Abstract
The study of bacteriophages (phages) and prophages has provided key insights into almost every cellular process as well as led to the discovery of unexpected new mechanisms and the development of valuable tools. This is exemplified for RNA-based regulation. For instance, the characterization and exploitation of the anti-phage CRISPR systems is revolutionizing molecular biology. Phage-encoded proteins such as the RNA binding MS2 protein, which is broadly used to isolate tagged RNAs, also have been developed as valuable tools. Hfq, the RNA chaperone protein central to the function of many base pairing small RNAs (sRNAs), was first characterized as a bacterial host factor required for Qβ phage replication. The ongoing studies of RNAs are continuing to reveal regulatory connections between infecting phages, prophages and bacteria and to provide novel insights. There are bacterial and prophage sRNAs that regulate prophage genes, which impact bacterial virulence as well as bacterial cell killing. Conversely, phage- and prophage-encoded sRNAs modulate the expression of bacterial genes modifying metabolism. An interesting subcategory of the prophage-encoded sRNAs are sponge RNAs that inhibit the activities of bacterial-encoded sRNAs. Phages also affect post-transcriptional regulation in bacteria through proteins that inhibit or alter the activities of key bacterial proteins involved in posttranscriptional regulation. However, what is most exciting about phage and prophage research, given the millions of phage-encoded genes that have not yet been characterized, is the vast potential for discovering new RNA regulators and novel mechanisms and for gaining insight into the evolution of regulatory RNAs.
INTRODUCTION
The impact that the study of phages, both in their lytic form and as prophages integrated into bacterial chromosomes, has had on molecular biology and microbiology is hard to overstate. The ease of phage manipulation helped establish several of the central dogmas in molecular biology. For example, characterization of various phage DNA polymerases contributed to the understanding of replication (1, 2), and models of transcription regulation were greatly influenced by studies of cI, the phage λ repressor (3, 4). Phages also have continually provided important tools such as transduction, the phage-assisted movement of DNA from one bacterium to another, which has been an essential tool since the early years of molecular biology (5, 6). As another example, the development of chain termination DNA sequencing approaches benefitted from single-stranded DNA cloning vectors derived from phage M13 (7).
Aside from their benefits as models and tools, the study of phages is important given their enormous impact on bacterial genome evolution, both as prophage integrated into the genomes, and through mechanisms related to their ability to transduce genes. For instance, ~10% of the genome of Streptococcus pyogenes, including several pathogenicity factors, is of prophage origin, whereas ~16% of the genetic information of Escherichia coli O157:H7 str. Sakai traces back to 18 prophages (8). Since some prophage sequences are similar, recombination between the integrated sequences can lead to chromosomal inversions and deletions. Phage sequences also can serve as precursors of new genes (9). Additionally, an estimated 1025 phage-infections occur worldwide every second (10). There are many mechanisms by which gene transfer takes place during these infections. These include a) specialized transduction whereby DNA located adjacent to an integrated prophage is transferred after imprecise excision of a prophage, b) gene transfer agents (GTAs), prophage-like elements that package random bacterial DNA but cannot package enough to enable the transmission of their own genes, and c) phage-inducible chromosomal islands (PICIs) that hijack helper phages to assist in their high-efficiency transfer to neighbouring bacteria (11, 12). In one medically-important example, the Staphylococcus aureus pathogenicity islands (SaPIs), which produce superantigens, utilize phage for effective transduction (13).
The transferred DNA can dramatically modify the recipient organism by encoding a wide range of genes including virulence factors, toxins, secretion systems and regulators. For bacteria to survive, the expression of the prophage or other foreign genes must be integrated into existing regulatory circuits (14–16). The diversity of the gene products encoded by phages, together with the rapidity by which these genes are integrated and transferred, results in great evolutionary pressure on the phages, prophages and bacteria leading to rapid changes in both the gene products and the regulatory circuits.
All of these concepts - the value of studying phage-prophage-bacterial interactions, the impact of rapid evolution, and the interwoven regulatory circuits - are applicable to RNA-based regulation as we will discuss in this chapter.
THE CONTRIBUTION OF PHAGE BIOLOGY TO THE DEVELOPMENT OF TOOLS AND NEW CONCEPTS IN RNA-BASED REGULATION
The study of phages and the cross-talk between phages and bacteria has led to a number of critical RNA-based tools for molecular biology. The most prominent tools come from the CRISPR phage defense systems, whose exploitation for genome engineering is changing molecular biology forever (17). A second class of important tools takes advantage of the high affinity RNA-binding proteins encoded by phages. Probably the most widely utilized protein is the MS2 coat protein of the RNA phage R17, which binds a 19 nucleotide RNA hairpin denoted MBS with nanomolar affinity (18). MS2 and other phage RNA-binding proteins are employed in techniques that rely on these proteins for detecting and isolating correspondingly tagged RNAs in complex with their associated molecules (19).
The study of phages has also led to the identification of key factors of sRNA-based regulation. For example, the OOP RNA, encoded by DNA phage λ, was one of the first characterized sRNAs (20, 21). Studies of this antisense sRNA showed that it base pairs with the cII-O mRNA leading to degradation in a process involving RNase III (an endoribonuclease that recognizes double-stranded RNA, also see chapter by Mohanty and Kushner) and possibly another ribonuclease, ultimately resulting in decreased levels of the cII activator (20, 22). As another example, Hfq, critical to the function of base pairing sRNAs in many bacteria was first identified as a host factor required for RNA phage Qβ replication (23). In the Qβ context, Hfq has been proposed to alter the structure of the phage RNA, possibly allowing the 3´end to be brought into the proximity of the replicase (24).
SRNAS REGULATING PROPHAGE-ENCODED VIRULENCE FACTORS
In pathogens, phage-mediated gene transfer has resulted in the acquisition of virulence genes. For example, the Vibrio cholerae toxin originates from the filamentous phage CTXΦ (25), and the emergence of new epidemic strains of Salmonella enterica has involved phages carrying virulence factors (26).
The role of regulatory sRNAs in the interplay of core genomic elements and the horizontally-acquired virulence genes has been particularly well studied in S. enterica, a model for enteric infections. This bacterium utilizes specialized protein secretion systems encoded within S. enterica pathogenicity islands 1 and 2 (SPI-1 and SPI-2) to deliver effector proteins that manipulate mammalian cell signaling cascades (27). Several effectors including SopE, SspH1, SseI and SopE2 are encoded by phages or phage remnants (8, 26).
Two core S. enterica genome-encoded sRNAs that impact both core-encoded and horizontally-acquired virulence genes are SgrS and RprA. SgrS induction is triggered by the accumulation of non-metabolizable, phosphorylated sugars in the cell. The SgrS role in responding to phosphosugar stress, repressing the synthesis of sugar transporters and increasing the level of a phosphatase, is conserved across several enteric bacteria (28). However, SgrS has broadened its regulatory repertoire in S. enterica to also repress expression of the horizontally-acquired SopD effector protein (29). Phosphosugar induction of SgrS, with the concomitant repression of SopD might help Salmonella adjust effector protein production to changes in carbon source availability during the infection process. The core-encoded sRNA RprA is induced by both the Rcs and Cpx two component systems in response to cell envelope stress and activates the expression of the coreencoded stationary sigma factor σS. Like SgrS, RprA has broadened its selection of targets to include two prophage-derived transcripts of S. enterica (SL2594 and SL2705) and several mRNAs encoded by the virulence plasmid pSLT (30). Interestingly, RprA activation of ricI, one of the pSLT encoded-targets that inhibits plasmid transfer, involves regulation of both core-encoded (indirectly through the stationary phase sigma factor σS which activates ricI transcription) and horizontally-acquired (directly through activation of ricI translation) genes.
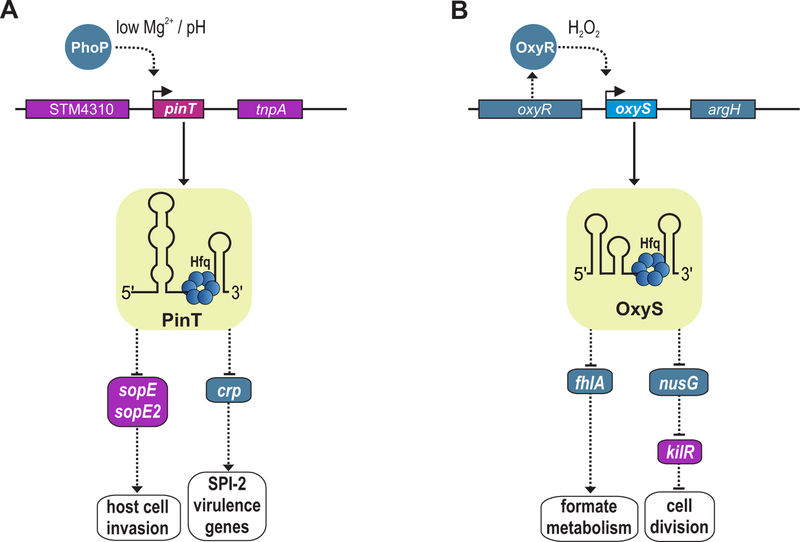
In contrast to SgrS and RprA, two other S. enterica sRNAs that control the synthesis of prophage-encoded virulence factors, are themselves encoded on horizontally-acquired genes specific to S. enterica. The first of these, PinT, is strongly induced by the PhoPQ two component system, a key regulator of S. enterica virulence, when the bacteria are internalized in the mammalian cells (31). By base pairing with the corresponding mRNAs, PinT blocks further synthesis of the prophage-encoded SopE and SopE2 effectors, which are expressed early in infection to facilitate bacterial invasion. PinT also base pairs with the mRNA that encodes Crp, the transcription factor that controls central carbon metabolism and activates transcription of genes encoding SPI-2 proteins. By regulating the temporal expression of both SPI-1 effectors and SPI-2 virulence genes, PinT facilitates Salmonella’s transition from an invasive state to a state capable of intracellular replication (Figure 1A). Another S. enterica island-encoded sRNA, IsrM, is also expressed during infection and inhibits the production of other horizontally-acquired genes, such as the SopA effector protein and HilE, a global regulator of SPI-1 transcription (32).
FIG 1.
Repression of both prophage- and bacterial-encoded mRNAs by sRNAs encoded by horizontally-acquired elements and the bacterial core genome. (A) Following host-cell invasion, the prophage-encoded sRNA, PinT (purple), is activated by the core genome-encoded transcription factor, PhoP (blue). PinT is an Hfq-binding sRNAs that regulates multiple target genes through direct base-pairing. These include the mRNAs of the two horizontally-acquired effector proteins, SopE and SopE2, as well as the core genome-encoded crp mRNA. The Crp protein acts as an activator of SPI-2 (intracellular) virulence genes of S. enterica. (B) The core genome-encoded (blue) OxyS sRNA is activated by the OxyR transcription factor under conditions of oxidative stress. OxyS associates with Hfq to regulate at least two targets: the mRNA encoding the FhlA transcription regulator of formate metabolism and the transcript encoding NusG, an important transcription termination factor. OxyS repression of NusG, which normally blocks expression of the prophage-encoded (purple) KilR protein together with the Rho termination factor, results in increased production of KilR, which transiently inhibits cell division.
In two examples from other bacteria, both the sRNAs and their target genes were acquired together within a single horizontally-transferred module (33). The AfaR sRNA of extra-intestinal pathogenic Escherichia coli (ExPEC) is expressed from the intergenic region between the afaABCD and afaE transcription units, adjacent to a prophage locus encoding a family of afimbrial adhesins (34). AfaR base pairs close to the afaD translational start site promoting cleavage by RNase E (an endoribonuclease that recognizes single-stranded RNA and is part of the degradosome, also see chapter by Bandyra and Luisi), thus reducing the production of AfaD VIII invasin protein while leaving afaABC unaffected (35). In another example from V. cholerae, the expression of the TarB sRNA from the horizontally-acquired Vibrio pathogenicity island (VPI) is activated by the master virulence regulator ToxT (36). Upon its induction, TarB inhibits the expression of the VPI-encoded tcpF mRNA, which codes for an essential colonization factor of V. cholerae (37). It has been speculated that, similar to PinT, TarB helps to coordinate the timing of steps in the infection process by repressing TcpF expression prior to penetration of the mucous barrier of the small intestine.
SRNAS REGULATING PROPHAGE GENES ENCODING TOXINS
Prophages also encode proteins that are toxic to bacteria when synthesized at high levels. sRNA-based mechanisms have evolved to modulate expression of some of these prophage toxins. One example of indirect induction of a toxin involves E. coli OxyS, a conserved sRNAs characterized early on ((38), also see chapter by Fröhlich and Gottesman). In fact, it was studies on OxyS that revealed Hfq functions to facilitate base pairing of trans-encoded sRNAs with their targets (39). Transcription of OxyS is induced by the OxyR transcription factor in response to hydrogen peroxide, and the sRNA was found to repress mutagenesis by an unknown mechanism (38). Only a limited number of OxyS targets such as fhlA were known (40) and none could explain the toxic phenotype of OxyS overexpression. To identify additional targets, a computational search for mRNAs encoded by essential genes that could base pair with a predominantly single stranded section of OxyS, a region encompassing point mutations known to exacerbate or suppress the toxic phenotype, was carried out (41). This approach led to the identification of nusG, encoding a vital transcription termination factor, as a target of OxyS. Mutational and probing experiments confirmed that OxyS base pairs with the transcript and blocks NusG synthesis.
NusG inhibits the production of toxic gene products encoded on horizontally-acquired genomic elements, including the rac prophage, which carries the kilR gene (42–44). The KilR protein blocks cell division by interfering with the function of FtsZ, which forms the tubulin-like ring required for division (45, 46). Consistent with the model that OxyS repression of NusG results in cell killing by KilR, the effects of OxyS overexpression on cell viability and cell elongation were decreased in a strain lacking the KilR toxin. Given that the OxyS-mediated antimutator phenotype was similarly lost in the kilR mutant strain, it seemed plausible that induction of OxyS results in a transient reduction of NusG production, which consequently increases KilR expression from the rac prophage and triggers temporary growth arrest. (Figure 1B). The growth arrest, like cell cycle checkpoints in response to DNA damage in eukaryotic cells, allows for DNA repair before normal growth is resumed.
An example of an sRNA that indirectly induces expression of a toxic protein is IsrK, encoded by Gifsy-1 prophage of Salmonella (47). The isrK promoter directs the synthesis of two distinct RNA species: a short IsrK sRNA and a long mRNA, which encodes an open reading frame of unknown function (orf45) and an antirepressor (anrP) but is translationally inactive. IsrK sRNA base pairs with the translationally-inactive orf45-anrP mRNA to increase translation of the antirepressor protein, AnrP. AnrP in turn activates the transcription of the prophage-encoded antiterminator AntQ. Increased levels of AntQ protein globally impact bacterial transcription termination, leading to growth arrest and ultimately cell death.
A more direct way of controlling toxin production and cell growth is exemplified by cis- encoded antisense RNAs that base pair directly with the toxin mRNAs transcribed from the opposite DNA strand. While a significant number of the classic so-called Type I toxin-antitoxin systems (see chapter by Darfeuille et al) are known to be encoded by plasmids and the core bacterial genome, they are also found in phage and prophage sequences (48). We speculate that large numbers of cis-encoded antitoxin sRNAs remain to be characterized. For example, a distinct antisense transcript detected in E. coli is encoded opposite the ymfL gene of the lambdoid prophage e14 (49) and may silence expression of the protein. While the function of YmfL is currently unknown, overexpression of this region of e14 leads to cell filamentation (50). In a slightly different variation, the RalA sRNA is encoded in trans, downstream of the toxic ralR gene of the rac prophage, but shares 16 nucleotides of complementarity and can block synthesis of the RalR endonuclease in an Hfq-dependent manner (51). Many other examples of trans-encoded sRNAs controlling toxin production also are anticipated.
PROPHAGE SRNAS REGULATING TRANSCRIPTS ENCODED ON THE CORE GENOME
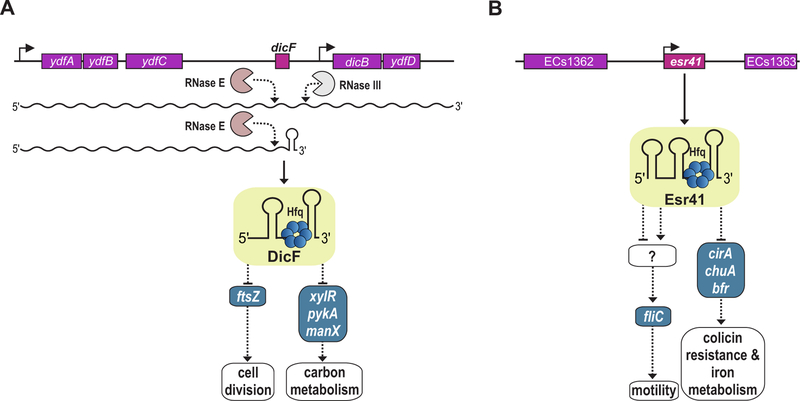
Given their sophisticated regulatory networks and the concise genomes, it is not surprising that phages and also prophages encode sRNAs (33, 52). These sRNAs are being found to not only regulate phage and prophage genes but also to modulate the expression of genes transcribed from the core genome (Table 1). One of the earliest sRNAs to be discovered in E. coli is DicF, an sRNA processed from a polycistronic transcript of the defective lambdoid prophage, Qin/Kim (53). Expression of the transcript, which also encodes five small proteins (YdfA, YdfB, YdfC, DicB and YdfD), is under the control of a cI-like repressor (54). The DicF sRNA accumulates as two isoforms in the cell (Figure 2A). RNase E-mediated processing of the polycistronic transcript generates the 5’ end of both, while alternative Rho-independent transcription termination and RNase III-mediated processing produce the 53-nucleotide and 72-nucleotide variants, respectively (55).
TABLE 1:
Examples of posttranscriptional cross-regulation between bacteria and phages
| Species | Phage | sRNA | Bacterial target | Bacterial/phase process affected | Reference |
|---|---|---|---|---|---|
| Prophage sRNAs regulating transcripts encoded on the core genome | |||||
| E. coli | Qin | DicF | ftsZ | Cell division | 58, 59 |
| xylR, pykA, manX | Metabolism | 58, 62 | |||
| E. coli | Degraded prophage | EcsR2 | ansB | Fumarate production | 65, 66 |
| EHEC | SpLE1 | Esr41 | fliC | Motility | 67, 68 |
| cirA, bfr, chuA | Iron metabolism | 69 | |||
| Prophage sRNAs regulating sRNAs encoded on the core genome | |||||
| EHEC | Sp5 | AgvB | GcvB | Niche colonization in cattle | 67 |
| Sp10 Sp11 | AsxR | FnrS | Iron release from heme | 67 | |
| Phage sRNAs regulating transcripts encoded on the core genome | |||||
| E. coli | PA-2 | IpeX | ompC | Porin regulation | 74 |
| E. coli | Φ24B | 24B_1 | d_ant | Phage production | 80 |
| P. aeruginosa | PAK_P3 | sRNA2 | TψC- tRNA loop | Translation | 83 |
| Phage proteins impacting host post-transcriptional regulation | |||||
| E. coli | T4 | SrD | RNase E | Bacterial RNA decay | 87 |
| E. coli | T7 | 0.7 | RNase E | Phage RNA stabilization | 88 |
| E. coli | T7 | 0.7 | RNase III | Phage RNA maturation | 89 |
| P. aeruginosa | ΦKZ | Dip | RNase E | Phage RNA stabilization | 90 |
FIG 2.
Prophage-encoded sRNAs that regulate the expression of host genes. (A) The prophage-encoded (purple) sRNA, DicF, is processed from a polycistronic transcript by RNase E, and for the second DicF isoform, by RNase III. DicF associates with Hfq to repress synthesis of the core genome-encoded (blue) FtsZ protein required for cell division, as well as XylR, PykA, and ManX, all involved in carbon metabolism. (B) Esr41 is a prophage-encoded (purple) sRNA, which binds Hfq to inhibit translation of the core genome-encoded (blue) cirA, chuA, and bfr mRNAs. The gene products of the mRNAs are involved in iron metabolism and repression of cirA results in colicin resistance. Esr41 also leads to increased motility by upregulation of FliC, however, the molecular mechanism underlying this process has not yet been determined.
Of the proteins encoded on the polycistronic RNA, only the DicB protein has been reported to have a biological function. The protein interacts with MinC and ZipA and thereby inhibits FtsZ polymerization and consequently cell division (56, 57). Overproduction of the DicF sRNA similarly inhibits cell division, in this case by pairing with the ftsZ mRNA to repress translation initiation (58, 59). Consistent with this base pairing role, DicF associates with the Hfq chaperone in vivo (60) and in vitro (61).
Two recent studies revealed possible metabolic functions for DicF. High levels of DicF inhibit the expression of metabolic genes encoding a transcription factor involved in D-xylose degradation (xylR), pyruvate kinase A (pykA), and a mannose transporter (manX) (58, 62). While repression of xylR requires the DicF 5´ end, repression of ftsZ and manX involves the very 3´ end of DicF (58, 62), which is unusual for Hfq-mediated base-pairing interactions (63) and might suggest an alternative mechanism of gene regulation. Indeed, rather than sequestering the manX ribosome binding site (RBS) directly, base pairing of DicF with the manX coding sequence recruits Hfq to the RBS to inhibit translation initiation (62). Whether DicF affects metabolic fluxes as a consequence is currently unknown; however, another link between DicF and metabolism comes from the report that DicF is stabilized by enolase under anaerobic conditions (64). Enolase is central to the glycolytic pathway and is also part of the degradosome complex, which is key for bulk RNA turnover in E. coli and related organisms (see chapter by Bandyra and Luisi).
Degraded prophage genes are the sources of the E. coli EcsR2 and Salmonella SesR2 sRNAs (65). Whereas very little is known about the biological function of SesR2, post-transcriptional gene regulation by EcsR2 has been studied in E. coli. The sRNA is expressed at low levels from the yagU-ykgJ intergenic region and phylogenetic analysis suggests that the sRNA originated from a vestigial phage gene (66). Two independent experimental approaches revealed that the mRNA for a periplasmic L-asparaginase (ansB) is a direct interaction partner of EcsR2 and suggested that base pairing with ansB requires an unstructured and conserved sequence element in the center of the sRNA. Because of its recent appearance in the E. coli genome, EcsR2 is considered a ‘young’ sRNA and could therefore serve as a model to study sRNA evolution in bacteria (see chapter by Dutcher and Raghavan).
As discussed above, horizontal gene transfer, especially through infecting phages and lysogens, has a major impact on the evolution of pathogenic microbes. The sRNAs encoded on these virulence-related prophages were previously often overlooked, however, are now being recognized as a source of post-transcriptional regulators. One such example is Esr41/EcOnc14 (67) from the Sakai prophage-like element (SpLE1) of enterohemorrhagic E. coli (EHEC) (Figure 2B). Esr41 is approximately 70 nucleotides long and was initially found to stimulate flagellin (fliC) expression, and consequently motility, when expressed from a multi-copy plasmid (68). It is currently not known if this phenotype requires base pairing of Esr41 with the fliC mRNA, or rather is an indirect effect involving additional factors.
A recent study applying CLASH (cross-linking, ligation, and sequencing of hybrids) to RNase E revealed multiple direct interaction partners of Esr41 in EHEC (69). Three mRNAs encoding an iron-siderophore complex uptake receptor (cirA), bacterioferritin (bfr), and outer membrane heme receptor (chuA) were confirmed to form duplexes with Esr41. In all three cases, base pairing with Esr41 involved the RBS of the target mRNA suggesting that Esr41 inhibits translation initiation (69). These findings were confirmed at the phenotypic level by showing that Esr41 overexpression renders E. coli resistant to pore-forming colicin 1A, which enters the cell through the CirA receptor. Additionally, EHEC cells deficient for esr41 gained a fitness advantage in iron-limited medium, which might be attributable to derepression of iron transporters in the mutant (69). It is interesting to note that some the Esr41 targets overlap with validated interaction partners of the core-encoded sRNA, RyhB (see chapter by Chareyre and Mandin) and that RyhB paralogs have been discovered in the horizontally acquired elements of other enteric pathogens such as S. enterica (70).
PROPHAGE SRNAS REGULATING THE ACTIVITY OF CORE GENOME-ENCODED SRNAS
As discussed in more detail in the chapter by Figueroa-Bossi and Bossi, it has become clear that the activities of sRNAs themselves can be regulated by other RNAs, often referred to as “sponge RNAs” that base pair with and block the activities of the target sRNA. Prophage examples of this type of sponge RNAs have now also been found through studies of Hfq-binding sRNAs in EHEC (67). In this study, transcripts that bound to Hfq were identified upon UV-induced crosslinking of the RNAs to affinity-tagged Hfq, followed by isolation of Hfq and deep sequencing. A surprising finding from this study was that very short sRNAs of 51–60 nt encoded by lambdoid prophages were among the most frequently recovered sRNAs. The sRNAs were found to be encoded in similar locations in eight different prophages, and the four most abundant transcripts carry variable 5’ regions of 14–18 together with highly conserved 3’ regions of 42 nt, which encompass a Rho-independent terminator.
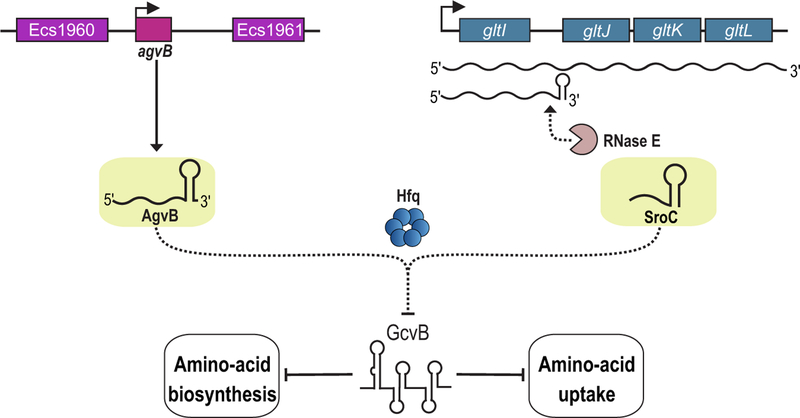
Two of the abundant sRNAs, AsxR and AgvB, were characterized in more detail and found to act as “anti-sRNAs” or sponges against the core genome-encoded FnrS and GcvB sRNAs, respectively (Table 1). Transcriptomic studies examining the consequences of short-term overexpression of AsxR, showed that AsxR increases the levels of the chuAS mRNA (encoding a heme outer membrane receptor and a heme oxygenase, respectively). These effects were at the post-transcriptional level, but the lack of homology between AsxR and the chuAS mRNA did not support a mechanism involving direct base pairing. Instead, complementarity was observed between the 5’ end of AsxR and the FnrS sRNA, whose expression is highest under anaerobic conditions (71). FnrS does base pair with and repress chuAS translation. Thus, by titrating the negative regulator FnrS and promoting its decay, AsxR indirectly promotes expression of chuAS, and potentially other FnrS target genes. AgvB, similarly was found to alleviate GcvB sRNA-mediated repression of the dpp mRNA (encoding a dipeptide transporter). GcvB is highly conserved among the enterobacteria, and controls a large regulon of genes coding for amino acid and peptide transporters (72). The 5’ end of AgvB is partially complementary to the conserved R1 seed region by which GcvB recognizes most of its targets, and base pairing of AgvB with this region antagonizes its function (67). Interestingly, the core-encoded sRNA SroC uses a similar mechanism to counteract GcvB function (Figure 3) (73). However, different from AgvB, SroC base pairs with two distinct sequence elements to achieve GcvB degradation. It is worth noting that E. coli O157 str. Sakai encodes two copies of agvB, which might act additively to curtail GcvB function.
FIG 3.
Prophage-encoded and core genome-encoded sRNAs that at as sponges to block the activities of core-encoded sRNAs. The prophage-encoded (purple) AgvB sRNA, as well as the core genome-encoded sRNA (blue), SroC, use Hfq to base-pair with the GcvB sRNA to inhibit the function of the GcvB global regulator of amino-acid uptake and metabolism. SroC is generated from RNase E-mediated endonucleolytic processing of a poly-cistronic transcript, while AgvB is transcribed from a free-standing gene.
As for other examples discussed thus far, expression of these prophage sponge sRNAs was proposed to impact EHEC virulence. Thus, repression of FnrS, which conceivably is upregulated in the microaerobic environments of the gastrointestinal tract, by AsxR (expressed from the same prophage as Shiga-toxin-2), would lead to increased synthesis of heme utilization proteins of benefit in the low iron environment of the infected mammalian cell.
PHAGE SRNAS REGULATING EXPRESSION OF TRANSCRIPTS ENCODED BY THE CORE GENOME
Similar to prophage sRNAs, sRNAs of infecting phages can control core genome expression (Table 1). One example is the IpeX sRNA transcribed downstream of the lc porin gene of phage PA-2. An identical sequence is also present on the genome of the cryptic phage qsr (DLP12). Infection of E. coli with PA-2 results in the reduction of OmpC production (74), which was attributed to IpeX activity since IpeX overexpression from a plasmid also inhibits OmpC and OmpF production in E. coli (75). However, the absence of convincing sequence complementarity between IpeX and ompC suggests that regulation might require additional factors. Similar to DicF, IpeX production was reported to require processing from a larger transcript, a feature that has now been reported for several other sRNAs from diverse microbes (76–79).
An unusual phage-derived sRNA is 24B_1, discovered on the genome of the Shiga toxin-converting phage Φ24B. Different from the bacterial sRNAs described above, 24B_1 is processed from a ~80 nucleotide precursor and accumulates as a transcript of only ~20 nucleotides in the cell (80). As such, 24B_1 has been suggested to resemble the ubiquitous eukaryotic microRNAs (81), a class of post-transcriptional regulators proposed to also exist in bacteria (82). Deletion of 24B_1 from the Φ24B genome has multiple effects on the physiology of the phage including more efficient prophage induction, increased phage production, and differential bacterial cell adsorption (80). The molecular mechanisms underlying these phenotypes are yet to be discovered, and it will be interesting to explore if these microRNA-sized transcripts also work through base pairing with mRNA targets or employ alternative regulatory mechanisms.
Unconventional types of gene regulation might also be employed by sRNA1 and sRNA2, which are expressed from the genome of the PAK_P3 phage infecting Pseudomonas aeruginosa (83). Both sRNAs accumulate as ~100 nucleotide transcripts and are differentially regulated during the infection process, with expression peaking during late stages of infection. Although complementary between sRNA2 and bacterial tRNAs has been noted, it again is not clear if sRNA1 and sRNA2 function by base pairing and whether the true targets are of bacterial and/or phage origin.
Phage- and prophage-encoded sRNA-sized transcripts have now been detected in the transcriptomes of other microbes including relevant pathogens, such as Mycobacterium and Listeria species (84, 85). These sRNAs await functional characterization, which will be key to understanding their biological functions during phage replication.
PHAGE PROTEINS IMPACTING HOST POST-TRANSCRIPTIONAL REGULATION
To promote their own proliferation, phages have evolved a multitude of mechanisms to exploit the core bacterial machineries; increasing the expression of phage genes while limiting the expression of bacterial genes. The mechanisms include using and modifying bacterial RNA polymerases and, as we will discuss next, the bacterial machinery for degrading RNA (Table 1). For example, infection of E. coli by T4 phage results in rapid degradation of bacterial mRNAs. Consequently, bacterial gene expression ceases while the associated generation of ribonucleotides and free ribosomes facilitates transcription and translation of T4 genes (86). A 29 kDa phage protein denoted Srd (due to its similarity with RpoD) was suggested to be responsible for the differential degradation. The association of Srd and RNase E in vivo, the involvement of Srd in the turnover of the unrelated bacterial mRNAs lpp and ompA, and the importance of Srd to phage proliferation led to the suggestion that Srd stimulates RNase E activity thus leading to rapid bacterial RNA degradation (87).
Unlike T4, the phage T7 achieves differential RNA stability by inhibiting RNase E. The protein kinase domain of gene 0.7 of T7 phage phosphorylates RNase E and the associated RNA helicase RhlB, which results in the stabilization of mRNAs that are synthesized by T7 RNA polymerase but not those synthesized by E. coli RNA polymerase, by mechanisms that to our knowledge are not yet understood (88). Phosphorylation by the phage T7 gp0.7 kinase conversely has been reported to stimulate the activity of RNase III (89).
A more recent study showed that the activity of the Pseudomonas aeruginosa RNA degradosome is inhibited by Gp37/Dip (degradosome interacting protein) encoded by the unusually large phage ϕKZ. Structural studies revealed that acidic patches on the convex outer surface of Dip contact two RNA binding sites on RNase E, thus preventing RNAs from being bound and degraded by the RNA degradosome (90). The three different phage proteins mentioned, Srd, gp0.7 and Dip, modulate RNase E by very different mechanisms. As the RNA degradation machinery is central to bacterial and phage growth and is broadly conserved, we propose that there are other phage proteins engaged in modulating RNase activity by additional mechanisms that remain to be identified.
VAST POTENTIAL TO IDENTIFY NEW RNA BINDING PROTEINS, REGULATORY RNAS AND UNIQUE FUNCTIONS
In general, one aspect of phage and prophage biology that is particularly exciting, also for investigators studying regulatory RNAs, is the vast numbers of unknown genes encoded by these elements. There are estimates of 1031 phage particles worldwide (91). Although some phage genes are similar and functionally conserved, the number of potential genes encoded by this many phages is incomprehensible. These worldwide estimates likely even underestimate the number of phages with either single-stranded RNA or double-stranded RNA genomes, or those that dominate unique environments, as these classes may not be identified by standard phage isolation and genomic sequencing approaches (92, 93), or are under-annotated because of their divergence from more well-characterized phage (94). One of the few characterized single-stranded RNA phage is the enterobacterial phage Qβ, whose study led to the discovery of Hfq (23). There is no doubt many of the as-yet uncharacterized genes and activities required for the proliferation of these viruses have RNA-related functions. It is worth noting that all of the examples mentioned in this chapter come from only three phage taxa (Caudovirales, Inoviridae and Leviviridae), while virtually nothing is known about RNA-mediated processes for Cystoviruses, Plasmaviruses, Tectiviruses and Microviruses as well as the viruses that infect archaeal cells.
Likely there are a number of uncharacterized protein families, encoded by phages or prophages or by bacterial genomes to modulate phage functions that carry out activities similar to those that are already known. For instance, for organisms that do not have recognizable RNA chaperone proteins such as Hfq, an analogous activity may be found among other proteins that are required for phage replication. Similarly, there is likely to be plethora of unidentified phage and prophage-encoded sRNAs with standard base pairing functions. Given the size of the phage metagenome, surprisingly few phage regulatory RNAs such as the λ OOP and PA-2 IpeX sRNAs (20) have been characterized. The expectation is that there are many such sRNA regulators, especially since sRNA regulators might provide a selective advantage given that phage genomes are small and genes are densely packed. Structured RNAs encoded by phages and prophages, such as the cis-acting BOXA and BOXB RNA structures of phage λ and the PUT RNAs of HK022 phage, have long been known to impact transcription elongation (95) (also see chapter by Winkler). Other structured regions of phage and prophage transcripts undoubtedly will be found to have roles in transcription antitermination or additional phage functions similar to the structured IRES (internal ribosome entry site) elements required for translation of eukaryotic RNA viruses (96). Some of these elements may bind proteins or tRNAs or molecules like T-box RNAs or riboswitches (also see chapters by Keiler and Henkin and Lotz and Suess). It is also possible that the structured sequences are sources for small RNA species cleaved from the longer transcripts.
In addition to uncovering different permutations of known mechanisms, the study of phage and prophage RNAs and their associated proteins, could lead to the discovery of unexpected new mechanisms. There still are no known homologs for many phage-encoded proteins. Proteins that are conserved across multiple phage species (core genes) are probably required for phage propagation and thus likely impact conserved bacterial processes, while those that are only present in a more limited number of species (accessory genes) may have predominantly regulatory roles. The characterization of proteins in both categories may uncover new RNA-based mechanisms. It is also worth noting that some of the largest noncoding RNAs of uncharacterized function such as the GOLLD and ROOL RNAs are encoded by phages and prophages (97, 98) (see chapter by Harris and Breaker). Since their size and predicted structural complexity rival those of ribosomes, there are expectations that these large RNAs may have novel ribozyme activities.
INSIGHTS INTO THE EVOLUTION OF RNA-BASED REGULATION FROM PHAGES AND PROPHAGES
Given the evolutionary constraints imposed on the interactions between bacteria, phages and prophages especially during phage-mediated horizontal gene transfer, the phage-bacteria interplay is an attractive system for studying the evolution of regulatory RNAs, a relatively unexplored topic (99) (see chapter by Dutcher and Raghavan). Interestingly, early studies noted that the genes encoding the E. coli CyaR and Salmonella SdsR/RyeB correspond to phage attachment or integration sites (100, 101) due to features that are not yet understood but are possibly shared with tRNA genes given that they also frequently overlap attachment sites (102). As already mentioned, the E. coli EcsR2 and Salmonella SesR2 sRNAs appear to have evolved from degraded prophage genes (66). Further comparisons among genomes should lead to additional examples and better understanding of the origin and evolution of regulatory RNAs.
The bacteria-phage/prophage systems are also useful for examining the evolution of base pairing between sRNAs and new mRNA targets. For example, if the main role of OxyS sRNA-mediated repression of nusG translation is to promote induction of the prophage KilR, which brings about cell stasis and allows DNA damage repair (41), did this base pairing only evolve in bacteria that carry the rac prophage? OxyS is fairly broadly conserved; did OxyS pairing with mRNAs encoding other proteins affecting cell stasis evolve in organisms without the rac prophage? Similar questions are relevant for horizontally-acquired targets such as sopD, which is repressed by SgrS. Interestingly, while SgrS effectively represses sopD, the sRNA does not repress the homolog sopD2, although the sequence with potential to base pair with SgrS only differs by one nucleotide (29). Regions of base pairing between sRNAs and their targets generally are relatively short and frequently single nucleotide differences can make or break an interaction and therefore gene regulation, but the rules for productive versus non-productive base pairing still are not well understood. Beyond base pairing-based regulation, comparisons of structured RNA elements across phage species should give insights into the evolution of these features. Given the strong selective forces, only those nucleotides or secondary or tertiary structures that are essential for regulation will be maintained.
Finally, phage and prophage-associated regulatory RNAs could be useful systems for exploring the evolution of the protein requirements for RNA function. While many bacterial sRNAs require Hfq for function, ProQ and other proteins with a FinO domain have recently been shown to facilitate sRNA-target mRNA pairing in some bacteria (103). Genes encoding potential FinO domain proteins have been discovered in genomes of many phages, and ligands of Salmonella ProQ include several phage-associated sRNAs (104). Future experiments will show if phage- or prophage-specific functions of ProQ exist and how these are integrated into the intrinsic gene regulatory networks orchestrated by Hfq and ProQ. A related issue is how many phage base pairing sRNAs even require RNA chaperones for function. Conceivably, some phage-associated sRNAs, which evolved as cis-acting regulators, subsequently adapted to control expression of bacterial genes in trans without a requirement for a chaperone protein. Indeed, the Gifsy-1 IsrK sRNAs does not require Hfq for function (47).
TAPPING INTO THE PHAGE AND PROPHAGE GOLD MINE OF RNA REGULATION
What are the best approaches to tap into the potential provided by phage and prophage genes? As the transcriptomes of more and more organisms are being determined, it will be critical to annotate the transcripts originating from phage or prophage sequences. This also is true for studies in which RNAs that associate with particular proteins or base pair with specific RNAs are determined by deep sequencing. Computational searches to identify phage and prophage genes that encode RNAs with predicted secondary structures similar to known regulatory RNAs or proteins with known RNA binding motifs as well as genes that are syntenic with other genes encoding RNAs or proteins with known functions in RNA metabolism could also be productive. Likely this will require interative searches with each newly identified homolog, as was carried out to find phage-encoded transcripts with Y-RNA-like structures and homologs of Ro60 RNA binding proteins ((105) see also chapter by Sim and Wolin). Regardless of the method for identification, detailed functional characterization of phage/prophage regulatory RNAs and RNA binding proteins will be required to uncover their roles in phage and host physiology.
The expanding and unforeseen biological functions and molecular mechanisms uncovered by the studies of phage/prophage regulatory RNAs are expected to lead to new tools in biotechnology as well as advances in synthetic biology and phage therapy. For example, the large burst sizes and sometimes promiscuous replication mechanisms have made phages useful tools for mutational analysis, and they are now being exploited for directed evolution experiments (106). Either alone or assisted by computational predictions, phages might be used to evolve RNAs with dedicated biological functions, or to construct synthetic gene regulatory circuits (107, 108). Finally, phages might be used as a potential treatment to change the human microbiome. In light of the rapidly worsening problem of multi-drug resistant bacterial pathogens, phage-directed antimicrobial therapies are currently experiencing a renaissance (109). Clinical applications of phage therapeutics require a thorough understanding of phage-controlled gene regulatory mechanisms including RNA-based regulation (110). Thus, the continued characterization of the intricate and sophisticated RNA-based regulatory systems controlling phages and their cross-talk with bacteria promises to be a fruitful direction for research for many years to come.
ACKNOWLEDGMENTS
We thank K. Fröhlich, S. Gottesman and S. Krishnamurthy for comments on this chapter. Work in the lab of S.A. is supported by GIF (G-1311–416.13/2015); the Israel Science Foundation founded by The Israel Academy of Sciences and Humanities (711/13); the Israel Centers of Research Excellence (ICORE), Chromatin and RNA (1796/12); and DIP (AM 441/1–1 SO 568/1–1), work in the lab of G.S. is supported by the Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and work in the lab of K.P. is supported by DFG (Exc114–2, GRK2062, SPP2002, PA2820/1 and TRR174), the Human Frontiers Science Program (CDA00024/2016-C), GIF (G-2411–416.13/2016), and the European Research Council (StG-758212).
REFERENCES
- 1.Rittie L, Perbal B. 2008. Enzymes used in molecular biology: a useful guide. J Cell Commun Signal 2:25–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benkovic SJ, Spiering MM. 2017. Understanding DNA replication by the bacteriophage T4 replisome. J Biol Chem 292:18434–18442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodd IB, Shearwin KE, Egan JB. 2005. Revisited gene regulation in bacteriophage lambda. Curr Opin Genet Dev 15:145–52. [DOI] [PubMed] [Google Scholar]
- 4.Herskowitz I 1973. Control of gene expression in bacteriophage lambda. Annu Rev Genet 7:289–324. [DOI] [PubMed] [Google Scholar]
- 5.Zinder ND, Lederberg J. 1952. Genetic exchange in Salmonella. J Bacteriol 64:679–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salmond GP, Fineran PC. 2015. A century of the phage: past, present and future. Nat Rev Microbiol 13:777–86. [DOI] [PubMed] [Google Scholar]
- 7.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103119. [DOI] [PubMed] [Google Scholar]
- 8.Brussow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daubin V, Ochman H. 2004. Start-up entities in the origin of new genes. Current Opinion in Genetics & Development 14:616–619. [DOI] [PubMed] [Google Scholar]
- 10.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR Jr., Hendrix RW, Hatfull GF. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–82. [DOI] [PubMed] [Google Scholar]
- 11.Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nat Rev Genet 16:472–82. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Aljaro C, Balleste E, Muniesa M. 2017. Beyond the canonical strategies of horizontal gene transfer in prokaryotes. Curr Opin Microbiol 38:95–105. [DOI] [PubMed] [Google Scholar]
- 13.Novick RP, Christie GE, Penades JR. 2010. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol 8:541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorman CJ. 2014. H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria. Plasmid 75:1–11. [DOI] [PubMed] [Google Scholar]
- 15.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. 2015. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol 13:641–50. [DOI] [PubMed] [Google Scholar]
- 16.Canchaya C, Fournous G, Brussow H. 2004. The impact of prophages on bacterial chromosomes. Mol Microbiol 53:9–18. [DOI] [PubMed] [Google Scholar]
- 17.Wright AV, Nuñez JK, Doudna JA. 2016. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164:29–44. [DOI] [PubMed] [Google Scholar]
- 18.Bardwell VJ, Wickens M. 1990. Purification of RNA and RNA-protein complexes by an R17 coat protein affinity method. Nucleic Acids Res 18:6587–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Varani G. 2013. Engineering RNA-binding proteins for biology. FEBS J 280:3734–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krinke L, Wulff DL. 1987. OOP RNA, produced from multicopy plasmids, inhibits l cII gene expression through an RNase III-dependent mechanism. Genes Dev 1:1005–1013. [DOI] [PubMed] [Google Scholar]
- 21.Hayes S, Szybalski W. 1973. Control of short leftward transcripts from the immunity and ori regions in induced coliphage lambda. Mol Gen Genet 126:275–290. [DOI] [PubMed] [Google Scholar]
- 22.Krinke L, Wulff DL. 1990. RNase III-dependent hydrolysis of l cII-O gene mRNA mediated by l OOP antisense RNA. Genes Dev 4:2223–2233. [DOI] [PubMed] [Google Scholar]
- 23.Franze de Fernandez MT, Eoyang L, August JT. 1968. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 219:588–590. [DOI] [PubMed] [Google Scholar]
- 24.Barrera I, Schuppli D, Sogo JM, Weber H. 1993. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J Mol Biol 232:512–21. [DOI] [PubMed] [Google Scholar]
- 25.Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–4. [DOI] [PubMed] [Google Scholar]
- 26.Ehrbar K, Hardt WD. 2005. Bacteriophage-encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium. Infect Genet Evol 5:1–9. [DOI] [PubMed] [Google Scholar]
- 27.LaRock DL, Chaudhary A, Miller SI. 2015. Salmonellae interactions with host processes.Nat Rev Microbiol 13:191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bobrovskyy M, Vanderpool CK. 2013. Regulation of bacterial metabolism by small RNAs using diverse mechanisms. Annu Rev Genet 47:209–232. [DOI] [PubMed] [Google Scholar]
- 29.Papenfort K, Podkaminski D, Hinton JC, Vogel J. 2012. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc Natl Acad Sci U S A 109:E757–E764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papenfort K, Espinosa E, Casadesús J, Vogel J. 2015. Small RNA-based feedforward loop with AND-gate logic regulates extrachromosomal DNA transfer in Salmonella. Proc Natl Acad Sci U S A 112:E4772–E4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westermann AJ, Forstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Muller L, Reinhardt R, Stadler PF, Vogel J. 2016. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529:496–501. [DOI] [PubMed] [Google Scholar]
- 32.Gong H, Vu GP, Bai Y, Chan E, Wu R, Yang E, Liu F, Lu S. 2011. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog 7:e1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frohlich KS, Papenfort K. 2016. Interplay of regulatory RNAs and mobile genetic elements in enteric pathogens. Mol Microbiol 101:701–13. [DOI] [PubMed] [Google Scholar]
- 34.Pichon C, du Merle L, Caliot ME, Trieu-Cuot P, Le Bouguénec C. 2012. An in silico model for identification of small RNAs in whole bacterial genomes: characterization of antisense RNAs in pathogenic Escherichia coli and Streptococcus agalactiae strains. Nucleic Acids Res 40:2846–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pichon C, du Merle L, Lequeutre I, Le Bouguénec C. 2013. The AfaR small RNA controls expression of the AfaD-VIII invasin in pathogenic Escherichia coli strains. Nucleic Acids Res 41:5469–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradley ES, Bodi K, Ismail AM, Camilli A. 2011. A genome-wide approach to discovery of small RNAs involved in regulation of virulence in Vibrio cholerae. PLos Pathog 7:e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirn TJ, Bose N, Taylor RK. 2003. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol Microbiol 49:81–92. [DOI] [PubMed] [Google Scholar]
- 38.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43–53. [DOI] [PubMed] [Google Scholar]
- 39.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell 9:11–22. [DOI] [PubMed] [Google Scholar]
- 40.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. 1998. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J 17:6069–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barshishat S, Elgrably-Weiss M, Edelstein J, Georg J, Govindarajan S, Haviv M, Wright PR, Hess WR, Altuvia S. 2017. OxyS small RNA induces cell cycle arrest to allow DNA damage repair. EMBO J:pii: e201797651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. 2008. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320:935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conter A, Bouché JP, Dassain M. 1996. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. J Bacteriol 178:5100–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burke C, Liu M, Britton W, Triccas JA, Thomas T, Smith AL, Allen S, Salomon R, Harry E. 2013. Harnessing single cell sorting to identify cell division genes and regulators in bacteria. PLoS One 8:e60964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernández-Rocamora VM, Alfonso C, Margolin W, Zorrilla S, Rivas G. 2015. Evidence That bacteriophage λ Kil peptide inhibits bacterial cell division by disrupting FtsZ protofilaments and sequestering protein subunits. J Biol Chem 290:20325–20335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haeusser DP, Hoashi M, Weaver A, Brown N, Pan J, Sawitzke JA, Thomason LC, Court DL, Margolin W. 2014. The Kil peptide of bacteriophage λ blocks Escherichia coli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLoS Genet 10:e1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hershko-Shalev T, Odenheimer-Bergman A, Elgrably-Weiss M, Ben-Zvi T, Govindarajan S, Seri H, Papenfort K, Vogel J, Altuvia S. 2017. Gifsy-1 prophage IsrK with dual function as small and messenger RNA modulates vital bacterial machineries. PLoS Genet 12:e1005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. 2010. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res 38:3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomason MK, Bischler T, Eisenbart SK, Förstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G. 2015. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol 197:18 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burke C, Liu M, Britton W, Triccas JA, Thomas T, Smith AL, Allen S, Salomon R, Harry E. 2013. Harnessing single cell sorting to identify cell division genes and regulators in bacteria. PLoS One 8:e60964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo Y, Quiroga C, Chen Q, McAnulty MJ, Benedik MJ, Wood TK, Wang X. 2014. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res 42:6448–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shinhara A, Matsui M, Hiraoka K, Nomura W, Hirano R, Nakahigashi K, Tomita M, Mori H, Kanai A. 2011. Deep sequencing reveals as-yet-undiscovered small RNAs in Escherichia coli. BMC Genomics 12:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouché F, Bouché JP. 1989. Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol Microbiol 3:991 994. [DOI] [PubMed] [Google Scholar]
- 54.Bejar S, Bouché F, Bouché JP. 1988. Cell division inhibition gene dicB is regulated by a locus similar to lambdoid bacteriophage immunity loci. Mol Gen Genet 212:11–19. [DOI] [PubMed] [Google Scholar]
- 55.Faubladier M, Cam K, Bouche JP. 1990. Escherichia coli cell division inhibitor DicFRNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E-dependent processing. J Mol Biol 212:461–71. [DOI] [PubMed] [Google Scholar]
- 56.Johnson JE, Lackner LL, Hale CA, de Boer PA. 2004. ZipA is required for targeting of DMinC/DicB, but not DMinC/MinD, complexes to septal ring assemblies in Escherichia coli. J Bacteriol 186:2418–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou H, Lutkenhaus J. 2005. MinC mutants deficient in MinD- and DicB-mediated cell division inhibition due to loss of interaction with MinD, DicB, or a septal component. J Bacteriol 187:2846–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balasubramanian D, Ragunathan PT, Fei J, Vanderpool CK. 2016. A prophage-encoded small RNA controls metabolism and cell division in Escherichia coli. mSystems 1:pii: e00021–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tetart F, Bouche JP. 1992. Regulation of the expression of the cell-cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol Microbiol 6:615–20. [DOI] [PubMed] [Google Scholar]
- 60.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50:1111–24. [DOI] [PubMed] [Google Scholar]
- 61.Olejniczak M 2011. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry 50:4427–40. [DOI] [PubMed] [Google Scholar]
- 62.Azam MS, Vanderpool CK. 2017. Translational regulation by bacterial small RNAs via an unusual Hfq-dependent mechanism. Nucleic Acids Res doi: 10.1093/nar/gkx1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9:578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murashko ON, Lin-Chao S. 2017. Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc Natl Acad Sci U S A 114:E8025–E8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raghavan R, Kacharia FR, Millar JA, Sislak CD, Ochman H. 2015. Genome rearrangements can make and break small RNA genes. Genome Biol Evol 7:557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kacharia FR, Millar JA, Raghavan R. 2017. Emergence of new sRNAs in enteric bacteria is associated with low expression and rapid evolution. J Mol Evol 84:204–213. [DOI] [PubMed] [Google Scholar]
- 67.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. 2014. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell 55:199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sudo N, Soma A, Muto A, Iyoda S, Suh M, Kurihara N, Abe H, Tobe T, Ogura Y, Hayashi T, Kurokawa K, Ohnishi M, Sekine Y. 2014. A novel small regulatory RNA enhances cell motility in enterohemorrhagic Escherichia coli. J Gen Appl Microbiol 60:44–50. [DOI] [PubMed] [Google Scholar]
- 69.Waters SA, McAteer SP, Kudla G, Pang I, Deshpande NP, Amos TG, Leong KW, Wilkins MR, Strugnell R, Gally DL, Tollervey D, Tree JJ. 2017. Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E. EMBO J 36:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. 2008. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res 36:1913–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Durand S, Storz G. 2010. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol 75:1215–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. 2011. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 81:1144–1165. [DOI] [PubMed] [Google Scholar]
- 73.Miyakoshi M, Chao Y, Vogel J. 2015. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J 34:1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schnaitman C, Smith D, de Salsas MF. 1975. Temperate bacteriophage which causes the production of a new major outer membrane protein by Escherichia coli. J Virol 15:1121 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Castillo-Keller M, Vuong P, Misra R. 2006. Novel mechanism of Escherichia coli porin regulation. J Bacteriol 188:576–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. 2012. An atlas of Hfq-bound transcripts reveals 3’ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31:4005–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chao Y, Vogel J. 2016. A 3’ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell 61:352–63. [DOI] [PubMed] [Google Scholar]
- 78.Papenfort K, Forstner KU, Cong JP, Sharma CM, Bassler BL. 2015. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci U S A 112:E766–E775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davis BM, Waldor MK. 2007. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol Microbiol 65:373–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nejman-Falenczyk B, Bloch S, Licznerska K, Dydecka A, Felczykowska A, Topka G, Wegrzyn A, Wegrzyn G. 2015. A small, microRNA-size, ribonucleic acid regulating gene expression and development of Shiga toxin-converting bacteriophage Phi24Beta. Sci Rep 5:10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–97. [DOI] [PubMed] [Google Scholar]
- 82.Bloch S, Wegrzyn A, Wegrzyn G, Nejman-Falenczyk B. 2017. Small and smaller-sRNAs and microRNAs in the regulation of toxin gene expression in prokaryotic cells: A mini- review. Toxins (Basel) 9:pii: E181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chevallereau A, Blasdel BG, De Smet J, Monot M, Zimmermann M, Kogadeeva M, Sauer U, Jorth P, Whiteley M, Debarbieux L, Lavigne R. 2016. Next-generation “-omics” approaches reveal a massive alteration of host RNA metabolism during bacteriophage infection of Pseudomonas aeruginosa. PLoS Genet 12:e1006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. 2011. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res 39:4235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dedrick RM, Marinelli LJ, Newton GL, Pogliano K, Pogliano J, Hatfull GF. 2013. Functional requirements for bacteriophage growth: gene essentiality and expression in mycobacteriophage Giles. Mol Microbiol 88:577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qi D, Alawneh AM, Yonesaki T, Otsuka Y. 2015. Rapid degradation of host mRNAs by stimulation of RNase E activity by Srd of bacteriophage T4. Genetics 201:977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qi D, Alawneh AM, Yonesaki T, Otsuka Y. 2015. Rapid Degradation of Host mRNAs by Stimulation of RNase E Activity by Srd of Bacteriophage T4. Genetics 201:977–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marchand I, Nicholson AW, Dreyfus M. 2001. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol Microbiol 42:767–776. [DOI] [PubMed] [Google Scholar]
- 89.Mayer JE, Schweiger M. 1983. RNase III is positively regulated by T7 protein kinase. J Biol Chem 258:5340–5343. [PubMed] [Google Scholar]
- 90.Van den Bossche A, Hardwick SW, Ceyssens PJ, Hendrix H, Voet M, Dendooven T, Bandyra KJ, De Maeyer M, Aertsen A, Noben JP, Luisi BF, Lavigne R. 2016. Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome. Elife 5:pii: e16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hatfull GF. 2015. Dark matter of the biosphere: the amazing world of bacteriophage diversity. J Virol 89:8107–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Decker CJ, Parker R. 2014. Analysis of double-stranded RNA from microbial communities identifies double-stranded RNA virus-like elements. Cell Rep 7:898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kauffman KM, Hussain FA, Yang J, Arevalo P, Brown JM, Chang WK, VanInsberghe D, Elsherbini J, Sharma RS, Cutler MB, Kelly L, Polz MF. 2018. A major lineage of nontailed dsDNA viruses as unrecognized killers of marine bacteria. Nature: doi: 10.1038/nature25474. [DOI] [PubMed] [Google Scholar]
- 94.Krishnamurthy SR, Janowski AB, Zhao G, Barouch D, Wang D. 2016. Hyperexpansion of RNA bacteriophage diversity. PLoS Biol 14:e1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weisberg RA, Gottesman ME. 1999. Processive antitermination. J Bacteriol 181:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martinez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Embarek AM. 2017. Insights into Structural and Mechanistic Features of Viral IRES Elements. Front Microbiol 8:2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR. 2017. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res 45:10811–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weinberg Z, Perreault J, Meyer MM, Breaker RR. 2009. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462:656–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Updegrove TB, Shabalina SA, Storz G. 2015. How do base-pairing small RNAs evolve? FEMS Microbiol Rev 39:379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15:16371651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Balbontín R, Figueroa-Bossi N, Casadesús J, Bossi L. 2008. Insertion hot spot for horizontally acquired DNA within a bidirectional small-RNA locus in Salmonella enterica. J Bacteriol 190:4075–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reiter WD, Palm P, Yeats S. 1989. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res 17:1907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Olejniczak M, Storz G. 2017. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers? Mol Microbiol 104:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smirnov A, Forstner KU, Holmqvist E, Otto A, Gunster R, Becher D, Reinhardt R, Vogel J. 2016. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci U S A 113:11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen X, Sim S, Wurtmann EJ, Feke A, Wolin SL. 2014. Bacterial noncoding Y RNAs are widespread and mimic tRNAs. RNA 20:1715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Esvelt KM, Carlson JC, Liu DR. 2011. A system for the continuous directed evolution of biomolecules. Nature 472:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodrigo G, Landrain TE, Jaramillo A. 2012. De novo automated design of small RNA circuits for engineering synthetic riboregulation in living cells. Proc Natl Acad Sci U S A 109:15271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brodel AK, Isalan M, Jaramillo A. 2017. Engineering of biomolecules by bacteriophage directed evolution. Curr Opin Biotechnol 51:32–38. [DOI] [PubMed] [Google Scholar]
- 109.Lu TK, Koeris MS. 2011. The next generation of bacteriophage therapy. Curr Opin Microbiol 14:524–31. [DOI] [PubMed] [Google Scholar]
- 110.Citorik RJ, Mimee M, Lu TK. 2014. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 32:1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]