Abstract
The nitric oxide (NO) secreted by vascular endothelium is required for the maintenance of cardiovascular homeostasis. Diminished release of NO generated by endothelial NO synthase contributes to endothelial dysfunction. Hypoxia and ischemia reduce endothelial eNOS expression via posttranscriptional mechanisms that result in NOS3 transcript destabilization. Here, we examine whether microRNAs contribute to this mechanism. We followed the kinetics of hypoxia-induced changes in NOS3 mRNA and eNOS protein levels in primary human umbilical vein endothelial cells (HUVECs). Utilizing in silico predictive protocols to identify potential miRNAs that regulate eNOS expression, we identified miR-200b as a candidate. We established the functional miR-200b target sequence within the NOS3 3′UTR, and demonstrated that manipulation of the miRNA levels during hypoxia using miR-200b mimics and antagomirs regulates eNOS levels, and established that miR-200b physiologically limits eNOS expression during hypoxia. Furthermore, we demonstrated that the specific ablation of the hypoxic induction of miR-200b in HUVECs restored eNOS-driven hypoxic NO release to the normoxic levels. To determine whether miR-200b might be the only miRNA that had this effect, we utilized Next Generation Sequencing (NGS) to follow hypoxia-induced changes in the miRNA levels in HUVECS and found 83 novel hypoxamiRs, with two candidate miRNAs besides miR-200b that could potentially influence eNOS levels. Taken together, the data establish miR-200b-eNOS regulation as a first hypoxamiR-based mechanism that limits NO bioavailability during hypoxia in endothelial cells, and show that hypoxamiRs could become useful therapeutic targets for cardiovascular diseases and other hypoxic-related diseases including various types of cancer.
Electronic supplementary material
The online version of this article (10.1007/s10456-018-9620-y) contains supplementary material, which is available to authorized users.
Keywords: eNOS, NOS3, MicroRNA 200b, Hsa-miR-200b-3p, Hypoxia, Nitric oxide bioavailability, Hypoxia-related diseases
Introduction
The vascular endothelium synthesizes and secrets a broad spectrum of substances including nitric oxide (NO) for the regulation of vascular tone and structure. This mediator is generated by endothelial NO synthase (eNOS encoded by NOS3 gene) in endothelial cells (ECs) and diffuses to the surrounding tissues in order to relax smooth muscle cells [1]. NO also prevents leukocyte adhesion and migration into the arterial wall, muscle cell proliferation, platelet adhesion and aggregation, and adhesion molecule expression [1]. Hence, maintaining the proper NO levels is cardioprotective.
Impaired eNOS activity contributes to endothelial dysfunction and is involved in the pathomechanisms of several cardiovascular diseases including atherosclerosis and hypertension [1, 2]. Although calcium/calmodulin binding and phosphorylation by serine/threonine-specific kinase (Akt) [3] are the major mechanisms governing eNOS activity, numerous pathological factors including hypoxia/ischemia modulate eNOS expression at both the transcriptional [4, 5] and posttranscriptional levels [5–7]. Although during the early stages of hypoxia, eNOS levels were shown to be induced transcriptionally due to hypoxia-inducible factor (HIF) activity [8, 9], numerous reports have linked the loss of NO bioavailability under prolonged hypoxia and ischemia to the reduction of endothelial eNOS levels [10, 11].
To date, an eNOS-specific posttranscriptional mechanism was proposed to be responsible for the hypoxic decline in the NOS3 mRNA half-life [12]. Under normoxic conditions, NOS3 mRNA is stabilized by heterogeneous nuclear ribonucleoprotein E1 (hnRNPE1) complex, while during hypoxia this interaction is lost, making the NOS3 transcript prone to destabilization by its natural antisense sONE RNA (also known as ATG9B, NOS3AS, and APG9L2). sONE binds to both the NOS3 coding sequence and the 3′UTR [11–13]. Interestingly, however, impairing the microRNA function via Dicer silencing resulted in eNOS upregulation during normoxia, and thus suggested that these non-coding RNAs (ncRNAs) can provide a general posttranscriptional epigenetic mechanism for the regulation of eNOS expression [14]. Indeed, miR-155 [15], miR-214 [16], and miR-24 [17] were reported to directly bind to NOS3 mRNA during normoxia, and a miR-200 family member, miR-200c, was shown to destabilize NOS3 transcript in response to oxidative stress [18].
Impairment of hnRNPE1 binding to NOS3 mRNA results in an increased susceptibility to downregulation by miR-765 in normoxic human umbilical vein endothelial cells (HUVECs) [12]; however, during hypoxia, access to this miRNA target site (TS) is impeded by sONE, and the physiological relevance of this interaction requires further clarification. Nevertheless, since sONE antisense only partially covers NOS3 3′UTR sequence [11], the hypoxic hnRNPE1 dissociation could reveal other miRNAs TSs at this 3′UTR, thus making this transcript sensitive for miRNA-based destabilization. Furthermore, hypoxia has been shown to specifically affect expression profiles of a large number of miRNAs [19].
Hence, it is very plausible that during hypoxia, specific miRNAs could directly reduce eNOS levels and consequently take a role in the physiological modulation of the bioavailability of NO [12], however, no such miRNA has been identified to date. In the present study, we identified a miR-200 family member, miR-200b, as a novel direct negative regulator of eNOS expression in human ECs in response to hypoxia. We also show that the specific ablation of hypoxic induction of miR-200b in HUVECs results not only in NOS3 mRNA and eNOS protein rescue, but also in an increase in hypoxic NO release in situ.
Methods
Cell culture
Primary human umbilical vein endothelial cells pooled from ten individual donors(HUVECs) were purchased from Cellworks (Caltag Medsystems Ltd, UK) and cultured in EGM-2 Bulletkit Medium (Lonza). All experiments were conducted between passage 2 and 6 at a confluence of 70–80%. Human embryonic kidney 293 cells (HEK-293) were obtained from ATCC (CRL-1573) and cultured in Minimal Essential Media (MEM) supplemented with 10% Fetal Bovine Serum (FBS), 2 mM l-glutamine (Sigma-Aldrich), and antibiotics (100 units/mL of penicillin, 100 µg/mL of streptomycin; Sigma-Aldrich).
Induction of hypoxia
Hypoxia was induced in a CO2/O2 incubator/chamber for hypoxia research (Invivo2 Baker Ruskin). Briefly, cells were cultured in 35 mm dishes at 0.9% O2 for the time periods specified. Control cells were maintained in normoxia in a CO2/O2 incubator (Binder).
miRNA analog transfections
Cells were seeded onto 6-well plates or 35 mm dishes and transfected at 70–80% confluence with Lipofectamine RNAiMax (Thermo Fisher Scientific) according to the manufacturer’s protocol. mirVana miRNA mimics and mirVana miRNA Inhibitors (Thermo Fisher Scientific) were used at final concentrations of 20 and 50 nM, respectively. mirVana mimics and inhibitors used in this study: miR-200b mimic (Assay ID: MC10492) and inhibitor (Assay ID: MH10492). In all experiments, cel-miR-67 was used as a scramble control since it has no homology to any known mammalian miRNA (Assay ID: MC22484) [20]. The degree of miRNA overexpression or knockdown was determined by qRT-PCR. Following the transfection, cells were cultured for 48 h prior to analysis.
Isolation of RNA and microRNA
Total RNA containing the microRNA fraction was isolated using miRNeasy kit (Qiagen). RNA concentrations were calculated based on the absorbance at 260 nm. RNA samples were stored at -70 °C until use.
Measurement of mRNA and miRNA levels using quantitative real-time PCR (qRT-PCR)
We used TaqMan One-Step RT-PCR Master Mix Reagents (Applied Biosystems) as described previously [21, 22] using the manufacturer’s protocol. The relative expressions were calculated using the method [23], with TATA-binding protein (TBP) and RNU44 genes as reference genes for the mRNA and miRNA, respectively. TaqMan probes ids used were NOS3 (Assay ID: Hs00176166_m1); TBP (Hs00427620_m1); hsa-miR-200b (Assay ID: 002251); hsa-miR-200c (Assay ID: 002300); hsa-miR-429 (Assay ID: 001024); hsa-miR-210 (Assay ID: 000512) and RNU44 (Assay ID: 001094).
Western Blots
Cells were lysed in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris–HCl, pH 8.0) supplemented with protease inhibitor [Complete Mini (Roche)] on ice for 15 min. The insoluble material was removed by centrifugation at 15,000×g for 15 min. Protein concentrations were determined by BioRad™ Protein Assay using bovine serum albumin (BSA) as a standard. Following the normalization of protein concentrations, lysates were mixed with an equal volume of 2× Laemmli sample buffer and incubated for 5 min at 95 °C prior to separation by SDS PAGE on stain-free TGX gradient gels (BioRad). Following SDS–PAGE, the proteins were transferred to polyvinylidene fluoride membranes (300 mA for 180 min at 4 °C). The membranes were then blocked with BSA (Sigma-Aldrich) dissolved in PBS/Tween-20 (3% BSA, 0.5% Tween-20 for 1–2 h), followed by immunoblotting with the primary antibody: anti-eNOS (1:1500, 610,297; BD Biosciences); anti-β-Actin (1:1000, ab1801; Abcam). After the washing steps, the membranes were incubated with goat anti-rabbit IgG (H+L chains) or with goat anti-mouse IgG (H+L) HRP-conjugated secondary antibodies (BioRad) and detected using ECL (Amresco). Densitometry was performed using Image Lab software v. 4.1 (BioRad).
Luciferase reporter assays
A human 3′UTR NOS3 firefly luciferase reporter construct (Vn) (HmiT088415-MT06) and its control vector (Vc) (CmiT000001-MT06) were purchased from GeneCopoeia. The reporter vector bearing the mutated NOS3 3′UTR (Vmut) was obtained by site-directed mutagenesis. Briefly, the template DNA (wild-type (Vn) vector) was amplified with mutation-introducing primers: Fwd: GTCTAATCTCTAAATCAgtcgacTATTATTGAAGATTTACC and Rev: GGTAAATCTTCAATAATAgtcgacTGATTTAGAGATTAGAC. The template DNA was then removed by digestion with DpnI, and the remaining mutated DNA was transformed into DH5α E.coli competent cells. Next, the designed Vmut vector was isolated from selected clones using GeneJET Plasmid Maxiprep Kit (Thermo Fisher Scientific). The correct mutagenesis clone was confirmed with Sanger sequencing. To test the posttranscriptional activity of the human NOS3 3′UTR regions, HEK293 cells were transfected with the constructs described above or with control plasmid. Twenty-four hours before the transfections, cells were seeded onto 24-well plates at ~80% confluency and then transfected using Lipofectamine 2000 (Thermo Fisher Scientific). Each well received 200 ng of total plasmid DNA and miR-200b/miR-200c mimics and or the cel-miR-67 scramble control at final concentration of 10 nM. 48 h after transfection, the cells were lysed using luciferase assay lysis buffer (Promega) and firefly/Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay (Promega) according to the manufacturer’s protocol. Results were plotted as a relative decrease in arbitrary light units compared with control cells.
NO measurements in cell culture
The concentration of NO was measured with an electrochemical microbiosensor. The microsensor was prepared as described previously [24]. To analyze the NO levels, the three-electrode system was used that consists of the sensor-working electrode, a platinum wire (0.1 mm) counter electrode, and a standard calomel reference electrode. The NO concentration was proportional to the current measured by a porphyrinic sensor in amperometric mode (EG&G PAR model 283 Potentiostat/Galvanostat) at constant potential of 0.65 V. A linear calibration curve was constructed from 500 nmol/L to 5 µmol/L before the measurements with aliquots of the NO standard solutions. eNOS was stimulated with 1 µmol/L of calcium ionophore (CaI; A23187). NO was measured as an increase of the current from its background level in the presence or absence of 300 µmol/L L-NAME (eNOS inhibitor) [24–26].
Next generation sequencing analyses of miRNAs
HUVECs at passage 3 were incubated under normoxic or hypoxic conditions for 16 h, and subsequently used for the RNA isolation and analyses. The induction of hypoxia was verified by accessing miR-210 levels prior to further analysis. MiRNA sequencing libraries were prepared using QIAseq miRNA library kit (Qiagen) following the manufacturer’s instructions and followed by sequencing on an Illumina NextSeq instrument. Using Qiagen’s Gene Globe Software, sequencing reads were aligned to the human reference genome assembly (hg19) followed by transcript assembly and estimation of the relative abundances. The analyses of the differential expression of small RNAs between the control (normoxia) and the experimental sample (16 h hypoxia) were performed with geNorm [27] the Gene Globe Software. Two independent pools of primary HUVECs were used in Next Generation Sequencing Experiments from two different laboratories: the UAB Heflin Center at UAB and the Medical University of Gdansk, Poland). The significant changes in miRNA levels (twofold changes), p value < 0.005) were only considered significant if they were seen in both experiments.
Statistical analysis
Results were expressed as mean ± standard deviation. Statistical significance was determined using the Student’s t test, with p < 0.05 considered significant. The correlation was accessed via Pearson product-moment correlation method.
Results
The hypoxic reduction in eNOS levels negatively correlates with miR-200b expression
Although previous studies have reported that eNOS protein and mRNA levels are reduced during hypoxia in human ECs [11, 13, 28], our goal was to understand the kinetics of hypoxia-induced changes in eNOS levels. To accomplish this, we performed time-course studies during hypoxia in HUVECs.
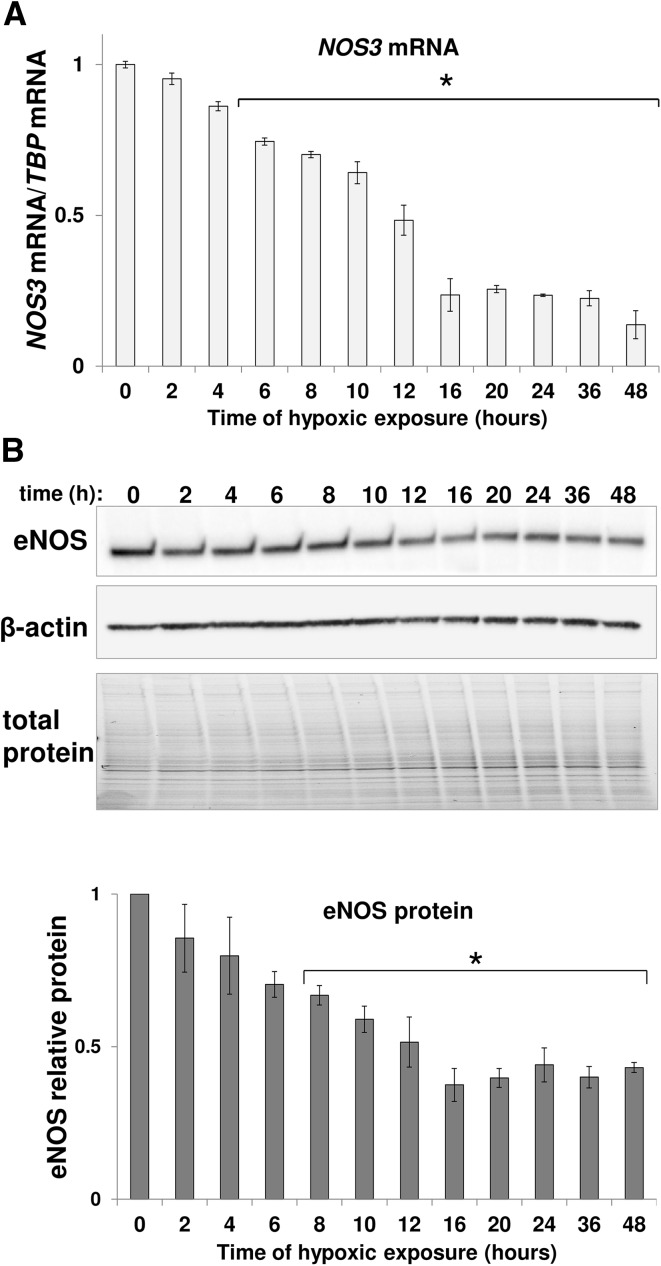
This analysis indicated that NOS3 mRNA is gradually reduced in response to hypoxia, as shown in Fig. 1a, reaching about 50 and 20% of its initial level after 12 and 24 h, respectively. The changes in NOS3 mRNA correlated well (correlation coefficient 0.965; p value 2.2 E-14) with the eNOS protein changes (Fig. 1b). During the early stages of hypoxia, the eNOS protein levels gradually decrease for the first 16 h, and then remain essentially unchanged up to 48 h (Fig. 1b).
Fig. 1.
Hypoxia reduces eNOS expression in HUVECs. HUVECs were exposed to hypoxia for the time periods specified and total RNA and protein lysates were collected. a NOS3 mRNA levels were quantified by qRT-PCR and normalized to TBP mRNA level. Data represent the mean ± SD of two independent experiments. b The corresponding changes in eNOS protein levels were evaluated by western blot normalized to β-Actin and total protein levels and related to normoxic control. *p < 0.05 was considered significant
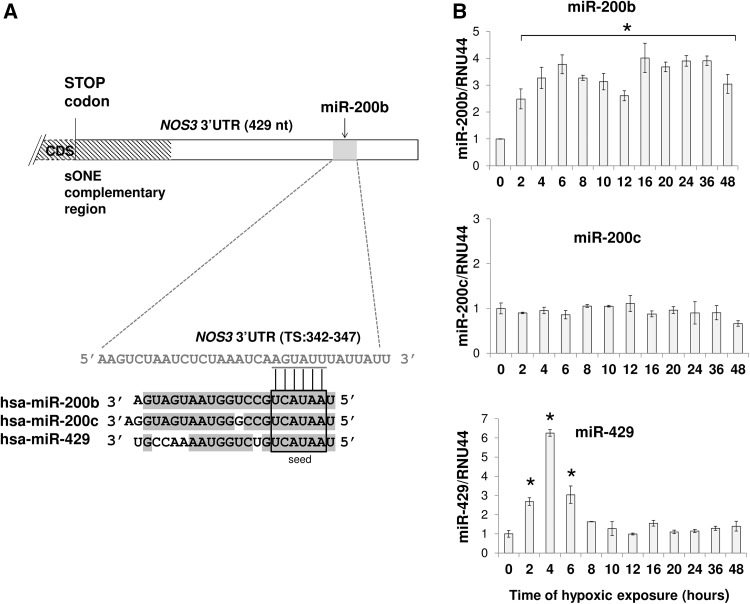
To test the hypothesis that hypoxia-induced miRNAs contribute to eNOS downregulation, we analyzed the NOS3 3′UTR sequence for potential hypoxamiR’s target sites using the miRANDA and TargetScan algorithms [29, 30]. Using this approach, we identified a potential 6mer target site for miR-429/200b/200c at position 342–347 from the stop codon in the 3′UTR of NOS3 mRNA (Fig. 2a).
Fig. 2.
miR-200b and miR-429 are upregulated in response to hypoxia. a The schematic representation of the interaction of the miR-200b/200c/429 seed sequence with the predicted target sequence (TS) in the NOS3 3′UTR sequence. As indicated by shading, the miR-200b, miR-200c, and miR-429 share high sequence homology. b HUVECs were exposed to hypoxia for the times indicated and total RNA enriched in miRNAs was isolated. miR-200b, mir-200c, and miR-429 levels were quantified by qPCR and normalized to RNU44. Data represent the mean ± SD of three independent experiments. *p < 0.05 was considered significant
Although the expression of these miRNAs was previously reported to be hypoxia-dependent in ECs [22, 31, 32], we verified their expression profiles during hypoxia in HUVECs. As shown in Fig. 2, miR-200b was induced up to fourfold and remained elevated throughout the 48-h test period, while miR-429 was induced only during acute hypoxia (2–6 h), and miR-200c was not elevated at all and therefore is not involved in eNOS regulation during hypoxia. More importantly, the miR-200b hypoxic expression profile correlated negatively with the corresponding changes in eNOS protein and NOS3 mRNA (correlation coefficients − 0.743 and − 0.639; p values of 0.000032 and 0.00077 for protein and mRNA, respectively), whereas no similar correlation was observed for miR-429 and miR-200c (correlation coefficients 0.423 and 0.262, respectively; p values of 0.039 and 0.214, respectively). This analysis supports miR-200b’s role in regulating eNOS expression during hypoxia.
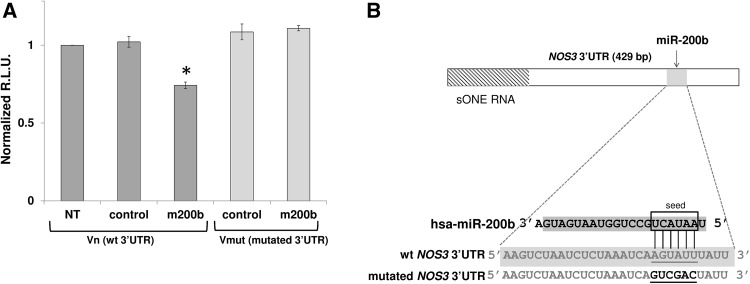
miR-200b binds to the NOS3 3′UTR
Although the miRNAs recognize specific target sequences, these sequences (6–8 nt) can be present in the 3′UTRs of many different genes. Hence, in order to exclude indirect effects of miR-200b on eNOS expression, we utilized a 3′UTR luciferase reporter. Briefly, a plasmid containing the 3′UTR of human NOS3 gene was tested in a luciferase gene construct that was expressed in HEK293 in the presence and absence of a miR-200b analog (mimic). As shown in Fig. 3a, miR-200b overexpression resulted in significantly reduced luciferase expression compared to control. Furthermore, a similar experiment with reporter vector containing the 3′ UTR of human NOS3 gene with a mutated miR-200b target site (Fig. 3b) did not result in a luciferase signal reduction (Fig. 3a), confirming a direct interaction between miR-200b and its target site at 3′UTR of NOS3 mRNA.
Fig. 3.
NOS3 3′UTR is a direct target for miR-200b, miR-200c, and miR-429. a Luciferase reporter constructs containing wild-type (Vn) or mutated (Vmut) NOS3 3′UTR sequence were transfected together with miR-200b or scramble control into HEK-293 cells. As a control reporter vector without 3′UTR (Vc) was used. The normalized R.L.U luciferase activities were calculated as a Vn/Vc or Vmut/Vc ratio (n = 2; *p < 0.05 vs. Vn/Vc). Data represent the mean ± SD of three experiments. *p < 0.05 was considered significant. b The schematic presentation of the mutations (bold) introduced into miR-200b TS in NOS3 3′UTR sequence
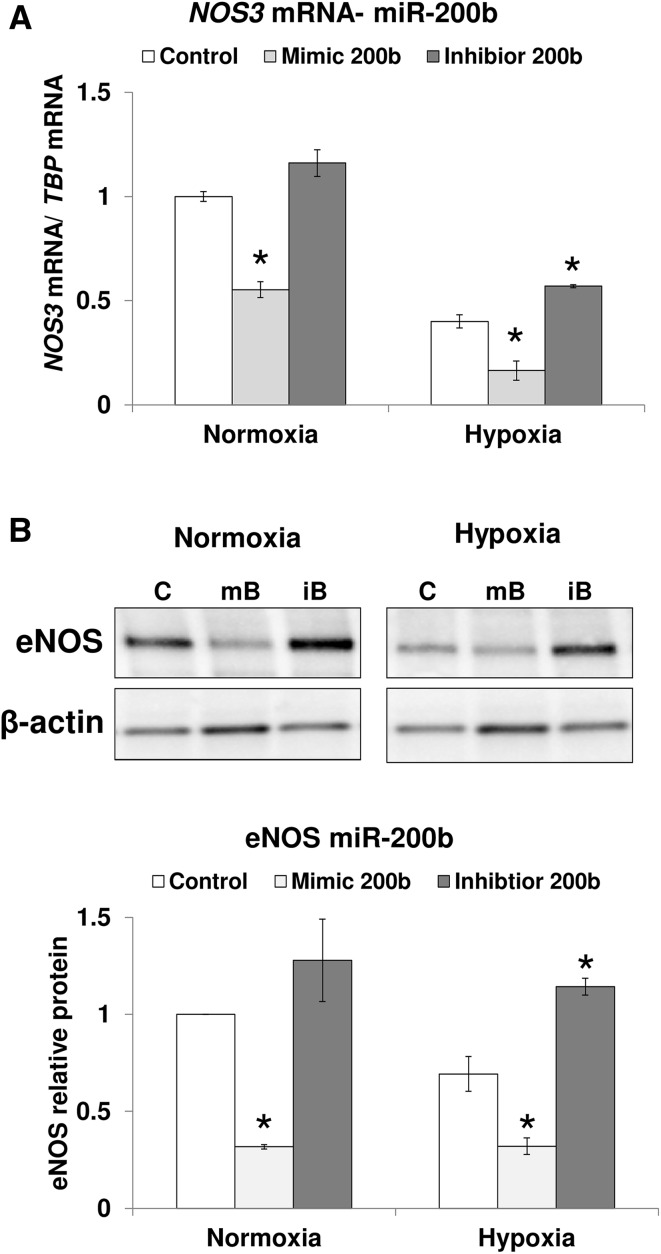
Next, we tested the effects of miR-200b overexpression and inhibition on NOS3 mRNA levels after 12 h of hypoxia in HUVECs. The miR-200b upregulation with mimic reduced NOS3 mRNA in both hypoxia and normoxia (Fig. 4a). The inhibition of miR-200b activity with antagomiR increased NOS3 mRNA significantly only during hypoxia (Fig. 4a), supporting the requirement of hnRNPE1 dissociation from NOS3 mRNA for the miR-200b-NOS3 functional interaction to occur. In parallel, we followed miR-200b analog effects on eNOS protein levels. As shown in Fig. 4b, both in normoxia and during hypoxia, miR-200b overexpression resulted in the reduction of eNOS protein levels in HUVECs. Notably, the antagomiR treatment during hypoxia had elevated eNOS protein, confirming the physiological effect of miR-200b on NOS3 expression during low oxygen levels (Fig. 4b). Importantly, despite the rather small miR-200b antagomiR effect on NOS3 mRNA during hypoxia, the attenuation of miR-200b expression restored eNOS protein to normoxic levels. This suggests that miR-200b effects on hypoxic eNOS expression are mainly caused by miRNA-mediated translational inhibition presumably due to low NOS3 mRNA stability during hypoxia.
Fig. 4.
miR-200b downregulates eNOS expression in response to hypoxia. HUVECs were transfected with miR-200b mimic, mir-200b antagomiR, or scramble control and cultured in normoxia or hypoxia for 12 h. a NOS3 mRNA levels were quantified by qRT-PCR and normalized to TBP mRNA. Data represent the mean ± SD of two independent experiments. b Corresponding changes in eNOS protein levels were monitored with western blot and normalized to β-Actin and related to the normoxic control. Data represent the mean ± SE of two independent experiments. *p < 0.05 was considered significant
Hypoxia miR-200b induction attenuates NO bioavailability
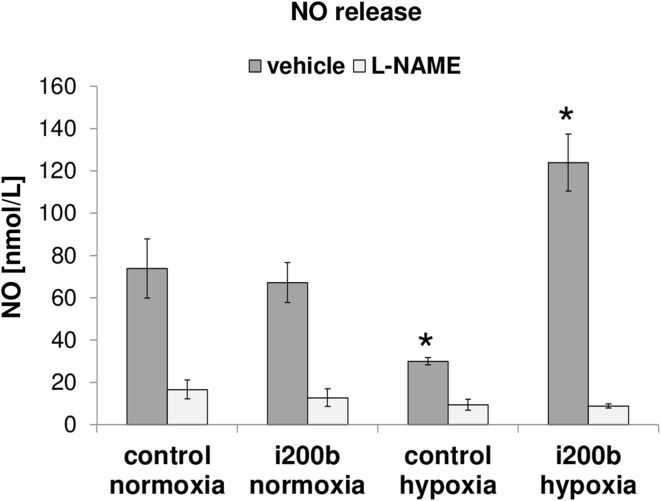
Given the effects on the eNOS RNA and protein, we next tested the effect of miR-200b antagomiR on NO release. The direct measurements of NO released from HUVECs cultured during hypoxia for 16 h indicate that eNOS produces approximately 50% less NO during hypoxia compared to normoxia (Fig. 5), confirming previous reports [33]. Furthermore, decreasing the physiological miR-200b levels with antagomiR had little effect on NO release during normoxia, but had a dramatic fourfold increase in eNOS-specific NO release (Fig. 5). This confirmed miR-200b as an important regulator limiting NO bioavailability during hypoxia. Furthermore, the antagomiR had no effect on NO release from normoxic cells, supporting the role of hnRNPE1 in stabilizing NOS3 transcript during normoxia. As expected, the release of NO after eNOS stimulation was dramatically inhibited by 300 μmol/L L-NAME in all sets of experiments.
Fig. 5.
Hypoxia-inducible miR-200b impairs NO release. HUVECs were transfected with miR-200b antagomiR or scramble control, the cells were then cultured in normoxia or hypoxia conditions for 16 h, and the NO release was measured. eNOS was stimulated by 1.0 µmol/L calcium ionophore. The specific eNOS inhibitor NG-nitro-l-arginine methyl ester (L-NAME) was used as a control. Results represent the mean ± SD of three measurements, *p < 0.05 was considered significant
Discussion
The mediator generated by eNOS, NO, is a key component of the vascular homeostasis [25, 34–37] and the loss of NO bioavailability provides an independent risk factor for cardiovascular diseases [35, 36]. Many of these disorders are associated with hypoxia or ischemia in different organs, while hypoxia and ischemia lower endothelial eNOS expression and lead to loss of NO bioavailability [10, 11]. Therefore, understanding the cellular pathways that regulate eNOS expression and consequently NO release during hypoxia is important.
The hypoxic loss of NO release is mediated by decreased eNOS activity brought about by altered eNOS phosphorylation [33], whereas posttranscriptional mechanisms that reduce the stability of NOS3 mRNA have been proposed to be another major contributing factor in the loss of NO bioavailability. The hnRNPE1 complex stabilizes NOS3 mRNA during normoxia by preventing binding factors that negatively affect the transcript half-life [12]. During hypoxia, however, hnRNPE1 protection is missing and this makes the NOS3 transcript accessible to destabilizing factors such as sONE [11–13]. However, attenuation of sONE in hypoxic ECs only partially restores NOS3 expression [11], and thus involvement of other posttranscriptional mechanisms must contribute to the observed reduction in the NOS3 mRNA levels during hypoxia.
Although the role of miRNAs in posttranscriptional gene regulation is clearly established, it is now evident that recent specific alterations in miRNA expression can also regulate eNOS levels [14–18] (reviewed in [6]). Moreover, differences in miRNA expression are also present in cardiovascular disorders [38], suggesting that miRNA network changes could influence disease pathogenesis. Despite the fact that inhibiting miRNAs function (through DICER silencing [12]) prevents eNOS downregulation, only five miRNAs that directly affect NOS3 expression have been identified that include miR-214 [16], miR-155 [15], miR-24 [39], miR-765 [12], and miR-200c [18]. However, previous studies did not examine whether miRNA-based mechanisms contribute to eNOS levels reduction during hypoxia.
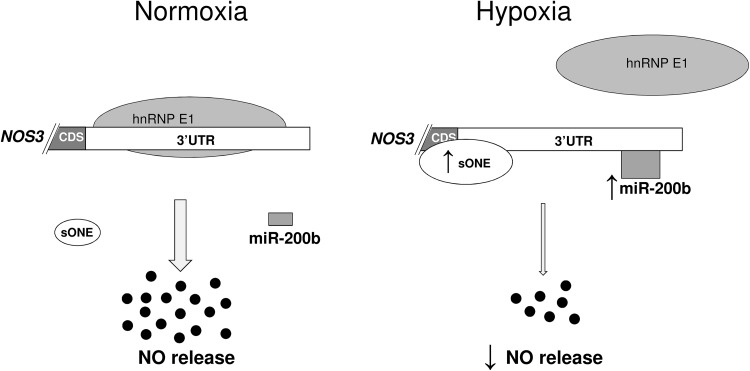
hnRPE1 dissociates and destabilizes NOS3 mRNA during hypoxia through sONE association (Fig. 6). It is also plausible that binding of sONE to NOS3 3′UTR would prevent miRNAs binding to this sequence. However, impairing sONE expression during hypoxic only partially attenuated the fall in both NOS3 mRNA and eNOS protein (~ one-half fold), whereas hypoxia results in a much more pronounced eNOS downregulation (~ fivefold drop) [11]. Notably, however, sONE occupies only one-third of NOS3 3′UTR (Fig. 2a), whereas the remaining sequence, due to hnRNPE1 dissociation, becomes available for miRNAs binding.
Fig. 6.
Hypoxia-inducible miR-200b reduces NO release in ECs by direct downregulation of eNOS expression. In normoxic ECs, NOS3 mRNA is actively stabilized by hnRNP E1 containing complex. Under hypoxia, hnRNP E1 dissociates from NOS3 3′UTR making it prone to destabilization by hypoxia-induced: miR-200b and sONE RNA. The hypoxic destabilization of NOS3 mRNA by miR-200b results in reduced eNOS protein and diminished NO release
This hypothesis is supported by a previous study in which siRNA-mediated impairment of hnRNP1 sensitized NOS3 transcript to destabilization by miR-765 in normoxic HUVECs [12]. However, miR-765 levels are reduced during hypoxia [40], and whether the hnRNPE1-dependent mechanism of NOS3 mRNA destabilization requires changes in miRNA levels is unclear. In contrast, numerous miRNAs are induced during hypoxia in ECs and the hnRNPE1-dependent assessment to these miRNA target sites at NOS3 3′UTR would allow dynamic and bidirectional modulation of NOS3 mRNA stability. Taken together, this suggests that hypoxamiRs contribute to the decline in NOS3 message and protein expression, and importantly provide intriguing targets for elevating NO levels during hypoxia.
Our in silico predictions indicate that miR-200b, miR-200c, and miR-429 were putative candidates for eNOS posttranscriptional regulation during hypoxia. Furthermore, the predicted target site for these miRNAs was shown to be target of miR-200c under oxidative stress [18]. To differentiate between miR-200b and miR-200c and miR-429, we utilized a hypoxia time course in HUVECs, and demonstrate a correlation between miR-200b induction and a corresponding decline in NOS3 mRNA and protein levels, whereas miR-200c and miR-429 changes did not correlate. Using the 3′UTR NOS3 luciferase constructs, we also demonstrated that miR-200b had a direct effect on luciferase expression, and this clearly established a direct effect on NOS3 message levels. Final support for the role of miR-200b comes from the negative and positive effects of miR-200b mimics and antagomirs on eNOS expression. Importantly, the miR-200b inhibition significantly increased eNOS levels under hypoxia exclusively, confirming that this miRNA regulates eNOS expression via an hnRNPE1-dependent mechanism. Taken together, the results suggest that during low oxygen conditions which could occur in various cardiovascular pathologies, miR-200b is upregulated and has a direct inhibitory effect on NOS3 message and eNOS protein expression in human ECs.
The removal of physiological miR-200b levels with antagomiR in hypoxic HUVECs resulted in the restoration of eNOS which released NO above normoxic levels and about fourfold above the hypoxic ones. This result shows that the hypoxic induction of miR-200b not only leads to reduced eNOS levels but also to reduced NO release as well. This was demonstrated in the most accurate method by using the NO-detecting porphyrinic biosensor, designed for cell cultures, that allows direct quantification of biologically active NO with high sensitivity in situ and in real time. This methodological approach for NO measurements provides the evidence on whether the changes in eNOS expression in endothelial cells can be truly translated into the increase of NO bioavailability [24–26].
This main finding of our study highlights the role of posttranscriptional mechanisms modulating eNOS expression for NO bioavailability. However, by inhibiting miR-200b function during hypoxia, we not only restored NO production, but we also increased it above normoxic levels. This can be explained by the network of miR-200 family targets involved in hypoxic response [22, 31, 32] that could mediate other mechanisms indirectly affecting eNOS activity during hypoxia.
The approach that we used for examining the miR-200b-eNOS interaction, however, does not answer the question about the potential role of other hypoxiamiRs in this process. To address this, we utilized Next Generation Sequencing (NGS) to follow hypoxia-induced changes in the miRNA levels in HUVECS. After 16 h of hypoxia, we observed a significant reduction of NOS3 mRNA and interestingly, 72 miRs were significantly induced, while 15 were reduced (Table 1). Among these 72 miRNAs, only 4 had been previously reported as hypoxiamiRs (including miR-210). Deregulation of many others, however, has been associated with various types of cancer (Table 1). In order to predict which miRNAs might govern hypoxic eNOS expression, we utilized the miRDIP database. This database effectively combines different target prediction algorithms in order to identify the most probable interactions [41]. The highest scores and thus the highest probability hits for binding to the NOS3 3′UTR were miR-424-5p and miR-503-5p. Their miRDIP scores were 0.322 and 0.265, respectively, and as a comparison, the score for miR-200b using this analysis was 0.212. Furthermore, both of these miRNAs were previously reported in ECs as hypoxiamiRs [42–44]. Whether or not they influence NOS3 mRNA expression levels, however, they will require experimental validation. Taken together, our deep sequencing analysis of the hypoxia effect on miRNAs expression in human primary ECs resulted in identification of 83 novel hypoxiamiRs. Among this group, 30 have previously been shown to be dysregulated in cancer cells.
Table 1.
Hypoxia-induced changes in miRNAs levels in HUVECs. Cells were exposed to hypoxia for 16 h and miRNA profiles determined with NGS analysis of two independent experiments
| MiRNA induced in HUVECs during 16 h of hypoxia | ||||
|---|---|---|---|---|
| miRNA | Fold change |
NOS3 target site (mirDIP integrated score) |
Process affected | References |
| hsa-miR-210-3p | 31.07 | 0.115 | HypoxiamiR (ECs and many other types of cells) | [45–47] |
| hsa-miR-1279 | 21.07 | 0.050 | – | – |
| hsa-miR-4256 | 18.52 | 0.008 | – | – |
| hsa-miR-4700-3p | 16.74 | 0.052 | – | – |
| hsa-miR-3131 | 14.72 | 0.011 | Dysregulated in gastric cancer | [48] |
| hsa-miR-33a-5p | 10.47 | 0.026 | Dysregulated in melanoma and hepatocellular carcinoma | [49, 50] |
| hsa-miR-4680-3p | 10.25 | 0.035 | Dysregulated in glioma | [51] |
| hsa-miR-6744-3p | 9.01 | 0.014 | – | – |
| hsa-miR-6789-5p | 6.95 | 0.011 | – | – |
| hsa-miR-4771 | 6.91 | 0.034 | – | – |
| hsa-miR-4710 | 6.64 | 0.010 | Upregulated in HUVECs exposed to estradiol | [52] |
| hsa-miR-3168 | 6.56 | 0.048 | – | – |
| hsa-miR-6828-3p | 6.31 | 0.014 | – | – |
| hsa-miR-4303 | 5.70 | 0.046 | – | – |
| hsa-miR-4433a-3p | 5.55 | 0.095 | – | – |
| hsa-miR-6820-3p | 5.48 | 0.014 | – | – |
| hsa-miR-6502-5p | 5.32 | 0.014 | – | – |
| hsa-miR-5095 | 5.19 | 0.070 | – | – |
| hsa-miR-6850-3p | 4.77 | 0.012 | – | – |
| hsa-miR-105-5p | 4.65 | 0.044 | Dysregulated in breast cancer, hepatocellular carcinoma, and glioma | [53–55] |
| hsa-miR-4659a-5p | 4.61 | 0.014 | – | – |
| hsa-miR-4468 | 4.54 | 0.067 | – | – |
| hsa-miR-649 | 4.44 | 0.024 | Dysregulated in bladder cancer | [56] |
| hsa-miR-200b-5p | 4.42 | 0.012 | Dysregulated in breast, head, and neck cancers | [57, 58] |
| hsa-miR-944 | 4.37 | 0.034 | Dysregulated in breast, gastric, and colorectal cancers | [59–61] |
| hsa-miR-6514-5p | 4.30 | 0.013 | – | – |
| hsa-miR-4322 | 4.25 | 0.012 | – | – |
| hsa-miR-8052 | 4.19 | 0.020 | – | – |
| hsa-miR-4450 | 4.08 | 0.096 | – | – |
| hsa-miR-206 | 4.02 | 0.086 | HypoxamiR (rat PASMCs, and rat cardiomyocytes) | [62] |
| hsa-miR-6131 | 3.90 | 0.066 | – | – |
| hsa-miR-5189-3p | 3.88 | 0.013 | – | – |
| hsa-miR-639 | 3.79 | 0.088 | Dysregulated in breast and tongue cancers | [63, 64] |
| hsa-miR-3916 | 3.79 | 0.091 | – | – |
| hsa-miR-4477a | 3.72 | 0.015 | – | – |
| hsa-miR-3679-3p | 3.71 | 0.009 | Dysregulated in non-small cell lung cancer | [65] |
| hsa-miR-503-3p | 3.62 | 0.031 | Dysregulated in breast cancer | [66] |
| hsa-miR-433-5p | 3.45 | 0.013 | – | – |
| hsa-miR-1266-5p | 3.44 | 0.031 | Dysregulated gastric in cancer | [67] |
| hsa-miR-6827-5p | 3.37 | 0.012 | – | – |
| hsa-miR-4285 | 3.35 | 0.012 | – | – |
| hsa-miR-3606-5p | 3.34 | 0.016 | – | – |
| hsa-miR-5590-5p | 3.31 | 0.016 | – | – |
| hsa-miR-215-3p | 3.28 | 0.015 | – | – |
| hsa-miR-505-5p | 3.27 | 0.013 | Dysregulated in breast cancer | [68] |
| hsa-miR-562 | 3.25 | 0.044 | Dysregulated in prostate cancer | [69] |
| hsa-miR-4687-5p | 3.23 | 0.028 | Dysregulated in colorectal cancer | [70] |
| hsa-miR-518d-3p | 3.13 | 0.020 | – | – |
| hsa-miR-5681a | 3.13 | 0.033 | – | – |
| hsa-miR-424-3p | 3.07 | 0.034 | Dysregulated colorectal in cancer | [71] |
| hsa-miR-4682 | 3.05 | 0.082 | – | – |
| hsa-miR-4772-5p | 3.05 | 0.013 | – | – |
| hsa-miR-3117-3p | 2.97 | 0.014 | Dysregulated in hepatocellular carcinoma | [72, 73] |
| hsa-miR-7162-3p | 2.95 | 0.013 | – | – |
| hsa-miR-891a-5p | 2.90 | 0.048 | – | – |
| hsa-miR-4725-3p | 2.89 | 0.012 | – | – |
| hsa-miR-617 | 2.84 | 0.044 | – | – |
| hsa-miR-3621 | 2.84 | 0.036 | Dysregulated in colorectal cancer | [74] |
| hsa-miR-503-5p | 2.71 | 0.266 | HypoxiamiR (rat MSCs) | [42] |
| hsa-miR-512-3p | 2.62 | 0.039 | Dysregulated in anaplastic large cell lymphoma and prostate cancer | [75, 76] |
| hsa-miR-144-3p | 2.50 | 0.045 | Dysregulated in papillary thyroid carcinoma and renal cell carcinoma | [77, 78] |
| hsa-miR-24-1-5p | 2.49 | 0.009 | Dysregulated in prostate cancer | [79] |
| hsa-miR-5007-3p | 2.46 | 0.009 | Dysregulated in gastric cancer | [80] |
| hsa-miR-8066 | 2.46 | 0.009 | – | – |
| hsa-miR-424-5p | 2.42 | 0.323 | Hypoxiamir (HUVECs and many other cell lines) | [43, 44] |
| hsa-miR-548bb-5p | 2.34 | 0.009 | – | – |
| hsa-miR-510-3p | 2.31 | 0.009 | – | – |
| hsa-miR-4500 | 2.27 | 0.015 | Dysregulated in colorectal cancer | [81] |
| hsa-miR-6751-5p | 2.26 | 0.017 | – | – |
| hsa-miR-4700-5p | 2.25 | 0.175 | – | – |
| hsa-miR-802 | 2.20 | 0.040 | Dysregulated in squamous cell carcinoma | [82] |
| hsa-miR-4735-5p | 2.18 | 0.009 | – | – |
| MiRNA reduced in HUVECs during 16 h of hypoxia | ||||
|---|---|---|---|---|
| hsa-miR-483-3p | 0.50 | 0.020 | Dysregulated in squamous cell carcinoma | [83] |
| hsa-miR-6761-5p | 0.45 | 0.041 | – | – |
| hsa-miR-4668-5p | 0.43 | 0.059 | – | – |
| hsa-miR-106a-3p | 0.42 | 0.035 | Dysregulated in breast cancer | [84] |
| hsa-miR-4461 | 0.40 | 0.033 | – | – |
| hsa-miR-6727-5p | 0.30 | 0.009 | – | – |
| hsa-miR-4472 | 0.28 | 0.031 | Dysregulated in breast cancer | [85] |
| hsa-miR-7706 | 0.27 | 0.006 | – | – |
| hsa-miR-922 | 0.25 | 0.163 | Dysregulated in hepatocellular carcinoma | [86] |
| hsa-miR-4484 | 0.25 | 0.033 | Dysregulated in glioblastoma | [87] |
| hsa-miR-6865-3p | 0.25 | 0.014 | – | – |
| hsa-miR-4483 | 0.20 | 0.067 | – | – |
| hsa-miR-449c-5p | 0.20 | 0.088 | Dysregulated in nasopharyngeal carcinoma | [88] |
| hsa-miR-6785-5p | 0.13 | 0.072 | – | – |
| hsa-miR-599 | 0.12 | 0.020 | Dysregulated in breast and non-small cell lung cancer | [89, 90] |
The table represents significant changes (fold change 2, p value < 0.005), that were consistent between the two independent experiments. The raw data are provided in Supplemental file 1. Potential miRNA binding to NOS3 3′UTR was predicted with mirDIP, and miRs that resulted in the prediction of high score are marked bold. Previously reported hypoxiamiRs are underlined
In summary, the studies show that hypoxia-induced physiological changes in miR-200b levels in human ECs contribute directly to eNOS downregulation during hypoxia and lead to diminished NO release. Hence, these results complement previous studies indicating the hnRNPE1-dependent sONE direct posttranscriptional effects on the downregulation of eNOS during hypoxia with a mechanism that involves a hypoxia-induced miRNA (Fig. 6). Furthermore, stabilization of eNOS protein levels during hypoxia through inhibition of miR-200b’s effects and the corresponding increase in NO release may provide a novel therapeutic opportunity for increasing NO bioavailability during various cardiovascular diseases. Moreover, in human ECs exposed to hypoxia, using Next Generation Sequencing, we identified 83 novel hypoxamiRs and proposed that miR-424-5p and miR-503-5p are candidate miRNAs that could potentially regulate eNOS expression during hypoxia. Although our initial analysis focused on eNOS-miRNA interactions, we also identified a number of hypoxamiRs many of which may be important in the cancer hypoxic microenvironment [91, 92].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- eNOS
Endothelial nitric oxide synthase
- NO
Nitric oxide
- HUVECs
Human umbilical vein endothelial cells
- HIF
Hypoxia-inducible factor
Author contributions
Conceived and designed the experiments: AJ-J, RB, and LK. Performed the experiments: AJJ, AS, SB, and MS. All authors analyzed the data. Contributed reagents/materials/analysis tools: RB and LK. All authors wrote, read, and revised the final version of the manuscript.
Funding
This work has been supported by National Science Center “OPUS” Program under contract UMO-2015/19/B/NZ7/03830 (L.K.) and NIH P30 DK072482 (J.F.C.).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing financial interests.
Contributor Information
Anna Janaszak-Jasiecka, Phone: 48 58 349 32 14, Email: ajanaszak@gumed.edu.pl.
Rafal Bartoszewski, Phone: 48 58 349 32 14, Email: rafalbar@gumed.edu.pl.
References
- 1.Versari D, Daghini E, Virdis A, Ghiadoni L, Taddei S. Endothelial dysfunction as a target for prevention of cardiovascular disease. Diabetes Care. 2009;32(Suppl 2):S314–S321. doi: 10.2337/dc09-S330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X, Chang Y, Wei W. Endothelial dysfunction and inflammation: immunity in rheumatoid arthritis. Mediat Inflamm. 2016;2016:6813016. doi: 10.1155/2016/6813016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fulton D, Gratton JP, McCabe TJ, Fontana J, Fujio Y, Walsh K, Franke TF, Papapetropoulos A, Sessa WC. Regulation of endothelium-derived nitric oxide production by the protein kinase Akt. Nature. 1999;399(6736):597–601. doi: 10.1038/21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziegler T, Silacci P, Harrison VJ, Hayoz D. Nitric oxide synthase expression in endothelial cells exposed to mechanical forces. Hypertension. 1998;32(2):351–355. doi: 10.1161/01.HYP.32.2.351. [DOI] [PubMed] [Google Scholar]
- 5.Searles CD. Transcriptional and posttranscriptional regulation of endothelial nitric oxide synthase expression. Am J Physiol Cell Physiol. 2006;291(5):C803–C816. doi: 10.1152/ajpcell.00457.2005. [DOI] [PubMed] [Google Scholar]
- 6.Kalinowski L, Janaszak-Jasiecka A, Siekierzycka A, Bartoszewska S, Wozniak M, Lejnowski D, Collawn JF, Bartoszewski R. Posttranscriptional and transcriptional regulation of endothelial nitric-oxide synthase during hypoxia: the role of microRNAs. Cell Mol Biol Lett. 2016;21:16. doi: 10.1186/s11658-016-0017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond GR, Cai H, Davis ME, Ramasamy S, Harrison DG. Transcriptional and posttranscriptional regulation of endothelial nitric oxide synthase expression by hydrogen peroxide. Circ Res. 2000;86(3):347–354. doi: 10.1161/01.res.86.3.347. [DOI] [PubMed] [Google Scholar]
- 8.Olson N, van der Vliet A. Interactions between nitric oxide and hypoxia-inducible factor signaling pathways in inflammatory disease. Nitric Oxide. 2011;25(2):125–137. doi: 10.1016/j.niox.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coulet F, Nadaud S, Agrapart M, Soubrier F. Identification of hypoxia-response element in the human endothelial nitric-oxide synthase gene promoter. J Biol Chem. 2003;278(47):46230–46240. doi: 10.1074/jbc.M305420200. [DOI] [PubMed] [Google Scholar]
- 10.McQuillan LP, Leung GK, Marsden PA, Kostyk SK, Kourembanas S. Hypoxia inhibits expression of eNOS via transcriptional and posttranscriptional mechanisms. Am J Physiol. 1994;267(5 Pt 2):H1921–H1927. doi: 10.1152/ajpheart.1994.267.5.H1921. [DOI] [PubMed] [Google Scholar]
- 11.Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, Ohh M, Marsden PA. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem. 2007;282(21):15652–15666. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- 12.Ho JJ, Robb GB, Tai SC, Turgeon PJ, Mawji IA, Man HS, Marsden PA. Active stabilization of human endothelial nitric oxide synthase mRNA by hnRNP E1 protects against antisense RNA and microRNAs. Mol Cell Biol. 2013;33(10):2029–2046. doi: 10.1128/mcb.01257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robb GB, Carson AR, Tai SC, Fish JE, Singh S, Yamada T, Scherer SW, Nakabayashi K, Marsden PA. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J Biol Chem. 2004;279(36):37982–37996. doi: 10.1074/jbc.M400271200. [DOI] [PubMed] [Google Scholar]
- 14.Suárez Y, Fernández-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100(8):1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 15.Sun HX, Zeng DY, Li RT, Pang RP, Yang H, Hu YL, Zhang Q, Jiang Y, Huang LY, Tang YB, Yan GJ, Zhou JG. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension. 2012;60(6):1407–1414. doi: 10.1161/HYPERTENSIONAHA.112.197301. [DOI] [PubMed] [Google Scholar]
- 16.Chan LS, Yue PY, Mak NK, Wong RN. Role of microRNA-214 in ginsenoside-Rg1-induced angiogenesis. Eur J Pharm Sci. 2009;38(4):370–377. doi: 10.1016/j.ejps.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Yan L, Li Y, Chen W, Hu N, Wang H, Ou H. Roles of miRNA-24 in regulating endothelial nitric oxide synthase expression and vascular endothelial cell proliferation. Mol Cell Biochem. 2015;405(1–2):281–289. doi: 10.1007/s11010-015-2418-y. [DOI] [PubMed] [Google Scholar]
- 18.Carlomosti F, D’Agostino M, Beji S, Torcinaro A, Rizzi R, Zaccagnini G, Maimone B, Di Stefano V, De Santa F, Cordisco S, Antonini A, Ciarapica R, Dellambra E, Martelli F, Avitabile D, Capogrossi MC, Magenta A. Oxidative stress-induced miR-200c disrupts the regulatory loop among SIRT1, FOXO1, and eNOS. Antioxid Redox Signal. 2017;27(6):328–344. doi: 10.1089/ars.2016.6643. [DOI] [PubMed] [Google Scholar]
- 19.Madanecki P, Kapoor N, Bebok Z, Ochocka R, Collawn JF, Bartoszewski R. Regulation of angiogenesis by hypoxia: the role of microRNA. Cell Mol Biol Lett. 2013;18(1):47–57. doi: 10.2478/s11658-012-0037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartoszewski R, Brewer JW, Rab A, Crossman DK, Bartoszewska S, Kapoor N, Fuller C, Collawn JF, Bebok Z. The unfolded protein response (UPR)-activated transcription factor X-box-binding protein 1 (XBP1) induces microRNA-346 expression that targets the human antigen peptide transporter 1 (TAP1) mRNA and governs immune regulatory genes. J Biol Chem. 2011;286(48):41862–41870. doi: 10.1074/jbc.M111.304956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartoszewski R, Rab A, Fu L, Bartoszewska S, Collawn J, Bebok Z. CFTR expression regulation by the unfolded protein response. Methods Enzymol. 2011;491:3–24. doi: 10.1016/B978-0-12-385928-0.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartoszewski R, Serocki M, Janaszak-Jasiecka A, Bartoszewska S, Kochan-Jamrozy K, Piotrowski A, Kroliczewski J, Collawn JF. miR-200b downregulates Kruppel Like Factor 2 (KLF2) during acute hypoxia in human endothelial cells. Eur J Cell Biol. 2017;96(8):758–766. doi: 10.1016/j.ejcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Kalinowski L, Dobrucki IT, Malinski T. Race-specific differences in endothelial function: predisposition of African Americans to vascular diseases. Circulation. 2004;109(21):2511–2517. doi: 10.1161/01.CIR.0000129087.81352.7A. [DOI] [PubMed] [Google Scholar]
- 25.Dobrucki LW, Marsh BJ, Kalinowski L. Elucidating structure-function relationships from molecule-to-cell-to-tissue: from research modalities to clinical realities. J Physiol Pharmacol. 2009;60(Suppl 4):83–93. [PubMed] [Google Scholar]
- 26.Domagala TB, Szeffler A, Dobrucki LW, Dropinski J, Polanski S, Leszczynska-Wiloch M, Kotula-Horowitz K, Wojciechowski J, Wojnowski L, Szczeklik A, Kalinowski L. Nitric oxide production and endothelium-dependent vasorelaxation ameliorated by N1-methylnicotinamide in human blood vessels. Hypertension. 2012;59(4):825–832. doi: 10.1161/HYPERTENSIONAHA.111.183210. [DOI] [PubMed] [Google Scholar]
- 27.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McQuillan LP, Leung GK, Marsden PA, Kostyk SK, Kourembanas S. Hypoxia inhibits expression of eNOS via transcriptional and posttranscriptional mechanisms. Am J Physiol. 1994;267(5):H1921–H1927. doi: 10.1152/ajpheart.1994.267.5.H1921. [DOI] [PubMed] [Google Scholar]
- 29.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janaszak-Jasiecka A, Bartoszewska S, Kochan K, Piotrowski A, Kalinowski L, Kamysz W, Ochocka RJ, Bartoszewski R, Collawn JF. miR-429 regulates the transition between Hypoxia-Inducible Factor (HIF)1A and HIF3A expression in human endothelial cells. Sci Rep. 2016;6:22775. doi: 10.1038/srep22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartoszewska S, Kochan K, Piotrowski A, Kamysz W, Ochocka RJ, Collawn JF, Bartoszewski R. The hypoxia-inducible miR-429 regulates hypoxia-inducible factor-1alpha expression in human endothelial cells through a negative feedback loop. FASEB J. 2015;29(4):1467–1479. doi: 10.1096/fj.14-267054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostergaard L, Stankevicius E, Andersen MR, Eskildsen-Helmond Y, Ledet T, Mulvany MJ, Simonsen U. Diminished NO release in chronic hypoxic human endothelial cells. Am J Physiol Heart Circ Physiol. 2007;293(5):H2894-2903. doi: 10.1152/ajpheart.01230.2006. [DOI] [PubMed] [Google Scholar]
- 34.Kalinowski L, Malinski T. Endothelial NADH/NADPH-dependent enzymatic sources of superoxide production: relationship to endothelial dysfunction. Acta Biochim Pol. 2004;51(2):459–469. [PubMed] [Google Scholar]
- 35.Landmesser U, Hornig B, Drexler H. Endothelial function: a critical determinant in atherosclerosis? Circulation. 2004;109(21 Suppl 1):II27–II33. doi: 10.1161/01.CIR.0000129501.88485.1f. [DOI] [PubMed] [Google Scholar]
- 36.Heitzer T, Schlinzig T, Krohn K, Meinertz T, Munzel T. Endothelial dysfunction, oxidative stress, and risk of cardiovascular events in patients with coronary artery disease. Circulation. 2001;104(22):2673–2678. doi: 10.1161/hc4601.099485. [DOI] [PubMed] [Google Scholar]
- 37.Channon KM, Guzik TJ. Mechanisms of superoxide production in human blood vessels: relationship to endothelial dysfunction, clinical and genetic risk factors. J Physiol Pharmacol. 2002;53(4 Pt 1):515–524. [PubMed] [Google Scholar]
- 38.Schulte C, Zeller T. microRNA-based diagnostics and therapy in cardiovascular disease—summing up the facts. Cardiovasc Diagn Ther. 2015;5(1):17–36. doi: 10.3978/j.issn.2223-3652.2014.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Yan L, Li Y, Chen W, Hu N, Wang H, Ou H. Roles of miRNA-24 in regulating endothelial nitric oxide synthase expression and vascular endothelial cell proliferation. Mol Cell Biochem. 2015;405(1):281–289. doi: 10.1007/s11010-015-2418-y. [DOI] [PubMed] [Google Scholar]
- 40.Ho JJ, Metcalf JL, Yan MS, Turgeon PJ, Wang JJ, Chalsev M, Petruzziello-Pellegrini TN, Tsui AK, He JZ, Dhamko H, Man HS, Robb GB, Teh BT, Ohh M, Marsden PA. Functional importance of Dicer protein in the adaptive cellular response to hypoxia. J Biol Chem. 2012;287(34):29003–29020. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, Lu R, Jurisica I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360-D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou BS, Ma RH, Si WX, Li SS, Xu Y, Tu X, Wang Q. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333(2):159–169. doi: 10.1016/j.canlet.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 43.Zhang D, Shi Z, Li M, Mi J. Hypoxia-induced miR-424 decreases tumor sensitivity to chemotherapy by inhibiting apoptosis. Cell Death Dis. 2014;5(6):e1301. doi: 10.1038/Cddis.2014.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y, Ramakrishnan S. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Investig. 2010;120(11):4141–4154. doi: 10.1172/jci42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283(23):15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voellenkle C, Rooij J, Guffanti A, Brini E, Fasanaro P, Isaia E, Croft L, David M, Capogrossi MC, Moles A, Felsani A, Martelli F. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA. 2012;18(3):472–484. doi: 10.1261/rna.027615.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bavelloni A, Ramazzotti G, Poli A, Piazzi M, Focaccia E, Blalock W, Faenza I. miRNA-210: a current overview. Anticancer Res. 2017;37(12):6511–6521. doi: 10.21873/anticanres.12107. [DOI] [PubMed] [Google Scholar]
- 48.Assumpcao MB, Moreira FC, Hamoy IG, Magalhaes L, Vidal A, Pereira A, Burbano R, Khayat A, Silva A, Santos S, Demachki S, Ribeiro-Dos-Santos A, Assumpcao P. High-throughput miRNA sequencing reveals a field effect in gastric cancer and suggests an epigenetic network mechanism. Bioinform Biol Insights. 2015;9:111–117. doi: 10.4137/BBI.S24066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Xu D, Xie H, Tang J, Liu R, Li J, Wang S, Chen X, Su J, Zhou X, Xia K, He Q, Chen J, Xiong W, Cao P, Cao K. miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1alpha. Cancer Biol Ther. 2015;16(6):846–855. doi: 10.1080/15384047.2015.1030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng W, Tai Y, Zhao H, Fu B, Zhang T, Liu W, Li H, Yang Y, Zhang Q, Feng Y, Chen G. Downregulation of miR-33a-5p in hepatocellular carcinoma: a possible mechanism for chemotherapy resistance. Med Sci Monit. 2017;23:1295–1304. doi: 10.12659/MSM.902692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shou JJ, Gu SX, Gu WT. Identification of dysregulated miRNAs and their regulatory signature in glioma patients using the partial least squares method. Exp Ther Med. 2015;9(1):167–171. doi: 10.3892/etm.2014.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vidal-Gomez X, Perez-Cremades D, Mompeon A, Dantas AP, Novella S, Hermenegildo C. MicroRNA as crucial regulators of gene expression in estradiol-treated human endothelial cells. Cell Physiol Biochem. 2018;45(5):1878–1892. doi: 10.1159/000487910. [DOI] [PubMed] [Google Scholar]
- 53.Zhou WY, Fong MY, Min YF, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Connor STF, Chin AR, Yen Y, Wang YF, Marcusson EG, Chu PG, Wu J, Wu XW, Li AX, Li Z, Gao HL, Ren XB, Boldin MP, Lin PC, Wang SE. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie RT, Yang HQ, Chang ZY, Sun R, Chai L, Cai MX, Zhong XJ, Zhu J, Fu D. High expression of miR-105-1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget. 2017;8(7):11896–11905. doi: 10.18632/oncotarget.14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan YL, Chen L, Bao YJ, Li ZP, Cui R, Li GY, Wang YJ. Identification of low miR-105 expression as a novel poor prognostic predictor for human glioma. Int J Clin Exp Med. 2015;8(7):10855–10864. [PMC free article] [PubMed] [Google Scholar]
- 56.Scheffer AR, Holdenrieder S, Kristiansen G, von Ruecker A, Muller SC, Ellinger J. Circulating microRNAs in serum: novel biomarkers for patients with bladder cancer? World J Urol. 2014;32(2):353–358. doi: 10.1007/s00345-012-1010-2. [DOI] [PubMed] [Google Scholar]
- 57.Rhodes LV, Martin EC, Segar HC, Miller DFB, Buechlein A, Rusch DB, Nephew KP, Burow ME, Collins-Burow BM. Dual regulation by microRNA-200b-3p and microRNA-200b-5p in the inhibition of epithelial-to-mesenchymal transition in triple-negative breast cancer. Oncotarget. 2015;6(18):16638–16652. doi: 10.18632/oncotarget.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hudcova K, Raudenska M, Gumulec J, Binkova H, Horakova Z, Kostrica R, Babula P, Adam V, Masarik M. Expression profiles of miR-29c, miR-200b and miR-375 in tumour and tumour-adjacent tissues of head and neck cancers. Tumor Biol. 2016;37(9):12627–12633. doi: 10.1007/s13277-016-5147-2. [DOI] [PubMed] [Google Scholar]
- 59.He HF, Tian W, Chen HL, Jiang K. MiR-944 functions as a novel oncogene and regulates the chemoresistance in breast cancer. Tumor Biol. 2016;37(2):1599–1607. doi: 10.1007/s13277-015-3844-x. [DOI] [PubMed] [Google Scholar]
- 60.Pan T, Chen WJ, Yuan XM, Shen J, Qin C, Wang LB. miR-944 inhibits metastasis of gastric cancer by preventing the epithelial-mesenchymal transition via MACC1/Met/AKT signaling. FEBS Open Bio. 2017;7(7):905–914. doi: 10.1002/2211-5463.12215. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Wen LQ, Li YR, Jiang ZP, Zhang YC, Yang B, Han FH. miR-944 inhibits cell migration and invasion by targeting MACC1 in colorectal cancer. Oncol Rep. 2017;37(6):3415–3422. doi: 10.3892/or.2017.5611. [DOI] [PubMed] [Google Scholar]
- 62.Yue J, Guan J, Wang X, Zhang L, Yang Z, Ao Q, Deng Y, Zhu P, Wang G. MicroRNA-206 is involved in hypoxia-induced pulmonary hypertension through targeting of the HIF-1alpha/Fhl-1 pathway. Lab Investig. 2013;93(7):748–759. doi: 10.1038/labinvest.2013.63. [DOI] [PubMed] [Google Scholar]
- 63.Li L, Qiu XG, Lv PW, Wang F. miR-639 promotes the proliferation and invasion of breast cancer cell in vitro. Cancer Cell Int. 2014;14(1):39. doi: 10.1186/1475-2867-14-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin ZY, Sun LJ, Chen WL, Liu BD, Wang YY, Fan S, Li YL, Li JS. miR-639 regulates transforming growth factor beta-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting FOXC1. Cancer Sci. 2014;105(10):1288–1298. doi: 10.1111/cas.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pu Q, Huang YC, Lu YR, Peng Y, Zhang J, Feng GL, Wang CG, Liu LX, Dai Y. Tissue-specific and plasma microRNA profiles could be promising biomarkers of histological classification and TNM stage in non-small cell lung cancer. Thorac Cancer. 2016;7(3):348–354. doi: 10.1111/1759-7714.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Z, Fan X, Jiang L, Xu Z, Xue L, Zhan Q, Song Y. miR-503-3p promotes epithelial-mesenchymal transition in breast cancer by directly targeting SMAD2 and E-cadherin. J Genet Genomics. 2017;44(2):75–84. doi: 10.1016/j.jgg.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 67.Chen L, Lu MH, Zhang D, Hao NB, Fan YH, Wu YY, Wang SM, Xie R, Fang DC, Zhang H, Hu CJ, Yang SM. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014;5(1):e1034. doi: 10.1038/Cddis.2013.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matamala N, Vargas MT, Gonzalez-Campora R, Minambres R, Arias JI, Menendez P, Andres-Leon E, Gomez-Lopez G, Yanowsky K, Calvete-Candenas J, Inglada-Perez L, Martinez-Delgado B, Benitez J. Tumor MicroRNA expression profiling identifies circulating MicroRNAs for early breast cancer detection. Clin Chem. 2015;61(8):1098–1106. doi: 10.1373/clinchem.2015.238691. [DOI] [PubMed] [Google Scholar]
- 69.Haldrup C, Kosaka N, Ochiya T, Borre M, Hoyer S, Orntoft TF, Sorensen KD. Profiling of circulating microRNAs for prostate cancer biomarker discovery. Drug Deliv Transl Res. 2014;4(1):19–30. doi: 10.1007/s13346-013-0169-4. [DOI] [PubMed] [Google Scholar]
- 70.Nagy ZB, Bartak BK, Kalmar A, Galamb O, Wichmann B, Dank M, Igaz P, Tulassay Z, Molnar B. Comparison of circulating miRNAs expression alterations in matched tissue and plasma samples during colorectal cancer progression. Pathol Oncol Res. 2017 doi: 10.1007/s12253-017-0308-1. [DOI] [PubMed] [Google Scholar]
- 71.Torres S, Garcia-Palmero I, Bartolome RA, Fernandez-Acenero MJ, Molina E, Calvino E, Segura MF, Casal JI. Combined miRNA profiling and proteomics demonstrates that different miRNAs target a common set of proteins to promote colorectal cancer metastasis. J Pathol. 2017;242(1):39–51. doi: 10.1002/path.4874. [DOI] [PubMed] [Google Scholar]
- 72.Cui X, Li QY, He YK. miR-3117 regulates hepatocellular carcinoma cell proliferation by targeting PHLPPL. Mol Cell Biochem. 2017;424(1–2):195–201. doi: 10.1007/s11010-016-2855-2. [DOI] [PubMed] [Google Scholar]
- 73.Neerincx M, Sie DLS, van de Wiel MA, van Grieken NCT, Burggraaf JD, Dekker H, Eijk PP, Ylstra B, Verhoef C, Meijer GA, Buffart TE, Verheul HMW. MiR expression profiles of paired primary colorectal cancer and metastases by next-generation sequencing. Oncogenesis. 2015;4(10):e170. doi: 10.1038/oncsis.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jung CK, Jung SH, Yim SH, Jung JH, Choi HJ, Kang WK, Park SW, Oh ST, Kim JG, Lee SH, Chung YJ. Predictive microRNAs for lymph node metastasis in endoscopically resectable submucosal colorectal cancer. Oncotarget. 2016;7(22):32902–32915. doi: 10.18632/oncotarget.8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu CL, Iqbal J, Teruya-Feldstein J, Shen YL, Dabrowska MJ, Dybkaer K, Lim MS, Piva R, Barreca A, Pellegrino E, Spaccarotella E, Lachel CM, Kucuk C, Jiang CS, Hu XZ, Bhagavathi S, Greiner TC, Weisenburger DD, Aoun P, Perkins SL, McKeithan TW, Inghirami G, Chan WC. microRNA expression profiling identifies molecular signatures associated with anaplastic large cell lymphoma. Blood. 2013;122(12):2083–2092. doi: 10.1182/blood-2012-08-447375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sadeghi M, Ranjbar B, Ganjalikhany MR, Khan FM, Schmitz U, Wolkenhauer O, Gupta SK. MicroRNA and transcription factor gene regulatory network analysis reveals key regulatory elements associated with prostate cancer progression. PLoS ONE. 2016;11(12):e0168760. doi: 10.1371/journal.pone.0168760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao W, Lou N, Ruan H, Bao L, Xiong Z, Yuan C, Tong J, Xu G, Zhou Y, Qu Y, Hu W, Gao Y, Ru Z, Liu L, Xiao H, Chen K, Yang H, Zhang X. miR-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A. Cell Physiol Biochem. 2017;43(6):2420–2433. doi: 10.1159/000484395. [DOI] [PubMed] [Google Scholar]
- 78.Liu C, Su C, Chen Y, Li G. miR-144-3p promotes the tumor growth and metastasis of papillary thyroid carcinoma by targeting paired box gene 8. Cancer Cell Int. 2018;18:54. doi: 10.1186/s12935-018-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coarfa C, Fiskus W, Eedunuri VK, Rajapakshe K, Foley C, Chew SA, Shah SS, Geng C, Shou J, Mohamed JS, O’Malley BW, Mitsiades N. Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene. 2016;35(18):2345–2356. doi: 10.1038/onc.2015.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang B, Jing C, Wang J, Guo X, Chen Y, Xu R, Peng L, Liu J, Li L. Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. Clin Transl Oncol. 2014;16(4):374–379. doi: 10.1007/s12094-013-1081-6. [DOI] [PubMed] [Google Scholar]
- 81.Zhang L, Qian JJ, Qiang Y, Huang HR, Wang CT, Li DM, Xu B. Down-regulation of miR-4500 promoted non-small cell lung cancer growth. Cell Physiol Biochem. 2014;34(4):1166–1174. doi: 10.1159/000366329. [DOI] [PubMed] [Google Scholar]
- 82.Wu XZ, Gong ZD, Sun LY, Ma L, Wang QB. MicroRNA-802 plays a tumour suppressive role in tongue squamous cell carcinoma through directly targeting MAP2K4. Cell Prolif. 2017;50(3):e12336. doi: 10.1111/cpr.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma JJ, Hong L, Xu GH, Hao JF, Wang R, Guo H, Liu JQ, Zhang YJ, Nie YZ, Fan DM. miR-483-3p plays an oncogenic role in esophageal squamous cell carcinoma by targeting tumor suppressor EI24. Cell Biol Int. 2016;40(4):448–455. doi: 10.1002/cbin.10585. [DOI] [PubMed] [Google Scholar]
- 84.Li M, Zhou Y, Xia T, Zhou X, Huang Z, Zhang H, Zhu W, Ding Q, Wang S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res Treat. 2018 doi: 10.1007/s10549-018-4757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang YW, Zhang WG, Ma R. Bioinformatic identification of chemoresistance-associated microRNAs in breast cancer based on microarray data. Oncol Rep. 2018;39(3):1003–1010. doi: 10.3892/or.2018.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu JP, Su Z, Zeng YJ, Zhang HY, Yang SL, Liu GJ. miR-922 regulates CYLD expression and promotes the cell proliferation of human hepatocellular carcinoma. Oncol Rep. 2017;37(3):1445–1450. doi: 10.3892/or.2017.5431. [DOI] [PubMed] [Google Scholar]
- 87.Nawaz Z, Patil V, Thinagararjan S, Rao SA, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Impact of somatic copy number alterations on the glioblastoma miRNome: miR-4484 is a genomically deleted tumour suppressor. Mol Oncol. 2017;11(8):927–944. doi: 10.1002/1878-0261.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang F, Lu J, Peng XH, Wang J, Liu X, Chen XM, Jiang YQ, Li XP, Zhang B. Integrated analysis of microRNA regulatory network in nasopharyngeal carcinoma with deep sequencing. J Exp Clin Canc Res. 2016;35(1):17. doi: 10.1186/s13046-016-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tian WJ, Wang GH, Liu YQ, Huang ZL, Zhang CQ, Ning K, Yu CX, Shen YJ, Wang MH, Li YT, Wang Y, Zhang BC, Zhao YR. The miR-599 promotes non-small cell lung cancer cell invasion via SATB2. Biochem Bioph Res Commun. 2017;485(1):35–40. doi: 10.1016/j.bbrc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 90.Wang YH, Sui YN, Zhu QW, Sui XM. Hsa-miR-599 suppresses the migration and invasion by targeting BRD4 in breast cancer. Oncol Lett. 2017;14(3):3455–3462. doi: 10.3892/ol.2017.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Beijnum JR, Giovannetti E, Poel D, Nowak-Sliwinska P, Griffioen AW. miRNAs: micro-managers of anticancer combination therapies. Angiogenesis. 2017;20(2):269–285. doi: 10.1007/s10456-017-9545-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Serocki M, Bartoszewska S, Janaszak-Jasiecka A, Ochocka RJ, Collawn JF, Bartoszewski R. miRNAs regulate the HIF switch during hypoxia: a novel therapeutic target. Angiogenesis. 2018;21:183–202. doi: 10.1007/s10456-018-9600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.