Abstract
Proximity ligation assay (PLA) achieves extremely low limits of detection, but requires the synthesis of antibody-DNA conjugates recognizing multiple target epitopes with appropriate proximity. In this work, we introduce a more generally-applicable method by replacing antibody-DNA conjugates with nanoparticles which create ultra-detectable PCR templates by capturing biotinylated oligonucleotides and catalyzing ligation. A competitive PCR readout was used to make the assay quantitative. We have demonstrated that NP-PLA can detect and quantitate human chorionic gonadotropin (hCG) levels as low as 2.6 fM (~ 0.1 pg/mL), nearly 1000 times more sensitive than the current state of the art ELISA.
Keywords: protein detection, nanoparticle, proximity ligation assay, competitive polymerase chain reaction, immunoassay
Graphical Abstract
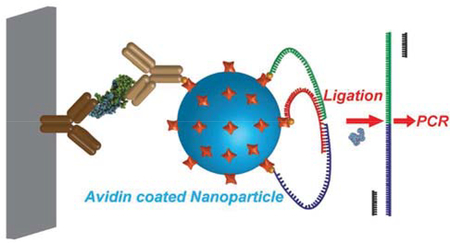
Measuring ultra-low levels of protein biomarkers is essential for early diagnosis, treatment, and management of many diseases1 and is critical for biomedical research. Exponential amplification processes, such as PCR, already allow the detection of specific nucleic acid sequences with exquisite sensitivity and specificity, but such direct amplification is not available for proteins. Efforts have been made to combine PCR and immunoassays for ultra-low-level protein detection. The first so-called immuno-PCR (iPCR), introduced in 1992, used a DNA PCR template as the reporter in an immunoassay to detect as few as several hundred protein molecules.3 Although decades have passed since iPCR was first introduced, it has not yet become very widely accepted due to two major challenges. First, false-positive signals are caused by the nonspecific binding of DNA to multiple surfaces4–6 and biomolecules7–9. Second, it requires complex and time-consuming preparation of specific DNA-antibody conjugated2,10. Several approaches2,10–11 have been developed to address these issues, and the proximity ligation assay (PLA) strategy is one of the most successful. PLA depends on the simultaneous recognition of target molecules by sets of two, three, or more antibodies conjugated to DNA oligonucleotides, followed by ligation of the DNA probes to produce PCR-amplifiable DNA products. In contrast with iPCR, the individual DNA probes in PLA cannot serve as PCR templates; thus, false-positive signals from single nonspecifically-bound DNAs are eliminated. However, PLA requires specific sets of two or more antibody-DNA conjugates that recognize different epitopes (with proper separation distances and angles) of the specific target protein. For later versions of PLA, including 3PLA12 (with higher sensitivity) and solid-phase PLA13 (with adjustable sample volume), sets of three or more antibody-DNA conjugates are required.
Therefore, PLAs are still limited by the availability of proper sets of antibodies to specific target proteins with appropriately-located epitopes.
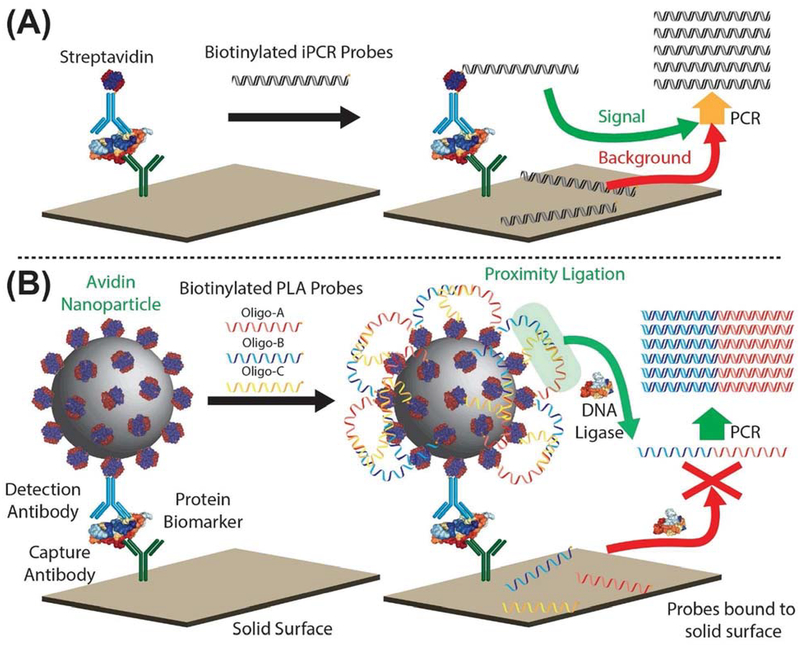
The past decade has witnessed extensive development of nanoparticles and their use in bioassays as molecular-level tools.14–17 In this paper, we report a nanoparticle-based PLA (NP-PLA) for protein detection, in which a nanoparticle serves as a universal labeling module to decouple the proximity ligation process from the molecular recognition of targets. In this proof-of-concept study, protein biomarkers were first captured on the surface of a microplate (Figure 1A). Biotinylated detection antibodies were bound to the microplate-captured protein of interest, avidin nanoparticles were bound to the detection antibody, and finally, the many unoccupied avidins on the bound nanoparticle catalyzed the ligation of a set of biotinylated DNA oligos. To our knowledge, this is the first time that the advantages of 3PLA and solid-phase PLA have been combined in one single assay, resulting in both higher detection sensitivity and adjustable sample volume as a result of the NP-PLA decoupling strategy.
Figure 1.
Schematic of nanoparticle-based proximity ligation assay (NP-PLA). (A) In traditional immuno-PCR (iPCR), a PCR-amplifiable template oligo is directly used as the reporter. Non-specifically bound oligos are PCR amplifiable, resulting in nonspecific background signal. (B) In NP-PLA, avidin-coated nanoparticles serve as reporters. The avidin nanoparticles bring the two split parts (biotinylated oligo-A and biotinylated oligo-B) of the PCR template and the biotinylated bridge oligo-C into proximity. Oligo-A and oligo-B are then ligated, with the resulting oligomer serving as the PCR template. Any non-specifically bound oligos in NP-PLA cannot be ligated into a PCR-amplifiable template, and nonspecific background is significantly decreased.
Although exponential amplification of PCR allows ultra-sensitivity with NP-PLA, it also limits the quantitative precision of the assay.18 To address this issue, we used competitive PCR (cPCR) to quantify the NP-PLA results. In cPCR, a known amount of internal standard competitor oligo using the same primers but with a different melting temperature due to its different internal sequence is co-amplified with the ligation products. The PCR efficiency of the competitor oligo is identical to that of the ligation products. Therefore, the ratio of the amount of competitor amplicon to that of ligation product amplicon is the same as their pre-PCR ratio (Figure SI)19 and the ratio of the their melting peak areas reflects their ratio in abundance (Figure S2). To our knowledge, this is the first application of melting curve-based cPCR (mp-cPCR) in quantitative immunoassays.
Nanoparticle screening.
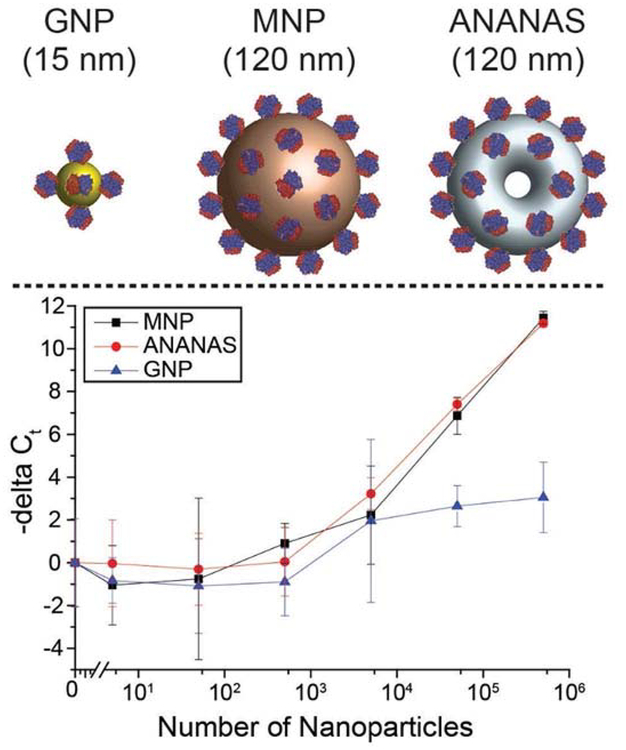
To test the feasibility of detecting avidin nanoparticles with PLA, three commercially available avidin/streptavidin-coated nanoparticles (streptavidin-coated spherical gold nanoparticles (EM.STP15, BBI Solutions) with a mean diameter of 15 nm, streptavidin-coated magnetic nanoparticles (Bio-Adembeads Streptavidin plus 0321) with a mean diameter of 120 nm, and ANANAS nanoparticles20 (ANANAS nanotech) with a mean DLS-effective diameter of 120 nm) (Figure 2) were tested using a set of optimized oligos (Table S1, SI). All three avidin/streptavidin-coated nanoparticles decreased the Ct values of the PLA products in a dose-responsive manner. While the 120-nm magnetic nanoparticles and 120-nm ANANAS nanoparticles showed similar limits of detection (LOD) (approximately 5000 particles) (Figure 2) and similar delta Ct slopes, as a function of the number of nanoparticles, the 15-nm gold nanoparticles showed a 10-fold higher LOD and a flatter delta Ct/nanoparticle concentration slope, indicating that PLA efficiency was higher with larger avidin/streptavidin-coated nanoparticles than smaller nanoparticles. Because ANANAS nanoparticles showed the highest detectability (and lowest variation) with PLA, they were selected for subsequent development of the NP-PLA. The ligation yield from avidin-coated nanoparticles was also analyzed (Figure S3). Based on a real-time quantitative PCR (qPCR) standard curve of the synthetic full-length ligation product, with the current settings, the ligation yield was constant at approximately 1.5 DNA templates per 120-nm avidin particle. Further optimization of the NP-PLA system may increase the ligation yield.
Figure 2.
Detectability of different nanoparticles using proximity ligation assay (PLA). Top, left to right: streptavidin-coated gold nanoparticles (GNP), streptavidin-coated magnetic nanoparticles (MNP), and ANANAS nanoparticles. Bottom: Dose-response curves for the three different nanoparticles by PLA. The -delta Ct values are calculated by subtracting the Ct value of samples from the Ct value of the blank control and are shown as the mean ± standard deviation; n = 3.
Human chorionic gonadotropin (hCG) detection.
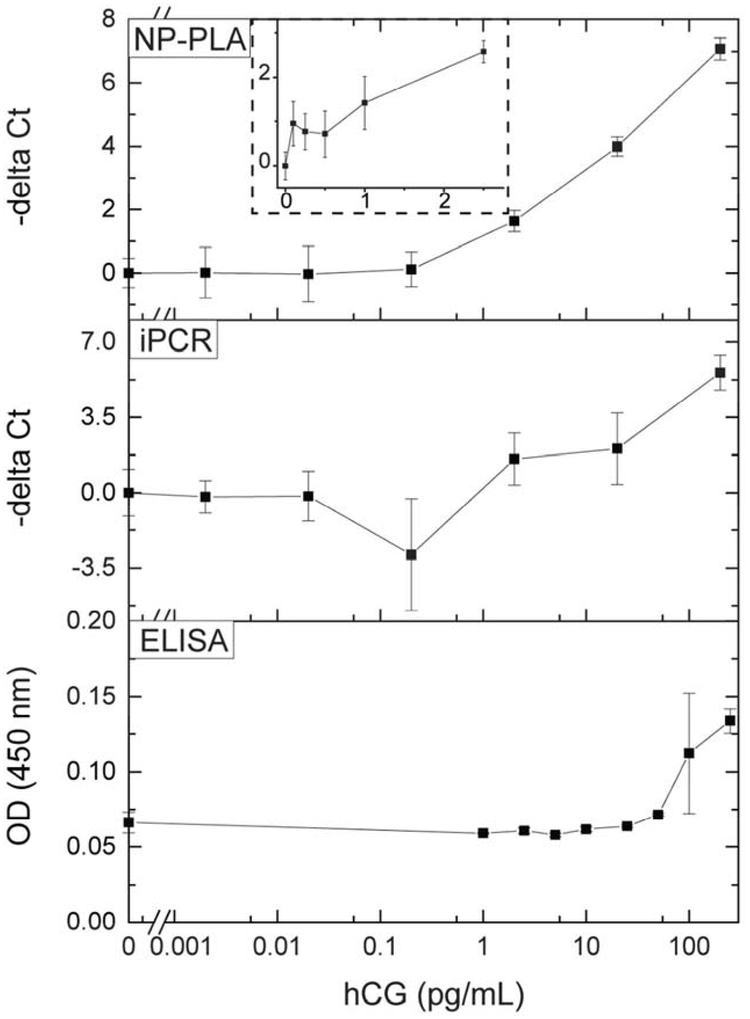
We further investigated the feasibility of NP-PLA by detecting human chorionic gonadotropin, the the most widely used pregnancy marker and also a biomarker for germ-cell ovarian and testicular cancers.21 To detect hCG, monoclonal antibodies recognizing the hCG β-subunit were immobilized on the surface of the wells of a microplate. Biotinylated polyclonal antibodies recognizing the hCG α-subunit were tagged with ANANAS nanoparticles by biotin-streptavidin linkage via a cleavable disulfide bond. The antibody was first covalently conjugated to NHS-SS-Biotin and then conjugated to the avidin-coated nanoparticle. As shown in Figure 3, -delta Ct increased logarithmically with the amount of hCG, with an LOD of 100 fg/mL (Figure S4; signal higher than the blank signal plus 3 standard deviations of the blank signal, 2.6 fM, 100 μL sample volume), thereby demonstrating the feasibility of NP-PLA. Compared to enzyme-linked immunosorbent assays (ELISA) using the same assay settings, NP-PLA was more than 100 times more sensitive.
Figure 3.
Detection of human chorionic gonadotropin (hCG) using nanoparticle-based proximity ligation assay (NP-PLA), immuno-PCR (iPCR) and ELISA. The -delta Ct values were calculated by subtracting the Ct value of samples from the Ct value of the blank control and are shown as the mean ± standard deviation; n = 3.; figure S5 includes data for the ELISA up to 1000 pg/mL.
Quantification by competitive PCR.
To precisely quantify protein biomarkers at ultra-low levels, we used melting peak-based competitive PCR (mp-cPCR) to quantify NP-PLA results. PCR product melting curves, obtained by monitoring fluorescence change while slowly heating to 95°C, are routinely used to characterize PCR products. Integration of the melting curve peaks is not used to quantify or compare the amounts of PCR products because of inevitable tube-to-tube variations in standard PCR, even when performed under the same PCR conditions. However, because species are amplified in parallel in the same reaction tube during competitive PCR, the ratio of the peak areas is expected to reflect the ratio of the amounts of the respective PCR products.19 In our mp-cPCR experiments, we designed a competitor sequence (Table S1) with the same primer-binding sites but with a melting temperature 10°C lower than that of the target sequence of the ligation products. Ligation products were co-amplified in the same PCR tubes with 30 copies of the competitor sequence added as an internal standard.
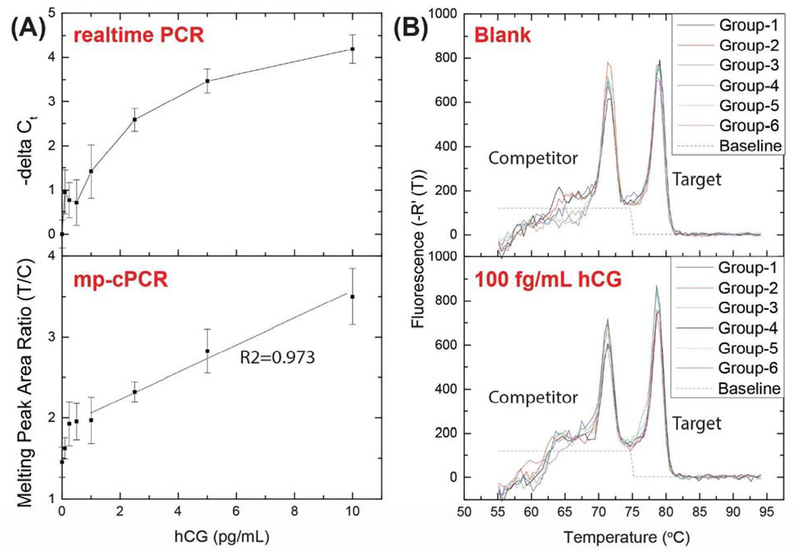
As shown in Figure 4A, instead of a logarithmic increase, the ratio between the peak areas of the target sequence and the competitor sequence (T/C) in mp-cPCR increased linearly with the concentration of hCG in the NP-PLA, demonstrating the utility of using mp-cPCR for quantification in NP-PLA. As shown in Figure 4B, with mp-cPCR, the LOD remained at 100 fg/mL of hCG (2.6 fM, 100 μL sample volume). At this hCG level, the ratio between the peak areas of the target and the competitor (1.62±0.13) was significantly higher than the blank controls (1.45±0.19), proving the feasibility of mp-cPCR for NP-PLA.
Figure 4.
Use of melting peak-based competitive PCR (mp-cPCR) to quantify the results of the nanoparticle-based proximity ligation assay (NP-PLA) for the detection of human chorionic gonadotropin (hCG). (A) Comparison of the results of NP-PLA for hCG detection with real-time PCR and mp-cPCR. The dashed lines indicate the linear fitting from 1 pg/mL to 10 pg/mL hCG. Mean ± standard deviation; n = 6. (B, top) Melting curves of six independent blank samples in the cPCR-based NP-PLA for hCG detection. (B, bottom) Melting curves of six independent samples with 0.1 pg/mL hCG in the cPCR-based NP-PLA for hCG detection. The dashed lines indicate the baselines used to integrate the peak areas.
In summary, we have demonstrated for the first time that avidin-coated nanoparticles can catalyze oligonucleotide ligation. We use this to decouple the proximity ligation process from the recognition of target proteins in PLA, reducing the background in nucleic acid amplification-based immunoassays, without being limited by the proximity of different antibody epitopes on the target proteins. Nanoparticle-based PLA also uses simple biotinylated PLA probes, avoiding the need to prepare antibody-DNA conjugates. The avidin-coated nanoparticles serve as a separate module in NP-PLA; the size, geometry, and spatial distribution of avidins can be freely adjusted for optimal assay performance. In this proof-of-concept study, as expected we found that larger nanoparticles were more detectable than smaller ones when used in similar numbers. In the first use of mp-cPCR in an immunoassay, we demonstrated that melting peak-based competitive PCR (mp-cPCR) is suitable for precise quantification of ultra-low protein biomarker levels using NP-PLA. Given their wide applicability to protein biomarkers, ultra-sensitivity, universal design and simple preparation of the NP-PLA probes, and the possibility of ultra-low-level protein quantification, combining NP-PLA with mp-cPCR creates a new approach for simple, sensitive, and quantitative detection of protein biomarkers at ultra-low levels.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Jay R. Adolacion for setting up the PCR hood, João Trabuco for fruitful discussions, and Mohammad Khodadady and Max Smith for helping characterize the nanoparticles.
FUNDING SOURCES
We gratefully acknowledge support from National Institute of Allergy and Infectious Diseases, National Institutes of Health, http://www.niaid.nih.gov, (Grant 1R21AI111120-01A1), National Science Foundation, www.nsf.gov, (Grant CBET-1511789) and Cancer Prevention & Research Institute of Texas, www.cprit.state.tx.us, (Grant RP150343).
ASSOCIATED CONTENT
Supporting Information.
Experimental section; table listing the sequences and modifications of oligos used for NP-PLA and cPCR (Table S1); optimization of the assay conditions for the detection of hCG with NP-PLA (Table S2); mp-cPCR to quantify the results of the NP-PLA for the detection of hCG (Table S3); working principle of cPCR (Figure S1); working principle of mp-cPCR (Figure S2); quantification of the ligation yield on the surface of nanoparticles (Figure S3); estimation of LOD for hCG using NP-PLA (Figure S4); detection of hCG up to 10ng/mL using ELISA (Figure S5); detection of hCG upto 10 pg/mL using NP-PLA, iPCR and ELISA. This material is available free of charge.
REFERENCES
- (1).Chen H; Hagstrom AE; Kim J; Garvey G; Paterson A; Ruiz-Ruiz F; Raja B; Strych U; Rito-Palomares M; Kourentzi K; Conrad JC; Atmar RL; Willson RC Flotation Immunoassay: Masking the Signal from Free Reporters in Sandwich Immunoassays. Sci Rep 2016, 6, 24297, DOI: 10.1038/srep24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Greenwood C; Ruff D; Kirvell S; Johnson G; Dhillon HS; Bustin SA Proximity Assays for Sensitive Quantification of Proteins. Biomol Detect Quantif 2015, 4, 10–16, DOI: 10.1016/j.bdq.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Sano T; Smith CL; Cantor CR Immuno-Pcr: Very Sensitive Antigen Detection by Means of Specific Antibody-DNA Conjugates. Science 1992, 258 (5079), 120. [DOI] [PubMed] [Google Scholar]
- (4).Shi B; Shin YK; Hassanali AA; Singer SJ DNA Binding to the Silica Surface. J Phys Chem B 2015,119 (34), 11030–11040, DOI: 10.1021/acs.jpcb.5b01983. [DOI] [PubMed] [Google Scholar]
- (5).Kan Y; Tan Q; Wu G; Si W; Chen Y Study of DNA Adsorption on Mica Surfaces Using a Surface Force Apparatus. Sci Rep 2015, 5, 8442, DOI: 10.1038/srep08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cai XE; Yang J Molecular Forces for the Binding and Condensation of DNA Molecules. Biophys J 2002, 82 (1 Pt 1), 357–365, DOI: 10.1016/S0006-3495(02)75400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Afek A; Lukatsky, David B. Nonspecific Protein-DNA Binding Is Widespread in the Yeast Genome. Biophysical Journal 102 (8), 1881–1888, DOI: 10.1016/j.bpj.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ganguly A; Rajdev P; Williams SM; Chatteiji D Nonspecific Interaction between DNA and Protein Allows for Cooperativity: A Case Study with Mycobacterium DNA Binding Protein. The Journal of Physical Chemistry B 2012,116 (1), 621–632, DOI: 10.1021/jp209423n. [DOI] [PubMed] [Google Scholar]
- (9).Sun L; Tabaka M; Hou S; Li L; Burdzy K; Aksimentiev A; Maffeo C; Zhang X; Holyst R The Hinge Region Strengthens the Nonspecific Interaction between Lac-Repressor and DNA: A Computer Simulation Study. PLOS ONE 2016,11 (3), e0152002, DOI: 10.1371/joumal.pone.0152002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhang H; Xu Y; Huang Q; Yi C; Xiao T; Li Q Natural Phage Nanoparticle-Mediated Real-Time Immuno-Pcr for Ultrasensitive Detection of Protein Marker. Chem Commun (Camb) 2013, 49 (36), 3778–3780, DOI: 10.1039/c3cc40688a. [DOI] [PubMed] [Google Scholar]
- (11).Litvinov J; Hagstrom AEV; Lopez Y; Adhikari M; Kourentzi K; Strych U; Monzon FA; Foster W; Cagle PT; Willson RC Ultrasensitive Immuno-Detection Using Viral Nanoparticles with Modular Assembly Using Genetically-Directed Biotinylation. Biotechnology letters 2014, 36(9), 1863–1868, DOI: 10.1007/sl0529-014-1555-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Schallmeiner E; Oksanen E; Ericsson O; Spangberg L; Eriksson S; Stenman UH; Pettersson K; Landegren U Sensitive Protein Detection Via Triple-Binder Proximity Ligation Assays. Nat Methods 2007, 4 (2), 135–137,DOI: 10.1038/nmeth974. [DOI] [PubMed] [Google Scholar]
- (13).Nong RY; Wu D; Yan J; Hammond M; Gu GJ; Kamali-Moghaddam M; Landegren U; Darmanis S Solid-Phase Proximity Ligation Assays for Individual or Parallel Protein Analyses with Readout Via Real-Time Per or Sequencing. Nature Protocols 2013, 8, 1234–1248, DOI: 10.1038/nprot.2013.070 [DOI] [PubMed] [Google Scholar]
- (14).Nguyen PD; Son SJ; Min J Upconversion Nanoparticles in Bioassays, Optical Imaging and Therapy. J Nanosci Nanotechnol 2014,14 (1), 157–174. [DOI] [PubMed] [Google Scholar]
- (15).Wang J Nanoparticle-Based Electrochemical Bioassays of Proteins. Electroanalysis 2007, 19 (7–8), 769–776, DOI: 10.1002/elan.200603789. [DOI] [Google Scholar]
- (16).Zhou W; Gao X; Liu D; Chen X Gold Nanoparticles for in Vitro Diagnostics. Chemical Reviews 2015,115 (19), 10575–10636, DOI: 10.1021/acs.chemrev.5b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kumar A; MazinderBoruah B; Liang X-J GoldNanoparticles: Promising Nanomaterials for the Diagnosis of Cancer and Hiv/Aids. Journal of Nanomaterials 2011, 2011, 17, DOI: 10.1155/2011/202187. [DOI] [Google Scholar]
- (18).Rutledge RG; Cote C Mathematics of Quantitative Kinetic Pcr and the Application of Standard Curves. Nucleic Acids Res 2003, 31 (16), e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Al-Robaiy S; Rupf S; Eschrich K Rapid Competitive Per Using Melting Curve Analysis for DNA Quantification. Biotechniques 2001, 31 (6), 1382–1386. [DOI] [PubMed] [Google Scholar]
- (20).Morpurgo M; Facchin S; Pignatto M; Silvestri D; Casarin E; Realdon N Characterization of Multifunctional Nanosystems Based on the Avidin-Nucleic Acid Interaction as Signal Enhancers in Immuno-Detection. Anal Chem 2012, 84 (7), 3433–3439, DOI: 10.1021/ac300276u. [DOI] [PubMed] [Google Scholar]
- (21).Fan R; Vermesh O; Srivastava A; Yen BK; Qin L; Ahmad H; Kwong GA; Liu CC; Gould J; Hood L; Heath JR Integrated Barcode Chips for Rapid, Multiplexed Analysis of Proteins in Microliter Quantities of Blood. Nat Biotechnol 2008, 26 (12), 1373–1378, DOI: 10.1038/nbt.l507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.