Abstract
The melon thrips, Thrips palmi is a serious pest and vector for plant viruses on a wide range of economically important crops. DNA barcoding evidenced the presence of cryptic diversity in T. palmi and that warrants exhaustive molecular studies. Our present study is on decoding the first complete mitochondrial genome of T. palmi (15,333 bp) through next-generation sequencing (NGS). The T. palmi mt genome contains 37 genes, including 13 Protein coding genes (PCGs), two ribosomal RNA (rRNAs), 22 transfer RNA (tRNAs), and two control regions (CRs). The majority strand of T. palmi revealed 78.29% A+T content, and 21.72% G+C content with positive AT skew (0.09) and negative GC skew (-0.06). The ATN initiation codons were observed in 12 PCGs except for cox1 which have unique start codon (TTG). The relative synonymous codon usage (RSCU) analysis revealed Phe, Leu, Ile, Tyr, Asn, Lys and Met were the most frequently used amino acids in all PCGs. The codon (CGG) which is assigned to Arginine in most insects but absent in T. palmi. The Ka/Ks ratio ranges from 0.078 in cox1 to 0.913 in atp8. We observed the typical cloverleaf secondary structure in most of the tRNA genes with a few exceptions; absence of DHU stem and loop in trnV and trnS, absence of DHU loop in trnE, lack of T-arm and loop in trnN. The T. palmi gene order (GO) was compared with ancestral GO and observed an extensive gene arrangement in PCGs, tRNAs and rRNAs. The cox2 gene was separated from the gene block ‘cox2-trnL2’ in T. palmi as compared with the other thrips mt genomes, including ancestor GO. Further, the nad1, trnQ, trnC, trnL1, trnV, trnF, rrnS, and rrnL were inversely transpositioned in T. palmi GO. The gene blocks ‘trnQ-trnS2-trnD’ and ‘trnN-trnE-trnS1-trnL1’ seems to be genus specific. The T. palmi mt genome contained 24 intergenic spacer regions and 12 overlapping regions. The 62 bp of CR2 shows the similarity with CR1 indicating a possible duplication. The occurrence of multiple CRs in thrips mt genomes seems to be a derived trait which needs further investigation. Although, the study depicted extensive gene rearrangements in T. palmi mt genome, but the negative GC skew reflects only strand asymmetry. Both the ML and BI phylogenetic trees revealed the close relationships of Thrips with Scirtothrips as compared to Frankliniella. Thus, more mt genomes of the diverse thrips species are required to understand the in-depth phylogenetic and evolutionary relationships.
Introduction
The members of insect order Thysanoptera (commonly known as thrips) are usually tiny, fringe winged and are classified into nine families within two suborders [1]. The family Thripidae is the most diverse family and further divided into four subfamilies (Dendrothripinae, Panchaetothripinae, Sericothripinae and Thripinae). Thrips are widely distributed throughout the world and known by 6154 species, of which 739 species are reported from India [2]. Thrips are one of the major sucking pests and sole transmitters of Tospoviruses (Family Bunyaviridae) on a wide number of agricultural and horticultural crops [3]. Fifteen species are reported as vectors of tospoviruses, of which six species are known from India (Ceratothripoides claratris, Frankliniella occidentalis, Frankliniella schultzei, Scirtothrips dorsalis, Thrips palmi and Thrips tabaci) [4–5]. Due to their economic importance, the accurate identification of these species is the basic necessity; for the study of their disease transmission efficiency in crops, and implementation of the effective management strategies. The morphological identification is time consuming and challenging because of their cryptic behaviour and overlapping geographical distributions. Therefore, modern molecular tools such as DNA barcoding have been applied for thrips identification and phylogenetic studies [6–8].
Thrips palmi is commonly known as melon thrips, one of the major pest on agricultural crops and has been reported as vector for four tospoviruses; Calla lily chlorotic spot virus (CCSV), Groundnut bud necrosis virus (GBNV), Melon yellow spot virus (MYSV), Watermelon silver mottle virus (WSMV) [4]. The species is widely distributed and highly polyphagous and is often confused with the Thrips flavus and Thrips alatus [9]. Recent DNA barcoding studies on thrips, revealed four molecular operations taxonomic units (MOTUs) in T. palmi (T. palmi Ia1, T. palmi IIa1, T. palmi Ib1, and T. palmi Ib2) representing multiple cryptic species [8]. Considering these hitches, more and in-depth molecular data on this species is required to understand the cryptic speciation and evolutionary affiliations.
Mitochondrial genome (mt genome) data have been widely used for phylogenetic, evolutionary studies, and population genetics in insects [10–11]. The mt genomes of insects were usually represented by 37 genes, including 13 PCGs, large and small ribosomal RNA genes (rrnL and rrnS), 22 transfer RNA genes (tRNAs), variable number of control regions (CRs) and chromosomes [12–14]. However, the availability of thrips mt genomes is limited in global database and six mt genomes of five species (Anaphothrips obscurus, Frankliniella intonsa, Frankliniella occidentalis, Scirtothrips dorsalis and Thrips imaginis) are available in the GenBank [15–19].
In the present study, we sequenced and characterized the complete mt genomes of the melon thrips, T. palmi under the family Thripidae using the next generation sequencing (NGS) technology and comparative analysis with other thrips mt genomes. The comparisons were based on gene arrangements, nucleotide composition, codon usage, evolutionary rates, and strand asymmetry etc. Further, to infer the phylogenetic relationships, 13 PCGs of T. palmi and other six thrips species mt genomes were analyzed using maximum likelihood (ML) and Bayesian inference (BI). This study will provide a better understanding of comparative mitochondrial genomics T. palmi with thysanopterans and other insects.
Materials and methods
Ethics statement
There is no need of specific permission for the collection and usage of the insects in this study because these insects are common pests of crops. Neither endangered nor protected species were involved in the study.
Sample collection, and DNA extraction
The specimens of T. palmi were collected from the Odisha state of India from Eggplant (Solanum melongena). The specimens were morphologically identified by the second author (K.T) with the available taxonomic keys [9], and preserved in absolute ethyl alcohol at −80°C in Centre for DNA Taxonomy, Molecular Systematics Division, Zoological Survey of India, Kolkata. The DNeasy DNA Extraction kit (Qiagen, Valencia, CA) was used to extract the genomic DNA and the concentration was measured on a quantified by Qubit fluorometer (Thermo Fisher Scientific, MA, USA) using a dsDNA high-sensitivity kit with the standard protocol.
Mitochondrial genome sequencing and assembly
The complete mt genome sequencing, assembly and annotation were carried out at the Genotypic Technology Pvt. Ltd. Bangalore, India (http://www.genotypic.co.in/). Whole genome sequencing (WGS) library was prepared with Illumina-compatible NEXTflex Rapid DNA sequencing kit (BIOO Scientific, Austin, Texas, U.S.A.). The DNA was sheared using a Covaris S2 sonicator (Covaris, Woburn, Massachusetts, USA) to generate approximate fragment size distribution of 200 bp to 600 bp. The fragment size distribution was checked on Agilent TapeStation and subsequently purified using Highprep magnetic beads (Magbio). Purified fragments were end-repaired, adenylated and ligated to Illumina multiplex barcode adaptors. Adapter-ligated DNA was purified using Highprep beads and the resulted fragments were amplified for eight cycles of PCR using Illumina-compatible primers. The final PCR product was purified, followed by library quality control check. Illumina-compatible sequencing library was quantified by Qubit fluorometer (Thermo Fisher Scientific, MA, USA) and its fragment size distribution were analyzed by Agilent 2200 Tapestation and Agilent Bioanalyzer. Libraries with adapter contamination were pooled and cleaned up with Highprep magnetic beads (Magbio) and then sequenced on Nextseq 500 150X2 chemistry. Raw Sequences were trimmed and filtered using NGS Toolkit. The adapter contamination and low-quality reads with base N’s or more than 70% of the bases with a quality score <20 also had been removed. The acquired high quality ~18 million reads were screened out using the Burrows-Wheeler Alignment (BWA) tool [20]. Out of 18 million reads, 0.10% (~1.8 million) of the reads got aligned, then assembled with SPAdes 3.9.0 [21], using default parameters considering T. imaginis mitochondrial genome (AF335993) as reference contig. The aligned reads were considered for the denovo mitochondrial genome of T. palmi.
Genome annotation, visualization, and comparative analysis
The assembled mt genome was annotated by using MITOS web-server (http://mitos.bioinf.uni-leipzig.de/index.py) to estimate the position of PCGs, tRNAs, rRNAs and their secondary structures. The boundaries of PCGs and rRNAs was confirmed manually by nucleotide-nucleotide BLAST (BLASTn), protein-protein BLAST (BLASTp), and open reading frame finder (ORF finder) in the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/orffinder/). The PCGs were translated into putative proteins using the invertebrate mitochondrial DNA genetic code. The ClustalX program was used to assign the initiation and termination codons in comparison with other thrips reference sequences [22]. MEGAX was used for alignment of the homologous sequences of T. palmi with other thrips species [23]. The complete annotated T. palmi mt genome was prepared using the Sequin submission tool (http://www.ncbi.nlm.nih.gov/Sequin/), for acquiring the accession number from GenBank database. The circular representation of T. palmi mt genome was drawn by CGView online server (http://stothard.afns.ualberta.ca/cgview_server/) with default parameters [24]. The assembled T. palmi mt genome was compared with the other thrips mt genomes to calculate the nucleotide composition, Relative Synonymous Codon Usage (RSCU), AT- GC skew, non-synonymous (Ka) and synonymous (Ks) substitutions, gene rearrangements, secondary structure of tRNAs and CRs etc. The nucleotide composition and RSCU were determined using MEGAX [23]. The following formula was used to calculate the skew: AT skew = (A−T) / (A+T) and GC skew = (G−C) / (G+C) [25]. The sequence substitution saturation analysis of PCGs was calculated by DAMBE5 software [26]. The overlapping and intergenic spacer regions of thrips mt genome genes were compared in terms of length and locations. Further the homology of CRs in T. palmi and other Thrips mt genomes were determined through sequence alignment using Clustal Omega [22]. The nucleotide sequences of each PCG were aligned based on the amino acid sequences in TranslatorX [27] with MAFFT algorithm using GBlocks parameters. The ratios of non-synonymous substitutions (Ka) and synonymous (Ks) substitutions were estimated in DnaSP6.0 [28]. The secondary structures of tRNAs were anticipated by the MITOS web server (http://mitos.bioinf.uni-leipzig.de) and confirmed by the tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/) [29] and ARWEN 1.2 [30]. The RNAstructure version 6.0.1 was used to predict the secondary structure of CRs [31]. The gene arrangements, pairwise comparisons of gene orders, and evolutionary pathways of T. palmi mt genome were evaluated by CREx web tool (Common Interval Rearrangement Explorer) [32]. This web-based program uses transpositions, reverse transpositions, reversals and tandem-duplication-random-loss (TDRL) events to find the parsimonious rearrangement events in a phylogenetic hypothesis. CREx analysis based on common intervals was implemented to determine the gene arrangement scenarios between the T. palmi gene order (GO) with the ancestral A. bakeri (Hemiptera) and other thrips species.
Phylogenetic analysis
Six complete mt genomes of five thrips species were retrieved from GenBank on April 1st, 2018 for phylogenetic inference (S1 Table). The A. bakeri mt genome was used in the dataset as an out-group [33]. The PCGs were individually aligned with the TranslatorX online platform using the MAFFT algorithm [27] with the GBlocks parameters and default settings. The dataset of all PCGs was concatenated using SequenceMatrix v1.7.845 to form 9696 bp dataset [34]. The optimal substitution model was estimated by jModelTest [35]. The dataset was analyzed using maximum likelihood (ML) implemented in RaxML, and Bayesian inference (BI) implemented in MrBayes 3.2 [36]. For likelihood analysis, the bootstrap analysis of 1,000 replicates was performed in RaxML [37] with GTR+G+I as a best fit model. For BI analysis, two simultaneous runs of 12 million generations were conducted for the dataset using GTR+G+I model and trees were sampled in every 1000 generations, with the first 25% discarded as burn-in. The BI analysis was stopped after reaching the stationary phase and average standard deviation of split frequencies below 0.01. The phylogenetic tree was visualized and edited using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) [38].
Results and discussion
Genome structure and nucleotide composition
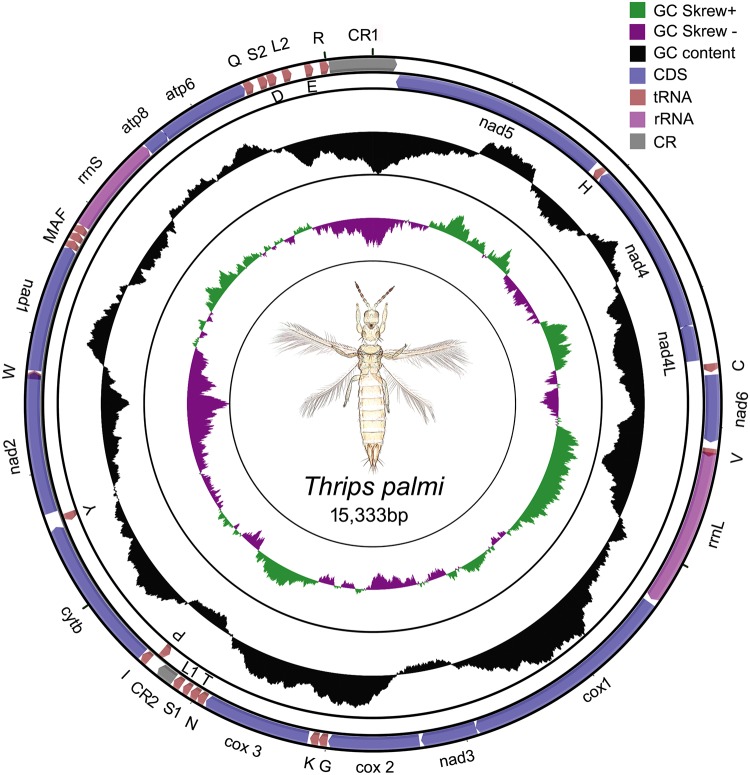
The complete mt genome of T. palmi (GenBank accession no. MH253898) was 15,333 base pairs (bp) in length. This is the second largest mt genome in size among all previously generated mt genomes of insect order Thysanoptera (S1 Table). The T. palmi mt genome was characterized by 37 genes, including 13 PCGs, large and small ribosomal RNA genes (rrnL and rrnS), 22 transfer RNA genes (tRNAs) and two control regions (CRs) (Fig 1). Among all genes, 31 genes were detected on the majority (light) strand and six genes on the minority (heavy) strand (Table 1). The nucleotide composition of T. palmi mt genome was 78.29% A+T content (42.71% A + 35.58% T) and 21.72% G+C content (10.14% G + 11.58% C) (Table 2). The A+T content was higher in control regions (85.81%) followed by tRNAs (80.26%), rRNAs (80.21%) and PCGs (77.43%). The sequence similarity searches of T. palmi mt genome showed the highest similarity (81%) with T. imaginis followed by F. occidentalis (74%), F. intonsa (73%), S. dorsalis SA1 (72%), S. dorsalis EA1 (71%) and A. obscurus (72%) in GenBank.
Fig 1. The circular representation of the complete mitochondrial genome of T. palmi.
Direction of gene transcription is indicated by arrows in entire complete genome. PCGs are shown as purple arrows, rRNA genes as pink arrows, tRNA genes as peach color arrows and CR regions as gray rectangles. The GC content is plotted using a black sliding window, as the deviation from the average GC content of the entire sequence. The GC-skew is plotted using a colored sliding window (green and orchid color), as the deviation from the average GC skew of the entire sequence. The figure was drawn using the CGView online server (http://stothard.afns.ualberta.ca/cgview_server/) with default parameters. The species photograph was taken by the second author (KT) using Leica Microscope DM1000 with Leica software application suite (LAS EZ) and edited manually in Adobe Photoshop CS 8.0.
Table 1. List of annotated mitochondrial genes of T. palmi and its characteristic features.
The PCGs and rRNA genes are represented by standard nomenclature, tRNAs are represented as trn followed by the IUPAC-IUB single letter amino acid codes. (+) values in strand represent as heavy (H) and (-) values represent as light (L). IGN represents (+) values as intergenic nucleotides and (-) values as overlapping regions. CR represents the control region.
| Gene | Strand | Location | Size (bp) | Anticodon | Start Codon | Stop Codon | IGN |
|---|---|---|---|---|---|---|---|
| nad5 | 176–1828 | 1653 | . | ATT | TAA | 29 | |
| trnH | - | 1858–1919 | 62 | CAC | . | . | 2 |
| nad4 | - | 1922–3241 | 1320 | . | ATT | TAA | -7 |
| nad4l | - | 3235–3510 | 276 | . | ATG | TAG | 34 |
| trnC | + | 3545–3606 | 62 | UGC | . | . | 19 |
| nad6 | + | 3626–4111 | 486 | . | ATT | TAA | 43 |
| trnV | + | 4155–4214 | 60 | GUA | . | . | 0 |
| rrnL | + | 4215–5325 | 1111 | . | . | . | 27 |
| cox1 | + | 5353–6915 | 1563 | . | TTG | TAA | -1 |
| nad3 | + | 6915–7319 | 405 | . | ATG | TAA | 12 |
| cox2 | + | 7332–7988 | 657 | . | ATA | TAA | 5 |
| trnG | + | 7994–8057 | 64 | GGA | . | . | -1 |
| trnK | + | 8057–8120 | 64 | AAA | . | . | 13 |
| cox3 | + | 8134–8916 | 783 | . | ATA | TAA | 4 |
| trnN | + | 8921–8983 | 63 | AAC | . | . | -3 |
| trnT | + | 8981–9044 | 64 | ACA | . | . | 7 |
| trnS1 | + | 9052–9108 | 57 | AGA | . | . | 15 |
| trnL1 | + | 9124–9188 | 65 | CUA | . | . | 3 |
| CR2 | + | 9192–9329 | 138 | . | . | . | 1 |
| trnP | - | 9331–9394 | 64 | CCA | . | . | 23 |
| trnI | + | 9418–9481 | 64 | AUC | . | . | 1 |
| cytb | + | 9483–10595 | 1113 | . | ATA | TAA | 8 |
| trnY | - | 10604–10666 | 63 | UAC | . | . | 37 |
| nad2 | + | 10704–11726 | 1023 | . | ATA | TAA | -50 |
| trnW | + | 11677–11738 | 62 | UGA | . | . | 0 |
| nad1 | + | 11739–12662 | 924 | . | ATA | TAA | -4 |
| trnM | + | 12659–12719 | 61 | AUG | . | . | 1 |
| trnA | + | 12721–12782 | 62 | GCA | . | . | -1 |
| trnF | + | 12782–12844 | 63 | UUC | . | . | -2 |
| rrnS | + | 12843–13570 | 728 | . | . | . | -1 |
| atp8 | + | 13570–13738 | 169 | . | ATA | T(TT) | -7 |
| atp6 | + | 13732–14391 | 660 | . | ATT | TAA | -1 |
| trnQ | + | 14391–14458 | 68 | CAA | . | . | 41 |
| trnS2 | + | 14500–14564 | 65 | UCA | . | . | 1 |
| trnD | + | 14566–14633 | 68 | GAC | . | . | 46 |
| trnL2 | + | 14680–14744 | 65 | CUA | . | . | 99 |
| trnE | + | 14844–14905 | 62 | GAA | . | . | 49 |
| trnR | + | 14955–15019 | 65 | CGA | . | . | 0 |
| CR1 | + | 15020–15333+175 | 489 | . | . | . | . |
Table 2. Nucleotide composition and skewness in different Thysanoptera mt genomes considered for comparative analysis.
| Species | Size(bp) | A% | G% | T% | C% | GC% | AT% | AT skew | GC skew |
|---|---|---|---|---|---|---|---|---|---|
| Whole mtgenome | |||||||||
| T. palmi | 15,333 | 42.71 | 10.14 | 35.58 | 11.58 | 21.72 | 78.28 | 0.09 | -0.07 |
| T. imaginis | 15,407 | 43.85 | 10.47 | 32.72 | 12.96 | 23.43 | 76.57 | 0.15 | -0.11 |
| F. intonsa | 15,215 | 41.24 | 11.06 | 34.68 | 13.01 | 24.07 | 75.93 | 0.09 | -0.08 |
| F. occidentalis | 14,889 | 40.98 | 11.35 | 36.62 | 11.06 | 22.41 | 77.59 | 0.06 | 0.01 |
| S. dorsalis EA1 | 15,343 | 39.12 | 11.61 | 36.62 | 12.64 | 24.26 | 75.74 | 0.03 | -0.04 |
| S. dorsalis SA1 | 15,204 | 39.83 | 11.18 | 37.56 | 11.42 | 22.60 | 77.40 | 0.03 | -0.01 |
| A. obscurus | 14,890 | 38.38 | 11.27 | 39.75 | 10.60 | 21.87 | 78.13 | -0.02 | 0.03 |
| PCG | |||||||||
| T. palmi | 11,030 | 41.64 | 10.52 | 35.32 | 12.52 | 23.04 | 76.96 | 0.08 | -0.09 |
| T. imaginis | 10,922 | 42.75 | 10.15 | 32.89 | 14.21 | 24.36 | 75.64 | 0.13 | -0.17 |
| F. intonsa | 11,009 | 39.95 | 11.39 | 34.58 | 14.08 | 25.47 | 74.53 | 0.07 | -0.11 |
| F. occidentalis | 10,852 | 39.82 | 11.62 | 36.72 | 11.84 | 23.46 | 76.54 | 0.04 | -0.01 |
| S. dorsalis EA1 | 10,954 | 38.06 | 11.92 | 36.53 | 13.48 | 25.41 | 74.59 | 0.02 | -0.06 |
| S. dorsalis SA1 | 10,973 | 38.94 | 11.36 | 37.67 | 12.03 | 23.38 | 76.62 | 0.02 | -0.03 |
| A. obscurus | 11,167 | 37.36 | 11.46 | 39.93 | 11.25 | 22.71 | 77.29 | -0.03 | 0.01 |
| tRNA | |||||||||
| T. palmi | 1,393 | 42.21 | 10.12 | 38.05 | 9.62 | 19.74 | 80.26 | 0.05 | 0.03 |
| T. imaginis | 1,492 | 43.83 | 9.45 | 36.66 | 10.05 | 19.50 | 80.50 | 0.09 | -0.03 |
| F. intonsa | 1,392 | 43.53 | 10.70 | 35.78 | 9.99 | 20.69 | 79.31 | 0.10 | 0.03 |
| F. occidentalis | 1,380 | 42.39 | 10.58 | 37.39 | 9.64 | 20.22 | 79.78 | 0.06 | 0.05 |
| S. dorsalis EA1 | 1,426 | 40.53 | 11.01 | 37.52 | 10.94 | 21.95 | 78.05 | 0.04 | 0.00 |
| S. dorsalis SA1 | 1,429 | 41.36 | 10.43 | 38.21 | 10.01 | 20.43 | 79.57 | 0.04 | 0.02 |
| A. obscurus | 1,430 | 39.79 | 10.63 | 39.86 | 9.72 | 20.35 | 79.65 | 0.00 | 0.04 |
| rRNA | |||||||||
| T. palmi | 1839 | 47.36 | 10.82 | 32.68 | 9.14 | 19.96 | 80.04 | 0.18 | 0.08 |
| T. imaginis | 1,876 | 47.65 | 10.77 | 32.14 | 9.43 | 20.20 | 79.80 | 0.19 | 0.07 |
| F. intonsa | 1,699 | 47.15 | 11.30 | 32.02 | 9.54 | 20.84 | 79.16 | 0.19 | 0.08 |
| F. occidentalis | 1,848 | 45.94 | 12.18 | 33.93 | 7.95 | 20.13 | 79.87 | 0.15 | 0.21 |
| S. dorsalis EA1 | 1,775 | 43.21 | 11.89 | 34.99 | 9.92 | 21.80 | 78.20 | 0.11 | 0.09 |
| S. dorsalis SA1 | 1,777 | 45.36 | 11.65 | 34.44 | 8.55 | 20.20 | 79.80 | 0.14 | 0.15 |
| A. obscurus | 1,812 | 43.16 | 11.70 | 36.59 | 8.55 | 20.25 | 79.75 | 0.08 | 0.16 |
| Control region | |||||||||
| T. palmi | 627 | 44.82 | 4.63 | 40.99 | 9.57 | 14.19 | 85.81 | 0.04 | -0.35 |
| T. imaginis | 900 | 47.56 | 16.67 | 25.22 | 10.56 | 27.22 | 72.78 | 0.31 | 0.22 |
| F. intonsa | 942 | 41.72 | 7.86 | 38.22 | 12.21 | 20.06 | 79.94 | 0.04 | -0.22 |
| F. occidentalis | 595 | 40.34 | 7.90 | 43.70 | 8.07 | 15.97 | 84.03 | -0.04 | -0.01 |
| S. dorsalis EA1 | 1,775 | 43.21 | 11.89 | 34.99 | 9.92 | 21.80 | 78.20 | 0.11 | 0.09 |
| S. dorsalis SA1 | 767 | 35.33 | 9.26 | 43.55 | 11.86 | 21.12 | 78.88 | -0.10 | -0.12 |
| A. obscurus | 145 | 25.52 | 8.97 | 62.76 | 2.76 | 11.72 | 88.28 | -0.42 | 0.53 |
Protein coding genes and Relative Synonymous Codon Usage
The T. palmi mt genome was represented by 13 PCGs (atp6, atp8, cox1, cox2, cox3, cytb, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6). The ATN initiation codons (six with ATA, four with ATT and two with ATG) was observed in 12 PCGs except cox1 (TTG). The TTG start codon for cox1 is unique in T. palmi as ATN start codon is observed for cox1 in all other thrips mt genomes (S2 Table). The PCGs were terminated with TAA stop codon except nad4L (TAG) and atp8 with an incomplete stop codon. The incomplete termination codons are common in other insect mt genomes which are assumed to be recovered by the post transcriptional polyadenylation [39–41]. The average A+T content of T. palmi PCGs was 77.43% with the highest of 84.06% in nad4L gene. However, the average G+C content of T. palmi PCGs was 22.57% with the highest of 27.9% in cox1 gene. The nucleotide composition in all T. plami PCGs showed 41.64% Adenine (A), 35.32% Thiamine (T), 10.52% Guanine (G), and 12.52% Cytosine (C) (S1 Fig, S3 Table).
Further, the comparative analysis of PCGs in all seven mt genomes of six thrips species showed that the adenine (A) and thiamine (T) composition was higher than guanine (G) and cytosine (C) (Table 2). The RSCU data analysis of 3,676 codons in 13 PCGs of T. palmi revealed that Phenylalanine (Phe), Leucine (Leu), Isoleucine (Ile), Tyrosine (Tyr), Asparagine (Asn), Lysine (Lys), and Methionine (Met) were the most frequently used amino acids (S2 Fig, S4 Table). Comparative RSCU analysis of seven thrips mt genomes showed that Phe, Leu, Ile, Tyr, Asn, Lys and Met were most frequent amino acids with TTT (Phe), TTA (Leu), ATT (Ile), TAT (Tyr), AAT (Asn) and AAA (Lys), ATA (Met) were the most frequently used codons. In contrast, almost all the frequently used codons were ended with A/T, which may lead to the A and T bias in thrips species mt genomes. The codon CGG (Arg) and GCG (Ala) were absent in T. palmi and S. dorsalis SA1 respectively, while these codons were present in other thrips mt genomes. Both of the missing codons were preferred to end with ‘G’ in the third codon position (S3 Fig). Sequence saturation analysis of PCGs of the thrips mt genomes exhibited an increasing rate of transitions and transversions ratio along with the divergence value (S4 Fig, S5 Table).
Non-synonymous and synonymous substitutions
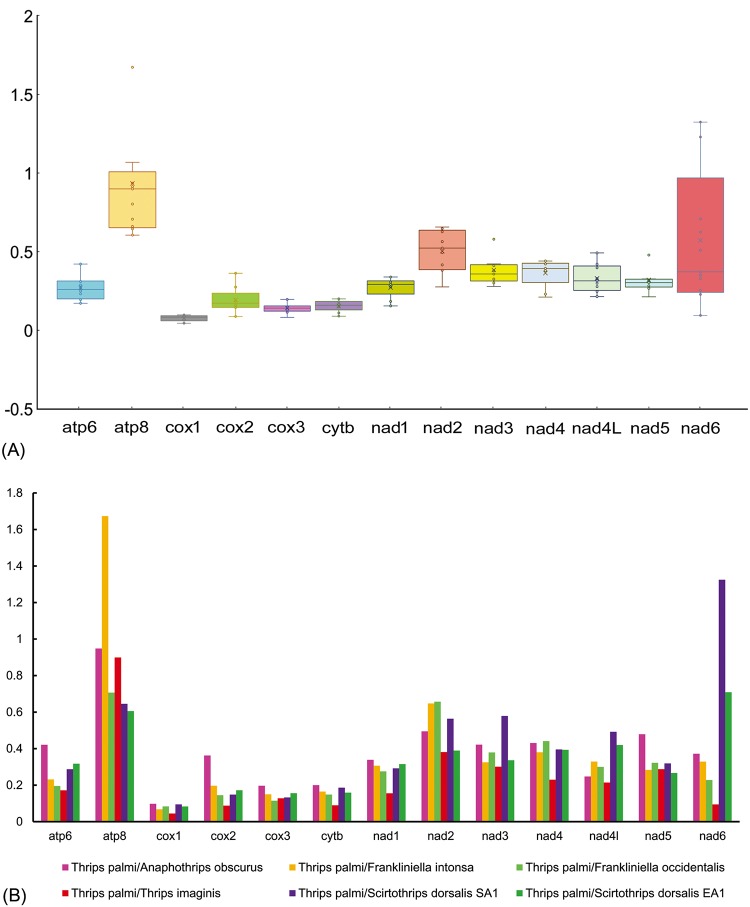
The non-synonymous and synonymous substitutions (Ka/Ks) ratio is an indicator for investigating the selective pressure and evolutionary relations of the homogenous or heterogeneous species [11]. It was reported that, (i) the Ka/Ks>1 for positive selection, (ii) Ka/Ks = 1 for neutral mutation, and (iii) Ka/Ks<1 for negative selection [42–44]. The Ka/Ks ratio ranges from 0.078±0.02 in cox1 to 0.913±0.40 in atp8 gene and the resulted following order: cox1<cox3<cytb<cox2<atp6<nad1<nad5<nad4L<nad4<nad3<nad6<nad2<atp8. This result indicated that the 13 PCGs of all thrips mt genomes including T. palmi were evolving under purifying selection (Fig 2A). Comparative analysis of the Ka/Ks ratio among 13 PCGs of thrips species showed that atp8 (1.7) and nad6 (1.3) genes are evolving under positive/relaxed selection with reference to F. intonsa and S. dorsalis SA1 respectively (Fig 2B). Further, the Ka/Ks was <1 for the remaining PCGs of T. palmi with reference to other thrips species, suggested that the mutations were replaced by synonymous substitutions. The lowest Ka/Ks ratio was observed in cox1 gene representing less variations in amino acids and hence had been evidenced as a potential molecular marker for thrips species identification and phylogenetic analysis [6–8].
Fig 2.
(A) Ratio estimation, box plot for pairwise divergence of Ka/Ks ratio for each one of the mitochondrial PCGs. (B) Evolutionary rates (Ka/Ks) of individuals PCGs of T. palmi with other thrips species.
Ribosomal RNA and transfer RNA
The T. palmi mt genome comprises two rRNA genes as observed in other insect mt genomes. The large ribosomal gene (rrnL) was 1111 bp long, and located between trnV and cox1. Further, the small ribosomal gene (rrnS) was 728 bp long, and located between trnF and atp8 gene. The A+T content of two rRNA was 80.04% in T. palmi which is the highest in comparison with other thrips mt genomes. Both AT skew (0.18) and GC skew (0.08) of T. palmi were positive, that is also similar to other previously sequenced thrips mt genomes. The locations of rrnL and rrnS were upstream of cox1 and atp8 gene, the arrangements seem to be conserved in insect order Thysanoptera. The T. palmi mt genome contained 22 tRNAs (ranging from 57 to 68 bp in length) with a total length of 1,393 bp. Nineteen tRNA genes were coded by the majority strand and three (trnY, trnP and trnH) by the minority strand. The A+T content of tRNAs was 85.81% with positive AT skew (0.05) and GC skew (0.3) (Table 2). Most of the tRNA showed the typical cloverleaf secondary structure; absence of DHU stem and loop was observed in trnV and trnS; absence of DHU loop in trnE; lack of TΨC loop in trnN; variation in TΨC arm and loop in other tRNAs (S5 Fig). The absence of DHU stem and loop in trnV was consistent in all the thrips mt genomes. The mismatched base pairs (G-U wobble pairs) were observed in seven tRNAs of T. palmi mt genome. These wobble mismatches were observed in trnL2 and trnL1 (in DHU arm), trnA, trnS1, and trnL2 (in acceptor arm), trnS2 and trnT (in TΨC arm).
Overlapping and intergenic spacer regions
The T. palmi mt genome had 24 intergenic spacer regions with a total of 520 bp, varying from 1 to 99 bp in length. There are 14 major intergenic spacers of >10 bp in length were observed (Table 1). The comparative analysis depicted highest intergenic spacer region in T. palmi in comparison with other thrips species; 17 intergenic spacer of 217 bp in T. imaginis, 14 intergenic spacer of 172 bp in F. intonsa, 14 intergenic spacer of 217 bp in F. occidentalis, 13 intergenic spacer of 436 bp in S. dorsalis EA1, 13 intergenic spacer of 342 bp, 19 intergenic spacer of 309 bp in A. obscurus. The longest intergenic spacer (99 bp) was observed between the trnL2 and trnE gene in T. palmi. However, the shortest intergenic spacer (1 bp) was observed in four positions: trnP and CR2, cytb and trnI, trnA and trnM, trnD and trnS2. Further, the comparative studies showed that the longest intergenic spacer 150 bp were between trnH and nad4 in S. dorsalis EA1. The T. palmi mt genome contained 11 overlapping regions with a total length of 78 bp. The comparative analysis showed that, the highest (15 overlapping regions of 86 bp) were observed in S. dorsalis SA1. The smallest overlapping region (1 bp) was observed in five positions in T. palmi: nad3 and cox1, trnK and trnG, trnF and trnA, atp8 and rrnS, trnQ and atp6. The largest overlapping region (50 bp) was observed between nad2 and trnW in T. palmi. However, the largest overlapping region (66 bp) was observed between rrnL and trnS2 in F. occidentalis in comparative studies (S6 Table).
Control regions
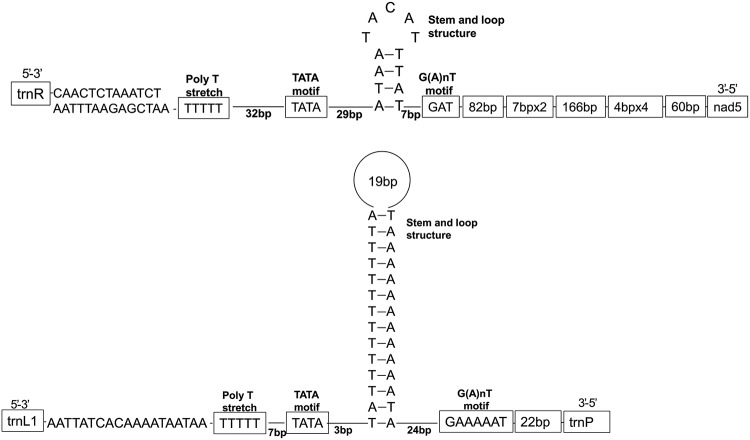
The CRs is usually characterized by five conserved elements, such as (i) a polyT stretch at the 5′ end; (ii) a [TA(A)]n-like stretch (iii) a stem and loop structure (iv) a TATA motif and a G(A)nT motif flanking the stem and loop structure (v) a G+A-rich sequence downstream of the stem and loop structure [45–47]. Both CRs of T. palmi were detected with four conserved elements except G+A-rich sequence (Fig 3). The control region (CR) in mt genomes is most crucial region, which regulates transcription and replication [48]. Duplicate CRs has been documented in many insect species, including thrips [15–18]. The study resulted in the duplication of CRs in T. palmi mt genome; 489 bp CR1 lied between trnR and nad5, 138 bp CR2 between the trnL1 and trnP. Further, we observed 62 bp sequence similarity of CR2 with CR1 indicating a possible duplication. The total length of CR1 and CR2 was 627 bp, which was higher than A. obscurus (145 bp) and F. occidentalis (595bp). T. palmi had two CRs, similar to T. imaginis, and S. dorsalis EA1. The presence of multiple CRs in thrips species that are serious pests and vectors for tospoviruses, seems to be a genomic apomorphy over other common thrips species and the ancestral species. However, more thrips mt genomes are needed to be studied to confirm, whether the duplication of CR in thrips species is linked with the association of tospoviruses.
Fig 3. Predicted structural elements for control regions (CRs) of T. palmi.
Gene arrangement
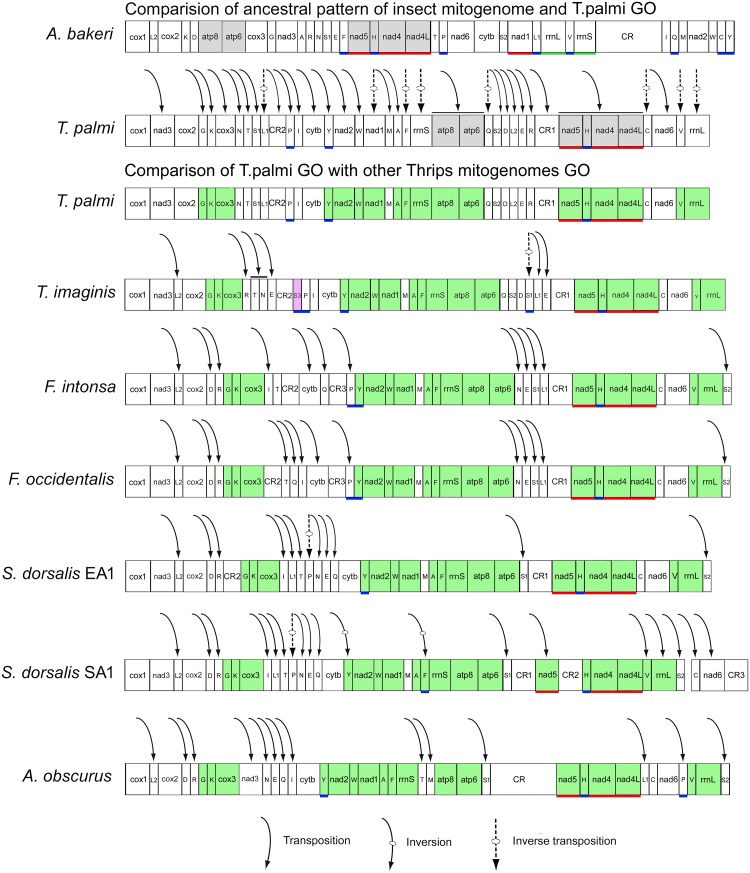
The gene arrangements can be categorized by following aspects, (i) transpositions, (ii) inversions, (iii) inverse transpositions, and (iv) the tandem duplication random loss operation (TDRL) [49–51]. The gene arrangement of T. palmi was evaluated by comparing the common intervals with A.bakeri GO, which was assumed to be the putative ancestor of hexapods [19, 33]. The CREx identified seven operations in the evolution of gene arrangement in T. palmi, including three inversions and four TDRLs with two sets of alternative scenarios (S6 Fig). The CREx analysis detected inversions of trnF, trnC and ‘nad1-rrnS’ gene block, in both scenarios. All the genes in T. palmi mt genome were encoded by the majority strand except three PCGs (nad5, nad4, nad4L) and three tRNAs (trnH, trnY and trnP). T. palmi GO showed intense gene rearrangements of 11 PCGs, 22tRNAs, and two rRNAs as compared with the ancestral GO. Most of the genes were transpositioned, while eight genes (nad1, trnL1, trnF, trnQ, trnC, trnV, rrnS, and rrnL) were inversely transpositioned as compared with the ancestral GO. Further, T. palmi GO was also compared other thrips mt genomes GO and resulted that nad2 was conserved between trnY and trnW in all thrips species. The cox2 was separated in T. palmi GO from ‘cox2-trnL2’ gene block, which was conserved in other thrips species including ancestral GO (Fig 4). The comparison of T. palmi GO with other thrips mt genomes further revealed trnD and trnR were translocated and separated from ‘trnD-cox3’ gene block in T. plami and T. imaginis. The gene blocks ‘cox3-nad2’ and ‘atp6-nad5’ have been identified as the most frequently rearranged position in thrips mt genomes, while ‘nad5-trnH-nad4-nad4L’ seems to be an ancestral gene block. The tRNAs gene blocks ‘trnQ-trnS2-trnD’ and ‘trnN-trnE-trnS1-trnL1’ might be specific to genus Thrips and Frankliniella respectively.
Fig 4. Linearized view of complete mitochondrial genome organization and gene rearrangement, transposition, inversion, and inverse transposition in T. palmi compared with the ancestral type of the insect (A. bakeri).
The gray color blocks show the conserve gene blocks of T. palmi and A. bakeri. The green color blocks show the conserve gene blocks of T. palmi and other Thysanoptera species. The pink color block shows the extra tRNA present in T. imaginis. Gene blocks with underline shows the position of genes in minority strand (red for PCGs, green for rRNAs, and blue for tRNAs). Different shapes of arrows are used for showing transposition, inversion, and inverse transposition. Gene nomenclature: atp6 and atp8; ATP synthase subunits 6 and 8; cytb: cytochrome b; cox1–3: cytochrome c oxidase subunits 1–3; nad1–6 and nad4L: NADH dehydrogenase subunits 1–6 and 4L; rrnS and rrnL: small and large subunit ribosomal RNA (rRNA) genes; Transfer RNA genes are denoted by a one-letter symbol according to the IPUCIUB single-letter amino acid codes. CR indicates the control region.
Strand asymmetry
The strand asymmetry is a remarkable feature of mitochondrial genomes denoted by the AT skew and GC skew on the majority strand (encoded with more genes) [52–54]. Positive AT skew (A>T) and negative GC skew (C>G) is usually observed in most of the insect’s mitochondrial genome with few exceptions; where strand asymmetry is reversed, indicating A<T and C<G on the majority strand [46–47, 50]. A recent study on the mechanism of strand asymmetry in insects confirmed that reversal of strand asymmetry was due to the inversion of replication origin in CRs [54]. We calculated and compared the AT and GC skews of T. palmi with other thrips mt genomes. The T. palmi mt genome showed positive AT skew (0.09) and negative GC skew (-0.07) found to be similar with most of the thrips mt genomes. The AT skew of complete mt genomes in other thrips species, ranged from -0.02 (A. obscurus) to 0.15 (T. imaginis), while the GC skew varies from -0.11 (T. imaginis) to 0.01 (F. occidentalis). The occurrence of Guanine (Gs) is more than Cytosine (Cs) with positive GC skew was observed in A. obscurus revealed the reversal of strand asymmetry (Table 2). We have also observed and compared the skew value of all PCGs in thrips mt genomes including T. palmi. The present study depicted that duplication of CRs and structural elements might regulate the extensive gene rearrangements in thrips mt genomes.
Phylogenetic analysis
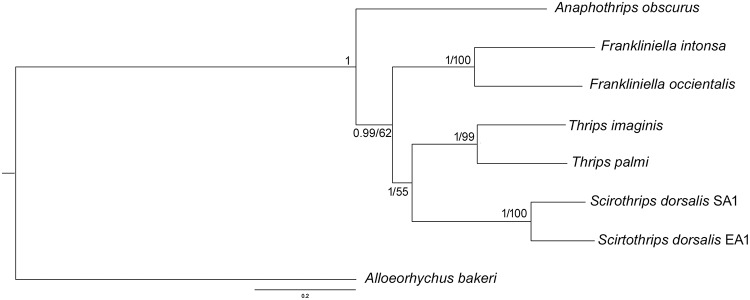
The Maximum likelihood (ML) and Bayesian Inference (BI) phylogenetic trees were constructed by using 13 PCGs. The A. bakeri mt genome was used as the out-group in the phylogenetic analysis. The phylogenetic trees generated using both the methods resulted similar topologies (Fig 5). The tree clustering revealed that species under genus Frankliniella and Thrips, were clustered under the respective genus clade. The phylogenetic analysis showed that genus Thrips is more closely related to genus Scirtothrips as compared to genus Frankliniella. The genus Thrips and Frankliniella forms two large genus groups ‘Thrips genus groups’ and ‘Frankliniella genus group’ respectively, which were supposed to be closely related based on the assumed homology for paired ctenidia on abdominal segments V-VIII [55]. However, Mound 2002 suggested that, these two genus-groups are not closely related based on the chaetotaxy of the abdomen and head [55]. Further, the close relationship between Scirtothrips and Thrips cannot be supported with their morphology. Till date, the comprehensive mt genomes data of thrips species is in its early stages and thus warrant generation of more mt genomes data of diverse thrips species from different hierarchical level to understand the phylogenetic and evolutionary relationships among them.
Fig 5. Bayesian phylogenetic tree inferred by 13 PCGs of thrips mt genomes.
The Bayesian posterior probabilities and Maximum likelihood bootstrap supports are superimposed with each node. The tree is drawn to scale with values indicated along with the branches (BI/ML).
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
The tRNAs are represented by full names and IUPAC-IUB single letter amino acid codes. The details of stem and loop is mentioned for one tRNA Serine which is applicable for all tRNAs secondary structures.
(TIF)
In total seven rearrangement operations occurred from the presumed ancestral gene order of A. bakeri to form the derived gene order of T. palmi GO. Two alternative sets of scenarios were found, i.e. operations 1–7 and operations 1a–7a.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors are thankful to the Director, Zoological Survey of India, Kolkata, for providing necessary facilities, constant support and encouragement throughout the study. This work is a part of the Ph. D thesis of the first author (RC).
Data Availability
All relevant data are available at NCBI GenBank (accession number MH253898).
Funding Statement
This research was funded by the Ministry of Environment, Forest and Climate Change: Zoological Survey of India, Core Funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.ThripsWiki. ThripsWiki—providing information on the World’s thrips. http://thrips.info/wiki/. (Accessed 1 May 2018).
- 2.Tyagi K, Kumar V. Thrips (Insecta: Thysanoptera) of India: An Updated Checklist. Halteres. 2016; 7: 64–98. [Google Scholar]
- 3.Tyagi K, Kumar V. Thrips of Economic importance in India: An identification Guide: 1–96. Published by the Director, Zoological Survey of India, Kolkata; 2017; ISBN: 978-81-8171-465-7. [Google Scholar]
- 4.Riley DG, Joseph SV, Srinivasan R, Diffie S. Thrips vectors of tospoviruses. J Int Pest Manage. 2011; 1: 1–10. [Google Scholar]
- 5.Zhou J, Tzanetakis IE. Epidemiology of Soybean vein necrosis-associated virus. Phytopathol. 2013; 103: 966–971, 10.1094/PHYTO-12-12-0322-R [DOI] [PubMed] [Google Scholar]
- 6.Buckman RS, Mound LA, Whiting MF. Phylogeny of thrips (Insecta: Thysanoptera) based on five molecular loci. Syst Entomol. 2013; 38: 123–133. 10.1111/j.1365-3113.2012.00650.x [DOI] [Google Scholar]
- 7.Iftikhar R, Ashfaq M, Rasool A, Hebert PDN. DNA Barcode Analysis of Thrips (Thysanoptera) Diversity in Pakistan Reveals Cryptic Species Complexes. PloS One 2016; 11: e0146014, 10.1371/journal.pone.0146014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyagi K, Kumar V, Singha D, Chandra K, Laskar BA, Kundu S, et al. DNA Barcoding studies on Thrips in India: Cryptic species, Species complexes. Sci Rep. 2017; 7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatti JS. Species of the genus Thrips from India (Thysanoptera). Syst Entomol. 1980; 5:109–166. [Google Scholar]
- 10.Singh D, Kabiraj D, Sharma P, Chetia H, Mosahari PV, Neog K, et al. The mitochondrial genome of Muga silkworm (Antheraea assamensis) and its comparative analysis with other lepidopteran insects. PloS One. 2017; 12: e0188077, 10.1371/journal.pone.0188077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao YJ, Zou YL, Ding YR, Xu WY, Yan ZT, Li XD, et al. Complete mitochondrial genomes of Anopheles stephensi and An. dirus and comparative evolutionary mitochondriomics of 50 mosquitoes. Sci Rep. 2017; 7: 7666 10.1038/s41598-017-07977-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999; 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992; 141: 173–216. [DOI] [PubMed] [Google Scholar]
- 14.Shao R, Kirkness EF, Barker SC. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 2009; 19: 904–912. 10.1101/gr.083188.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao R, Barker SC. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol Biol Evol. 2003; 20: 362–370. 10.1093/molbev/msg045 [DOI] [PubMed] [Google Scholar]
- 16.Yan D, Tang Y, Xue X, Wang M, Liu F, Fan J. The complete mitochondrial genome sequence of the western flower thrips Frankliniella occidentalis (Thysanoptera: Thripidae) contains triplicate putative control regions. Gene. 2012; 506: 117–124. 10.1016/j.gene.2012.06.022 [DOI] [PubMed] [Google Scholar]
- 17.Yan D, Tang Y, Xue X, Wang M, Liu F, Fan J, et al. The mitochondrial genome of Frankliniella intonsa: insights into the evolution of mitochondrial genomes at lower taxonomic levels in Thysanoptera. Genomics. 2014; 104: 306–312. 10.1016/j.ygeno.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 18.Dickey AM, Kumar V, Morgan JK, Cavieres AJ, Shatters RG Jr, McKenzie CL, et al. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genomics. 2015; 16: 439 10.1186/s12864-015-1672-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Li H, Song F, Gu W, Feng J, Cai W, et al. Novel insights into mitochondrial gene rearrangement in thrips (Insecta: Thysanoptera) from the grass thrips, Anaphothrips obscurus. Sci Rep. 2017; 7: 4284 10.1038/s41598-017-04617-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25: 1754–1760. 10.1093/bioinformatics/btp324 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012; 19: 45519:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson JD, Gibson TJ, Higgins DG. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002; 2.3.1–2.3.22. 10.1002/0471250953.bi0203s00 354. [DOI] [PubMed] [Google Scholar]
- 23.Kumar S, Stecher G, Li M, Knyaz C, and Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018; 35: 1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008; 36: W181–W184. 10.1093/nar/gkn179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perna NT, Kocher TD. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 1995; 41: 353–358. [DOI] [PubMed] [Google Scholar]
- 26.Xia X. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 2013; 30: 1720–1728. 10.1093/molbev/mst064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010; 38: W7–W13. 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozas J, Rozas R. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci. 1995; 11: 621–625. [DOI] [PubMed] [Google Scholar]
- 29.Lowe TM, Chan PP. tRNAscan-SE On-line: Search and Contextual Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016; 44: W54–57. 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laslett D. Canbäck B. ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008; 24: 172–175. 10.1093/bioinformatics/btm573 [DOI] [PubMed] [Google Scholar]
- 31.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11: 129 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernt M, Merkle D, Ramsch K, Fritzsch G, Perseke M, Bernhard D, et al. CREx: Inferring Genomic Rearrangements Based on Common Intervals. Bioinformatics. 2007; 23: 2957–2958. 10.1093/bioinformatics/btm468 [DOI] [PubMed] [Google Scholar]
- 33.Li H, Liu H, Cao L, Shi A, Yang H, Cai W. The complete mitochondrial genome of the damsel bug Alloeorhynchus bakeri (Hemiptera: Nabidae). Int J Biol Sci. 2012; 8: 93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2010; 27: 171–180. [DOI] [PubMed] [Google Scholar]
- 35.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998; 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 36.Ronquist F, Huelsenbeck JP. MrBayes 3.2: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003; 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 37.Stamatakis A. RAxML Version 8: a tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. In Bioinformatics. 2014; 30: 1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rambaut A. FigTree. Version 1.4.2, Inst Evol Biol Univ. Edinburgh. 2014.
- 39.Clary DO, Wolstenholme DR. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985; 22: 252–271. 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Li Y, Pan M, Dai F, Zhu X, Lu C, et al. The complete mitochondrial genome of the Chinese oak silkmoth, Antheraea pernyi (Lepidoptera: Saturniidae). Acta Biochim Biophys Sin (Shanghai). 2008; 40: 693–703. 10.1111/j.1745-7270.2008.00449.x. [DOI] [PubMed] [Google Scholar]
- 41.Kim SR, Kim MI, Hong MY, Kim KY, Kang PD, Hwang JS, et al. The complete mitogenome sequence of the Japanese oak silkmoth, Antheraea yamamai (Lepidoptera: Saturniidae). Mol Biol Rep. 2009; 36: 1871–1880. 10.1007/s11033-008-9393-2. [DOI] [PubMed] [Google Scholar]
- 42.Shen X, Ren J, Cui Z, Sha Z, Wang B, Xiang J, et al. The complete mitochondrial genomes of two common shrimps (Litopenaeus vannamei and Fenneropenaeus chinensis) and their phylogenomic considerations. Gene. 2007; 403: 98–109. 10.1016/j.gene.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Huang Y, Lei F. Comparative mitochondrial genomics and phylogenetic relationships of the Crossoptilon species (Phasianidae, Galliformes). BMC Genomics. 2015; 16: 42 10.1186/s12864-015-1234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meiklejohn CD, Montooth KL, Rand DM. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007; 23: 259–263. 10.1016/j.tig.2007.03.008 [DOI] [PubMed] [Google Scholar]
- 45.Cameron SL, Johnson KP, Whiting MF. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): effects of extensive gene rearrangements on the evolution of the genome. J Mol Evol. 2007; 65: 589–604. 10.1007/s00239-007-9042-8 [DOI] [PubMed] [Google Scholar]
- 46.Hassanin A, Leger N, Deutsch J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst Biol. 2005; 54: 277–298. [DOI] [PubMed] [Google Scholar]
- 47.Hassanin A. Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenetics Evol. 2006; 38: 100–116. [DOI] [PubMed] [Google Scholar]
- 48.Zhang DX, Hewitt GM. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 1997; 25: 99–120. [Google Scholar]
- 49.Dowton M, Castro LR, Austin AD. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: The examination of genome ‘‘morphology”. Invert Syst. 2002; 16: 345–356. [Google Scholar]
- 50.Cameron SL, Johnson KP, Whiting MF. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): effects of extensive gene rearrangements on the evolution of the genome. J Mol Evol. 2007; 65, 589–604. 10.1007/s00239-007-9042-8 [DOI] [PubMed] [Google Scholar]
- 51.Chen L, Chen PY, Xue XF, Hua HQ, Li YX, Zhang F, et al. Extensive gene rearrangements in the mitochondrial genomes of two egg parasitoids, Trichogramma japonicum and Trichogramma ostriniae (Hymenoptera: Chalcidoidea: Trichogrammatidae). Sci Rep. 2018; 8: 7034 10.1038/s41598-018-25338-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikolaou C, Almirantis Y. Deviations from Chargaff’s second parity rule in organellar DNA—insights into the evolution of organellar genomes. Gene. 2006; 381: 34–41. 10.1016/j.gene.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 53.Albrecht-Buehler G. Asymptotically increasing compliance of genomes with Chargaff’s second parity rules through inversions and inverted transpositions. Proc Natl Acad Sci USA. 2006; 103: 17828–17833. 10.1073/pnas.0605553103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei SJ, Shi M, Chen XX, Sharkey MJ, van Achterberg C, Ye GY, et al. New Views on Strand Asymmetry in Insect Mitochondrial Genomes. PLoS ONE. 2010; 5: e12708 10.1371/journal.pone.0012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mound LA. The Thrips and Frankliniella genus groups: the phylogenetic significance of ctenidia. 2002; Pp. 379–386 in Marullo R & Mound LA [eds] Thrips and Tospoviruses: Proceedings of the 7th International Symposium on Thysanoptera. Australian National Insect Collection, Canberra.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
The tRNAs are represented by full names and IUPAC-IUB single letter amino acid codes. The details of stem and loop is mentioned for one tRNA Serine which is applicable for all tRNAs secondary structures.
(TIF)
In total seven rearrangement operations occurred from the presumed ancestral gene order of A. bakeri to form the derived gene order of T. palmi GO. Two alternative sets of scenarios were found, i.e. operations 1–7 and operations 1a–7a.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are available at NCBI GenBank (accession number MH253898).