Abstract
Genotype imputation estimates unobserved genotypes from genome-wide makers, to increase genome coverage and power for genome-wide association studies. Imputation has been successful for European ancestry populations in which very large reference panels are available. Smaller subsets of African descent populations are available in 1000 Genomes (1000G), the Consortium on Asthma among African-Ancestry Populations in the Americas (CAAPA) and the Haplotype Reference Consortium (HRC). We compared the performance of these reference panels when imputing variation in 3,747 African Americans (AA) from 2 cohorts (HCV and COPDGene) genotyped using Illumina Omni microarrays. The haplotypes of 2,504 (1000G), 883 (CAAPA) and 32,470 individuals (HRC) were used as reference. We compared number of variants, imputation quality, imputation accuracy and coverage between panels. In both cohorts, 1000G imputed 1.5−1.6× more variants than CAAPA and 1.2× more than HRC. Similar findings were observed for variants with imputation R2>0.5 and for rare, low frequency, and common variants. When merging imputed variants of the three panels the total number was 62M-63M with 20M overlapping variants imputed by all three panels, and a range of 5 to 15M variants imputed exclusively with one of them. For overlapping variants, imputation quality was highest for HRC, followed by 1000G, then CAAPA, and improved as the minor allele frequency increased. 1000G, HRC and CAAPA provided high performance and accuracy for imputation of African American individuals, increasing the number of variants available for subsequent analyses. These panels are complementary and would benefit from the development of an integrated African reference panel.
Keywords: Imputation, African Americans, Performance, 1000G, HRC, CAAPA
Introduction
Over the past 10 years, genome-wide association studies (GWAS) have uncovered a large number of replicated associations for many complex human diseases (Marchini and Howie 2010; McRae 2017; Visscher et al. 2017). These studies have used different genotyping arrays with 300,000 to 2.5 million single nucleotide polymorphisms (SNPs), varied genomic coverage, and a wide range of allelic frequencies across populations. In general, these arrays provide excellent genomic coverage and density for European ancestry populations. Despite efforts at enrichment, imputation remains modest at best for other ancestral populations, especially populations of African ancestry. Genotype imputation is a cost-effective method for statistically predicting un-typed genotypes not directly assayed in a sample of individuals based on a dense reference panel of haplotypes. Imputation methods estimate haplotypes of observed genotypes shared between genotyped individuals and a sequenced reference panel, and use this information to infer alleles at un-typed SNPs (Marchini and Howie 2010). This process can increase the overall genome coverage of an array by increasing the number of testable single nucleotide variants (SNVs) across the entire genome and can improve fine-mapping of a targeted region of interest. Imputation also facilitates the comparison and meta-analyses of studies originally done on different microarrays (Marchini and Howie 2010; Hancock et al. 2012; Das et al. 2016; McRae 2017), potentially bridging the gap in coverage between various genome-wide SNP platforms (Anderson et al. 2008).
Imputation of rare SNVs is more challenging since rare alleles are often ethnicity or population-specific and reflect fine-scale linkage disequilibrium (LD) structure impacted by recent demographic events (Wojcik et al. 2017). Options for imputing low-frequency and rare variants more accurately in any specific population include increasing the size of the imputation reference panel to capture more reference haplotypes, or increasing the sequencing depth in the reference samples to minimize error rates inherent in low-coverage sequencing (Browning and Browning 2009). Recently admixed populations, which have higher degrees of LD and greater heterogeneity in their haplotype block structure (reflect the dynamics of admixture), may also benefit from using more diverse or larger reference populations.
Earlier available reference panels include the Human Genome Diversity Project (Cavalli-Sforza 2005), the HapMap Consortium (The International HapMap 3 Consortium et al. 2010) and the 1000 Genomes Project (1000G) (Sudmant et al. 2015). More recently, the Haplotype Reference Consortium (HRC) (McCarthy et al. 2016) was constructed via a predominantly European ancestry consortium currently comprised of 32,611 individuals with whole genome or exome sequences available. The HRC includes the Genome of The Netherlands (GoNL), 250 Dutch parent-offspring families sequenced at 12× depth (Genome of the Netherlands Consortium et al. 2014), the UK10K project with nearly 10,000 individuals whose whole genome was sequenced at 7×, or exome sequenced at 80× (Walter et al. 2015) and 1000G subjects among other cohorts (http://www.haplotype-reference-consortium.org/participating-cohorts). Another project, funded by the UK government, plans to sequence 100,000 whole genomes from patients registered and treated by the National Health Service (http://www.genomicsengland.co.uk/the-100000-genomes-project/). These dense reference panels will allow better imputation of low frequency and rare variants (Deelen et al. 2014) and the discovery of new variants (Walter et al. 2015; Warren et al. 2017), but are generally focused on populations of European descent.
There are a only few reference panels available for imputation in African Americans, those include the 1000 Genomes Project (1000G) (Sudmant et al. 2015) and the Consortium on Asthma among African ancestry Populations in the Americas (CAAPA) (Mathias et al. 2016). The 1000G includes 661 individuals with African ancestry from Esan, Gambian, Luhya, Mende, Yoruba, Barbadian and African-American populations (Sudmant et al. 2015). The CAAPA panel is an additional resource completed on populations of African ancestry from the Americas (Mathias et al. 2016). CAAPA included 883 unrelated individuals of African descent from 15 locations in North, Central, and South America, the Caribbean, and Yoruba-speaking individuals from Nigeria. Their relatively small size of these panels compared to the references populations for European ancestry, limits the ability to discover new variants beyond those already present on the commercially available chips for subjects of African descent. Other projects assessing genetic diversity through dense genotyping and at the WGS level in African populations include the African Genome Variation Project (AGVP) (Gurdasani et al. 2014) and the African Genome Resources (AGR) reference panel (https://www.apcdr.org/).
In this paper, we compared imputation performance using publicly available reference panels to evaluate imputation accuracy and quality in African or admixed populations of African descent. Imputation performance has been evaluated for African American populations comparing the 1000G, HapMap and the Exome Sequencing Project (Chanda et al. 2012; Hancock et al. 2012; Sung et al. 2012; Duan et al. 2013; Roshyara et al. 2016), and also using several combinations of populations from 1000G. Previous analyses suggest multi-ethnic panels in 1000G (primarily European (EUR) and African (AFR)) improve imputation performance compared to a reference panel from any single population (AFR) (Chanda et al. 2012). In previous studies of African Americans imputed with several combinations of 1000G populations, imputation accuracy (based on concordance and imputation quality score) was comparable across the reference panels. Imputation quality for SNPs with MAF between 0.02–0.50 was better when using more distantly related reference panels containing several continental African populations (AFR+EUR or ALL populations) in comparison with more closely related populations (Yoruba (YRI), CEPH European (CEU), and African Americans from the Southwest US (ASW)), but when analyzing all ranges of MAF including those with MAF < 0.02, the most closely related (YRI+CEU+ASW) panel produced better imputation results. On the other hand, genotype concordance was similar for both distant and closely related reference panels from 1000G (Hancock et al. 2012).
Imputation is standard part of all array-based genome-wide association analyses. However, the relative performance of these newer imputation reference panels – with varying total sample size and number of African individuals – is unknown. In this study, we extend these prior imputation comparisons beyond the 1000G populations by evaluating genotype imputation performance using CAAPA, HRC, and 1000G reference panels in two independent populations of African Americans (Regan et al. 2010; Duggal et al. 2013; Wojcik et al. 2014).
Methods
The current study includes a total of 3,747 African Americans participating in previous genome-wide association studies of spontaneous resolution of Hepatitis C viral infection (HCV cohort) and Chronic Obstructive Pulmonary Disease (COPD) from the COPDGene cohort, a multi-site study of heavy smokers (Regan et al. 2010; Duggal et al. 2013; Wojcik et al. 2014). Metrics of imputation performance, accuracy, genome coverage and annotation of variants were calculated in these two cohorts, separately.
Study subjects
African Americans from the HCV cohort:
A genome-wide marker panel from 447 African Americans was used, as previously described (Duggal et al. 2013; Wojcik et al. 2014). Briefly, 2,401 individuals participating in a longitudinal cohort study or identified through blood repositories as having HCV infection (spontaneously resolved or persistent) were enrolled, and were genotyped as part of the HCV Genetics Consortium. African American subjects are part of a multicenter cohort composed of several study groups including ALIVE (AIDS Link to the Intravenous Experience, Baltimore, MD) (Vlahov et al. 1990); BBAASH (Baltimore Before and After Acute Study of Hepatitis, Baltimore, MD) (Cox et al. 2005); BAHSTION (Boston Acute HCV Study: Transmission, Immunity and Outcomes Network, Boston, MA) (Kim et al. 2011); Cramp and colleagues’ study at King’s College Hospital, London, UK (Cramp et al. 1998); HGDS (Hemophilia Growth and Development Study in 14 hemophilia treatment centers in USA) (Hilgartner et al. 1993); Mangia and colleagues’ study at San Giovanni Rotondo, Italy (Mangia et al. 1999); MHCS (Multicenter Hemophilia Cohort Study) and MHCS-II recruited in 16 sites located in the United States, Greece, Germany, and Austria (Goedert et al. 2007); REVELL study (Correlates of Resolved Versus Low-Level Viremic Hepatitis C Infection in Blood Donors recruited at 17 blood banks in Western and Southern USA) (Tobler et al. 2010); the Swan Project recruited at the Lower East Side of Manhattan, NY (Edlin et al. 2009); the Toulouse cohort form the south of France (Alric et al. 1997); WIHS (Women’s Interagency HIV Study), a multicenter study with 10 recruitment sites across United states (Kuniholm et al. 2011); and the United Kingdom Drug Use cohort, London, UK (Khakoo et al. 2004). Each individual study obtained consent for genetic testing from their governing Institutional Review Board (IRB) and the Johns Hopkins School of Medicine Institutional Review Board.
African Americans from the COPDGene cohort.
This study included 3,300 African Americans participants in the COPDGene study. A complete study protocol for COPDGene had been described elsewhere (Regan et al. 2010). Briefly, 10,280 self-identified Non-Hispanic Whites and African Americans between the ages of 45 and 80 years with a minimum of 10 pack-years smoking history were enrolled at 21 centers across the US with the goal of identifying genetic causes of COPD. The majority of were recruited from: Temple University, Philidelphia, PA (n=798), Morehouse School of Medicine, Atlanta, GA (n=454), Harbor-UCLA Hospital, Los Angeles, CA (n=368), University of California San Diego, San Diego, CA (n=319), Columbia University, New York, NY (n=309), Johns Hopkins University, Baltimore, MD (n=251). Each study site has obtained local IRB approval to enroll participants in this project, and all subjects provide informed consent (Regan et al. 2010).
Genotyping and Quality Control
African Americans from the HCV cohort:
Genetic variants and their locations for the genotypic data, reference panels and whole genome sequencing data were specified based on The Genome Reference Consortium Human build 37 (GRCh37) (Lander et al. 2001). A total of 774,792 SNPs genotyped on the Illumina Omni Quad array (IIlumina, Inc. San Diego) met quality control criteria and were used for imputation. SNPs with MAF < 0.01, those with missing call rate ≥ 5% and those deviating from Hardy Weinberg equilibrium at p < 1×10−5 were removed from the analysis for quality control. Individuals cryptically related, duplicated replicates, and individuals with sex discrepancies were excluded (Duggal et al. 2013).
African Americans from the COPDGene cohort:
A total of 624,564 SNPs genotyped on the Illumina Omni Express array (IIlumina Inc. San Diego, CA) were used for imputation. All SNPs with MAF < 0.05, those with missing call rate ≥ 2%, those deviating from Hardy Weinberg equilibrium at p < 1×10−3 and individuals cryptically related, duplicated replicates and individuals with sex discrepancies were excluded (Cho et al. 2012).
Whole-Genome Sequencing: Library Preparation and Bioinformatic Analysis
Whole genome sequencing data in a subgroup of 17 subjects of the HCV cohort was performed at the New York Genome Center. In brief, libraries of 350-bp fragments were generated from 1 μg sheared genomic DNA using the TruSeq PCR-Free library preparation kit (Illumina, San Diego, CA). WGS was performed at a coverage of 30×. Base calling and filtering were performed using current Illumina software; sequences were aligned to NCBI genome (build 37) using Burrows-Wheeler Aligner (Li and Durbin 2009); Picard was used to remove duplicate reads (http://broadinstitute.github.io/picard/); base quality scores were recalibrated using GATK (DePristo et al. 2011). Assessment of reads not aligning fully to the reference genome was performed, locally realigning around indels to identify putative insertions or deletions in the region. Variants were called using GATK HaplotypeCaller tool, which generates single-sample Genomic VCF (GVCF) files. To improve variant call accuracy, multiple single-sample GVCF files were jointly genotyped using GATK Genotype GVCFs, which generates a multi-sample VCF. Variant Quality Score Recalibration (VQSR) was performed on the multi-sample VCF, which adds quality metrics to each variant that can be used in downstream variant filtering (Van der Auwera et al. 2013). Quality control of all variants included filtering out based on genotyping quality score < 20, read depth < 10 and removing variants in genomic duplicated segments.
Estimation of Genetic African American Ancestry
Genetic ancestry for both cohorts was determined by principal components using the smartpca program in EIGENSOFT (Price et al. 2006). A subset of independent SNPs across the genome were selected by pruning the full dataset for markers with an r2 < 0.01 to insure independence between SNPs. Chromosomal regions known to be associated with ethnicity were removed (including the lactase regions on chromosomes 2, 8, and the HLA region on chromosome 6). African-American ancestry groups were determined based on their distribution over the first 2 principal components (Supplementary Figure 1). Outliers were removed based on heterozygosity, and if the subjects were 6 standard deviations from either of the 2 first principal components (Regan et al. 2010; Duggal et al. 2013). We also have performed local ancestry inference on both the HCV and COPDGene cohorts using the algorithm implemented in the Local Ancestry in adMixed Populations (LAMPLD) software (Baran et al. 2012; Parker et al. 2014; Wojcik et al. 2014). Averaging local ancestry estimates over all sites, COPDGene and HCV subjects have an average of 80.7% and 79.5% African ancestry respectively, with a range of 29.5% – 99.6% African ancestry in both cohorts.
Phasing and Imputation
For both cohorts, Eagle v2.3, a reference-based phasing algorithm was used to phase genotypes prior to imputation (Durbin 2014; Loh et al. 2016). Imputation was performed for chromosomes 1 to 22 using the Minimac3 software through the publicly available Michigan Imputation Server (Das et al. 2016). Minimac3 is a Markov Chain based haplotyper that can resolve long haplotypes or infer missing genotypes in samples of unrelated individuals (Li et al. 2010). Imputation of genotypes was performed using 3 different reference panels:
-
a)
1000 Genomes Phase 3, Version 5 (referred to here as “1000G”) included 49,143,605 sites located in chromosomes 1 to 22 for the complete set of 2,504 individuals representing 5 continental and sub-continental populations: East Asian (EAS= 504), European (EUR=503), South Asian (SAS=489), African (AFR= 661) and Mixed American (AMR= 347) (Sudmant et al. 2015). The 1000G uses a combination of low-coverage whole-genome sequencing (WGS) (mean depth of 7.4×), deep exome sequencing (mean depth of 65.7×) and dense microarray genotyping;
-
b)
The CAAPA reference panel (“CAAPA”) comprising 883 individuals from 19 case-control studies of asthma with 31,163,897 variants identified on chromosomes 1–22 by high coverage WGS (30×). The populations for this panel include individuals from Barbados (N=39), Jamaica (N=45), Dominican Republic (N=47), Honduras (N=41), Colombia (N=31), Puerto Rico (N= 53), Brazil (N=33) and Nigeria (N=25) and African Americans (N=328) (Mathias et al. 2016);
-
c)
The Haplotype Reference Consortium (“HRC”) reference panel combining data sets from 20 different studies with low-coverage WGS (4–8× coverage) of subjects with predominantly European ancestry. For this analysis we used the version HRC r1.1,2016 (http://imputationserver.readthedocs.io/en/latest/reference-panels/) of the first release of the consortium (http://www.haplotype-reference-consortium.org/) consisting of 32,470 individuals with 64,940 haplotypes including 39,635,008 SNPs, each with a minor allele count (MAC) greater or equal to 5 (McCarthy et al. 2016).
Imputation Performance Metrics
Evaluation of imputed variants by reference panel:
For each reference panel and cohort, we assessed imputation performance using the following criteria: 1) the total number of imputed variants; 2) the distribution of all variants based on MAF ranges; and 3) the relationship between imputation quality and MAF. Imputation quality was determined using the R2 score, or the estimated value of the squared correlation between imputed genotypes and true, unobserved genotypes basing its calculation in the population allelic frequencies (Howie et al. 2012a).
Comparison of imputed variants between reference panels:
To compare imputation results between panels, we analyzed variants imputed by all three panels (“overlapping” variants) and exclusively by each panel (“unique” variants). For overlapping variants, the imputation quality and genotype concordance between the panels was compared. Unique variants with R2 >0.5, were evaluated by their number and MAF and for its presence and MAF in the CEU, YRI and CHB populations from1000G as a method to evaluate the potential ancestral origin of them.
Imputation accuracy
Imputation accuracy is defined as the proportion of correctly imputed SNPs among all successfully imputed SNPs (Zhao et al. 2008; Huang et al. 2009; Nothnagel et al. 2009; Zhang et al. 2011). We calculated imputation accuracy using three separate approaches:
-
a)
A “masked analysis” where we removed genotypes of a subset of 25,000 SNPs and then imputed them as though they were not genotyped. Imputed genotypes of these SNPs then were compared back to their original genotypes (Huang et al. 2009; Shriner et al. 2010; Hancock et al. 2012). The MAF of these SNPs ranged from 0.01 to 0.5;
-
b)
A comparison of the allelic dosage of the original genotypic data with the allelic dosage of imputed data. Given three genotypes AA, AB, and BB for each SNP, the allelic dosage for each individual can be calculated as probabilities (P) of each of three genotypes via 2∗P (AA) + 1∗P(AB) + 0∗P(BB) to obtain the expected allelic dosage from the original genotypic data and from the observed allelic dosage for masked and imputed genotype at each SNP (Verma et al. 2014). The metric EmpRsq obtained in Minimac3 is the correlation between the true genotyped values and the imputed dosages calculated by hiding all known genotypes for a given SNP (Howie et al. 2012a), similar to the masked analysis described above. We calculated the mean of this EmpRsq by bins of 0.001–0.01 value of frequency of the minor allele.
-
c)
A comparison of the imputed genotypes with whole genome sequencing genotypes in a subgroup of 17 individuals from the HCV study. This analysis was restricted to variants located on chromosome 22, and was done independently for all the variants imputed with each reference panel.
Genomic Coverage and Density of Imputed Variants
The total proportion of genomic variation captured by an array, either directly or indirectly, is referred to as “genomic coverage.” Assessments of imputation-based genomic coverage leverages observed array SNPs which imputed from a more densely genotyped or sequenced reference panel, such as the HapMap Project3 or 1000 Genomes Project (Abecasis et al. 2012; Auton et al. 2015). In this study we based our calculations on the imputation R2 (calculated as squared correlation between the actual (discrete) allelic dosage at a SNP and the imputed (continuous) allelic dosage, over a defined set of samples). Genomic coverage was quantified as the proportion of variants with an imputation R2 ≥ 0.8, and the reference set of variants used to determine imputation-based genomic coverage was the total number of variants described in each imputation reference panel. This method has been described and used previously as one assessment of genomic coverage in imputation performance studies (Hoffmann et al. 2011; Nelson et al. 2013).
We also calculated the density of imputed variants (represented as the number of variants with R2>0.8 per Kb) across all autosomes, by chromosomes and in chromosomal regions harboring known genes. We compared the results obtained with the three panels. Variants genotyped on the arrays were given imputation R2 = 1 for all coverage and density calculations; chromosome sizes in base pairs were obtained from the UCSC Known Gene Human Annotation (GRCh37). Coordinates of the regions containing genes were obtained from the RefSeq database via UCSC genome browser (Kent et al. 2002) (http://genome.ucsc.edu/). Plink version 1.90 beta (Purcell et al. 2007), bcftools (Danecek et al. 2011) and customized scripts in R (R Core Team 2013) were used for the analyses with both cohorts.
Results
Imputation Performance Metrics
Evaluation of imputed variants by reference panel:
The total number of imputed variants and their distribution by MAF was similar for both the HCV and COPDGene cohorts (Table 1 and Supplementary Table 1). In both cohorts, 1000G imputed approximately 1.5× more variants than did CAAPA, and 1.2× more variants than did HRC regardless of imputation quality. However, It is important to note the 1000G imputation includes small insertions/deletions (INDELS) that are not currently available in the HRC and CAAPA panels. These INDELS corresponded approximately 7.0 % of total imputed variants in both cohorts. The actual values of this and other metrics obtained in the COPDGene cohort are described in detail in Supplementary Results.
Table 1.
Number of variants imputed by reference panel, minor allele frequency ranges and imputation quality for the HCV cohort.
| CAAPA | 1000G | HRC | ||||
|---|---|---|---|---|---|---|
| Minor allele frequency | Number of variants | R2 > 0.5 (%) | Number of variants | R2 > 0.5 (%) | Number of variants | R2 > 0.5 (%) |
| 0–0.0001 | 12,164 | 0 | 396,072 | 0 | 3,568,020 | 0 |
| 0.0001–0.01 | 15,481,830 | 36.8 | 29,683,849 | 33.9 | 21,736,936 | 32.1 |
| 0.01–0.05 | 6,343,792 | 83.9 | 7,050,809 | 97.4 | 6,055,818 | 99.3 |
| >0.05 | 8,012,926 | 94.2 | 9,495,981 | 98.6 | 7,658,238 | 99.7 |
| Total | 29,850,712 | 62.3 | 46,626,711 | 56.4 | 39,019,012 | 52.9 |
For variants imputed with R2 > 0.5, 1000G imputed nearly 1.4× and 1.3× more variants in both cohorts. In the HCV cohort they were 26,310,578 vs. 18,584,433 and 20,643,333 for CAAPA and HRC, respectively. All three reference panels had a similar percent of variants imputed with R2 > 0.5 in both cohorts being slightly high for the COPDGene cohort. For HCV cohort, HRC had 53%; 1000G, 56% and CAAPA, 62%. For the three panels panels, the percentage of variants imputed with R2 > 0.5 increased with increasing MAF (Table 1 and Supplementary Table 1).
Regardless of allele frequencies, the number of imputed variants was greater in 1000G than for CAAPA: ~1.8× more rare variants (MAF = 0.0001–0.01), ~1.3× more low MAF variants (MAF=0.01–0.05) and ~1.2× more common variants (MAF>0.05). These numbers were also higher for 1000G compared to HRC being 1.4×, 1.1× and 1.2×, respectively. The distribution of the number of variants with R2 > 0.5 by MAF for each panel was similar between all reference panels with a high number of low frequency SNPs (i.e. those with MAF < 0.1) in the three panels.
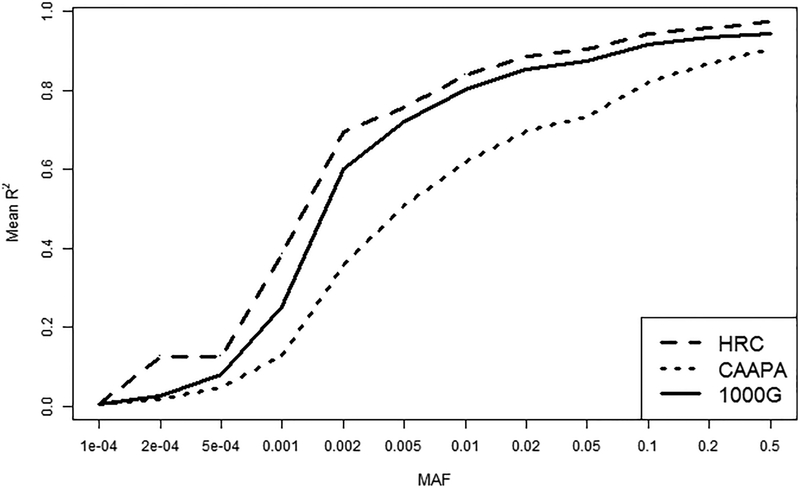
For both cohorts and all three panels, imputation quality improved as the MAF increased, reaching a mean quality score or R2 of 0.6 or higher for common variants (MAF>0.05). CAAPA imputed with slightly lower quality across all MAF followed by 1000G and HRC (Figure 1 and Supplementary Figure 2). The higher imputation quality observed with HRC and 1000G was particularly evident in low frequency variants (i.e. those with MAF from 0.002 to 0.05). In the COPDGene cohort, HRC had a better performance compared to 1000G for very rare variants (MAF < 0.001, Supplementary Figure 2) but this was not observed in the HCV cohort, likely due to sample size differences in the two target populations.
Figure 1.
Relationship between imputation quality and minor allele frequency for all variants imputed with 1000G, CAAPA and HRC in the HCV cohort. The graph represents the mean of imputation R2 in each minor allele frequency (MAF) bin (intervals of 0.001 for variants with MAF < 1% and intervals of 0.01 for for variants with MAF > 1%).
Comparison of imputed variants between reference panels:
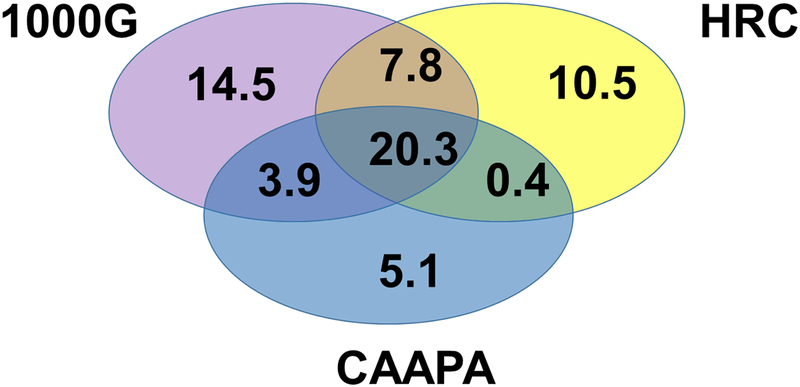
When merging the variants imputed independently with each reference panel, the total number of imputed variants was 62–63 millions, representing an increase of 20 – 30 million variants compared to the imputation of each panel separately (Figure 2, Supplementary Figure 3 and Supplementary Table 2). There were approximately 20 million overlapping variants imputed with all three reference panels and a range of 5 to 15 million unique variants imputed exclusively within one of the three panels.
Figure 2.
Number of overlapping and unique variants imputed with 1000G, CAAPA and HRC for the HCV cohort. The intersection shows the number of variants (in millions) imputed with the three reference panels and the non-overlapping sections of the circles show the variants unique to each panel.
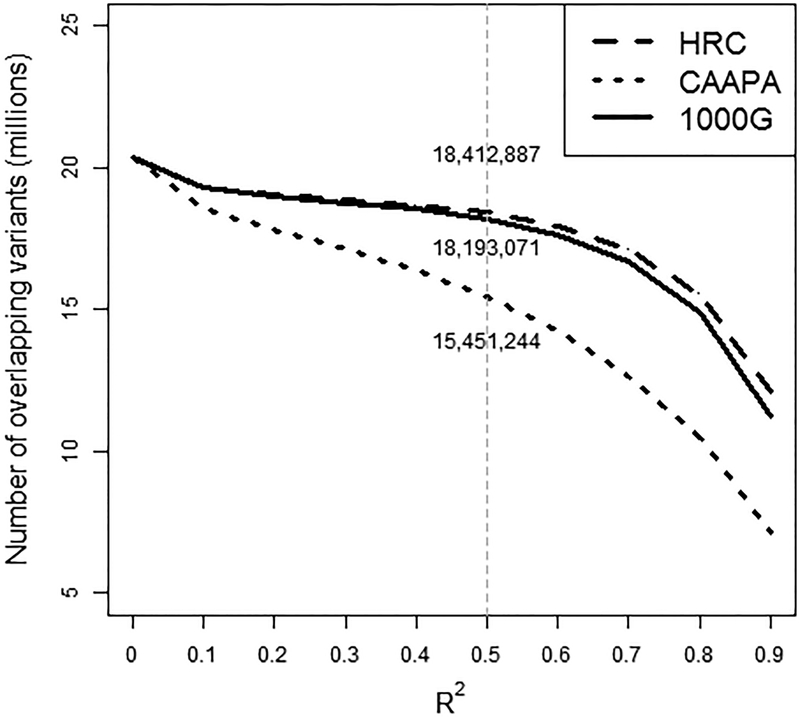
For overlapping variants, we compared the imputation quality obtained with each of the three panels (Figure 3 and Supplementary Figure 4). For the same variants, the imputation quality was higher for HRC and 1000G compared to imputation run against CAAPA. From approximately 20 million overlapping variants, HRC and 1000G imputed ~18–19 million variants with R2 > 0.5, whereas CAAPA imputed ~15 million (Figure 3 and Supplementary Figure 4). Genotype calls of overlapping variants were 98–99% concordant between pairs of panels in both cohorts.
Figure 3.
Number of variants by imputation quality (R2) for all overlapping variants imputed with CAAPA, 1000G and HRC for the HCV cohort. The values on the gray line at imputation R2=0.5 correspond to the number of overlapping variants imputed with R2>= 0.5 with each panel.
Unique variants corresponded to 17%, 31% and 27% of all variants imputed with in CAAPA, 1000G and HRC, respectively. Most of them had MAF < 0.01 (75%–90%) in both cohorts for the three panels (Supplementary Tables 3 and 4), 5–24% of these rare variants were imputed with R2 > 0.5. There was a lower percentage (0.2–5%) of low frequency variants (MAF between 0.01–0.05) that were imputed with better quality (> 45% had R2> 0.5). The percentage of all unique variants imputed with R2 > 0.5 in the HCV cohorts was 27% for 1000G; 24% for CAAPA and 4% for HRC and slighlyt higher percentages were observed for the COPDGene cohort (Supplementary Results).
We interrogated the three parental populations of 1000G (CEU, YRI and CHB) to estimate allele frequencies of unique variants imputed with R2 > 0.5 in each population. 31% of the variants imputed with CAAPA were present in parental populations of 1000G. Only a small percentage of those were polymorphic in the CEU and CHB (0.3%–6% in both cohorts) as compared to YRI (12–22%), Supplementary Figure 5. Of those unique variants imputed using HRC, 34–35% were present in the parental populations; from those, 17–18% were polymorphic in CEU, in contrast with a 0.5–4% and 2% of variants that were polymorphic in YRI and CHB in both the HCV and COPDGene cohorts. All the unique variants with R2 > 0.5 imputed with 1000G in both cohort were also present in the parental populations, and 30–60% were actually polymorphic (i.e. MAF > 0) in YRI, CEU, and CHB (Supplementray Results).
Imputation accuracy
The concordance of genotype calls between the original genotype data and imputed data was quite high (96–97%) across all three panels using masked SNPs from both cohorts. The correlations between dosages of the imputed genotypes and actual genotypes ranged from 0.80–0.94, with higher correlations occurring when the MAF was greater than 1% (Supplementary Figure 6). In addition, we evaluated the concordance between whole genome sequenced and imputed data using variants on chromosome 22 in the HCV cohort. The genotype concordance for the three panels was 99% for 213,467, 190,005, and 195,591 variants overlapping between sequenced genotypes and imputed genotypes with 1000G, CAAPA and HRC, respectively.
Genomic Coverage and Density of Imputed Variants
In the HCV cohort, we imputed 20,222,182 variants with R2> 0.8 using the 1000G panel, 11,684,700 with CAAPA and 16,941,215 with HRC. The genomic coverage was 0.41, 0.37 and 0.43 for 1000G, CAAPA and HRC, respectively. Genomic density of markers included in the genotype array was estimated to be at 0.3 marker/Kb. In contrast, imputation with 1000G, CAAPA and HRC increased the genome density to 7, 4.1 and 5.9 markers per Kb, respectively. Similar values were observed in the COPDGene cohort (Supplementary Results). For both cohorts, the average density across chromosomes was ~6.8 variants/Kb for 1000G, ~3.9 variants/Kb for CAAPA and ~5.7 variants/Kb for HRC. The density was considerably lower in gene regions with an average ~1.4, ~2.3 and ~1.9 variants/Kb for CAAPA, 1000G and HRC, respectively (Supplementary Table 5 and 6).
Discussion
In the current study, we used three reference panels to impute GWAS genotyping data in two independent cohorts of African American individuals with remarkably consistent results between the two studies. Imputation to three reference panels increased coverage and density of markers across all autosomal chromosomes and facilitated the accurate imputation of both rare and common alleles with R2 > 0.5. Somewhat surprisingly, despite the smaller size of the reference panel and number of African-Americans, the 1000G reference panel resulted in a higher number of imputed variants (even after removing INDELS) than either the HRC or CAAPA cohorts alone. Additionally, while all three panels led to accurate estimated genotypes, the imputation quality was highest for HRC across all MAFs, but especially for low frequency and rare variants.
A greater number of variants were imputed with 1000G as compared to CAAPA. The substantially larger sample size of the 1000G panel may explain this difference by itself. Previous studies comparing references panels have shown larger reference panels considerably increase the number of imputed variants, as well as their imputation quality and accuracy, particularly for low-frequency variants (Browning and Browning 2009; Deelen et al. 2014; McCarthy et al. 2016). However, the composition of the reference population and similarity to the target population is also very important. Shriner et al. (Shriner et al. 2010) imputed variants on chromosome 22 in African Americans from the Washington, D.C. metropolitan area participating in the Howard University Family Study (Adeyemo et al. 2009) and concluded the YRI reference panel outperformed other HapMap reference panels, including ASW, as well as a combination of the CEU and YRI. Previous studies in European populations have indicated population specific panels can improve imputation quality and coverage (Pistis et al. 2015; Mitt et al. 2017) compared to broader panels. This improvement in the number of variants imputed and the accuracy using population specific panels argues that LD patterns of ethnic-specific variants may not be captured by different ethnic groups with distinct ancestral genetic background (Chou et al. 2016), which might include haplotypes from irrelevant populations. We would expect a population specific panel like CAAPA genotyped at a high depth, where ~50% are African Americans with African ancestry estimates of 76% or higher (Mathias et al. 2016), would be optimal for imputing more rare and common variants with higher quality in a target sample with similar high proportion of African ancestry (African ancestry estimates averaging 79.5%) (Duggal et al. 2013; Wojcik et al. 2014). However, in our study, the higher number of individuals from populations from continental Africa contained in 1000G compared to CAAPA (N=504 vs. N=25) may have outweighed the larger number of total African ancestry individuals in CAAPA, and provided more information on parental haplotype diversity (Jorde et al. 2000; Campbell and Tishkoff 2008) improving the chances of a rare variant being effectively tagged by a characteristic haplotype in admixed individuals (Auton et al. 2015).
1000G imputed also more variants than HRC in both cohorts. The difference is primarily because of the imputation of INDELs by 1000G, but even after accounting for this, there is still a smaller total number of variants in HRC (even though 1000G is contained within this larger reference panel). The predominantly European ancestry haplotypes of the HRC panel might impair the selection of optimal haplotypes for imputation of these populations with high proportion of African ancestry and consequently explain why there was a less of an increase in imputation success in them. Our sensitivity analysis investigating the potential ancestral origin of the unique variants indicated 1000G was able to impute variants of European and African origin compared to the HRC panel alone where the unique variants were present mostly in the CEU population of 1000G indicating an exclusive European origin. This may also explain why HRC imputed more variants than obtained with the CAAPA reference panel alone, if the higher number of imputed variants obtained with HRC compared to CAAPA reflects underlying European haplotypes in admixed African Americans. It is reasonable to suggest that the inclusion of more European and African parental populations in the CAAPA panel will improve its performance.
Our results are remarkably consistent in the two datasets even though they were imputed based on two diverse genotyping platforms and have different sample sizes. Previous analysis have demonstrated that differences in the density and distribution of markers in diverse genotyping array plays a role in the results of imputation (Marchini and Howie 2010; Nelson et al. 2017); however, our findings suggest that a difference in 150K genotyped markers in these two platforms does not alter the imputation performance. On the other hand, some studies indicate that merging the individual datasets before imputation performs slightly better than combining datasets after imputation(van Iperen et al. 2017). Since the actual analysis was not done here, it is unknown if, for these two particular studies, the results of the imputation of the datasets separately are comparable to those potentially obtained with the imputation of the intersection of the two datasets, as required and recommended (Johnson et al. 2013) for analyzing combined datasets in association studies.
Regardless of reference panel, imputation yielded accurate genotypes as shown in the analysis of correlation between true genotypes and imputed genotypes, the masked analysis and the concordance analysis comparing imputed SNPs and sequencing data in the HCV cohort. The accuracy reflected in the estimated correlation of true genotypes and imputed genotypes was comparable (but slightly higher) for 1000G when studied as a function of minor allele frequency. The lower accuracy found using the CAAPA and HRC reference panels separately might be due to the inclusion of admixed populations without a large African and European ancestral panel in CAAPA or the predominance of European haplotypes in HRC. This could limit selection of the best reference haplotypes. Imputation accuracy increases with the number of haplotypes in the reference panel of sequenced genomes (Howie et al. 2012a; Fuchsberger et al. 2015; Pistis et al. 2015), particularly for rare and low-frequency variants. The estimation of the effect of effective sample size (and ancestral composition) of a reference panel has been the subject of analysis of several papers for populations of diverse ancestry and demographic history (Chanda et al. 2012; Krithika et al. 2012; Duan et al. 2013; Huang and Tseng 2014; Huang et al. 2015). Previous studies determined that reference panels as small as 60 European ancestry individuals were sufficient for imputation in a set of 2,759 European Americans with imputation accuracy of 0.91 for variants with MAF > 5% using Minimac (Li et al. 2010). Similarly for a MAF >5 %, 60 individuals of European ancestry and 59 of African ancestry (CEU/YRI of 1000G) were sufficient to obtain an accuracy of 0.83 in a sample of 8,421 African Americans from the Women’s Health Initiative (WHI) study (Howie et al. 2012b). The authors note that the accuracy improvement is directly related to the sample size of the panel for these common variants. Therefore, we believe that the expansion of the number of sequenced individuals from African ancestral populations will be necessary for more accurate imputation, especially for rare variants. Additional studies evaluating the effective number of sequence based haplotypes for admixed populations are necessary to determine target goals for these reference panels for less frequent and rare variants.
In this study we used global concordance as a measure of imputation accuracy excluding variants with MAF < 0.01. Accuracy can be inflated when calculations of concordance rate include rare and low frequency variants, due to chance concordance or chance agreement (Lin et al. 2010; Ramnarine et al. 2015). Due to the baseline low allele frequency, there is a low probability of any rare allele being present in any imputed sample; therefore, when the major allele is assigned in imputation, this inference would be almost uniformly “correct” by chance alone. This inflation is increasingly problematic whenever studies are more interested in evaluating low frequency and rare variants. Since our study didn’t include rare variants in the estimate, we consider our obtained values reliable. Our global estimates of accuracy were higher than previous results obtained in a group of 40 African Americans imputed with MACH using CEU, YRI, MEX and JPT+CHB HapMap populations. Accuracy values (measured as percentage of most likely genotypes agreeing with the original genotypes) of 88.8, 87.9 and 87.2 were found when masking 50%, 70% and 100% of all high quality SNPs (Roshyara and Scholz 2015). The differences between this study and the current analysis are likely due to our larger sample size and type of reference population (Hapmap vs 1000G, HRC and CAAPA, separately).
Imputation resulted in a considerable number of rare and common variants unique to CAAPA, although they are present in the 1000G database. These variants are predominantly of African origin even though a great number are monomorphic in YRI subjects and possibly for this reason they were not imputed in the 1000G panel. It is likely these unique variants may be derived from African genomes not included in the 1000G, and may be unique to African descent populations in the Americas (where there is also a small percentage of Native American genes included). Similarly, the unique variants imputed with HRC may correspond to European derived polymorphisms not captured by the haplotype structure of the other reference panels.
In this study, the total number of imputed variants increased when merging imputed variants obtained from each reference panel individually. However, although all panels are publicly available for imputation on the server and 1000G and a subset of CAAPA raw sequencing data are publically available and in dbGap under request, respectively; we were not able to evaluate the imputation of a fully integrated reference panel given that the full HRC is not available for offline imputation. Previously integration of the 1000G and African Genome Variation Project panels markedly improved imputation accuracy across the entire allele frequency spectrum for populations poorly represented in the 1000G panel (Gurdasani et al. 2014). Similar results were found when merging the Estonian Biobank of the Estonian Genome Center, University of Tartu (EGCUT) and 1000G datasets (Mitt et al. 2017) and the GoNL and 1000G (Pistis et al. 2015) and when using a combined reference panel of 1,092 subjects from 1000G and 3,781 from UK10K Project for imputing rare variants in the Framingham Heart Study and the North Chinese Study (Chou et al. 2016). We encourage the development of more publically available combined reference panels, like HRC, for African ancestry populations. The African Genome Resources (AGR) data is a great example with data merged with 1000G and available via the Sanger Imputation Center. The AGR incorporates ~2000 Ugandan samples and ~100 samples each from Ethiopia, Egypt, Namibia and South African populations. These joint panels should also include the NHLBI funded Trans-Omics for Precision Medicine (TOPMed) project (Brody et al. 2017). The TOPMed panel includes over 60,000 WGS samples (125,568 haplotypes), sequenced to a mean depth of 30×. Unlike the HRC panel, TOPMed sample are ~ 50% non-white, including ~25% (or ~15,000 samples) of African ancestry. The African Ancestry samples in the TOPMed reference include the COPDGene subjects presented in this manuscript. We expect this huge sample of both African and European ancestry individuals will result in improved overlap of haplotypes represented in the reference and our African Americans subjects. Therefore, we expect more variants of higher quality will be imputed using the TOPMed panel. Additionally, unlike the HRC and CAAPA panels, TOPMed imputation through the Michigan Imputation Server includes small INDELs, affording investigators the opportunity to analyze these in addition to single nucleotide variants.
In addition, The 1000 Genomes Project will soon become “The International Genome Sample Resource” with all sequenced reads being re-mapped to the GRCh38 map producing new variants calls specific to this assembly. It will also expand the global catalogue of freely available sequence information by incorporating Russian samples, new African populations and whole genome sequences from the Simons Genome Diversity Project (Zheng-Bradley and Flicek 2016). Data from the CAAPA project is also available at the database of Genotypes and Phenotypes (dbGaP) and can be used to explore the option of “custom reference panels” for imputation in African Americans and other admixed populations from Latin America and the Caribbean. But as we show in this study, there is still a need for characterizing large, diverse parental populations such as those from sub-Saharan African to better capture populations such as those in the Americas.
In summary, we found the 1000G, HRC and CAAPA reference panels provide high performance and accuracy for imputing dense marker panels for admixed African American individuals, increasing the total number of high quality imputed variants available for subsequent analyses. The 1000G panel also showed higher performance compared to the HRC and CAAPA reference populations in terms of number of imputed variants with high accuracy likely because it included more diverse parental populations. Finally, there are a large number of variants unique to these three reference panels, making them complementary to each other. We recommend directing efforts to the construction of an integrated African panel including data from multiple resources and populations.
Supplementary Material
Acknowledgments
Funding
This project was funded in part with federal funds from the office of AIDS Research through the Center for Inherited Diseases at Johns Hopkins University, the National Institutes of Drug Abuse R01013324. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Liliana Franco was supported by COLCIENCIAS’s (Administrative Department of Science, Technology and Innovation -Departamento Administrativo de Ciencia, Tecnología e Innovación-) scholarship program for PhD students and the Epidemiology Group of National School of Public Health of the University of Antioquia. The COPDGene project (NCT00608764) was supported by Award Number R01HL089897 and Award Number R01HL089856 from the National Heart, Lung, And Blood Institute. Margaret M. Parker was supported by T32HL007427. The COPDGene project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, Sunovion, and GlaxoSmithKline.
Footnotes
Conflict of Interest: M.H.C. has received grant support from GSK. The remaining authors declare that they have no conflict of interest.
References
- Abecasis GR, Auton A, Brooks LD, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. doi: 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo A, Gerry N, Chen G, et al. (2009) A Genome-Wide Association Study of Hypertension and Blood Pressure in African Americans. PLoS Genet 5:e1000564. doi: 10.1371/journal.pgen.1000564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alric L, Fort M, Izopet J, et al. (1997) Genes of the major histocompatibility complex class II influence the outcome of hepatitis C virus infection. Gastroenterology 113:1675–81 [DOI] [PubMed] [Google Scholar]
- Anderson CA, Pettersson FH, Barrett JC, et al. (2008) Evaluating the Effects of Imputation on the Power, Coverage, and Cost Efficiency of Genome-wide SNP Platforms. Am J Hum Genet 83:112–119. doi: 10.1016/j.ajhg.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Abecasis GR, Altshuler DM, et al. (2015) A global reference for human genetic variation. Nature 526:68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran Y, Pasaniuc B, Sankararaman S, et al. (2012) Fast and accurate inference of local ancestry in Latino populations. Bioinformatics 28:1359–1367. doi: 10.1093/bioinformatics/bts144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody JA, Morrison AC, Bis JC, et al. (2017) Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology. Nat Genet 49:1560–1563. doi: 10.1038/ng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR (2009) A Unified Approach to Genotype Imputation and Haplotype-Phase Inference for Large Data Sets of Trios and Unrelated Individuals. Am J Hum Genet 84:210–223. doi: 10.1016/j.ajhg.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL (2005) The Human Genome Diversity Project: past, present and future. Nat Rev Genet 6:333–40. doi: 10.1038/nrg1596 [DOI] [PubMed] [Google Scholar]
- Chanda P, Yuhki N, Li M, et al. (2012) Comprehensive evaluation of imputation performance in African Americans. J Hum Genet 57:411–421. doi: 10.1038/jhg.2012.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Castaldi PJ, Wan ES, et al. (2012) A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet 21:947–957. doi: 10.1093/hmg/ddr524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou W-C, Zheng H-F, Cheng C-H, et al. (2016) A combined reference panel from the 1000 Genomes and UK10K projects improved rare variant imputation in European and Chinese samples. Sci Rep 6:39313. doi: 10.1038/srep39313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AL, Netski DM, Mosbruger T, et al. (2005) Prospective evaluation of community-acquired acute-phase hepatitis C virus infection. Clin Infect Dis 40:951–8. doi: 10.1086/428578 [DOI] [PubMed] [Google Scholar]
- Cramp ME, Carucci P, Underhill J, et al. (1998) Association between HLA class II genotype and spontaneous clearance of hepatitis C viraemia. J Hepatol 29:207–13 [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, et al. (2016) Next-generation genotype imputation service and methods. Nat Genet 48:1284–1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen P, Menelaou A, van Leeuwen EM, et al. (2014) Improved imputation quality of low-frequency and rare variants in European samples using the “Genome of The Netherlands.” Eur J Hum Genet 22:1321–1326. doi: 10.1038/ejhg.2014.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–8. doi: 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Liu EY, Auer PL, et al. (2013) Imputation of coding variants in African Americans: Better performance using data from the exome sequencing project. Bioinformatics 29:2744–2749. doi: 10.1093/bioinformatics/btt477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal P, Thio CL, Wojcik GL, et al. (2013) Genome wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann Intern Med 158:235–245. doi: 10.7326/0003-4819-158-4-201302190-00003.Genome [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R (2014) Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT). Bioinformatics 30:1266–72. doi: 10.1093/bioinformatics/btu014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlin BR, Shu MA, Winkelstein E, et al. (2009) More rare birds, and the occasional swan. Gastroenterology 136:2412–4. doi: 10.1053/j.gastro.2009.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger C, Abecasis GR, Hinds DA (2015) minimac2: faster genotype imputation. Bioinformatics 31:782–4. doi: 10.1093/bioinformatics/btu704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome of the Netherlands Consortium LC, Menelaou A, Pulit SL, et al. (2014) Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 46:818–25. doi: 10.1038/ng.3021 [DOI] [PubMed] [Google Scholar]
- Goedert JJ, Chen BE, Preiss L, et al. (2007) Reconstruction of the hepatitis C virus epidemic in the US hemophilia population, 1940–1990. Am J Epidemiol 165:1443–53. doi: 10.1093/aje/kwm030 [DOI] [PubMed] [Google Scholar]
- Gurdasani D, Carstensen T, Tekola-Ayele F, et al. (2014) The African Genome Variation Project shapes medical genetics in Africa. Nature 517:327–332. doi: 10.1038/nature13997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Levy JL, Gaddis NC, et al. (2012) Assessment of Genotype Imputation Performance Using 1000 Genomes in African American Studies. PLoS One 7:e50610. doi: 10.1371/journal.pone.0050610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgartner MW, Donfield SM, Willoughby A, et al. (1993) Hemophilia growth and development study. Design, methods, and entry data. Am J Pediatr Hematol Oncol 15:208–18 [DOI] [PubMed] [Google Scholar]
- Hoffmann TJ, Zhan Y, Kvale MN, et al. (2011) Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics 98:422–430. doi: doi: 10.1016/j.ygeno.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, et al. (2012a) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44:955–9. doi: 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, et al. (2012b) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44:955–959. doi: 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GH, Tseng YC (2014) Genotype imputation accuracy with different reference panels in admixed populations. BMC Proc 8:S64. doi: 10.1186/1753-6561-8-s1-s64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Howie B, McCarthy S, et al. (2015) Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun 6:8111. doi: 10.1038/ncomms9111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Li Y, Singleton AB, et al. (2009) Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet 84:235–250. doi: 10.1016/j.ajhg.2009.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EO, Hancock DB, Levy JL, et al. (2013) Imputation across genotyping arrays for genome-wide association studies: Assessment of bias and a correction strategy. Hum Genet 132:509–522. doi: 10.1007/s00439-013-1266-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM (2002) The Human Genome Browser at UCSC. Genome Res 12:996–1006. doi: 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakoo SI, Thio CL, Martin MP, et al. (2004) HLA and NK Cell Inhibitory Receptor Genes in Resolving Hepatitis C Virus Infection. Science (80- ) 305:872–874. doi: 10.1126/science.1097670 [DOI] [PubMed] [Google Scholar]
- Kim AY, Kuntzen T, Timm J, et al. (2011) Spontaneous control of HCV is associated with expression of HLA-B 57 and preservation of targeted epitopes. Gastroenterology 140:686–696.e1. doi: 10.1053/j.gastro.2010.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krithika S, Valladares-Salgado A, Peralta J, et al. (2012) Evaluation of the imputation performance of the program IMPUTE in an admixed sample from Mexico City using several model designs. BMC Med Genomics 5:12. doi: 10.1186/1755-8794-5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuniholm MH, Gao X, Xue X, et al. (2011) The relation of HLA genotype to hepatitis C viral load and markers of liver fibrosis in HIV-infected and HIV-uninfected women. J Infect Dis 203:1807–14. doi: 10.1093/infdis/jir192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, et al. (2010) MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34:816–834. doi: 10.1002/gepi.20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P, Hartz SM, Zhang Z, et al. (2010) A new statistic to evaluate imputation reliability. PLoS One 5:e9697. doi: 10.1371/journal.pone.0009697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P-R, Danecek P, Palamara PF, et al. (2016) Reference-based phasing using the Haplotype Reference Consortium panel. bioRxiv 48:52308. doi: 10.1101/052308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangia A, Gentile R, Cascavilla I, et al. (1999) HLA class II favors clearance of HCV infection and progression of the chronic liver damage. J Hepatol 30:984–9 [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B (2010) Genotype imputation for genome-wide association studies. Nat Rev Genet 11:499–511. doi: 10.1038/nrg2796 [DOI] [PubMed] [Google Scholar]
- Mathias RA, Taub MA, Gignoux CR, et al. (2016) A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat Commun 7:12522. doi: 10.1038/ncomms12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, et al. (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48:1279–1283. doi: 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae AF (2017) Analysis of Genome-Wide Association Data In: Keith JM (ed) Bioinformatics, Second. Humana Press, Melbourne, pp 161–174 [Google Scholar]
- Mitt M, Kals M, Pärn K, et al. (2017) Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur J Hum Genet. doi: 10.1038/ejhg.2017.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SC, Doheny KF, Pugh EW, et al. (2013) Imputation-Based Genomic Coverage Assessments of Current Human Genotyping Arrays. G3 3:1795–1807. doi: 10.1534/g3.113.007161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SC, Romm JM, Doheny KF, et al. (2017) Imputation-Based Genomic Coverage Assessments of Current Genotyping Arrays: Illumina HumanCore, OmniExpress, Multi-Ethnic global array and sub-arrays, Global Screening Array, Omni2.5M, Omni5M, and Affymetrix UK Biobank. bioRxiv. doi: 10.1101/150219 [DOI] [Google Scholar]
- Nothnagel M, Ellinghaus D, Schreiber S, et al. (2009) A comprehensive evaluation of SNP genotype imputation. Hum Genet 125:163–171. doi: 10.1007/s00439-008-0606-5 [DOI] [PubMed] [Google Scholar]
- Parker MM, Foreman MG, Abel HJ, et al. (2014) Admixture mapping identifies a quantitative trait locus associated with FEV1/FVC in the COPDGene Study. Genet Epidemiol 38:652–659. doi: 10.1002/gepi.21847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistis G, Porcu E, Vrieze SI, et al. (2015) Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. Eur J Hum Genet 23:975–83. doi: 10.1038/ejhg.2014.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909. doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013) R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing [Google Scholar]
- Ramnarine S, Zhang J, Chen L-S, et al. (2015) When Does Choice of Accuracy Measure Alter Imputation Accuracy Assessments. PLoS One 10:e0137601. doi: 10.1371/journal.pone.0137601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan EA, Hokanson JE, Murphy JR, et al. (2010) Genetic Epidemiology of COPD (COPDGene) Study Design. COPD J Chronic Obstr Pulm Dis 7:32–43. doi: 10.3109/15412550903499522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshyara NR, Horn K, Kirsten H, et al. (2016) Comparing performance of modern genotype imputation methods in different ethnicities. Sci Rep 6:34386. doi: 10.1038/srep34386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshyara NR, Scholz M (2015) Impact of genetic similarity on imputation accuracy. BMC Genet 16:90. doi: 10.1186/s12863-015-0248-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D, Adeyemo A, Chen G, Rotimi CN (2010) Practical considerations for imputation of untyped markers in admixed populations. Genet Epidemiol 34:258–265. doi: 10.1002/gepi.20457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, et al. (2015) An integrated map of structural variation in 2,504 human genomes. Nature 526:75–81. doi: 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Gu CC, Tiwari HK, et al. (2012) Genotype Imputation for African Americans Using Data From HapMap Phase II Versus 1000 Genomes Projects. Genet Epidemiol 36:508–516. doi: 10.1002/gepi.21647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap 3 Consortium, Altshuler DM, Gibbs RA, et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58. doi: 10.1038/nature09298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler LH, Bahrami SH, Kaidarova Z, et al. (2010) A case-control study of factors associated with resolution of hepatitis C viremia in former blood donors (CME). Transfusion 50:1513–23. doi: 10.1111/j.1537-2995.2010.02634.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G, Carneiro M, Hartl C, et al. (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–33. doi: 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iperen E, Hovingh G, Asselbergs F, Zwinderman A (2017) Extending the use of GWAS data by combining data from different genetic platforms. PLoS One 12:e0172082. doi: 10.1371/journal.pone.0172082. eCollection 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SS, de Andrade M, Tromp G, et al. (2014) Imputation and quality control steps for combining multiple genome-wide datasets. Front Genet 5:370. doi: 10.3389/fgene.2014.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, et al. (2017) 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet 101:5–22. doi: 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahov D, Muñoz A, Anthony J, et al. (1990) Association of drug injection patterns with antibody to human immunodeficiency virus type 1 among intravenous drug users in Baltimore, Maryland. Am J Epidemiol 132:847–856 [DOI] [PubMed] [Google Scholar]
- Walter K, Min JL, Huang J, et al. (2015) The UK10K project identifies rare variants in health and disease. Nature 526:82–90. doi: 10.1038/nature14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren HR, Evangelou E, Cabrera CP, et al. (2017) Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet 49:403–415. doi: 10.1038/ng.3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik GL, Fuchsberger C, Taliun D, et al. (2017) Imputation aware tag SNP selection to improve power for multi-ethnic association studies. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik GL, Thio CL, Kao WHL, et al. (2014) Admixture Analysis of Spontaneous Hepatitis C Virus Clearance in Individuals of African-Descent. Genes Immun 15:241–246. doi: 10.1038/gene.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhi D, Zhang K, et al. (2011) Practical Consideration of Genotype Imputation: Sample Size, Window Size, Reference Choice, and Untyped Rate. Stat Interface 4:339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Timofeev N, Hartley SW, et al. (2008) Imputation of missing genotypes: an empirical evaluation of IMPUTE. BMC Genet 9:85. doi: 10.1186/1471-2156-9-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng-Bradley X, Flicek P (2016) Applications of the 1000 Genomes Project resources. Brief Funct Genomics 16:elw027. doi: 10.1093/bfgp/elw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.