et al. Using transcriptomics to improve butanol tolerance of Synechocystis sp. strain PCC 6803. Appl Environ Microbiol. 2013;79:7419–7427. doi: 10.1128/AEM.02694-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Wu T-Y, Machado IMP, et al. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol. 2010;6:449. doi: 10.1038/msb.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao G, Dong H, Zhu Y, et al. Comparative genomic and proteomic analyses of Clostridium acetobutylicum Rh8 and its parent strain DSM 1731 revealed new understandings on butanol tolerance. Biochem Biophys Res Commun. 2014;450:1612–1618. doi: 10.1016/j.bbrc.2014.07.052. [DOI] [PubMed] [Google Scholar]
- Belkin S, Smulski DR, Vollmer AC, et al. Oxidative stress detection with Escherichia coli harboring a katG: lux fusion. Appl Environ Microbiol. 1996;62:2252–2256. doi: 10.1128/aem.62.7.2252-2256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Carty CE, Ingram LO. Alcohol-induced changes in the phospholipid molecular species of Escherichia coli. J Bacteriol. 1980;142:1040–1044. doi: 10.1128/jb.142.3.1040-1044.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden JR, Papoutsakis ET. Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl Environ Microbiol. 2007;73:3061–3068. doi: 10.1128/AEM.02296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette JL, Russel M, Weiner L, Model P. Phage shock protein, a stress protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:862–866. doi: 10.1073/pnas.87.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsen MP, Liao JC. An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol Syst Biol. 2009;5:277. doi: 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury-Moné S, Nomane Y, Reymond N, et al. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009;5:e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Wang J, Zeng L, et al. Significant rewiring of the transcriptome and proteome of an Escherichia coli strain harboring a tailored exogenous global regulator irre. PLoS One. 2012;7:e371126. doi: 10.1371/journal.pone.0037126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Y, Chen TH, et al. Identification and manipulation of a novel locus to improve cell tolerance to short-chain alcohols in Escherichia coli. J Ind Microbiol Biotechnol. 2018;45:589–598. doi: 10.1007/s10295-017-1996-y. [DOI] [PubMed] [Google Scholar]
- Chong H, Huang L, Yeow J, et al. Improving ethanol tolerance of Escherichia coli by rewiring Its global regulator cAMP receptor protein (CRP) PLoS One. 2013;8:e57628. doi: 10.1371/journal.pone.0057628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong H, Geng H, Zhang H, et al. Enhancing E. coli isobutanol tolerance through engineering its global transcription factor cAMP receptor protein (CRP) Biotechnol Bioeng. 2014;111:700–708. doi: 10.1002/bit.25134. [DOI] [PubMed] [Google Scholar]
- Clomburg JM, Gonzalez R. Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl Microbiol Biotechnol. 2010;86:419–434. doi: 10.1007/s00253-010-2446-1. [DOI] [PubMed] [Google Scholar]
- Conrad TM, Frazier M, Joyce AR, et al. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci USA. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida A, Catone MV, Rhodius VA, et al. Unexpected stress-reducing effect of PhaP, a poly(3-hydroxybutyrate) granule-associated protein, in Escherichia coli. Appl Environ Microbiol. 2011;77:6622–6629. doi: 10.1128/AEM.05469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits M, Mattanovich D. Adaptive laboratory evolution—principles and applications for biotechnology. Microb Cell Fact. 2013;12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits M, Mozhayskiy V, Quinones-Soto S, et al. Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol Syst Biol. 2013;9:643. doi: 10.1038/msb.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop MJ, Dossani ZY, Szmidt HL, et al. Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol. 2011;7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaida SM, Sandoval NR, Nicolaou SA, et al. Expression of heterologous sigma factors enables functional screening of metagenomic and heterologous genomic libraries. Nat Commun. 2015;6:7045. doi: 10.1038/ncomms8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe TR, Yukawa H. Common solvent toxicity: autoxidation of respiratory redox-cyclers enforced by membrane derangement. Zeitschrift fur Naturforsch - Sect C J Biosci. 2001;56:483–491. doi: 10.1515/znc-2001-7-801. [DOI] [PubMed] [Google Scholar]
- Gonzalez R, Tao H, Purvis JE, et al. Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: comparison of KO11 (parent) to LY01 (resistant mutant) Biotechnol Prog. 2003;19:612–623. doi: 10.1021/bp025658q. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Bennett BD, Amini S, et al. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol. 2010;6:378. doi: 10.1038/msb.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandvalet C, Assad-García JS, Chu-Ky S, et al. Changes in membrane lipid composition in ethanol- and acid-adapted Oenococcus oeni cells: characterization of the cfa gene by heterologous complementation. Microbiology. 2008;154:2611–2619. doi: 10.1099/mic.0.2007/016238-0. [DOI] [PubMed] [Google Scholar]
- Gronenberg LS, Marcheschi RJ, Liao JC. Next generation biofuel engineering in prokaryotes. Curr Opin Chem Biol. 2013;17:462–471. doi: 10.1016/j.cbpa.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft RJF, Keating DH, Schwaegler T, et al. Correcting direct effects of ethanol on translation and transcription machinery confers ethanol tolerance in bacteria. Proc Natl Acad Sci. 2014;111:E2576–E2585. doi: 10.1073/pnas.1401853111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai T, Atsumi S, Liao JC. Engineered synthetic pathway for isopropanol production in Escherichia coli. Appl Environ Microbiol. 2007;73:7814–7818. doi: 10.1128/AEM.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann M, Fayolle F, Marchal R, et al. Isolation and characterization of butanol-resistant mutants of Clostridium acetobutylicum. Appl Environ Microbiol. 1985;50:1238–1243. doi: 10.1128/aem.50.5.1238-1243.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa Y, Kanesaki Y, Arai S, et al. Mutations responsible for alcohol tolerance in the mutant of Synechococcus elongatus PCC 7942 (SY1043) obtained by single-cell screening system. J Biosci Bioeng. 2018;125:572–577. doi: 10.1016/j.jbiosc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Horinouchi T, Tamaoka K, Furusawa C, et al. Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genom. 2010;11:579. doi: 10.1186/1471-2164-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi T, Suzuki S, Hirasawa T, et al. Phenotypic convergence in bacterial adaptive evolution to ethanol stress. BMC Evol Biol. 2015;15:180. doi: 10.1186/s12862-015-0454-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi T, Sakai A, Kotani H, et al. Improvement of isopropanol tolerance of Escherichia coli using adaptive laboratory evolution and omics technologies. J Biotechnol. 2017;255:47–56. doi: 10.1016/j.jbiotec.2017.06.408. [DOI] [PubMed] [Google Scholar]
- Horinouchi T, Suzuki S, Kotani H, et al. Prediction of cross-resistance and collateral sensitivity by gene expression profiles and genomic mutations. Sci Rep. 2017;7:14009. doi: 10.1038/s41598-017-14335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram LO. Adaptation of membrane lipids to alcohols. J Bacteriol. 1976;125:670–678. doi: 10.1128/jb.125.2.670-678.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Lee SW, Kim SH, et al. Global functional analysis of butanol-sensitive Escherichia coli and its evolved butanol-tolerant strain. J Microbiol Biotechnol. 2017;27:1171–1179. doi: 10.4014/jmb.1702.02021. [DOI] [PubMed] [Google Scholar]
- Jia K, Zhang Y, Li Y. Identification and characterization of two functionally unknown genes involved in butanol tolerance of clostridium acetobutylicum. PLoS One. 2012;7:e388115. doi: 10.1371/journal.pone.0038815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Li S, Hu Y, et al. Adaptive evolution for fast growth on glucose and the effects on the regulation of glucose transport system in Clostridium tyrobutyricum. Biotechnol Bioeng. 2012;109:708–718. doi: 10.1002/bit.23346. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Van Der Westhuizen A, Long S. Solvent production and morphological changes in Clostridium acetobutylicum. Appl Environ Microbiol. 1982;43:1434–1439. doi: 10.1128/aem.43.6.1434-1439.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasling JD, Chou H. Metabolic engineering delivers next-generation biofuels. Nat Biotechnol. 2008;26:298–299. doi: 10.1038/ng.85. [DOI] [PubMed] [Google Scholar]
- Koppolu V, Vasigala VK. Role of Escherichia coli in biofuel production. Microbiol Insights. 2016;9:MBI.S10878. doi: 10.4137/MBI.S10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau MW, Gunawan C, Balan V, Dale BE. Comparing the fermentation performance of Escherichia coli KO11, Saccharomyces cerevisiae 424A(LNH-ST) and Zymomonas mobilis AX101 forcellulosic ethanol production. Biotechnol Biofuels. 2010;3:11–20. doi: 10.1186/1754-6834-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NS, Matsui M, Abdullah AAA. Cyanobacteria: photoautotrophic microbial factories for the sustainable synthesis of industrial products. Biomed Res Int. 2015;2015:1–9. doi: 10.1155/2015/754934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Yang KS, Jang SA, et al. Engineering butanol-tolerance in Escherichia coli with artificial transcription factor libraries. Biotechnol Bioeng. 2011;108:742–749. doi: 10.1002/bit.22989. [DOI] [PubMed] [Google Scholar]
- Lenski RE. Bacterial evolution and the cost of antibiotic resistance. Int Microbiol. 1998;1:265–270. doi: 10.2436/im.v1i4.27. [DOI] [PubMed] [Google Scholar]
- Lepage C, Fayolle F, Hermann M, Vandecasteele J-P. Changes in membrane lipid composition of Clostridium acetobutylicum during acetone–butanol fermentation: effects of solvents, growth temperature and pH. Microbiology. 1987;133:103–110. doi: 10.1099/00221287-133-1-103. [DOI] [Google Scholar]
- Lopes da Silva T, Passarinho PC, Galriça R, et al. Evaluation of the ethanol tolerance for wild and mutant Synechocystis strains by flow cytometry. Biotechnol Rep. 2018;17:137–147. doi: 10.1016/j.btre.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LH, Seo PS, Seo JW, et al. Improved ethanol tolerance in Escherichia coli by changing the cellular fatty acids composition through genetic manipulation. Biotechnol Lett. 2009;31:1867–1871. doi: 10.1007/S10529-009-0092-4. [DOI] [PubMed] [Google Scholar]
- Lupino KM, Romano KA, Simons MJ, et al. A recurrent silent mutation implicates fecA in ethanol tolerance by Escherichia coli. BMC Microbiol. 2018;18:36. doi: 10.1186/s12866-018-1180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S, Luo Y, Zhang T, et al. Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield. J Proteome Res. 2010;9:3046–3061. doi: 10.1021/pr9012078. [DOI] [PubMed] [Google Scholar]
- Matsusako T, Toya Y, Yoshikawa K, Shimizu H. Identification of alcohol stress tolerance genes of Synechocystis sp. PCC 6803 using adaptive laboratory evolution. Biotechnol Biofuels. 2017;10:307. doi: 10.1186/s13068-017-0996-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzina MP, Wetzler DE, de Almeida A, et al. A phasin with extra talents: a polyhydroxyalkanoate granule-associated protein has chaperone activity. Environ Microbiol. 2015;17:1765–1776. doi: 10.1111/1462-2920.12636. [DOI] [PubMed] [Google Scholar]
- Mezzina MP, Álvarez DS, Egoburo DE, et al. A new player in the biorefineries field: phasin PhaP enhances tolerance to solvents and boosts ethanol and 1,3-propanediol synthesis in Escherichia coli. Appl Environ Microbiol. 2017 doi: 10.1128/AEM.00662-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty JJ, Lesnefsky A, Lin F, et al. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb Cell Fact. 2011;10:18. doi: 10.1186/1475-2859-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, VanBogelen RA, Vaughn V. The genetics and regulation of heat-shock proteins. Annu Rev Genet. 1984;18:295–329. doi: 10.1146/annurev.ge.18.120184.001455. [DOI] [PubMed] [Google Scholar]
- Nicolaou SA, Gaida SM, Papoutsakis ET. Exploring the combinatorial genomic space in Escherichia coli for ethanol tolerance. Biotechnol J. 2012;7:1337–1345. doi: 10.1002/biot.201200227. [DOI] [PubMed] [Google Scholar]
- Niu X, Zhu Y, Pei G, et al. Elucidating butanol tolerance mediated by a response regulator Sll0039 in Synechocystis sp. PCC 6803 using a metabolomic approach. Appl Microbiol Biotechnol. 2015;99:1845–1857. doi: 10.1007/s00253-015-6374-y. [DOI] [PubMed] [Google Scholar]
- Nozzi NE, Oliver JWK, Atsumi S. Cyanobacteria as a platform for biofuel production. Front Bioeng Biotechnol. 2013;1:7. doi: 10.3389/fbioe.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei G, Sun T, Chen S, et al. Systematic and functional identification of small non-coding RNAs associated with exogenous biofuel stress in cyanobacterium Synechocystis sp. PCC 6803. Biotechnol Biofuels. 2017;10:57. doi: 10.1186/s13068-017-0743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta-Yahya PP, Keasling JD. Advanced biofuel production in microbes. Biotechnol J. 2010;5:147–162. doi: 10.1002/biot.200900220. [DOI] [PubMed] [Google Scholar]
- Pósfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J, Wang J, Chen L, et al. Quantitative iTRAQ LC-MS/MS proteomics reveals metabolic responses to biofuel ethanol in cyanobacterial Synechocystis sp. PCC 6803. J Proteome Res. 2012;11:5286–5300. doi: 10.1021/pr300504w. [DOI] [PubMed] [Google Scholar]
- Rau MH, Calero P, Lennen RM, et al. Genome-wide Escherichia coli stress response and improved tolerance towards industrially relevant chemicals. Microb Cell Fact. 2016;15:176. doi: 10.1186/s12934-016-0577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes LH, Almario MP, Kao KC. Genomic library screens for genes involved in n-butanol tolerance in Escherichia coli. PLoS One. 2011;6:e17678. doi: 10.1371/journal.pone.0017678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes LH, Almario MP, Winkler J, et al. Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metab Eng. 2012;14:579–590. doi: 10.1016/j.ymben.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Reyes LH, Abdelaal AS, Kao KC. Genetic determinants for n-butanol tolerance in evolved Escherichia coli mutants: cross adaptation and antagonistic pleiotropy between n-butanol and other stressors. Appl Environ Microbiol. 2013;79:5313–5320. doi: 10.1128/AEM.01703-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbold K, Van Buijsen H, Overkamp K, et al. Microbial production host selection for converting second generation feedstocks into bioproducts. Microb Cell Fact. 2009;8:64–64. doi: 10.1186/1475-2859-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford BJ, Dahl RH, Price RE, et al. Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl Environ Microbiol. 2010;76:1935–1945. doi: 10.1128/AEM.02323-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si HM, Zhang F, Wu AN, et al. DNA microarray of global transcription factor mutant reveals membrane-related proteins involved in n-butanol tolerance in Escherichia coli. Biotechnol Biofuels. 2016;9:114. doi: 10.1186/s13068-016-0527-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Chen L, Wang J, et al. A transcriptional regulator sll0794 regulates tolerance to biofuel ethanol in photosynthetic Synechocystis sp. PCC 6803. Mol Cell Proteom. 2014;13:3519–3532. doi: 10.1074/mcp.M113.035675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos G. Metabolic engineering: enabling technology for biofuels production. Metab Eng. 2008;10:293–294. doi: 10.1016/j.ymben.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Swings T, Weytjens B, Schalck T, et al. Network-based identification of adaptive pathways in evolved ethanol-tolerant bacterial populations. Mol Biol Evol. 2017;34:2927–2943. doi: 10.1093/molbev/msx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas CA, Alsaker KV, Bonarius HPJ, et al. DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J Bacteriol. 2003;185:4539–4547. doi: 10.1128/JB.185.15.4539-4547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas CA, Welker NE, Papoutsakis ET. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell’s transcriptional program. Appl Environ Microbiol. 2003;69:4951–4965. doi: 10.1128/AEM.69.8.4951-4965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas CA, Beamish J, Papoutsakis ET. Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J Bacteriol. 2004;186:2006–2018. doi: 10.1128/JB.186.7.2006-2018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen L, Tian X, et al. Global metabolomic and network analysis of Escherichia coli responses to exogenous biofuels. J Proteome Res. 2013;12:5302–5312. doi: 10.1021/pr400640u. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shi M, Niu X, et al. Metabolomic basis of laboratory evolution of butanol tolerance in photosynthetic Synechocystis sp. PCC 6803. Microb Cell Fact. 2014;13:151. doi: 10.1186/s12934-014-0151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YF, Tian J, Ji ZH, et al. Intracellular metabolic changes of Clostridium acetobutylicum and promotion to butanol tolerance during biobutanol fermentation. Int J Biochem Cell Biol. 2016;78:297–306. doi: 10.1016/j.biocel.2016.07.031. [DOI] [PubMed] [Google Scholar]
- Winkler JD, Kao KC. Recent advances in the evolutionary engineering of industrial biocatalysts. Genomics. 2014;104:406–411. doi: 10.1016/j.ygeno.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Woodruff LBA, Pandhal J, Ow SY, et al. Genome-scale identification and characterization of ethanol tolerance genes in Escherichia coli. Metab Eng. 2013;15:124–133. doi: 10.1016/j.ymben.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Xu M, Zhao J, Yu L, Yang ST. Comparative genomic analysis of Clostridium acetobutylicum for understanding the mutations contributing to enhanced butanol tolerance and production. J Biotechnol. 2017;263:36–44. doi: 10.1016/j.jbiotec.2017.10.010. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Suda M, Niimi S, et al. Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol Bioeng. 2013;110:2938–2948. doi: 10.1002/bit.24961. [DOI] [PubMed] [Google Scholar]
- Zhao J, Xu L, Wang Y, et al. Homofermentative production of optically pure L-lactic acid from xylose by genetically engineered Escherichia coli B. Microb Cell Fact. 2013;12:1. doi: 10.1186/1475-2859-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pei G, Niu X, et al. Metabolomic analysis reveals functional overlapping of three signal transduction proteins in regulating ethanol tolerance in cyanobacterium Synechocystis sp. PCC 6803. Mol Biosyst. 2015;11:770–782. doi: 10.1039/C4MB00651H. [DOI] [PubMed] [Google Scholar]
- Zingaro KA, Nicolaou SA, Yuan Y, Papoutsakis ET. Exploring the heterologous genomic space for building, stepwise, complex, multicomponent tolerance to toxic chemicals. ACS Synth Biol. 2014;3:476–486. doi: 10.1021/sb400156v. [DOI] [PubMed] [Google Scholar]