Abstract
Purpose
The methods (IHC/FISH) typically used to assess ER, PR, HER2, and Ki67 in FFPE specimens from breast cancer patients are difficult to set up, perform, and standardize for use in low and middle-income countries. Use of an automated diagnostic platform (GeneXpert®) and assay (Xpert® Breast Cancer STRAT4) that employs RT-qPCR to quantitate ESR1, PGR, ERBB2, and MKi67 mRNAs from formalin-fixed, paraffin-embedded (FFPE) tissues facilitates analyses in less than 3 h. This study compares breast cancer biomarker analyses using an RT-qPCR-based platform with analyses using standard IHC and FISH for assessment of the same biomarkers.
Methods
FFPE tissue sections from 523 patients were sent to a College of American Pathologists-certified central reference laboratory to evaluate concordance between IHC/FISH and STRAT4 using the laboratory’s standard of care methods. A subset of 155 FFPE specimens was tested for concordance with STRAT4 using different IHC antibodies and scoring methods.
Results
Concordance between STRAT4 and IHC was 97.8% for ESR1, 90.4% for PGR, 93.3% for ERBB2 (IHC/FISH for HER2), and 78.6% for MKi67. Receiver operating characteristic curve (ROC) area under the curve (AUC) values of 0.99, 0.95, 0.99, and 0.85 were generated for ESR1, PGR, ERBB2, and MKi67, respectively. Minor variabilities were observed depending on the IHC antibody comparator used.
Conclusion
Evaluation of breast cancer biomarker status by STRAT4 was highly concordant with central IHC/FISH in this blinded, retrospectively analyzed collection of samples. STRAT4 may provide a means to cost-effectively generate standardized diagnostic results for breast cancer patients in low- and middle-income countries.
Electronic supplementary material
The online version of this article (10.1007/s10549-018-4889-5) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer biomarker assays, STRAT4, Estrogen receptor, Progesterone receptor, Human epidermal growth factor receptor 2, Tumor proliferation rate, IHC, FISH
Introduction
Breast cancer is becoming increasingly recognized as a major health problem in low- and middle-income countries (LMIC) [1–5]. Although the impact of cancer diagnoses overall has often been overshadowed in these settings by infectious diseases like tuberculosis, malaria, and HIV, the numbers of patients affected by breast cancer is already substantial, and is likely to increase among LMIC in regions where populations are growing the fastest [6].
Currently, however, the treatment of breast cancer in LMIC is fraught with difficulty. In recent years, effective treatments like tamoxifen have become available at low or no cost for women with estrogen receptor (ER) positive breast cancer, accounting for approximately two-thirds of cases. Unfortunately, access to high-quality diagnostic technologies capable of identifying a tumor as ER-positive have been difficult to set up and maintain in a standardized and cost-effective manner (personal communication, John Flanigan, Senior Advisor, Center for Global Health, National Cancer Institute), owing largely to their reliance on antibody-based methods requiring significant expertise to perform and interpret. As lower cost biosimilars of trastuzumab become available [7, 8], breast cancer patients with tumors that overexpress the human epidermal growth factor receptor 2 (HER-2) may find themselves struggling to access a highly effective drug because diagnostic tests that are standard of care for every breast cancer patient in the United States and Europe are unavailable to women in LMIC.
Stimulated by several studies that showed an association between quantitative measurements of mRNA for the transcripts encoding ER and HER-2 (ESR1 and ERBB2) and clinical outcomes on tamoxifen and trastuzumab, respectively [9–11], we anticipated that an assay based on quantitative, real-time, polymerase chain reaction (RT-qPCR) methodology would be highly concordant with central measurements of ER and HER-2 using IHC and/or FISH, and might, therefore, be extremely useful in LMIC. A critical consideration in the genesis of this idea was the fact that such RT-qPCR assays could be developed and run on a distributed diagnostic platform called the GeneXpert®, [(Cepheid, Sunnyvale, CA, USA), (http://www.cepheid.com/us/cepheid-solutions/systems/genexpert-systems/genexpert-i)], which performs automated sample preparation and multiplexed RT-qPCR assays in approximately 2 h. The platform is designed for ease of use, and is already widely distributed throughout the world with more than 17,000 instruments running in 182 countries. Moreover, the platform has been adapted to extract nucleic acids from formalin-fixed, paraffin-embedded tissue (FFPE), the most common tissue-type employed by pathologists for the analysis of breast cancer specimens.
Thus, we aimed to demonstrate that the measurement of mRNAs for the analytes ESR1 and ERBB2 were concordant with high-quality central laboratory assessments by immunohistochemistry (IHC) for ER and HER2 protein expression and fluorescence-in-situ-hybridization (FISH) for HER-2 gene amplification. A multiplexed assay was built that included, in addition to ESR1 and ERBB2, primers and probes to detect and quantitate mRNAs for the progesterone receptor (PR, PGR) and the cell proliferative antigen identified by monoclonal antibody Ki-67 (Ki67, MKi67). The panel is referred to as Xpert® Breast Cancer STRAT4 (STRAT4). Once constructed, the assay was analytically validated by demonstrating linearity and dynamic range, analytical sensitivity (minimal sample input), analytical specificity (tests for interfering substances), prevention of carryover contamination, and assay kit stability (Chu et. al. manuscript submitted for publication). Additional studies were performed to examine the impact of pre-analytical sample handling (selection of invasive carcinoma for testing, macro-dissection techniques, and STRAT4 assay performance by different pathologists) on assay result variability, as well as assess concordance with central IHC/FISH and define preliminary cutoff values (Wong et al. manuscript submitted for publication).
The current study is designed to investigate concordance between STRAT4 and standard IHC and FISH performed at a central laboratory using a large cohort of FFPE specimens tested in a blinded, retrospective manner, and interpreted according to the 2013–2014 ASCO/CAP guidelines. In addition, we examined the concordance between STRAT4 and several different antibodies commonly used in IHC assays performed at different central laboratories.
STRAT4 is a CE-IVD (Conformité Européene In-vitro Medical Device) product that is available in some, but not all, European countries, and is not available in the United States. Where the STRAT4 assay is not available under CE-IVD, evaluations of its performance using specimens prepared under local pre-analytical sample handling procedures can be supported under collaborative research agreements using a Research Use Only version.
Materials and methods
Specimen collection, IHC and HER-2 FISH analysis
523 surgically excised breast tumors prepared as FFPE specimens, ranging from 6 months to 22 years in block age, were sourced from five institutes worldwide. Tumor blocks were selected based on what was available at each site. For each specimen, one sectioned slide was stained with hematoxylin and eosin (H&E) and used by pathologists to mark tumor areas, estimate tumor size, and estimate percentage tumor content. Serial unstained tumor samples (4 µm in thickness) were delivered to Cepheid for STRAT4 testing and to the University of Southern California (USC, Los Angeles, CA) Breast Cancer Analysis Laboratory for ER, PR, HER2, and Ki67 IHC and HER-2 FISH analyses. More sections were cut from a subset of tumor blocks (155 out of 523 total blocks) and sent to USC, Molecular Pathology Laboratory Network, Inc. (MPLN, Maryville, TN), and LabCorp (previously Pathology Inc., Torrance, CA) where different antibodies and scoring methods were used to generate IHC results for each analyte. Each reference laboratory generated its own H&E slides for each sample. All IHC, HER-2 FISH and STRAT4 testing was performed within two weeks after block sectioning. Only a subset of samples was tested for concordance for Ki67/MKi67. The immunohistochemical assay methods used in the USC central laboratory for assessment of ER, PR, Ki67 and HER2 are described elsewhere [21–24].
Sample processing and testing for STRAT4
FFPE samples were processed according to the package insert instructions of the STRAT4 assay kit. For each specimen, one unstained slide was overlaid onto the H&E slide which had been marked by a pathologist to select the invasive carcinoma, and then was used to choose the material to be macro-dissected into a 1.5 mL eppendorf tube using a razor blade. Macro-dissected tumor material was then mixed with 1.2 mL of FFPE lysis reagent and 20 µL of proteinase K. The tubes containing the sample lysate were placed in heat blocks for incubation at 80 °C for 30 min. Sample lysate was then mixed with 1.2 mL of ethanol (molecular biology grade, Sigma-Aldrich). For each sample, 520 µL of the lysate was transferred to the sample chamber of a STRAT4 cartridge and placed into a GeneXpert module for RNA extraction, purification, and RT-qPCR analysis.
GeneXpert DX software analysis settings
ESR1, PGR, ERBB2, and MKi67 mRNA measurements were normalized against the mRNA measurement of the internal reference target Cytoplasmic FMR1-Interacting Protein 1 (CYFIP1). Optical readouts of PCR amplifications and cycle threshold (Ct) determination for all targets and CYFIP1 in STRAT4 test runs were analyzed with settings specified in the GeneXpert DX software. Delta Ct assay cutoffs for ESR1 and ERBB2 were set at “− 1” and dCt cutoffs for PGR and MKi67 were set at “− 3.5” and “− 4”, respectively. Preliminary assay cutoffs were determined in a previous study involving 32 FFPE breast cancer samples that were tested by both STRAT4 and central IHC/FISH in a reference laboratory. The delta Ct numerical limits were set to maximize the concordance with IHC (IHC/FISH for HER2). To minimize the rate of false negatives for PGR and MKi67, a minimum assay input value of 31 for the CYFIP Ct was implemented. If the dCt for PGR or MKi67 was lower than the pre-specified cutoffs (dCt = − 3.5 for PGR or − 4 for MKi67) and the CYIFIP Ct was greater than 31, the result was reported as “INDETERMINATE” instead of “NEGATIVE” indicating that the minimum assay input criteria had not been met (CYFIP1 Ct ≥ 31), and the test should be repeated adding more lysate to the cartridge to achieve a CYFIP1 Ct of at least 31.
Results
Among 523 specimens tested with the STRAT4 assay, 503 samples yielded valid test results (“POSITIVE” or “NEGATIVE”) for at least one assay target. 20 specimens had no or insufficient PCR amplification signal for the reference RNA CYFIP1 (CYFIP1 Ct > 35). Most of these 20 specimens came from FFPE blocks that were more than 10-years old (data not shown).
Agreement rates between ESR1 mRNA and ER protein by IHC
The overall concordance rate of the STRAT4 ESR1 results compared with central IHC results was 97.8% using either a 1% or 10% immunostaining level for positivity (Table 1).
Table 1.
Comparison of mRNA and protein status for ESR1 (ER), PGR (PR), ERBB2 (HER2), and MKI67 (Ki67) determined by IHC and RT-qPCR
| Analyte | IHC+/RTqPCR+ | IHC−/RTqPCR+ | IHC+/RTqPCR− | IHC−/RTqPCR− | Total number | Sensitivity (PPA) (95% CI) | Specificity (NPA) (95% CI) | Kappa statistic (95% CI) | Concordance rate (OPA) (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| ESR1 (ER)* | 407 | 4 | 7 | 75 | 493 | 98.3% (96.5–99.3%) | 94.9% (87.5–98.6%) | 91.8% (87.1–96.6%) | 97.8% (96.0–98.9%) |
| ESR1 (ER)x | 404 | 7 | 4 | 78 | 493 | 99.0% (97.5–99.7%) | 91.8% (83.8–96.9%) | 92.1% (87.4–96.7%) | 97.8% (96.0–98.9%) |
| PGR (PR)* | 333 | 23 | 23 | 99 | 478 | 93.5% (90.5–95.9%) | 81.1% (73.1–87.7%) | 74.7% (67.8–81.6%) | 90.4% (87.4–92.9%) |
| PGR (PR)x | 320 | 36 | 7 | 115 | 478 | 97.9% (95.6–99.1%) | 76.2% (68.8–82.7%) | 78.1% (71.9–84.2%) | 91.0% (88.1–93.4%) |
| ERBB2 (HER2)** | 66 | 29 | 4 | 391 | 490 | 94.3% (86.0–98.4%) | 93.1% (90.2% − 95.3%) | 76.1% (68.4–83.8%) | 93.3% (90.7–95.3%) |
| ERBB2 (HER2)xx | 42 | 16 | 2 | 361 | 421 | 95.5% (84.5–99.4%) | 95.8% (93.2% − 97.6%) | 80.0% (71.1–88.9%) | 95.7% (90.7–95.3%) |
| ERBB2 (HER2)xxx | 56 | 23 | 6 | 242 | 327 | 90.3% (80.1–96.4%) | 91.3% (87.3% − 94.4%) | 73.9% (65.0–82.8%) | 91.1% (87.5–94.0%) |
| ERBB2 (HER2)*** in ER+ | 39 | 28 | 1 | 341 | 389 | 97.5% (86.8–99.9%) | 92.4% (89.2–94.9%) | 69.1% (58.8–79.5%) | 92.9% (90.0–95.2%) |
| ERBB2 (HER2)**** in ER− | 27 | 1 | 3 | 48 | 79 | 90.0% (73.5–97.9%) | 98.0% (89.1–99.9%) | 89.1% (78.7–99.5%) | 94.9% (87.5–98.6%) |
| MKI67 (Ki67)xxxx | 117 | 63 | 15 | 94 | 289 | 88.6% (82.0%–93.5%) | 59.9% (51.8%–67.6%) | 47.1% (37.6–56.6%) | 73% (67.5–78%) |
| MKi67 (Ki67)xxxxx | 155 | 25 | 37 | 72 | 289 | 80.7% (74.5–85.7%) | 74.2% (64.7–81.9%) | 53.3% (43.2–63.5%) | 78.6% (73.4–82.9%) |
*Using the IHC cut-off of 1% as recommended by ASCO-CAP (2010)
xUsing the IHC cut-off of 10% cut-off, as described elsewhere
**Using central IHC and central FISH for resolution of IHC 2+ to either FISH-negative or FISH-positive, as recommended by the ASCO-CAP guidelines for HER2 testing (2013/2014)
xxComparison of STRAT4 ERBB2 dCt result and HER2 result with IHC 2+ equivocals excluded from analysis using the Herceptest
***In the ER-positive subset only (as determined by IHC), overall percent agreement (OPA), positive percent agreement (PPA), and negative percent agreement (NPA) are shown for the comparison of STRAT4 ERBB2 dCt result and HER2 result by central IHC and central FISH where IHC 2+ equivocals were resolved to positive or negative calls by the FISH assay
xxxComparison of STRAT4 ERBB2 dCt result and HER2 result by central FISH
****In the ER-negative subset only (as determined by IHC), overall percent agreement (OPA), positive percent agreement (PPA), and negative percent agreement (NPA) are shown for the comparison of STRAT4 ERBB2 dCt result and HER2 result by central IHC and central FISH where IHC 2+ equivocals were resolved to positive or negative calls by the FISH assay
xxxxComparison of STRAT4 MKi67 dCt result and Ki67 result by central IHC using a Ki67 IHC cutoff of 20% to discriminate “high proliferation rate” from “low proliferation rate”
xxxxxComparison of STRAT4 MKi67 dCt result and Ki67 result by central IHC using a Ki67 IHC cutoff of 10% to discriminate “high proliferation rate” from “low proliferation rate”
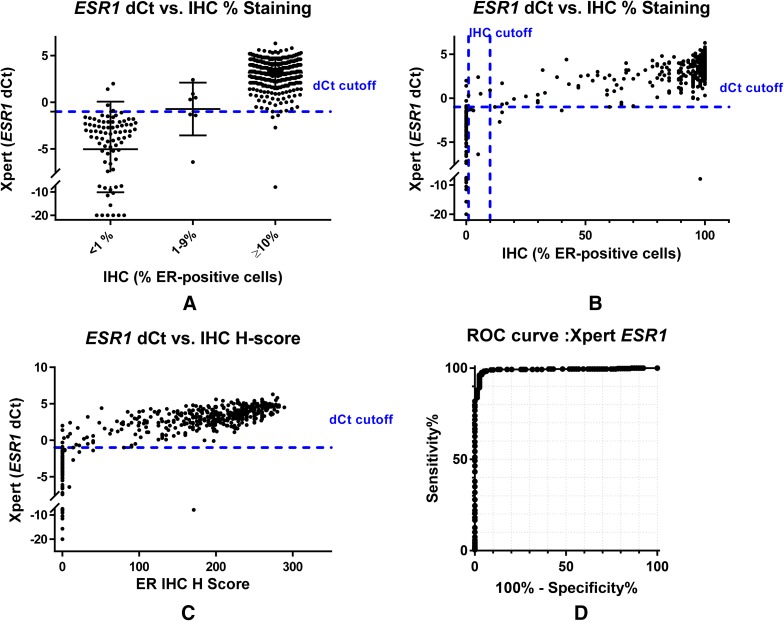
ESR1 dCt values were plotted against percent positive staining treated as categorical or continuous variables and H score for the same samples (Fig. 1a–c). These data suggest high levels of concordance between STRAT4 and central IHC for ESR1/ER, and demonstrate that the discordant samples are nearly all close to the ESR1 dCt cutoff.
Fig. 1.
Comparison of estrogen receptor status determined by immunohistochemistry and RT-qPCR (STRAT4 or [STRAT4 (Xpert)]) assays. a Graph of STRAT4 ESR1 dCt values by ER IHC result categorized as negative (0%), low positive (1–9%), or positive (≥ 10%). Among ER-positive and ER-negative breast cancers according to IHC assessment, there is also a clear separation by ESR1 mRNA by RT-qPCR into high and low expression subgroups. In contrast, those breast cancers with from 1 to 9% ER-positive carcinoma cells have predominantly ESR1 mRNA quantities near the RT-qPCR cut-off separating “positive” from “negative”. b Comparison of STRAT4 ESR1 dCt values according to ER IHC % staining alone. The plot of ER IHC percentage positive tumor cell immunohistochemical staining demonstrated a strong correlation with ESR1 mRNA quantity. c Graph of STRAT4 ESR1 dCt values by ER IHC H-Score. H-Score is defined as [3(% of tumor staining 3+)] + [2(% of tumor staining 2+)] + [1(% of tumor staining 1+)]. Quantitative stratification of the IHC protein assessment by combining percentage of immunostained tumor cells with intensity of immunohistochemical staining demonstrated an improved correlation with ESR1 mRNA determined by RT-qPCR. d The ROC curve for STRAT4 ESR1 including all samples in the analysis. The area under the curve (AUC) is 0.99
Agreement rates between PGR mRNA and PR protein by IHC
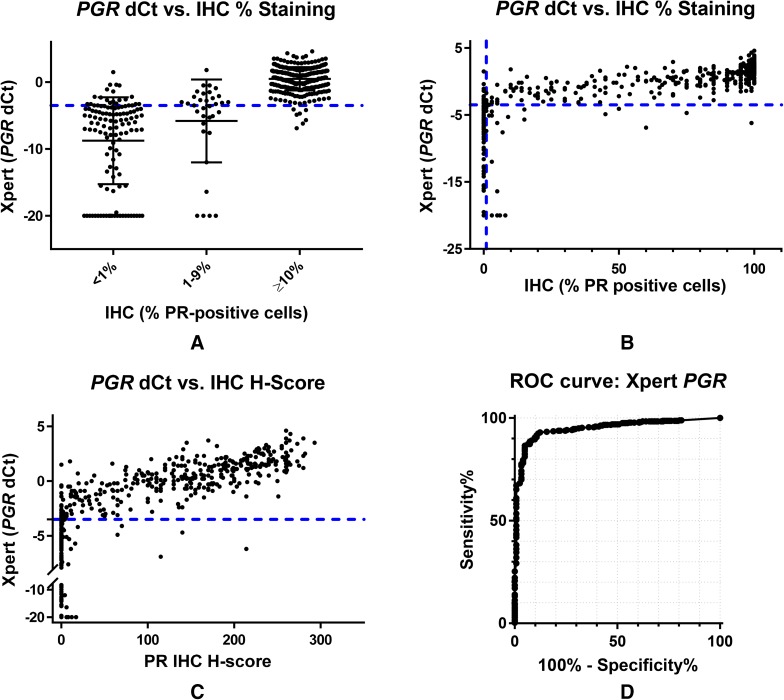
The overall concordance rate between the STRAT4 PGR results and the central PR IHC results with the PGR 636 antibody was in the 90–91% range whether 1 or 10% staining was used to determine PR-positive status (Table 1). Fifteen samples with “indeterminate” STRAT4 PGR results (delta Ct < − 3.5 and CYFIP Ct > 31) were excluded from the concordance analysis. The correlation between PGR dCt values and PR IHC results considered as a categorical variable suggests that there are more samples close to the PGR dCt cutoff with different IHC staining results, explaining the lower overall percent agreement for PGR (Fig. 2a). Of samples considered as low positives by IHC (1–9% PR staining), roughly half were called positive by STRAT4 and half were called negative. Scatterplots of STRAT4 PGR dCt values compared with PR IHC percent staining and H Score suggest a positive correlation between the absolute level of PGR transcript detected and the absolute amount of PR staining observed by IHC (Fig. 2b, c).
Fig. 2.
Comparison of progesterone receptor status determined by immunohistochemistry (IHC) and RT-qPCR (STRAT4) assays. a Graph of STRAT4 PGR dCt values by PR IHC result categorized as negative (0%), low positive (1–9%), or positive (≥ 10%). Among PR-positive and PR-negative breast cancers according to IHC assessment, there is also a relatively good separation by PGR mRNA by RT-qPCR into high and low expression subgroups. In contrast, the majority of those breast cancers with 1–9% PR-positive carcinoma cells have predominantly PGR mRNA quantities near the RT-qPCR cut-off separating “positive” from “negative”. b Graph of STRAT4 PGR dCt values by PR IHC % staining. The plot of the percentage of PR-positive tumor cells by immunohistochemical staining demonstrated a strong correlation with PGR mRNA quantity which was improved only slightly by consideration of tumor cell IHC staining intensity as shown in c. c Graph of STRAT4 PGR dCt values by PR IHC H-Score. H-Score is defined as [3(% of tumor staining 3+)] + [2(% of tumor staining 2+)] + [1(% of tumor staining 1+)]. Quantitative stratification of the IHC protein assessment by combining percentage of immunostained tumor cells with intensity of immunohistochemical staining demonstrated a slightly improved correlation with PGR mRNA determined by RT-qPCR. d The ROC curve for STRAT4 PGR including all samples in the analysis. The area under the curve (AUC) is 0.95
Agreement rates between ERBB2 mRNA and HER2 protein by IHC (and FISH)
The overall concordance rate between STRAT4 ERBB2 and HER-2 IHC was 95.7%, excluding samples with staining results of “2+” (Table 1). Concordance between STRAT4 and FISH alone was 91.1% (Table 1). When STRAT4 was compared to IHC plus FISH, where IHC 2+ samples were tested by FISH and categorized as either ISH-positive or ISH-negative, the overall positive agreement rate was 93.3% (Table 1). Finally, similar concordance rates were obtained when the population was stratified first by ER status (Table 1).
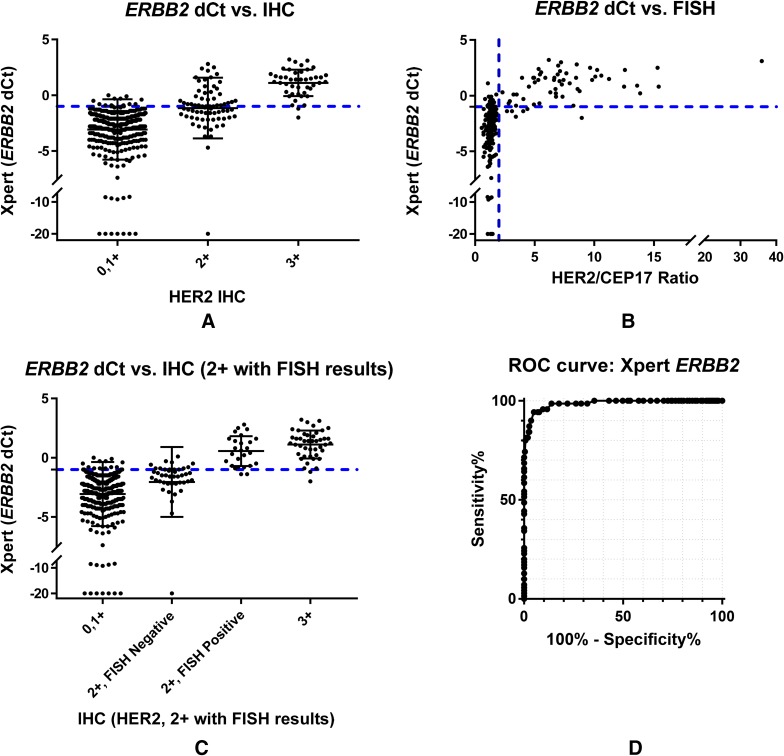
Comparison of STRAT4 ERBB2 dCt values and IHC results demonstrated minimal overlap in the IHC negative (0–1+) and IHC positive (3+) populations (Fig. 3a), while the IHC equivocal group (2+) is almost perfectly bisected by the ERBB2 dCt cutoff. A comparison between ERBB2 dCt values and the HER-2/CEP17 ratios determined by FISH demonstrates an apparent correlation between increasing HER-2/CEP17 ratios and the amount of ERBB2 transcript detected by STRAT4 (Fig. 3b).
Fig. 3.
Comparison of human epidermal growth factor receptor 2 determined by either RT-qPCR or by immunohistochemistry with or without FISH assessment of IHC2+. a Graph of STRAT4 ERBB2 dCt values by HER2 IHC result categorized as negative (0–1+), equivocal (2+), or positive (3+). There is a significant correlation between HER2 status determined by IHC and ERBB2 mRNA determined by RT-qPCR with IHC 0/1+ showing low-level expression of ERBB2 mRNA and IHC 3+ showing high-level ERBB2 mRNA expression. The IHC 2+ breast cancers appear to have a level of ERBB2 mRNA intermediate between that of the IHC 0/1+ and IHC 3+ subgroups. b Graph of STRAT4 ERBB2 dCt values by HER2/CEP17 Ratio by the FISH assay. Increasing levels of gene amplification determined by FISH (increasing HER2/CEP17 ratio) are associated with increasing levels of ERBB2 mRNA expression, as expected. c Graph of STRAT4 ERBB2 dCt values by IHC plus FISH where FISH was used to resolve the IHC 2+ equivocals into HER2-positive or HER2-negative status. Although the IHC 2+ breast cancers appear to have a level of ERBB2 mRNA intermediate between that of the IHC 0/1+ and IHC 3+ subgroups (as illustrated in A above), further stratification can be achieved by the use of FISH in this group to determine which cases are HER2-amplified and which are not. Those IHC 2+ breast cancers with HER2-amplification by FISH have ERBB2 mRNA expression levels similar to IHC 3+ breast cancers. In contrast, those IHC 2+ breast cancers that are HER2-not-amplified by FISH have ERBB2 mRNA expression levels similar to IHC 1+ breast cancers. d The ROC curve for STRAT4 ERBB2 including all samples in the analysis. The area under the curve (AUC) is 0.99
Figure 3c shows the correlation between ERBB2 dCt values by STRAT4 and the combined results generated when IHC and FISH are used together to determine HER-2 over-expression. Among those patients who were IHC 2+, STRAT4 for ERBB2 overwhelmingly agreed with FISH when it came to resolving those patients into HER-2 positive and HER-2 negative groups.
Stratifying the population into ER-positive and ER-negative sub-populations and then examining the correlation between the ERBB2 dCt values and HER-2 results by IHC + FISH suggests that the assay is slightly better at discriminating over-expressors in the ER-negative than the ER-positive sub-population, but the difference is small (Supplementary Fig. 1).
Agreement rates between MKi67 mRNA and Ki67 proliferation rates determined by IHC
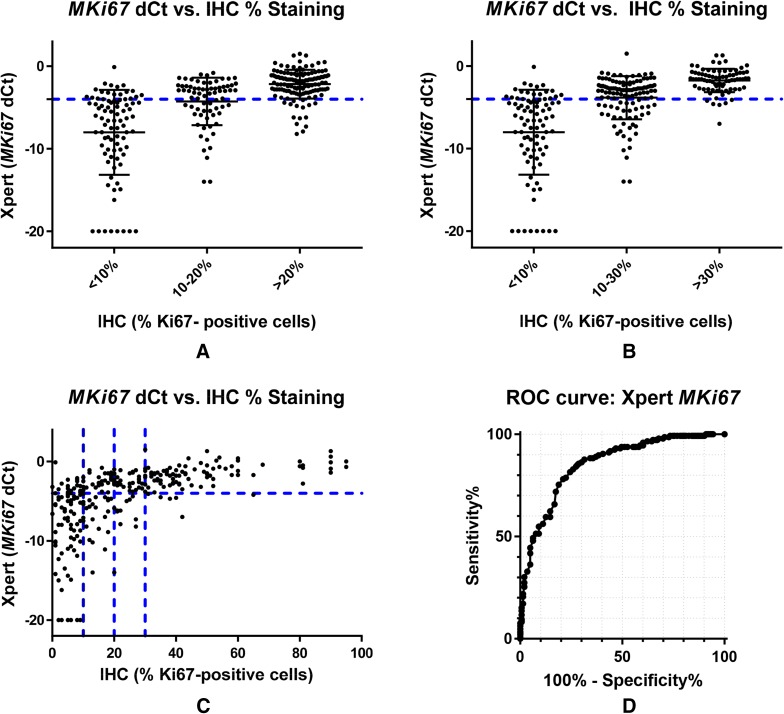
For MKi67/Ki67 concordance, twenty-four samples with indeterminate STRAT4 results were excluded from the analysis. We examined the correlation between MKi67 by RT-qPCR and Ki67 by IHC using cutoffs of 10% and 20% for the determination of high proliferation rate. Although many laboratories consider < 10% to indicate low proliferation rate, > 20% to indicate high proliferation rate, and use an intermediate zone between 10 and 20%, we elected to choose single cutoffs for these analyses because we had not previously defined an intermediate zone for the MKi67 dCt distribution. The overall positive agreement rate for STRAT4 MKi67 results with Ki67 by IHC using a 20% cutoff was 73%, and using a 10% cutoff was 78.6%. (Table 1). Comparison of MKi67 dCt values with Ki67 IHC results categorized as low (< 10%), intermediate (10–20%), or high (> 20%) offers a possible explanation for these lower concordance rates. Rather than distinct subsets, the distribution appears as a continuum with the large intermediate population showing substantial overlap with both the low and high populations, although the median values of each sub-population are clearly different, and correlate positively with increasing MKi67 dCt values by STRAT4 (Fig. 4a). Expanding the intermediate zone to include patients with Ki67 IHC values of 10–30% leads to the same conclusion (Fig. 4b). Comparison of MKi67 dCt values with a continuous measure of IHC % immunostaining for Ki67 demonstrates a weak correlation with STRAT4 and significant scatter at low proliferation rates below 30% (Fig. 4c).
Fig. 4.
Comparison of Ki67 proliferation rate determined by either RT-qPCR or immunohistochemistry. a Graph of STRAT4 MKi67 dCt values by Ki67 IHC % staining where the IHC high proliferation rate cutoff is defined as 20% and the intermediate proliferation rate is defined as 10–20% with < 10% considered a low proliferation rate. There is some overlap in MKi67 mRNA values between the high (> 20%) and low (< 10%) proliferation rate groups, with the intermediate group (10–20%) showing intermediate mRNA values by RT-qPCR, but with substantial overlap with both the high and low proliferation rate groups. MKi67 as measured by RT-qPCR appears as a continuum without a clear cutoff evident from the distributions when compared to Ki67 levels measured by IHC. b Graph of STRAT4 MKi67 dCt values by Ki67 IHC % staining where the IHC positivity cutoff is defined as 30% and the equivocal zone is defined as 10–30%. Raising the IHC cutoff for the determination of high proliferation rate has no appreciable impact on the correlation between RT-qPCR and IHC methods. The MKi67 distribution still shows a continuum of values without a clear cutoff. c Graph of STRAT4 MKi67 dCt values by Ki67 IHC % staining. There appears to be a discernable correlation between the percentage of tumor cells with immunochemical staining for Ki67 with MKi67 mRNA levels by RT-qPCR, especially at levels above 40%. d The ROC curve for STRAT4 MKi67 where all samples were included in the analysis. The area under the curve (AUC) is 0.85
Receiver-operator characteristic analyses
STRAT4 results were compared with central IHC/FISH using receiver-operator characteristic (ROC) analyses for all four analytes (Figs. 1d, 2d, 3d, 4d). The ROC area under the curve (AUC) values for each of the four analytes were 0.99 for ESR1 and ERBB2, 0.95 for PGR, and 0.85 for MKi67. For MKi67, the AUC value improved to 0.92 when the equivocal samples (IHC Ki67 staining of 10–20%) were excluded from the analysis (Supplementary Fig. 2). The shape of the curves for ESR1 and ERBB2, the two most important analytes for LMIC use, demonstrated that the STRAT4 assay is highly correlated with results generated using conventional IHC and FISH assays. While the PGR assay results were highly correlated according to the ROC curve, they were not as highly correlated as ESR1 and ERBB2. The ROC curve for MKi67 STRAT4 and conventional IHC was the weakest of the four correlations. Whether all patients were included in the analysis, or the 10–20% intermediate group were excluded, the data suggest that it is more difficult to identify a clear cutoff when the distribution appears to define a continuum, an observation that has been suggested even when using IHC to measure this analyte [19].
Variability of STRAT4 concordance with different IHC antibodies and scoring methods
The antibodies selected for the primary concordance analysis in this study were the antibodies used at the central reference laboratory, but not all reference laboratories used the same antibodies to perform IHC for these four analytes. In order to get a sense of how disparate the concordance between STRAT4 and IHC might be when compared across a variety of central reference laboratories, we selected a subset of samples (n = 155) and tested them at three different central reference laboratories that used different IHC antibodies and scoring methods. The concordance rates for STRAT4 ESR1 to IHC ER range from 97.8% for the 6F11 antibody scored manually to 91.9% for the SP1 antibody using an automated scoring system (Supplementary Fig. 3). Concordance across different IHC methods for ER were in the 94–95% range (data not shown).
For STRAT4 PGR to PR IHC comparisons, the concordance ranged from 94.4 to 89% while the IHC to IHC comparisons were similar at 94.3 and 93.6% (Supplementary Fig. 4).
For the STRAT4 ERBB2 to IHC HER2 comparisons, with equivocal breast cancers (IHC 2+ samples) excluded, concordance ranged from 94.3 to 92.8%, and from 93.3 to 91.6% if FISH was employed to resolve the IHC 2+ samples (Supplementary Table 1). IHC to IHC comparisons using different antibodies were not performed.
For the STRAT4 MKi67 to IHC Ki67 comparisons, excluding samples with IHC staining in the 10–20% range, concordance was quite variable depending on the antibody/method comparator. Using the MIB1 antibody and manual scoring, concordance was 84.6%, while the same antibody used in a different central laboratory and employing an automated scoring system yielded only a 63.7% concordance. Another automated scoring system using antibody 30-9 was intermediately concordant at 76%. A comparison of the STRAT4 MKi67 dCt values to % staining with these three antibodies/methods is shown in Supplementary Fig. 5.
Discussion
The measurement of mRNAs that encode protein biomarkers like ER, PR, HER2, and Ki67 (ESR1, PGR, ERBB2, and MKi67, respectively) on an automated, broadly distributed diagnostic platform is feasible and carries certain advantages for LMIC applications, including ease-of-use, accessibility, standardization, reproducibility, and a short time-to-result. These features suggest that such an approach has real potential for clinical benefit in the management of patients with breast cancer in low- and middle-income countries, where accessing more standard diagnostic methods like IHC and FISH is difficult. The ASCO/CAP guidelines currently recommend IHC and/or FISH for the determination of hormone receptor or HER-2 status for the purpose of selecting therapy, therefore, it is important that we determine the degree of concordance between STRAT4 and these standard methods [12–14].
In prior studies, we have shown that STRAT4 is analytically valid and capable of generating reproducible results even when variability in pre-analytical sample handling exists across laboratories performing the test. Our goal in this study was to determine the degree of concordance between measurements of the mRNAs ESR1, PGR, ERBB2, and MKi67 by STRAT4 and central laboratory IHC and FISH results for ER, PR, HER2, and Ki67.
The results demonstrate high concordance for ESR1/ER and ERBB2/HER2 with ROC AUC values of 0.99 for both, and very good concordance for PGR/PR (ROC AUC = 0.95). For MKi67/Ki67, including all samples in the analysis, the ROC AUC was reduced to 0.85. This may not be surprising, however, due to the significant challenges already described in achieving standardization of Ki67 result reporting by IHC across laboratories [15–18]. Denkert and colleagues [19] have shown that Ki67 results by IHC are highly variable unless at least 500, and better yet 1000, cells are carefully counted for each sample. Multicenter studies to address improved methods of standardization for Ki67 measurement by IHC are in progress. For these markers, examination of direct correlations between the RNA measurements and clinical outcomes is desirable, but this is particularly important for Ki67, where the establishment of MKi67 dCt cutoffs derived directly from clinical outcomes would obviate the need for further concordance studies and perhaps offer pathologists an easier method of reliably measuring this proliferation marker.
The results of the antibody comparison studies demonstrate some minor variabilities in concordance rates depending on the laboratory, antibody, and scoring method used for ESR1/ER, PGR/PR, and ERBB2/HER2, suggesting that the STRAT4 assay results are generally concordant across central laboratories using different methods. For MKi67/Ki67, there was significant variability in concordance, but once again, this is not unexpected given the challenges associated with Ki67 assessments by IHC.
The dCt cutoffs used for all four of the STRAT4 analytes were pre-specified based on prior testing in small datasets. However, we learned during the performance of this study that reliable negative results for both PGR and MKi67 depended upon having a larger amount of sample available for analysis by the cartridge than it did for ESR1 or ERBB2. Based on these observations, the requirement was established that for each of these two analytes, the CYFIP1 internal control Ct value needed to be ≤ 31 rather than 35, which is the CYFIP1 Ct cutoff for ESR1 and ERBB2. Thus, select samples were classified as indeterminate if the PGR or MKi67 values were below the cutoff for positivity and also had a CYFIP1 Ct of greater than 31. Moving forward, such samples would be re-run using the lysate from the entire FFPE section rather than only 520 µL, which represents 25% of the final lysate volume.
The current study suggests that the determination of ESR1, PGR, ERBB2, and MKi67 RNA levels by RT-qPCR on the GeneXpert automated diagnostic platform is not only feasible, but also generates results from FFPE tumor sections that are highly concordant with high quality central laboratory measurements of ER, PR, and HER2 using standardized IHC and FISH assays. For Ki67, the continuous nature of the distribution of values we observed as well as known difficulties in standardizing IHC assessments across laboratories presents unique challenges, but one we hope to address more fully by using clinical outcomes from appropriately designed studies to define the STRAT4 MKi67 dCt cutoff(s) that will provide the clearest and most clinically informative interpretation.
STRAT4 has already been shown to be highly concordant with automated quantitative analysis of IHC (AQUA) [20]. From a LMIC perspective, ESR1 and ERBB2 measurements have the greatest relevance currently. As such, given these results, the STRAT4 assay could be considered a potential solution to the problem of limited access to breast cancer diagnostics that currently exists for patients in low resource countries, and should move forward to a prospective concordance study without delay, paying particular attention to the impact of local sample handling and fixation methods on STRAT4 results. Assuming the STRAT4 assay can be validated for use in those geographies, prior experience with the GeneXpert system can be leveraged for rapid progress toward a workable diagnostic solution for patients with breast cancer in countries with limited healthcare resources, particularly LMIC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Dr. John Flanigan (Center for Global Health, National Cancer Institute, Bethesda, MD), Dr. Jane Brock (Department of Pathology, Brigham and Women’s Hospital, Boston MA), Dr. Dan Milner (American Society for Clinical Pathology, Chicago, IL), and Dr. Carol Harris (Department of Medicine, Albert Einstein College of Medicine, New York, NY) for helpful discussions and insightful observations. The authors would also like to thank Roberta Guzman (USC) and Angela Santiago (USC) for laboratory technical assistance with FISH and IHC and Ivonne Villalobos (USC) for data management.
Funding
This study was funded by Cepheid, Sunnyvale, CA.
Conflict of interest
Authors NCW, WW, KEH, VCC, and AR declared themselves to be employees of Cepheid and have received remuneration from Cepheid. Author SD declared no conflict of interest. Author DK declared no conflict of interest. Author RM declared no conflict of interest. Author JJ declared being a consultant for Bristol-Myers-Squib and G1 Therapeutics. Author RD declared no conflict of interest. Author WB declared no conflict of interest. Author BR declared no conflict of interest. Author TF declared no conflict of interest. Author JLK and MS are employees of Molecular Pathology Laboratory Network, Inc. and declared receiving remuneration from Molecular Pathology Laboratory Network, Inc. Author SC declared no conflict of interest. Author HJ declared remuneration and stock ownership from Indivumed. Authors JMW and MB declared receiving remuneration from Cepheid and stock ownership from Danaher. Author MFP declared consulting for Brogent International LLC and his institution has received research grants for his laboratory from Cepheid, Eli Lilly and Company, Novartis Pharmaceuticals Corporation, Pfizer Inc, F. Hoffmann-La Roche Ltd, and Zymeworks.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the respective institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from individual participants included in the study by Indivumed.
References
- 1.Akuoko CP, Armah E, Sarpong T, Quansah DY, Amankwaa I, Boateng D. Barriers to early presentation and diagnosis of breast cancer among African women living in sub-Saharan Africa. PLoS ONE. 2017;12(2):e0171024. doi: 10.1371/journal.pone.0171024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jedy-Agba E, McCormack V, Adebamowo C, dos-Santos-Silva I. Stage at diagnosis of breast cancer in sub-Saharan Africa: a systematic review and meta-analysis. Lancet Glob Health. 2016;4(12):e923–e935. doi: 10.1016/s2214-109x(16)30259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefan DC. Cancer care in Africa: an overview of resources. J Glob Oncol. 2015;1(1):30–36. doi: 10.1200/jgo.2015.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson BO. Breast cancer–thinking globally. Science. 2014;343(6178):1403. doi: 10.1126/science.1253344. [DOI] [PubMed] [Google Scholar]
- 5.Edge J, Buccimazza I, Cubasch H, Panieri E. The challenges of managing breast cancer in the developing world: a perspective from sub-Saharan Africa. S Afr Med J. 2014;104(5):377–379. doi: 10.7196/SAMJ.8249. [DOI] [PubMed] [Google Scholar]
- 6.Bray F, Moller B. Predicting the future burden of cancer. Nat Rev Cancer. 2006;6(1):63–74. doi: 10.1038/nrc1781. [DOI] [PubMed] [Google Scholar]
- 7.Rugo HS, Linton KM, Cervi P, Rosenberg JA, Jacobs I. A clinician’s guide to biosimilars in oncology. Cancer Treat Rev. 2016;46:73–79. doi: 10.1016/j.ctrv.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Rugo HS, Barve A, Waller CF, Hernandez-Bronchud M, Herson J, Yuan J, Sharma R, Baczkowski M, Kothekar M, Loganathan S, Manikhas A, Bondarenko I, Mukhametshina G, Nemsadze G, Parra JD, Abesamis-Tiambeng ML, Baramidze K, Akewanlop C, Vynnychenko I, Sriuranpong V, Mamillapalli G, Ray S, Yanez Ruiz EP, Pennella E. Effect of a proposed trastuzumab biosimilar compared with trastuzumab on overall response rate in patients with ERBB2 (HER2)-positive metastatic breast cancer: a randomized clinical trial. Jama. 2017;317(1):37–47. doi: 10.1001/jama.2016.18305. [DOI] [PubMed] [Google Scholar]
- 9.Loi S, Dafni U, Karlis D, Polydoropoulou V, Young BM, Willis S, Long B, de Azambuja E, Sotiriou C, Viale G, Ruschoff J, Piccart MJ, Dowsett M, Michiels S, Leyland-Jones B. Effects of estrogen receptor and human epidermal growth factor receptor-2 levels on the efficacy of trastuzumab: a secondary analysis of the HERA trial. JAMA Oncol. 2016;2(8):1040–1047. doi: 10.1001/jamaoncol.2016.0339. [DOI] [PubMed] [Google Scholar]
- 10.Pogue-Geile KL, Kim C, Jeong JH, Tanaka N, Bandos H, Gavin PG, Fumagalli D, Goldstein LC, Sneige N, Burandt E, Taniyama Y, Bohn OL, Lee A, Kim SI, Reilly ML, Remillard MY, Blackmon NL, Kim SR, Horne ZD, Rastogi P, Fehrenbacher L, Romond EH, Swain SM, Mamounas EP, Wickerham DL, Geyer CE, Jr, Costantino JP, Wolmark N, Paik S. Predicting degree of benefit from adjuvant trastuzumab in NSABP trial B-31. J Natl Cancer Inst. 2013;105(23):1782–1788. doi: 10.1093/jnci/djt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim C, Tang G, Pogue-Geile KL, Costantino JP, Baehner FL, Baker J, Cronin MT, Watson D, Shak S, Bohn OL, Fumagalli D, Taniyama Y, Lee A, Reilly ML, Vogel VG, McCaskill-Stevens W, Ford LG, Geyer CE, Jr, Wickerham DL, Wolmark N, Paik S. Estrogen receptor (ESR1) mRNA expression and benefit from tamoxifen in the treatment and prevention of estrogen receptor-positive breast cancer. J Clin Oncol. 2011;29(31):4160–4167. doi: 10.1200/jco.2010.32.9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Poznak C, Somerfield MR, Bast RC, Cristofanilli M, Goetz MP, Gonzalez-Angulo AM, Hicks DG, Hill EG, Liu MC, Lucas W, Mayer IA, Mennel RG, Symmans WF, Hayes DF, Harris LN. Use of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2015;33(24):2695–2704. doi: 10.1200/JCO.2015.61.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF, American Society of Clinical O, College of American P Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 14.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG, Viale G, Zabaglo LA, Penault-Llorca F, Bartlett JM, Gown AM, Symmans WF, Piper T, Mehl E, Enos RA, Hayes DF, Dowsett M, Nielsen TO. An international Ki67 reproducibility study. J Natl Cancer Inst. 2013;105(24):1897–1906. doi: 10.1093/jnci/djt306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polley MY, Leung SC, Gao D, Mastropasqua MG, Zabaglo LA, Bartlett JM, McShane LM, Enos RA, Badve SS, Bane AL, Borgquist S, Fineberg S, Lin MG, Gown AM, Grabau D, Gutierrez C, Hugh JC, Moriya T, Ohi Y, Osborne CK, Penault-Llorca FM, Piper T, Porter PL, Sakatani T, Salgado R, Starczynski J, Laenkholm AV, Viale G, Dowsett M, Hayes DF, Nielsen TO. An international study to increase concordance in Ki67 scoring. Mod Pathol. 2015;28(6):778–786. doi: 10.1038/modpathol.2015.38. [DOI] [PubMed] [Google Scholar]
- 17.Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, McShane L, Paik S, Penault-Llorca F, Prudkin L, Regan M, Salter J, Sotiriou C, Smith IE, Viale G, Zujewski JA, Hayes DF. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103(22):1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11(2):174–183. doi: 10.1016/s1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 19.Denkert C, Budczies J, von Minckwitz G, Wienert S, Loibl S, Klauschen F. Strategies for developing Ki67 as a useful biomarker in breast cancer. Breast. 2015;24(Suppl 2):S67–S72. doi: 10.1016/j.breast.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 20.Wasserman BE, Carvajal-Hausdorf DE, Ho K, Wong W, Wu N, Chu VC, Lai EW, Weidler JM, Bates M, Neumeister V, Rimm DL. High concordance of a closed-system, RT-qPCR breast cancer assay for HER2 mRNA, compared to clinically determined immunohistochemistry, fluorescence in situ hybridization, and quantitative immunofluorescence. Lab Investig. 2017;97(12):1521–1526. doi: 10.1038/labinvest.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang JE, Wecsler JS, Press MF, Tripathy D. Molecular markers for breast cancer diagnosis, prognosis and targeted therapy. J Surg Oncol. 2015;111(1):81–90. doi: 10.1002/jso.23732. [DOI] [PubMed] [Google Scholar]
- 22.Press M, Spaulding B, Groshen S, Kaminsky D, Hagerty M, Sherman L, Christensen K, Edwards DP. Comparison of different antibodies for detection of progesterone receptor in breast cancer. Steroids. 2002;67(9):799–813. doi: 10.1016/S0039-128X(02)00039-9. [DOI] [PubMed] [Google Scholar]
- 23.Press MF, Sauter G, Buyse M, Fourmanoir H, Quinaux E, Eiermann W, Robert N, Pienkowski T, Crown J, Martin M, Valero V, Mackey JR, Bee V, Ma Y, Villalobos I, Campeau A, Mirlacher M, Lindsay MA, Slamon DJ. HER2 gene amplification testing by fluorescent in situ hybridization (FISH): comparison of the (ASCO)-College of American Pathologists (CAP) Guidelines with FISH Scores used for enrollment in Breast Cancer International Research Group (BCIRG) Clinical Trials. Journal of Clinical Oncology. 2016;34(29):3518–3528. doi: 10.1200/JCO.2016.66.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Press MF, Villalobos I, Santiago A, Guzman R, Estrada MC, Gasparyan A, Campeau A, Ma Y, Tsao-Wei D, Groshen SL. Assessing the new American Society of Clinical Oncology/College of American Pathologists guidelines for HER2 testing by fluorescence in situ hybridization: Experience of an academic consultation practice. Archiv Patholo Lab Med. 2016;140(11):1250–1258. doi: 10.5858/arpa.2016-0009-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.