SUMMARY
The intestinal tract is constantly exposed to various stimuli. Group 3 innate lymphoid cells (ILC3s) reside in lymphoid organs and in the intestinal tract and are required for immunity to enteric bacterial infection. However, the mechanisms that regulate the ILC3s in vivo remain incompletely defined. Here, we show that GPR183, a chemotactic receptor expressed on murine and human ILC3s, regulates ILC3 migration toward its ligand 7α,25-dihydroxycholesterol (7α,25-OHC) in vitro, and GPR183 deficiency in vivo leads to a disorganized distribution of ILC3s in mesenteric lymph nodes and decreased ILC3 accumulation in the intestine. GPR183 functions intrinsically in ILC3s, and GPR183-deficient mice are more susceptible to enteric bacterial infection. Together, thes1e results reveal a role for the GPR183-7α,25-OHC pathway in regulating the accumulation, distribution, and anti-microbial and tissue-protective functions of ILC3s and define a critical role for this pathway in promoting innate immunity to enteric bacterial infection.
In Brief
Chu et al. demonstrate that GPR183 and its ligand 7α,25-OHC regulate the accumulation, distribution, and antimicrobial and tissue-protective functions of group 3 innate lymphoid cells, thus revealing a critical role for this pathway in promoting innate immunity against enteric bacterial infection.
Graphical Abstract
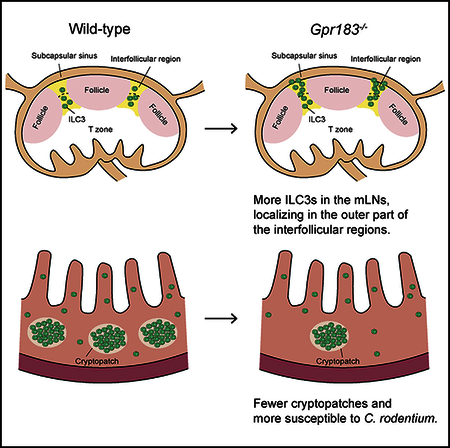
INTRODUCTION
The intestinal mucosal barrier surface is constantly exposed to food antigens, beneficial microbes, pathogens, and a multitude of other environmental stimuli (Turner, 2009). Innate lymphoid cells (ILCs) are known to contribute to innate and adaptive immune responses against these stimuli and play a critical role in maintaining barrier function and intestinal homeostasis (Artis and Spits, 2015; Diefenbach et al., 2014; Eberl et al., 2015; Klose and Artis, 2016; Spits et al., 2013, 2016). ILCs are lineage-negative (Lin-), interleukin-7 (IL-7) receptor α-positive (CD127+), CD90+ innate immune cells that are widely distributed throughout the body, particularly enriched at the mucosal barriers (Artis and Spits, 2015; Diefenbach et al., 2014; Eberl et al., 2015; Klose and Artis, 2016). Group 3 ILCs (ILC3s) express the transcription factor RORγt and play pivotal roles in protecting against bacterial, viral, and fungal infections in the intestine by fortifying the epithelial barrier via rapid secretion of soluble factors, such as IL-22, lymphotoxin α, and IL-17A, as well as regulating CD4+ T cell responses toward intestinal commensal bacteria (Fernandes et al., 2014; Gladiator et al., 2013; Hepworth et al., 2013, 2015; Kim et al., 2012; Klose et al., 2013; Satoh-Takayama et al., 2008). ILC3s are enriched in lymphoid tissues and at mucosal barrier surfaces, such as the intestinal tract, protecting against hazardous environmental stimuli together with other immune cells (Artis and Spits, 2015; Diefenbach et al., 2014; Eberl et al., 2015; Klose and Artis, 2016). Immune cells express various G-protein-coupled receptors (GPRs), including C-C motif chemokine receptors (CCRs), C-X-C motif chemokine receptors, and other GPRs, such as GPR183 and sphingosine-1-phosphate receptors, which regulate cell migration, accumulation, and distribution in tissues. Several chemokine receptors have been reported to control the accumulation of a subset of the ILC3s (Ivanov et al., 2006; Kim et al., 2015; Mackley et al., 2015; Satoh-Takayama et al., 2014); however, the molecular mechanisms that regulate the accumulation, distribution, and function of the entire ILC3 population in lymphoid and mucosal tissues and their effects on anti-bacterial responses and tissue protection are incompletely defined.
GPR183 (also known as EBI2) is a Gα-coupled seven-transmembrane chemotactic receptor. It is highly expressed on follicular B cells, CD4+ dendritic cells (DCs), and CD4+ T cells but is downregulated on germinal center (GC) B cells in secondary lymphoid organs and controls cell migration to achieve efficient antibody responses and CD4+ T cell responses (Gatto et al., 2009, 2013; Li et al., 2016; Pereira et al., 2009; Yi and Cyster, 2013; Yi et al., 2012). GPR183 ligand, 7α,25-dihydroxycholesterol (7α,25-OHC), is produced by stromal cells residing in the interfollicular regions of lymph nodes (LNs) and the bridging channels of the spleen (Hanne-douche et al., 2011; Liu et al., 2011; Yi et al., 2012). GPR183 expressed on CD4+ ILC3s (also termed as lymphoid tissue inducer cells [LTis]) controls their migration and the formation of colonic tertiary lymphoid organs (Emgård et al., 2018). However, whether GPR183 and 7α,25-OHC control the accumulation, distribution, and tissue-protective function of ILC3s in the gut-associated lymphoid tissues and in the intestinal lamina propria (LP) has not been examined.
In this study, we demonstrate that ILC3s isolated from the mesenteric LNs (mLNs) and intestinal LP express GPR183 and intestinal ILC3s migrate toward 7α,25-OHC in vitro. Quantitative PCR (qPCR) analysis indicated 7α,25-OHC production by gut stromal cells, and genetic deletion of GPR183 or 7α,25-OHC resulted in a disorganized accumulation of ILC3s in the subcap-sular sinus of the mLNs and reduced ILC3 accumulation in the intestine. The regulation of ILC3 accumulation in the intestine by GPR183 was ILC3 intrinsic and was required for optimal IL-22 production and protective immunity against the enteric bacterium, Citrobacter rodentium (C. rodentium). Taken together, these data reveal a previously unrecognized role of the GPR183-7α,25-OHC pathway in regulating ILC3-dependent immunity to enteric bacterial infection.
RESULTS
GPR183 Is Constitutively Expressed on ILC3s and Mediates Migration toward 7α,25-OHC
To study the pathways that regulate the distribution and accumulation of ILC3s in lymphoid and mucosal tissues in vivo, we undertook an unbiased analysis of molecules associated with these processes by RNA sequencing (RNA-seq). Among the genes listed in the gene ontology (GO) term “lymphocyte chemo-taxis,’’ Gpr183 was the most abundant gene expressed in intestinal LP ILC3s (Figure S1A). To further examine this, we sorted ILC subsets from intestinal LP cells as well as mLN cells harvested from Rorc(γt)Gfp mice and analyzed the expression of Gpr183 mRNA by quantitative PCR. Consistent with the RNA-seq data, intestinal CCR6+ ILC3s and CCR6- lLC3s and mLN ILC3s had high Gpr183 mRNA expression, and the expression in intestinal CCR6+ ILC3s was higher than in ILC1s and ILC2s (Figure 1A). To examine GPR183 protein expression, we employed Gpr183LacZ/+ reporter mice, which were generated by replacing the Gpr183 coding region with the LacZ gene, and we could detect LacZ expression in non-GC B cells, but not in GC B cells (Figure S1B). We confirmed the LacZ expression in mLN and small intestinal (SI) ILC3s (Figure 1B; gating strategy in Figure S1C). We also detected GPR183 protein staining on CD45+Lin-CD127+CD117+CRTH2- cells, which are ILC3s in human ileal LP and ILC precursors in peripheral blood mononuclear cells (PBMCs) (Lim et al., 2017) from healthy donors (Figures 1C and 1D; gating strategy in Figure S1D). Collectively, these data show that murine and human ILC3s express GPR183.
Figure 1. GPR183 Is Expressed on ILC3s and Regulates ILC3 Migration.
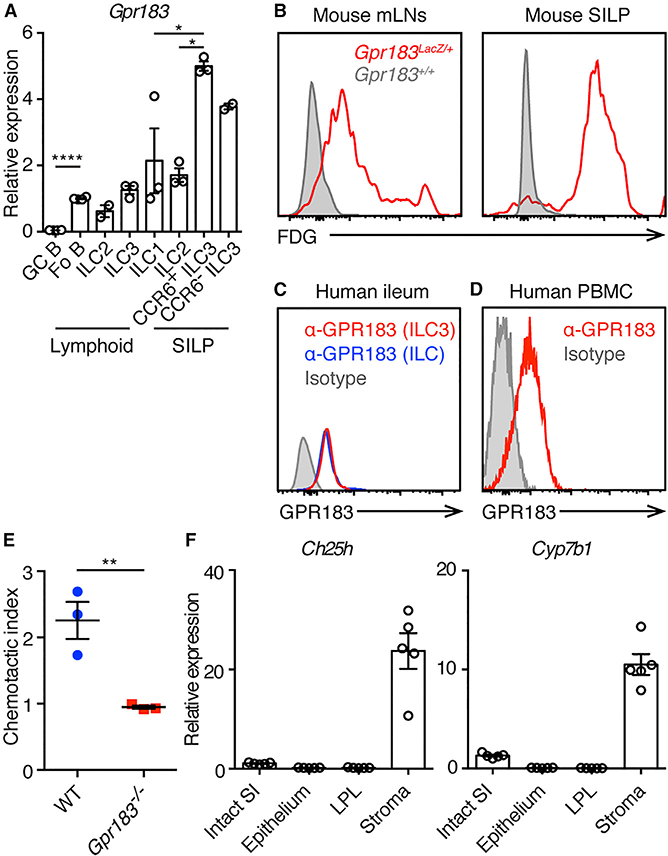
(A) qPCR analysis of Gpr183 transcript abundance in indicated sort-purified cell populations presented relative to Hprt1. Each symbol represents sample from one mouse or pooled from 2 or 3 mice. Fo B, follicular B cells; GC B, germinal center B cells. *p < 0.05 and ****p < 0.0001 by one-way ANOVA with Dunnett’s multiple comparison.
(B) LacZ staining (conversion of the fluorescent LacZ substrate fluorescein di-β-D-galactopyrano-side [FDG]) indicating GPR183 expression on ILC3s from mLNs and SILP. Data are representative of 2 independent experiments.
(C) Flow cytometry histograms of anti-GPR183 staining on ILC3s (red line), total ILCs (blue line), and isotype control staining on total ILCs (shaded gray) from human terminal ileum. Data are representative of 5 donors.
(D) Flow cytometry histograms of anti-GPR183 staining (red line) and isotype control staining (shaded gray) on ILC precursors from human PBMCs. Data are representative of 4 donors.
(E) In vitro migration of ILC3s toward 7α,25-OHC. Data are representative of 2 independent experiments. **p < 0.01.
(F) qPCR analysis of 7α,25-OHC-producing enzymes Ch25h and Cyp7b1 transcript abundances in fractionated SI samples from WT mice, presented relative to Hprt1. LPL, lamina propria leukocyte.
Each symbol represents one mouse unless specifically indicated. Data are mean ± SEM. See also Figure S1.
Given that GPR183 is a chemotactic receptor expressed on follicular B cells, CD4+ T cells, and DCs, mediating their migration toward the GPR183 ligand 7α,25-OHC (Gatto et al., 2009, 2013; Li et al., 2016; Pereira et al., 2009; Yi and Cyster, 2013; Yi et al., 2012), we tested whether ILC3s migrate toward 7α,25-OHC in vitro. We performed transwell migration assays with SILP cells from wild-type (WT) mice and Gpr183−/−mice. WT ILC3s migrated toward 7α,25-OHC in vitro, whereas Gpr183−/− ILC3s did not, indicating that GPR183 regulates ILC3s migration toward 7α,25-OHC (Figure 1E).
CH25H and CYP7B1 Are Expressed in Intestinal Stromal Cells
Cholesterol 25-hydroxylase (CH25H) and oxysterol 7α-hydroxy-lase (CYP7B1), which are enzymes required for the biosynthesis of GPR183 ligand 7α,25-OHC, are highly expressed in spleen and LN stromal cells (Hannedouche et al., 2011; Liu et al., 2011; Yi et al., 2012). To determine whether CH25H and CYP7B1 are also expressed in intestinal stromal cells, we fractionated mouse SI into epithelial, LP leukocyte (LPL), and stromal compartments and measured the relative expression of Ch25h and Cyp7b1 mRNA in each compartment by qPCR. Expression of Ch25h and Cyp7b1 was higher in the stromal compartment compared to epithelial and LPL compartments of the SI (Figure 1F), suggesting 7α,25-OHC production by intestinal stromal cells. Together with previous reports (Emgård et al., 2018; Yi et al., 2012), these data indicate that the interaction of GPR183 expressed on ILC3s and 7α,25-OHC produced by stromal cells in mLNs and the intestine may control the accumulation of ILC3s in these organs.
GPR183 and 7α,25-OHC Control the Distribution of ILC3s in mLNs
To examine the effect of GPR183 deficiency on ILC3s at steady state in vivo, we analyzed ILC3s from WT mice and Gpr183−/− mice using flow cytometry. Percentages of RORγt+ ILC3s within the total ILC population (CD45+Lin-CD90+CD127+; refer to the gating strategy in Figure S2A) were significantly increased in the mLNs of Gpr183−/− mice, along with increased ILC3 numbers compared to WT mice (Figures 2A and 2B). The frequencies and numbers of NKp46+RORγt- ILC1s and GATA-3+ ILC2s were reduced in Gpr183−/− mice compared to WT mice (Figures S2B and S2C). Ch25h−/− mice that have defective 7α,25-OHC production (Hannedouche et al., 2011; Liu et al., 2011) exhibited increased ILC3s, reduced ILC1s, and comparable ILC2s in the mLNs compared to WT mice at steady state (Figures 2C, 2D, S2D, and S2E).
Figure 2. Disorganized ILC3 Distribution in the mLNs of Gpr183−/− and Ch25h−/− Mice.
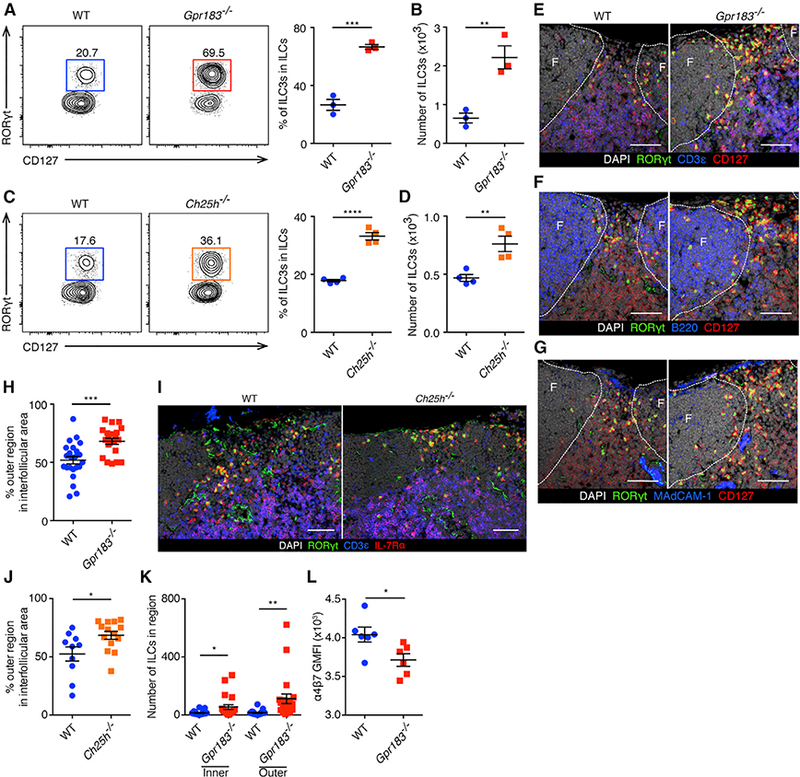
(A and B) Population frequencies (A) and numbers
(B) of RORγt+ ILC3s in the mLNs of WT and Gpr183−/− mice at steady state, gated on CD45+Lin-CD127+CD90+ cells. Data are representative of 3 independent experiments. **p < 0.01, ***p < 0.001.
(C and D) Population frequencies (C) and numbers (D) of RORγt+ ILC3s in the mLNs of WT and Ch25h−/− mice at steady state, gated on CD45+Lin-CD127+CD90+ cells. Data are representative of 3 independent experiments. **p < 0.01, ****p < 0.0001.
(E-G) Representative images of mLN sections of WT and Gpr183−/− mice stained for RORγt, CD127 and DAPI together with CD3ε (E), B220(F) or MAdCAM-1 (G). Follicles, or F, are designated by dashed lines. The scale bars represent 50 μm.
(H) Percentages of ILC3s localized in the outer regions over ILC3s localized in the interfollicular areas between follicles in mLN sections of WT and Gpr183−/− mice. Twenty-three interfollicular areas in15 mLNs from 3 WT mice and 21 interfollicular areas in 15 mLNs from 3 Gpr183−/− mice were examined. ***p < 0.001. (I and J) Representative images of ILC3 distribution (I) and percentages of ILC3s localized in the outer regions (J) in mLN sections of WT and Ch25h−/− mice. Ten interfollicular areas in 10 mLNs from 2 WT mice and 14 interfollicular areas in 10 mLNs from 2 Ch25h−/− mice were examined. The scale bars represent 100 μm. In
(J), *p < 0.05.
(K) Numbers of ILC3s in interfollicular areas (inner and outer regions) in mLN sections of WT and Gpr183−/− mice. See Figures 2E-2H legend for details. *p <0.05, **p < 0.01.
(L) The enumeration of anti-a4b7 geometric mean fluorescence intensity (GMFI) on mLN ILC3s of WT and Gpr183−/− mice. Data are representative of 3 independent experiments. *p < 0.05.
Each symbol represents one mouse (A-D and L) or one region (H, J, and K). Data are mean ± SEM. See also Figure S2.
Previous studies identified that the localization of GPR183-expressing T cells, B cells, and DCs within lymphoid tissues is regulated by 7α,25-OHC produced from tissue stromal cells (Gatto et al., 2009, 2013; Hannedouche et al., 2011; Li et al., 2016; Liu et al., 2011; Pereira et al., 2009; Yi and Cyster, 2013; Yi et al., 2012). To examine whether GPR183 regulates the localization of ILC3s in mLNs, we stained mLN sections from WT mice and Gpr183−/− mice with anti-RORγt, CD3ε, and CD127 antibodies to visualize ILC3s (Figure 2E). Serial sections were stained for B220 or MAdCAM-1 to examine the position of follicles and the subcapsular sinuses (Figures 2F and 2G). There was no overt difference in the structure of follicles and T cell areas in mLNs from Gpr183−/− mice compared to WT mice, which is consistent with the previous report (Pereira et al., 2009). RORγt+CD3ε-CD127+ ILC3s were found predominantly in the interfollicular areas of WT mLN sections, which is consistent with published results (Hepworth et al., 2015; Mackley et al., 2015). A significantly higher proportion of the Gpr183−/− ILC3s were found in the outer regions of the in-terfollicular areas rather than their normal localization in mLNs compared to that of the WT ILC3s (Figures 2E-2H; refer to Figure S2F for the definition of inner and outer regions). Similarly, staining of mLN sections from WT mice and Ch25h−/− mice showed more ILC3s localized in the outer regions of the interfollicular areas of Ch25h−/− mLNs compared to WT mLNs (Figures 2I, 2J, and S2F). Consistent with the flow cytometry data (Figures 2A and 2B), more ILC3s were found in the interfollicular areas in the Gpr183−/− mLNs compared to WT (Figure 2K). We also examined the Peyer’s patches (PPs) and found increased ILC3s in the Gpr183−/− PPs compared to WT (Figure S2G). Immunofluorescent staining of PP sections revealed accumulation of ILC3s in between the follicle and the T cell area in the luminal side of Gpr183−/− mice (Figure S2H). Collectively, these results suggest that GPR183 and 7α,25-OHC control the distribution and accumulation of ILC3s in the mLNs and PPs. Notably, ILC3s from Gpr183−/− mice had comparable expression of CCR7 and CCR9 but significantly lower expression of integrin α4β7, which mediates cell trafficking to the intestine (Kim et al., 2015), compared to ILC3s from WT mice (Figures 2L and S2I).
GPR183 and 7α,25-OHC Regulate ILC3 Accumulation in the Intestine
Given that GPR183 appears to regulate migration of intestinal ILC3s (Figure 1E) and expression of gut-tropic integrin expression (Figure 2L), we analyzed the accumulation of ILC3s in the SILP of Gpr183−/− mice and WT mice. In contrast to mLNs, RORγt+ ILC3s, including both the NKp46+ and CCR6+ ILC3 subsets, were significantly reduced in Gpr183-l- SILP compared to WT at steady state (Figures 3A-3D). As intestinal stromal cells are potential producers of 7α,25-OHC (Figure 1F; Emgård et al., 2018), we also examined the SILP of Ch25−/− mice and found fewer ILC3s in Ch25−/− mice compared to WT mice (Figures 3E-3H). To examine whether GPR183 regulates the localization of ILC3s in the SI, we crossed Gpr183−/− mice with Rorc(γt)Gfp mice and stained the SI sections with anti-GFP, CD3ε, and B220 antibodies (Figure 3I). WT and Gpr183−/− GFP +CD3ε- ILC3s were similarly scattered in the LP, but Gpr183−/− mice had fewer cryptopatches (CPs) compared to WT mice, which is consistent with the flow cytometry data (Figures 3A, 3B, and 3J). When we analyzed the colon LP at steady state, the numbers and proportions of ILCs were comparable between WT and Gpr183−/− mice or Ch25h−/− mice (Figures S3A-S3D), but Gpr183−/− mice had fewer CPs and isolated lymphoid follicles in the colon compared to WT mice (Figures S3E and S3F). Taken together, these results suggest that, through binding to 7α,25-OHC produced by intestinal stromal cells, GPR183 promotes the accumulation of ILC3s in the intestine.
Figure 3. Defective ILC3 Accumulation in the SI of Gpr183−/− and Ch25h−/− Mice.
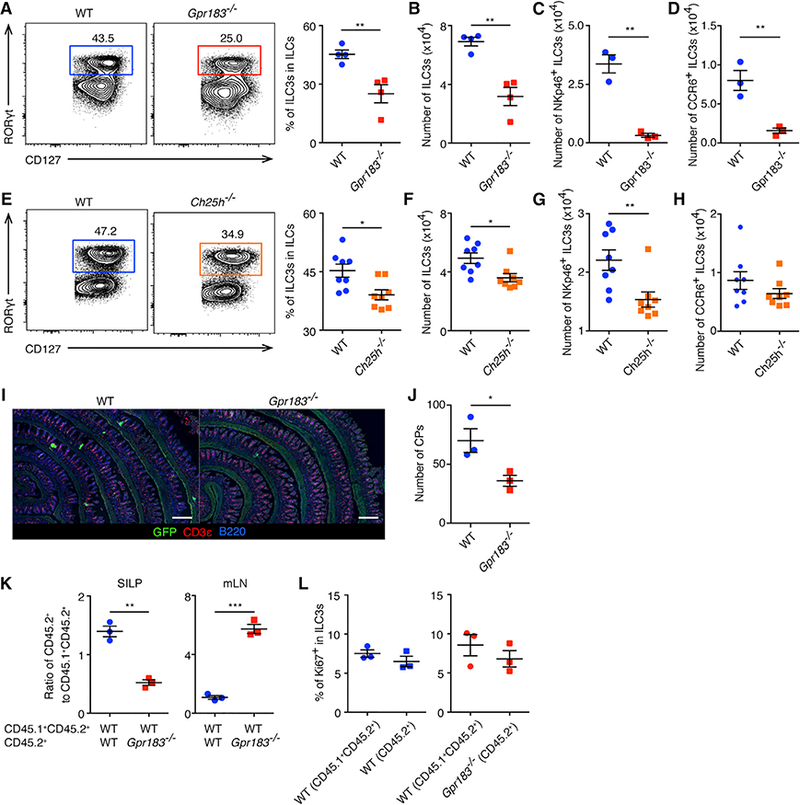
(A-D) Population frequencies (A) and numbers (B) of RORγt+ ILC3s and numbers of NKp46+RORγt+ ILC3s (C) and CCR6+RORγt+ ILC3s (D) in the SI of WT and Gpr183−/− mice at steady state, gated on CD45+Lin- CD127+CD90+ cells. Data are representative of 3 independent experiments. **p < 0.01.
(E-H) Population frequencies (E) and numbers (F) of RORγt+ ILC3s and numbers of NKp46+RORγt+ ILC3s (G) and CCR6+RORγt+ ILC3s (H) in the SI of WT and Ch25h−/− mice at steady state, gated on CD45+Lin-CD127+CD90+ cells. Data are pooled from 2 experiments. In (E) and (F), *p < 0.05. In (G), **p < 0.01.
(I)Representative images of ILC3 distribution and CPs in SI sections of WT and Gpr183−/− mice. The scale bars represent 500 μm.
(J) Numbers of CPs in SI sections of WT and Gpr183−/− mice. *p < 0.05.
(K) Ratios of CD45.2+ ILC3s to CD45.1+CD45.2+ ILC3s in the SI and mLNs of mixed BM chimera mice with indicated genotypes. Data are representative of 3 independent experiments. **p < 0.01, ***p < 0.001. See also Figure S3G.
(L) Frequencies of Ki67-expressing ILC3s in the SI of WT:WT (left) and Gpr183−/−:WT (right) BM chimera mice. Data are representative of 3 independent experiments.
Each symbol represents one mouse. Data are mean ± SEM. See also Figure S3.
ILC3-Intrinsic Expression of GPR183 Controls Their Accumulation in the Intestine and mLNs
To test whether the regulation of ILC3 accumulation in the intestine and mLNs by GPR183 is ILC3-intrinsic, we generated mixed bone marrow (BM) chimera mice by transferring congenically labeled WT or Gpr183−/− cells (CD45.2+) mixed with WT BM cells (CD45.1+CD45.2+) into lethally irradiated recipient mice (CD45.1+). After 8 weeks of reconstitution, we examined the ratios of WT or Gpr183−/− CD45.2+ cells to internal control WT CD45.1+CD45.2+ cells within ILC3 populations. Compared to WT CD45.2+ BM mixed with WT CD45.1+CD45.2+ BM chimera (WT:WT mice), Gpr183−/− CD45.2+ BM mixed with WT CD45.1+CD45.2+ BM chimera (Gpr183−/−:WT mice) had significantly reduced ratios of CD45.2+ ILC3s to CD45.1+CD45.2+ ILC3s in the SI (Figures 3K and S3G). We also observed smaller ILC3 percentages in Gpr183−/− CD45.2+ cells compared to that in WT CD45.1+CD45.2+ cells in total SI ILCs within each Gpr183−/−:WT mice (Figure S3H; 8.3 ± 0.1 versus 13.4 ± 0.9; mean ± SEM; p < 0.01). Meanwhile, Gpr183−/−:WT mice exhibited significantly increased ratios of CD45.2+ ILC3s to CD45.1+CD45.2+ ILC3s in the mLNs compared to WT:WT mice (Figure 3K).
The expression of the proliferation marker Ki67 was comparable in CD45.1+CD45.2+ ILC3s (WT) and CD45.2+ ILC3s (WT or Gpr183−/−) in both chimeras (Figure 3L). Therefore, GPR183 appears to regulate the accumulation, but not the proliferation, of ILC3s in the intestine and mLNs in a cell-intrinsic manner. Moreover, Gpr183−/−:WT mice exhibited increased ratios of CD45.2+ cells in the ILC1 gate and comparable ratios of ILC2s compared to WT:WT mice, suggesting that the reduced ILC1s and ILC2s in Gpr183−/− mLNs was secondary to the cell-intrinsic effect on ILC3s (Figure S3I).
GPR183 Is Required for ILC3-Mediated Protective Immunity following Enteric Bacterial Infection
ILC3s are critical in promoting innate immunity to C. rodentium infection through producing IL-22, which triggers the secretion of anti-microbial peptides from intestinal epithelial cells (Satoh-Takayama et al., 2008; Sawa et al., 2011; Sonnenberg et al., 2011; Zheng et al., 2008). Considering that GPR183 deficiency hampers ILC3 accumulation in the intestine, we sought to test whether GPR183 regulates ILC3-mediated innate immunity against C. rodentium infection. Similar to the SI of Gpr183−/− mice (Figures 3A-3D), percentages and numbers of ILC3s were significantly reduced in the SI and the colon of Rag1−/−Gpr183−/− mice compared to Rag1−/− mice at steady state (Figures S4A-S4D). Numbers of NKp46+ ILC3s and CCR6+ ILC3s were both significantly reduced in Rag1−/−Gpr183−/− mice (Figures S4E and S4F). At 8 days after C. rodentium infection, Rag1−/−Gpr183−/− mice also exhibited significantly reduced frequencies and numbers of ILC3s in the SI and the colon compared to Rag1−/− mice (Figures 4A, 4B, S4G, and S4H). Similar to steady state, numbers of NKp46+ ILC3s and CCR6+ ILC3s in the SI were both significantly reduced in infected Rag1−/−Gpr183−/− mice compared to Rag1−/− mice (Figures S4I and S4J). The percentages of Ki67+ cells in intestinal ILC3s were comparable between Rag1−/− mice and Rag1−/− Gpr183−/− mice, suggesting that the proliferation of ILC3s during infection is not regulated by GPR183 (Figures 4C and S4K). We next examined whether the IL-22-producing capacity of ILC3s was regulated by GPR183 expression. Notably, both Rag1−/− and Rag1−/−Gpr183−/− ILC3s contained similar percentages of IL-22+ cells, suggesting that GPR183 deficiency does not affect the capability of IL-22 production in ILC3s (Figures 4D, S4L, and S4M). However, numbers of IL-22-producing ILC3 were significantly reduced in Rag1−/−Gpr183−/− compared to Rag1−/− mice, due to lower total numbers of ILC3 in Rag1−/−Gpr183−/− mice (Figures 4E and S4N).
Figure 4. GPR183 Is Required for ILC3-Mediated Protection against C. rodentium Infection.
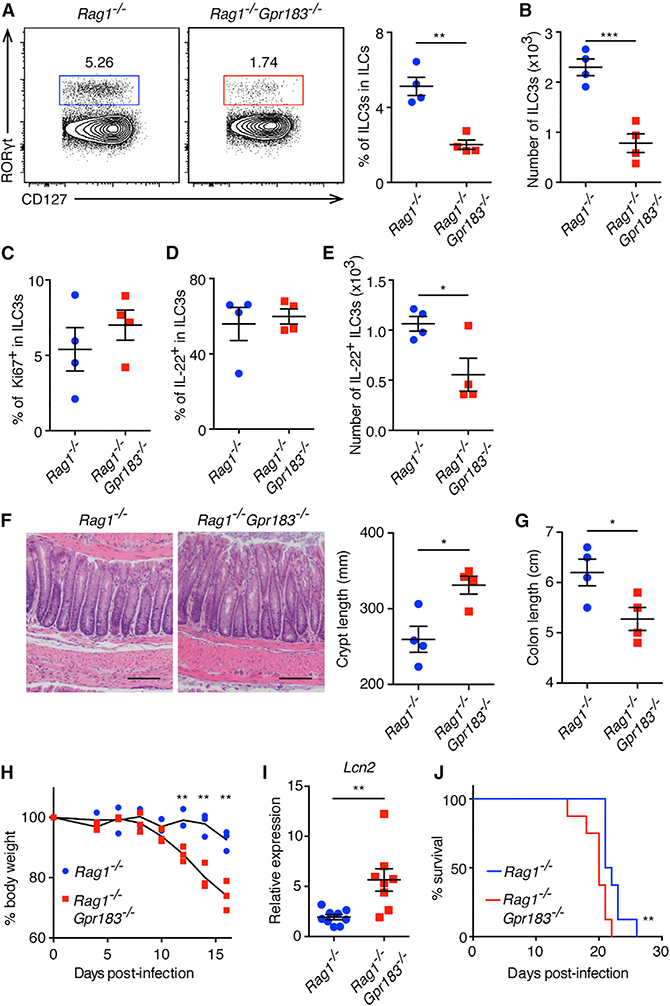
(A-H) Rag1−/− and Rag1−/−Gpr183−/− mice were infected with C. rodentium and were analyzed 8 (A-E and I) and 10(F-H) days post-infection (d.p.i.).
(A and B) Population frequencies (A) and numbers (B) of ILC3s in the colon LP, gated on CD45+Lin-CD127+CD90+ cells. **p < 0.01, ***p < 0.001.
(C) Frequencies of Ki67-expressing colonic ILC3s. (D and E) Frequencies (D) and numbers (E) of IL-22-producing colonic ILC3s following IL-23 restimulation. In (E), *p < 0.05.
(F) Representative H&E staining sections of colon (left) and enumeration of the crypt length (right). The scale bars represent 100 μm.
(G-J) Colon length (G), changes in body weight (H), Lcn2 mRNA expression (I), and survival (J) of infected Rag1−/− and Rag1−/−Gpr183−/− mice. Body weight is presented relative to initial weight, set as 100%. Survival data are pooled from 2 experiments (8 mice per group). *p < 0.05, **p < 0.01. All data are representative of 2 independent experiments unless stated. Each symbol represents one mouse. Data are mean ± SEM. See also Figure S4.
In accordance with reduced numbers of IL-22-producing ILC3s, Rag1−/−Gpr183−/− mice exhibited significantly more severe colonic crypt hyperplasia, colonic shortening, and more severe loss of body weight compared to Rag1−/− mice, indicating impaired ILC3-dependent innate immunity and tissue protection in Rag1−/−Gpr183−/− mice (Figures 4F-4H). Lipocalin 2 (Lcn2) mRNA expression was higher in Rag1−/−Gpr183−/− mice, which is consistent with exacerbated inflammation compared to Rag1−/− mice (Figure 4I). Moreover, Rag1−/−Gpr183−/− mice exhibited a reduced survival rate following infection compared to Rag1−/− mice (Figure 4J). Given the importance of IL-22 in tissue protection following bacterial infection (Satoh-Takayama et al., 2008; Sawa et al., 2011; Sonnenberg et al., 2011; Zheng et al., 2008), these results suggest that GPR183 regulates anti-bacterial responses and tissue protection through facilitating the accumulation of IL-22-producing ILC3s in the intestine. Taken together, these data identify a crosstalk between GPR183-expressing ILC3s and intestinal stromal cells that express 7α,25-OHC, which is required for optimal ILC3 responses and host protective immunity against enteric bacterial infection.
DISCUSSION
Mucosal barriers are constitutively challenged by various stimuli, and the homeostasis of mucosal barriers both at steady state and upon challenge are maintained by tissue-resident immune cells (Kurashima et al., 2013; Okumura and Takeda, 2016). ILC3s are found in lymphoid tissues and are enriched in the intestine, where they play critical roles in regulating adaptive immune responses against commensal bacteria, as well as in innate immunity against enteric bacterial infections (Hepworth et al., 2013, 2015; Rankin et al., 2016; Satoh-Takayama et al., 2008; Sawa et al., 2011; Song et al., 2015; Sonnenberg et al., 2011). Although the mechanisms ILC3s employ to control infections and promote tissue repair continue to be defined (Satoh-Takayama et al., 2008; Sawa et al., 2011; Sonnenberg et al., 2011), our understanding of how the accumulation, distribution, and tissue-protective function of ILC3s in the intestine and its associated lymphoid organs are controlled remained limited. Emgård et al. (2018) recently reported that CD4+ LTi-like ILC3s express GPR183 that controls cell migration and formation of solitary intestinal lymphoid tissues in the colon and enhances IL-22 production by ILC3s in the colon at steady state. In the current study, we demonstrate that GPR183 is expressed on murine and human ILC3s and that GPR183 and its ligand 7α,25-OHC regulate the accumulation and distribution of ILC3s in lymphoid tissues and the intestine, and consequently, GPR183 controls ILC3-dependent innate immunity and tissue protection following enteric bacterial infection. We also identify GPR183-dependent accumulation of IL-22-producing ILC3s in the intestine following C. rodentium infection. Of note, enhanced IL-22 production by ILC3s was not detectable, possibly due to heightened inflammation elicited by the bacterial infection.
ILC3s reside in the interfollicular areas of the mLNs, where they present commensal bacterial antigen through major histocompatibility complex class II and prevent CD4+ T cell-induced chronic intestinal inflammation toward commensal bacteria (Hepworth et al., 2015). In this study, we show that GPR183 controls the distribution of ILC3s in mLNs. GPR183-deficient ILC3s accumulated in the outer regions of the inter-follicular areas, which are close to the subcapsular sinuses. DCs migrate into the LNs via the lymph through subcapsular sinuses and then move to the paracortex where they interact with helper T cells (Lian and Luster, 2015). This pathway is regulated by CCR7, a molecule that also controls the accumulation of ILC3s to LNs (Lian and Luster, 2015; Mackley et al., 2015). Similarly, ILC3s migrate from other organs, such as the intestine (Mackley et al., 2015), and enter the LNs through subcapsular sinuses. In the context of GPR183 deficiency, ILC3s cannot migrate into the interfollic-ular areas because they fail to respond to the ligand expressed in the inner regions of the interfollicular areas and hence are sequestered in the subcapsular sinuses. As GPR183 plays such important roles in regulating the distribution and function of ILC3s in both lymphoid and non-lymphoid tissues, GPR183 itself and its oxysterol ligand-producing pathway could be potential therapeutic targets for controlling and regulating ILC3 functions in multiple infectious and inflammatory diseases.
EXPERIMENTAL PROCEDURES
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures.
Mice
C57BL/6 (Jax 664), Gpr183LacZ/+ (Jax 26443), Ch25h−/− (Jax 16263), CD45.1 (Jax 2014), and Rag1−/− (Jax 2216) mice were purchased from The Jackson Laboratory. Rorc(γt)Gfp was provided by Dr. G. Eberl (Institut Pasteur, France). Breeding of Gpr183LacZ/+ to homozygosity resulted in Gpr183-deficient (Gpr183LacZ/LacZ) mice referred to as Gpr183−/− throughout the manuscript. Sex-and age-matched WT and transgenic mice between 6 and 16 weeks of age were co-housed and used for experiments. All mice were maintained under specific pathogen-free conditions and were used in accordance with the Institutional Animal Care and Use Committee guidelines at Weill Cornell Medical College.
Isolation and Flow Cytometry Staining of Human ILC3s
Intestinal biopsies from the terminal ileum were obtained, processed, and viably cryopreserved as previously described (Hepworth et al., 2013,2015). Following thawing, cells were stained for CD3 (UCHT1), CD19(HIB19),CD11c(S-HCL-3), CD11b (M1/70), CD14 (M5E2) as line age markers, and CD45 (HI30), CRTH2 (BM16), CD127 (A019D5), CD117 (104D2), GPR183 (SA313E4), or mouse immunoglobulin G2a (IgG2a), κ (MOPC-173) was used as isotype control. Dead cells were excluded with the Live/Dead Fixable Aqua Dead Cell Stain Kit. ILCs were gated as CD45+Lin-CD127+, and ILC3s were gated as CD45+Lin-CD127+CRTH2-CD117+. PBMCs were isolated from buffy coats (New York Blood Center) with a Ficoll-Paque PLUS (GE Healthcare) gradient. All samples were cryopreserved and stored in liquid nitrogen. For staining ILC precursorsfrom PBMCs, cells were incubated with FcR Blocking Reagent (Miltenyi Biotec) and subsequentlystained forCD3(UCHT1), CD123(6H6), CD5 (UCHT2), FcεRI (AER-37), CD11c (S-HCL-3), CD11b(M1/70), CD34 (581), CD14 (HCD14) as lineage markers and CD19 (HIB19), CD45 (HI30), CRTH2 (BM16), CD94 (DX22), CD127 (A019D5), and CD117 (104D2). For surface receptor expression analysis, anti-GPR183 (SA313E4) or mouse IgG2a, κ isotype control (MOPC-173) was used. Dead cells were excluded with the Live/Dead Fixable Aqua Dead Cell Stain Kit. ILC precursors were gated as CD45+Lin-CD19-CD94-CD127+CRTH2-CD117+. All antibodies were purchased from eBioscience, BioLegend, or BD Bioscience.
Quantification and Statistical Analysis
Statistical tests were performed with Prism (GraphPad). Unless specifically indicated otherwise, Student’s t tests were used to compare endpoint means of different groups. Error bars depict the SEM. For the comparison of Kaplan-Meier survival curves, log rank (mantel-Cox) test was used. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Supplementary Material
Highlights.
ILC3s from mouse mesenteric lymph nodes and mouse and human intestine express GPR183
GPR183 and its ligand 7α,25-OHC promote ILC3 migration in vitro
GPR183 and 7α,25-OHC regulate the accumulation and function of ILC3s in vivo
GPR183 is required for ILC3-mediated immunity against enteric bacterial infection
ACKNOWLEDGMENTS
We thank the Artis lab members and the Sonnenberg lab members for discussion and critical reading of the manuscript. We also thank Dr. G. Eberl for Rorc(γt)Gfp mice and Dr. B. Vallance for Citrobacter rodentium. This work was supported by the grants from the Japan Society for the Promotion of Science Overseas Research Fellowships (to S.M.), the German Research Foundation (KL 2963/1-1 to C.S.N.K.), the Novo Nordic Foundation (14052 to J.B.M.), the Jill Roberts Institute (to G.G.P.), the Wellcome Trust (Senior Research Fellowship 110199/Z/15/Z to D.R.W.), Cure for IBD (to D.A.), the Crohn’s and Colitis Foundation of America (to D.A. and G.F.S.), the Searle Scholars Program (to G.F.S.), an American Asthma Foundation Scholar Award (to G.F.S.), the Burroughs Wellcome Fund (to D.A.), and the NIH (DP5OD012116, AI123368, DK110262, and AI095608 to G.F.S.; AI074878, AI083480, AI095466, AI095608, AI102942, AI097333, and AI106697 to D.A.).
Footnotes
AUTHOR CONTRIBUTIONS
C.C. and S.M. carried out most experiments and analyzed the data with help from Z.L., L.Z., A.-L.F., C.S.N.K., J.B.M., D.R.W., and G.F.S. G.G.P. preformed RNA-seq analysis. C.C., S.M., and D.A. conceived the project and wrote the manuscript with input from all co-authors.
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.05.099.
REFERENCES
- Artis D, and Spits H (2015). The biology of innate lymphoid cells. Nature 517, 293–301. [DOI] [PubMed] [Google Scholar]
- Diefenbach A, Colonna M, and Koyasu S (2014). Development, differentiation, and diversity of innate lymphoid cells. Immunity 41, 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Colonna M, Di Santo JP, and McKenzie AN (2015). Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science 348, aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emgård J, Kammoun H, García-Cassani B, Chesné J, Parigi SM, Jacob JM, Cheng HW, Evren E, Das S, Czarnewski P, et al. (2018). Oxysterol Sensing through the Receptor GPR183 Promotes the Lymphoid-Tissue-Inducing Function of Innate Lymphoid Cells and Colonic Inflammation. Immunity 48, 120–132.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes SM, Pires AR, Ferreira C, Foxall RB, Rino J, Santos C, Correia L, Pocas J, Veiga-Fernandes H, and Sousa AE (2014). Enteric mucosa integrity in the presence of apreserved innate interleukin 22 compartment in HIV type 1-treated individuals. J. Infect. Dis 210, 630–640. [DOI] [PubMed] [Google Scholar]
- Gatto D, Paus D, Basten A, Mackay CR, and Brink R (2009). Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity 31, 259–269. [DOI] [PubMed] [Google Scholar]
- Gatto D, Wood K, Caminschi I, Murphy-Durland D, Schofield P, Christ D, Karupiah G, and Brink R (2013). The chemotactic receptor EBI2 regulates the homeostasis, localization and immunological function of splenic dendritic cells. Nat. Immunol 14, 446–453. [DOI] [PubMed] [Google Scholar]
- Gladiator A, Wangler N, Trautwein-Weidner K, and LeibundGut-Land-mann S (2013). Cutting edge: IL-17-secreting innate lymphoid cells are essential for host defense against fungal infection. J. Immunol 190, 521–525. [DOI] [PubMed] [Google Scholar]
- Hannedouche S, Zhang J, Yi T, Shen W, Nguyen D, Pereira JP, Guerini D, Baumgarten BU, Roggo S, Wen B, et al. (2011). Oxysterols direct immune cell migration via EBI2. Nature 475, 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, et al. (2013). Innate lymphoid cells regulate CD4+T-cell responses to intestinal commensal bacteria. Nature 498, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, Withers DR, Hugues S, Farrar MA, Reith W, et al. (2015). Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4+ T cells. Science 348, 1031–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- sIvanov II, Diehl GE, and Littman DR (2006). Lymphoid tissue inducer cells in intestinal immunity. Curr. Top. Microbiol. Immunol 308, 59–82. [DOI] [PubMed] [Google Scholar]
- Kim CJ, Nazli A, Rojas OL, Chege D, Alidina Z, Huibner S, Mujib S, Benko E, Kovacs C, Shin LY, et al. (2012). A role for mucosal IL-22 production and Th22 cells in HIV-associated mucosal immunopathogenesis. Mucosal Immunol. 5, 670–680. [DOI] [PubMed] [Google Scholar]
- Kim MH, Taparowsky EJ, and Kim CH (2015). Retinoic Acid Differentially Regulates the Migration of Innate Lymphoid Cell Subsets to the Gut. Immunity 43, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CS, and Artis D (2016). Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol 17, 765–774. [DOI] [PubMed] [Google Scholar]
- Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, Göppert N, Croxford AL, Waisman A, Tanriver Y, and Diefenbach A (2013). A T-bet gradient controlsthe fate and function of CCR6-RORγt+ innate lymphoid cells. Nature 494, 261–265. [DOI] [PubMed] [Google Scholar]
- Kurashima Y, Goto Y, and Kiyono H (2013). Mucosal innate immune cells regulate both gut homeostasis and intestinal inflammation. Eur. J. Immunol 43, 3108–3115. [DOI] [PubMed] [Google Scholar]
- Li J, Lu E, Yi T, and Cyster JG (2016). EBI2 augmentsTfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature 533, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J, and Luster AD (2015). Chemokine-guided cell positioning in the lymph node orchestrates the generation of adaptive immune responses. Curr. Opin. Cell Biol 36, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, Serafini N, Puel A, Bustamante J, Surace L, et al. (2017). ). Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell 768, 1086–1100.e10. [DOI] [PubMed] [Google Scholar]
- Liu C, Yang XV, Wu J, Kuei C, Mani NS, Zhang L, Yu J, Sutton SW, Qin N, Banie H, et al. (2011). Oxysterols direct B-cell migration through EBI2. Nature 475, 519–523. [DOI] [PubMed] [Google Scholar]
- Mackley EC, Houston S, Marriott CL, Halford EE, Lucas B, Cerovic V, Filbey KJ, Maizels RM, Hepworth MR, Sonnenberg GF, et al. (2015). CCR7-dependent trafficking of RORγ+ ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat. Commun 6, 5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura R, and Takeda K (2016). Maintenance of gut homeostasis by the mucosal immune system. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci 92,423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JP, Kelly LM, Xu Y, and Cyster JG (2009). EBI2 mediates B cell segregation between the outer and centre follicle. Nature 460, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A, Wieduwild E, Putoczki T, Mondot S, Lantz O, et al. (2016). Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat. Immunol 77, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Loch-ner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, et al. (2008). Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970. [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama N, Serafini N, Verrier T, Rekiki A, Renauld JC, Frankel G, and Di Santo JP (2014). The chemokine receptor CXCR6 controls the functional topography of interleukin-22 producing intestinal innate lymphoid cells. Immunity 47, 776–788. [DOI] [PubMed] [Google Scholar]
- Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Klein-schek M, Cua D, Di Santo JP, and Eberl G (2011). RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol 72, 320–326. [DOI] [PubMed] [Google Scholar]
- Song C, Lee JS, Gilfillan S, Robinette ML, Newberry RD, Stappen- beck TS, Mack M, Cella M, and Colonna M (2015). Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med 272, 1869–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, and Artis D (2011). CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. (2013). Innate lymphoid cells-a proposal for uniform nomenclature. Nat. Rev. Immunol 73, 145–149. [DOI] [PubMed] [Google Scholar]
- Spits H, Bernink JH, and Lanier L (2016). NK cells and type 1 innate lymphoid cells: partners in host defense. Nat. Immunol 77, 758–764. [DOI] [PubMed] [Google Scholar]
- Turner JR (2009). Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol 9, 799–809. [DOI] [PubMed] [Google Scholar]
- Yi T, and Cyster JG (2013). EBI2-mediated bridging channel positioning supports splenic dendritic cell homeostasis and particulate antigen capture. eLife 2, e00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T, Wang X, Kelly LM, An J, Xu Y, Sailer AW, Gustafsson JA, Russell DW, and Cyster JG (2012). Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity 37, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, and Ouyang W (2008). Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med 74, 282–289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


