Abstract
Aconitum carmichaelii, commonly known as Fuzi, is a typical traditional Chinese medicine (TCM) herb that has been grown for more than one thousand years in China. Although root rot disease has been seriously threatening this crop in recent years, few studies have investigated root rot disease in Fuzi, and no pathogens have been identified. In this study, fungal libraries from rhizosphere soils were constructed by internal transcribed spacer (ITS) sequencing using the HiSeq 2500 high-throughput platform. A total of 948,843 tags were obtained from 17 soil samples, and these corresponded to 195,583,495 nt. At 97% identity, the libraries yielded 12,266 operational taxonomic units (OTUs), of which 97.5% could be annotated. In sick soils, Athelia, Mucor and Mortierella were the dominant fungi, comprising 10.3%, 10.1% and 7.7% of the fungal community, respectively. These fungi showed 2.6-, 1.53- to 6.31- and 1.38- to 2.65-fold higher enrichment in sick soils compared with healthy soils, and their high densities reduced the fungal richness in the areas surrounding the rotted Fuzi roots. An abundance analysis suggested that A. rolfsii and Mucor racemosus, as the dominant pathogens, might play important roles in the invading Fuzi tissue, and Phoma adonidicola could be another pathogenic fungus of root rot. In contrast, Mortierella chlamydospora, Penicillium simplicissimum, Epicoccum nigrum, Cyberlindnera saturnus and Rhodotorula ingeniosa might antagonize root rot pathogens in sick soils. In addition, A. rolfsii was further verified as a main pathogen of Fuzi root rot disease through hypha purification, morphological observation, molecular identification and an infection test. These results provide theoretical guidance for the prevention and treatment of Fuzi root rot disease.
Introduction
Aconitum carmichaelii Debx. is a typical TCM herb that has been clinically used for almost two thousand years [1, 2]. Fuzi, the lateral roots of A. carmichaelii Debx., has been extensively used as a cardiotonic, analgesic, anti-inflammatory, and diuretic agent for the treatment of colds, polyarthralgia, diarrhoea, heart failure, beriberi, and oedema [3]. As a result, the government of China as well as various Chinese companies have highly invested in A. carmichaelii (Fuzi). More than two thousand hectares of Fuzi, which is considered an important daodi herb and economic crop, were cultivated in Jiangyou and Butuo in Sichuan Province in 2015. Some East Asian countries, such as Japan, Korea, Mongolia and India, import a large quantity of this traditional medicine from China.
However, large-scale planting encourages mortal threats, and the Fuzi crop yield per unit area has been reducing each year. For instance, ten years ago, 15,000 kg of Fuzi could be harvested per hectare, but only approximately 11,000 kg per hectare can be harvested at present. This decrease is due to root rot disease, which is difficult to remedy using traditional methods. Once one Fuzi plant is infected by the root rot pathogen, its leaves turn brown (or reddish purple) and droop, and its stems eventually wilt. In severe cases, strands of white hyphae are visible on the ground, and a reddish brown sclerotium forms approximately a week later. Underground lateral roots eventually show skin lesions and tissue decay. The normal infection speed of this disease can reach 0.5–1 metres per day. Plants in the same farm or farm located near the disease sources soon become sick, and this resulted in the infection of more than 50% of the area of Fuzi farmlands by root rot fungus in 2016.
Root rot is generally regarded as the primary disease caused by soil-borne pathogens, such as the genus Fusarium in cereal plants, Rhizoctonia bataticola in chickpea plants [4], Phytophthora nicotianae in pepper crops [5], and Cylindrocarpon destructans in Panax ginseng [6]. Other pathogens, such as Magnaporthe grisea, M. oryzae, Colletotrichum graminicola, Leptosphaeria maculans and Cercospora beticola, also infect the roots of their hosts to cause root rot [7]. Crop plants can display dry root rot, collar rot or black root rot disease under different environmental conditions [4], but the causes of root rot appear to vary. On the one hand, pathogens have been shown to exhibit great adaptability to various soils and hosts. For instance, the host range of P. nicotianae includes 255 plant genera in 90 families [5], and this pathogen can cause not only root but also collar rot in pepper plants [5, 8]. C. destructans can also cause disease symptoms in the roots of species other than P. ginseng, such as strawberry [6, 9] and grapevines [10]. On the other hand, root rot might derive from microbial imbalances in rhizosphere soils. Rotted chickpea roots exhibit a different microbial community structure compared with control roots [4], indicating that a balanced microbial community is extremely important in the maintenance of healthy plants.
Few studies have investigated the pathogens of Fuzi root rot, and neither the invasion process nor prevention and control methods are known. Moreover, the pathogens leading to Fuzi root rot have not yet been reported; therefore, the encouragement of fundamental research studies on Fuzi root rot is crucial for solving the problem of pathogen invasion. In this study, we used high-throughput sequencing technology to screen the fungal community in sick soils and identify potential pathogens for Fuzi root rot disease. In addition, the fungal colonies of sick roots were isolated, purified and identified based on ITS sequences. Additionally, a pathogen infection test was performed to determine which fungi could cause the disease. Through this research, we aim to provide theoretical guidance for the prevention and treatment of Fuzi root rot disease.
Materials and methods
Infected soils and Fuzi
In this study, sick soils refer to soils attached to Fuzi roots around which sclerotia emerge. Samples of rot-infected Fuzi and sick soils were harvested from Jiangyou in Sichuan Province, which is the core area of the Aconitum Good Agricultural Practice of Medicinal Plants and Animals (GAP) planting bases in China. In Jiangyou, the Da-Hua leaf-type Fuzi is the main type of Fuzi plants cultivated, and the Xiao-Hua leaf type was cultivated to a lesser degree. Compared with Xiao-Hua, Da-Hua leaf-type Aconitum has a higher yield, larger root tubers and weaker resistance to root rot disease.
The following four groups of root surface soils were obtained: healthy soils (T1-T5, combined to form G1), healthy Da-Hua Fuzi (D1-D4, combined to form G2), diseased Da-Hua Fuzi (DV1, DV2 and DV4, combined to form G3) and healthy Xiao-Hua Fuzi (X1-X5, combined to form G4). Each group of dry soil was approximately 300–500 g (100 g for each replicate), and 17 soil replicates were prepared respectively as described by Miao [11]. The samples of diseased Fuzi materials without soil were stored at 4°C prior pathogen isolation and purification. All backup samples used in the study were frozen at the State Bank of Chinese Drug Germplasm Resources, which is located at Chengdu University of Traditional Chinese Medicine.
Soil genomic DNA extraction and PCR amplification
Genomic DNA was extracted from each soil using the CTAB reagent, and PCR was then performed using specific barcode primers. The reaction contained 15 μl of PCR Mix (2× Phusion® High-Fidelity PCR Master Mix with GC Buffer), 3 μl of each primer (2 μM), 2 μl of gDNA (1 ng/μl) and 10 μl of H2O. The reaction conditions were 98°C for 1 min, 30 cycles of 98°C for 10 s, 50°C for 30 s and 72°C for 30 s, and 72°C for 5 min, and the ITS primer sequences used were ITS5-1737F (5’-GGA AGT AAA AGT CGT AAC AAG G-3’) and ITS2-2043R (5’-GCT GCG TTC TTC ATC GAT GC-3’).
cDNA library construction, species annotation and analysis
The TruSeq® DNA PCR-Free Sample Preparation Kit was used to construct cDNA libraries of the fungal communities in the soil samples. After the libraries were quantified by qRT-PCR and Qubit 2.0, a HiSeq 2500 PE250 instrument was used to sequence their ITS nucleotides. The high-throughput data was analysed using Qiime (V1.7.0, http://qiime.org/scripts/). After removing primers, tags, adaptors and low-quality nucleotides, the MUSCLE programme (Version 3.8.31, http://www.drive5.com/muscle/) was used for sequence alignments to identify the phylogenetic relationships and representative sequences of all OTUs. The OTUs were assigned at the 97% identity level. The community composition of each soil sample was calculated at each classification level, including kingdom, phylum, class, order, family, genus and species. The OTU counts were normalized into relative abundances (%). Statistical analyses were conducted with SPSS (v19, http://www.spss.com.cn/), and curve fitting was performed with SigmaPlot (v. 12.5, www.sigmaplot.com/). Unless otherwise stated, P values ≤0.05 were considered significant.
Diversity and community structure analysis
To evaluate the species richness of rhizosphere fungi in each soil sample, an α-diversity analysis was performed using Qiime software (Version 1.7.0) with the Shannon index.
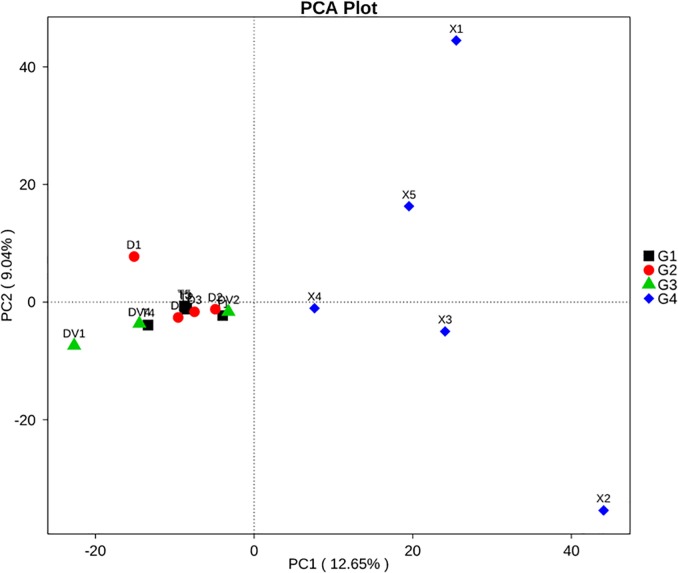
The focus of this study was the differences in fungal diversity between sick and healthy soils. Qiime software was used for an unweighted pair-group method with arithmetic means (UPGMA) analysis based on the UniFrac distance. The soil groups were clustered in a weighted UniFrac tree based on their relative fungal abundances at the phylum level. A principal component analysis (PCA) of the soil fungi in the four groups of soils was performed using the ade4 function in R software (Version 2.15.3).
Isolation and identification of rot-disease pathogens in Fuzi
Fresh samples of rotted Fuzi were washed and sterilized with 75% ethanol. The interior milky tissue was sliced into 0.5-cm2 pieces, and the square blocks of Fuzi were incubated on PDA medium (200 g of potato, 30 g of glucose, 5 g of agar, 1 g of MgSO4, and 5 g of KH2PO4) at 32°C. Once the hyphae grew to approximately 2–3 cm, they were purified three times.
ITS barcode PCR, sequencing and nucleotide blast analysis () were performed to identify the pathogens isolated from rotted Fuzi tissues. The DNA of the pathogens was extracted using a Plant Fungal DNA Extraction Kit with the ITS sequence primers ITSlF (5’-CTT GGT CAT TTA GAG GAA GTA A-3’) and ITS4B (5’-CAG GAG ACT TGT ACA CGG TCC AG-3’). The PCR reactions contained 25 μl of PCR Mix (SsoFast TM EvaGreen® Supermix), 2 μl of ITS1F, 2 μl of ITS4B, 1 μl of gDNA and 20 μl of ddH2O. The reaction programme was 94°C for 3 min, 35 cycles of 94°C for 40 s, 55°C for 40 s, and 72°C for 90 s, and 72°C for 5 min. Sequencing was finished by the Chengdu branch of Biological Science and Technology Co., Ltd.
After ITS sequence alignment with the NCBI database, the pathogens in rotted Fuzi tissue were identified with homology >90%. A phylogenetic tree was then constructed based on the neighbour-joining (NJ) method using MEGA 4.0 software (http://www.megasoftware.net/) with the default parameters.
Pathogenic infection
To detect the invasion effects of candidate rot pathogens, 0.5 cm2 of healthy fresh Fuzi was inoculated onto candidate mycelia of purified pathogens and cultured in the dark at 32°C. The infection status of the Fuzi materials and their morphology and colours were observed and compared with those of diseased aconite in the field.
Results and discussion
Fuzi root rot disease
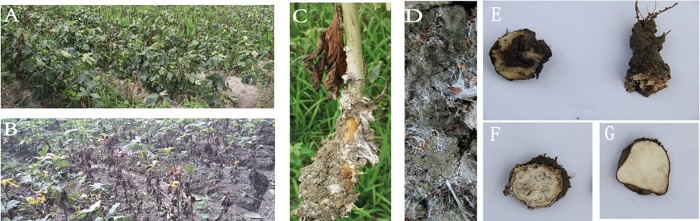
Diseased Aconitum carmichaelii plants were observed in this study (Fig 1). Once the plants were infected with root rot disease, which is also called “southern blight”, the lateral root tissues of Fuzi plants became soft from the outside to the inside, with the root skins rotting first and the hairy roots falling off. At the later stages of the disease, the roots emitted a foul smell, and white hyphae were observed on their surfaces. Moreover, the upper parts of A. carmichaelii plants wilted, and the leaves turned brown (or reddish purple) and dropped from the plant starting from the bottom. In addition, white hyphae were distributed unevenly in the soil, usually in visible clusters, and reddish-brown sclerotia arose and gathered near the soil surface (this type of soil is denoted “sick soil” in the manuscript). The series of symptoms observed was similar to that described in previous reports [12, 13].
Fig 1. Diseased Aconitum carmichaelii (Fuzi) plants.
Construction of the soil fungal database
To explore the pathogenesis of Fuzi root rot disease, 17 soil replicates from four group of samples were used to construct fungal databases. In total, high-throughput sequencing produced 996,653 reads from the 17 soil samples. After removing the barcode primers, these were combined into 975,213 reads, and 948,843 ITS tags (95.25%) were obtained after the QC and Nochime analyses, yielding a total of 195,583,495 nt and an average length of 206 nt for each tag (S1 Table.). The Q30 value was 99.36% after filtrating the chimeric sequences in effective tags, indicating that the fungal databases were of high quality. The high-throughput sequencing data were uploaded to the NCBI database under Accession Number SRP142412.
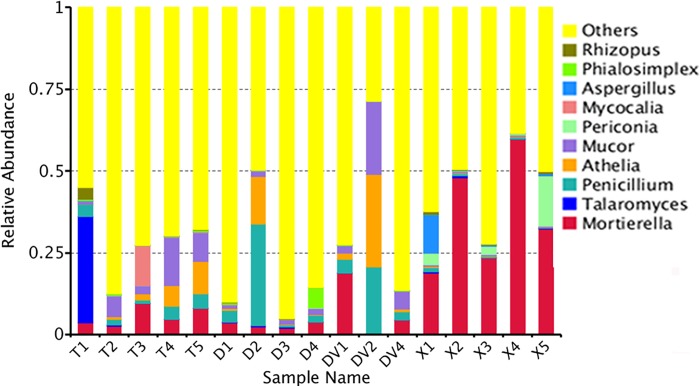
At 97% identity, the fungal libraries yielded 12,266 clusters, also known as OTUs, of which 9198 (75%) were unique, and each soil sample provided an average of 722 OTUs (S1 Fig). For annotation analysis, 39.5–97.5% of fungi were classified into seven taxonomic levels (species, genus, family, order, class, phylum and kingdom) (S2 Fig). The soil fungi were mainly distributed in the Zygomycota (38.3%), Ascomycota (31.3%), Basidiomycota (28.2%) and Chytridiomycota (0.5%) phyla, and the genus-level analysis classified most of the soil fungi as Mortierella, Talaromyces, Penicillium, Athelia and Mucor (Fig 2 and Table 1).
Fig 2. Relative abundance of rhizosphere fungi around Fuzi roots at the genus level.
Table 1. Relative abundance of rhizosphere fungi at the genus level.
| Genus | Relative abundance in rhizosphere soils (%) | |||
|---|---|---|---|---|
| G1 | G2 | G3 | G4 | |
| Mortierella | 0.05627±0.0294 | 0.029317±0.0089 | 0.077484±0.0977 | 0.364652±0.0172 |
| Talaromyces | 0.066607±0.0015 | 0.00192±0.0013 | 0.000812±0.0005 | 0.002565±0.0011 |
| Penicillium | 0.029211±0.0085 | 0.094064±0.0145 | 0.09101±0.0784 | 0.006624±0.0045 |
| Athelia | 0.038747±0.0084 | 0.038703±0.0072 | 0.10287±0.0856 | 0.001846±0.0008 |
| Mucor | 0.066228±0.0554 | 0.015694±0.0044 | 0.100743±0.1006 | 0.003539±0.0005 |
| Periconia | 0.000752±0.0003 | 0.000996±0.0001 | 0.000048±0.0000 | 0.044616±0.0231 |
| Mycocalia | 0.025143±0.0238 | 0.000268±0.0002 | 0.000014±0.0000 | 0.000244±0.0002 |
| Aspergillus | 0.00038±0.0002 | 0.000243±0.0002 | 0.000124±0.0001 | 0.026304±0.0112 |
| Phialosimplex | 0.002945±0.0018 | 0.016984±0.0089 | 0.001149±0.0010 | 0.000219±0.0002 |
| Rhizopus | 0.007578±0.0016 | 0.000294±0.0001 | 0.000076±0.0000 | 0.003705±0.0033 |
| Others | 0.706139±0.1167 | 0.801516±0.2072 | 0.62567±0.3017 | 0.545687±0.1307 |
Fungal diversity of four soil groups
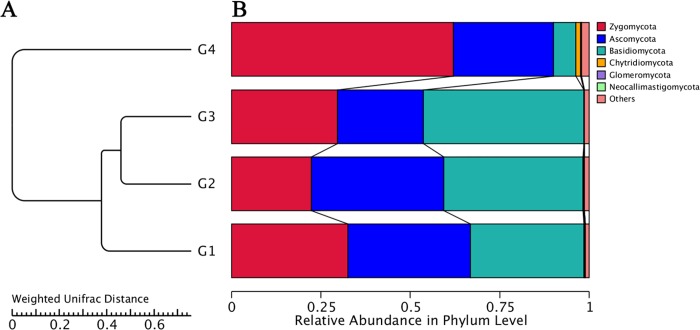
For the diversity analysis, a visualized UPGMA tree was constructed with the fungi in sick (G3) and healthy soils (G1, G2 and G4). The results revealed various diversities at every taxonomic level. At the phylum level, G3 contained the least abundances of Chytridiomycota, Ascomycota and Zygomycota fungi but the highest abundance of Basidiomycota fungi. The fungi in the G4 samples, which were obtained from rot-resistant Fuzi, mostly included Chytridiomycota, Zygomycota and Glomeromycota, with a lower abundance of Basidiomycota (Fig 3).
Fig 3.
UPGMA cluster tree (A) and relative abundance (B) of rhizosphere fungi around Fuzi roots at the phylum level.
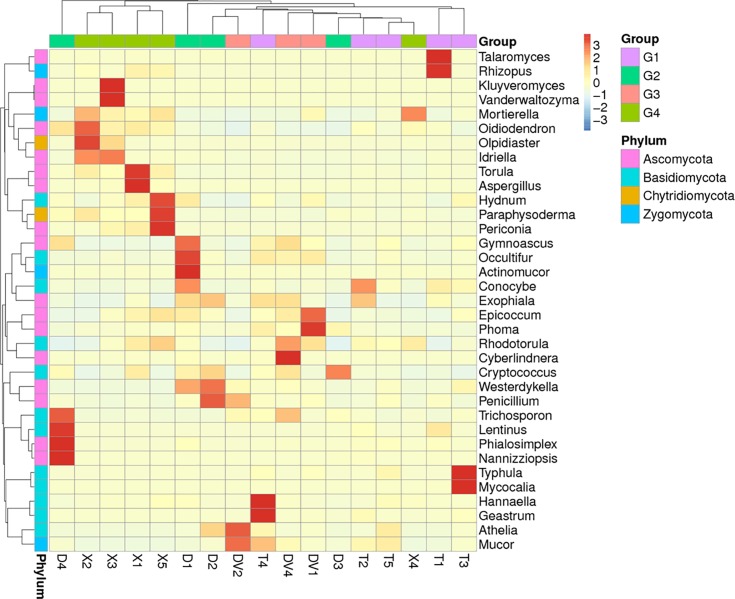
At the genus level, the healthy soils (G1, healthy) showed the highest species richness (Shannon value = 5.32), and the fungal community was mainly composed of Talaromyces (6.66%), Mucor (6.62%), Mortierella (5.63%), and Athelia (3.87%) (Fig 4 and Table 2). The rhizosphere soils (G2 and G3) collected from areas surrounding Da-Hua Fuzi plants shared similar richness values (4.91 and 4.76, respectively) and some common dominant fungi, such as Penicillium (9.4% in G2 and 9.1% in G3). However, the Athelia content in sick soils (G3) contributed 10.3% of the fungal biomass, which is 2.6-fold higher than that found for G2. In addition, the abundances of Mucor and Mortierella in sick soils were 1.6- and 5.41-fold higher than those in G2, respectively (Fig 4 and Table 2). High densities of dominant fungi reduced the biodiversity of G3 in comparison to G2 and G1. Mortierella made up 36.4% of the fungal community in Xiao-Hua Fuzi plants (Fig 4 and Table 2), which decreased the richness value (Shannon value = 4.46) of G4 soils. The results were supported by the PCA results of the four groups of soil fungi (Fig 5). In particular, the PCA results showed that the fungal structure of G4 soils was far from those of G1-G3, echoing the results of the phylum-level diversity analysis of rhizosphere fungi around Fuzi roots (Fig 3). Based on the rot-resistant feature of Xiao-Hua Fuzi, an in-depth study of the fungal community of G4 soils might shed light on the biological prevention and control for Fuzi root-rot disease.
Fig 4. Heat map of the abundance of rhizosphere fungi around Fuzi roots at the genus level.
Table 2. Relative abundance of rhizosphere fungi around Fuzi roots.
| Fungus species | Relative abundance in rhizosphere soils (%) | |||
|---|---|---|---|---|
| G1 | G2 | G3 | G4 | |
| Mortierella alpina | 0.003721±0.0008 | 0.005126±0.0049 | 0.001115±0.0005 | 0.289409±0.1870 |
| Talaromyces purpureogenus | 0.065368±0.0443 | 0.001136±0.0010 | 0.000138±0.0001 | 0.001379±0.0013 |
| Penicillium simplicissimum | 0.024462±0.0145 | 0.086455±0.0778 | 0.073272±0.0680 | 0.004989±0.0038 |
| Athelia rolfsii | 0.038677±0.0221 | 0.038367±0.0209 | 0.102828±0.0956 | 0.001797±0.0008 |
| Mucor racemosus | 0.059967±0.0479 | 0.014398±0.0045 | 0.093584±0.0734 | 0.001925±0.0007 |
| Mortierella chlamydospora | 0.041382±0.0312 | 0.010645±0.0058 | 0.07352±0.0580 | 0.001879±0.0006 |
| Periconia byssoides | 0.000752±0.0003 | 0.000996±0.0006 | 0.000048±0.0000 | 0.044616±0.0331 |
| Mycocalia denudata | 0.025143±0.0138 | 0.000268±0.0002 | 0.000014±0.0000 | 0.000244±0.0002 |
| Aspergillus tamarii | 0.000211±0.0001 | 0.000134±0.0001 | 0.000007±0.0000 | 0.02601±0.0110 |
| Phialosimplex caninus | 0.002912±0.0018 | 0.016984±0.0099 | 0.001149±0.0010 | 0.000219±0.0002 |
| Mortierella ambigua | 0.000756±0.0002 | 0.001482±0.0004 | 0.000213±0.0002 | 0.03273±0.0244 |
| Mortierella oligospora | 0.001161±0.0004 | 0.000661±0.0008 | 0 | 0.026704±0.0222 |
| Rhizopus arrhizus | 0.007529±0.0016 | 0.000263±0.0001 | 0.000076±0.0000 | 0.002817±0.0027 |
| Conocybe macrospora | 0.002577±0.0011 | 0.008151±0.0045 | 0.000255±0.0001 | 0.000496±0.0002 |
| Olpidiaster brassicae | 0.000322±0.0001 | 0.000196±0.0001 | 0.000096±0.0000 | 0.009115±0.0073 |
| Conocybe coprophila | 0.008541±0.0053 | 0.000780±0.0005 | 0.000028±0.0000 | 0.000058±0.0000 |
| Hannaella oryzae | 0.006088±0.0041 | 0.000909±0.0006 | 0.000847±0.0006 | 0.001747±0.0011 |
| Mortierella hypsicladia | 0.005555±0.0020 | 0.008900±0.0017 | 0.002154±0.0011 | 0.000525±0.0004 |
| Mortierella capitata | 0.000376±0.0003 | 0.000160±0.0001 | 0 | 0.007603±0.0034 |
| Kluyveromyces hubeiensis | 0.000045±0.0000 | 0.00001±0.0000 | 0.000014±0.0000 | 0.004328±0.0016 |
| Penicillium oxalicum | 0.000946±0.0005 | 0.001223±0.0010 | 0.006925±0.0018 | 0.001218±0.0010 |
| Lentinus squarrosulus | 0.001627±0.0009 | 0.005152±0.0011 | 0.000021±0.0000 | 0.000021±0.0000 |
| Penicillium solitum | 0.000206±0.0002 | 0.000284±0.0002 | 0.006814±0.0011 | 0.000025±0.0000 |
| Mortierella exigua | 0.000644±0.0002 | 0.000191±0.0000 | 0.000034±0.0000 | 0.005513±0.0039 |
Fig 5. Variation in the fungal composition of the four soil groups determined by PCA.
Candidate pathogens of fuzi root rot disease
To identify pathogens of Fuzi root rot disease, the fungal species enriched in sick soils were targeted. In the rotted Fuzi rhizosphere soils (G3), A. rolfsii, Mucor racemosus, Mortierella chlamydospora and P. simplicissimum were detected at levels higher than 5% (Table 2 and Fig 4). A. rolfsii, also called Sclerotium rolfsii, is a facultative plant pathogen and the causal agent of “southern blight” disease in crops such as potato, soybean, sunflower and some ornamental plants [14, 15, 16]. This pathogen is confined to warm and moist territories (such as Jiangyou in China) with daily average air temperatures of 30–33°C in mid-summer. In this study, the obvious rot morphology of DV2 Fuzi (Fig 1) and its dominant fungus in rhizosphere soil (Fig 4) demonstrated that A. rolfsii is one of the pathogens of Fuzi root rot disease. Mucor racemosus has been reported to be a causative pathogen of soft rot in cherry tomato [17]. Mortierella is ubiquitous in rhizosphere soils from a range of temperate and tropical forests and agricultural habitats [11]. This fungus usually plays a role in the maintenance of the micro-ecological balance by suppressing soil-borne pathogens through competition for nutrients and by assisting host plants with phosphorus/nitrogen absorption. P. simplicissimum is a plant growth-promoting fungus that induces plant resistance through the activation of multiple defence signals [18].
Some fungi that were not identified as the main constituent species in sick soils (G3) were nevertheless found to be enriched in these soils compared with healthy soils. For example, Phoma adonidicola and Epicoccum nigrum in DV1 and Cyberlindnera saturnus and Rhodotorula ingeniosa in DV4 are shown in Fig 4. Many fungi belonging to the Phoma genus are the causal agents of brown root rot, which causes severe root lesions in alfalfa, soybean and other perennial forage legumes [14]. This phenomenon might also be true for Fuzi plants, whose rotted-root annexed soil (DV1) had 20- to 40-fold higher amounts of P. adonidicola than healthy soils (T1-T5). It is noteworthy that the other three fungi that were enriched in sick soils appear to play competitive and antagonistic roles against the rot pathogens in Fuzi. A few E. nigrum isolates have been reported to significantly reduce the radial growth of both Pythium debaryanum and P. ultimum [19]. A Cyberlindnera fungus has been proven to exhibit antagonistic activities against a range of pathogenic and saprophytic filamentous fungi [20]. Additionally, Rhodotorula can protect sugar beets against the soil pathogen Rhizoctonia solani [21, 22]. Thus, these fungi might have potential as a novel bio-control agent against Fuzi pathogenic fungi.
These findings suggest that A. rolfsii and Mucor racemosus, as the dominant pathogens, play important roles in the invasion of Fuzi tissue and the induction of root rot symptoms, and increases in the P. adonidicola abundance would worsen the disease. In contrast, M. chlamydospora, P. simplicissimum, E. nigrum, C. saturnus and R. ingeniosa might antagonize the pathogens in sick soils around Fuzi roots.
Separation and identification of the root rot pathogen
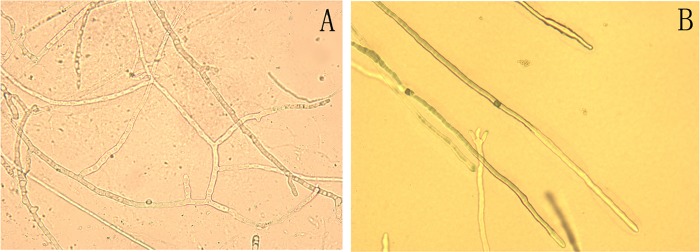
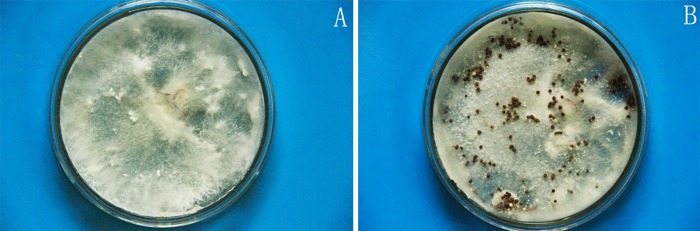
In this study, the rotten tissue of diseased Da-Hua Fuzi plants was used for pathogen isolation, and one strain of white fungus was obtained after purification. Its white, slender and crossed hyphae spread rapidly in the PDA medium, as a rate of approximately 2–3 cm per day (Fig 6). The fungal morphology observed under a microscope closely matched that observed in other studies [23]. After one week, brown and black sclerotia appeared in the culture dishes, and the hyphae showed slight shrinkage (Fig 7).
Fig 6. Hyphal morphology of suspected pathogens causing Fuzi root rot disease.
Fig 7.
Cultivation of suspected pathogenic fungus (A) and its sclerotium (B) in PDA medium.
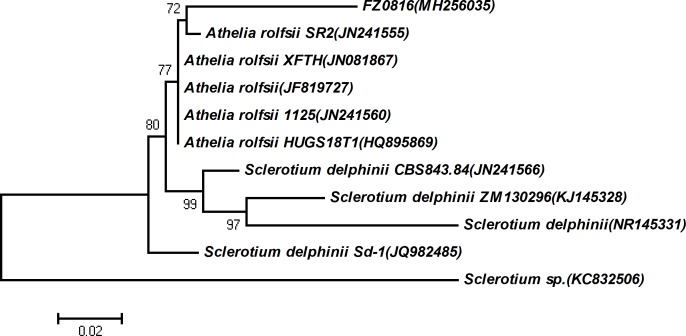
In addition to morphological verification, the ITS barcode method was used for the identification of fungal species. The ITS fragment of fungus isolated from rotten Fuzi plants was found to be highly similar to sequences of known A. rolfsii strains in the NCBI database (S3 Fig). Then, the construction of a phylogenetic tree revealed that the fungus was closely related with other strains of A. rolfsii (Fig 8), indicating that it differs from known strains. These results verified previous inferences that A. rolfsii is the main pathogen causing Fuzi root rot disease. The newly discovered A. rolfsii was named FZ0816 and submitted to the NCBI database under Accession Number MH256035.
Fig 8. Phylogenetic tree of A. rolfsii strains.
Fuzi Infection by the suspected pathogen
To detect the invasion effects of the suspected pathogenic fungi FZ0816, fresh blocks of healthy Fuzi tissue were inoculated onto candidate mycelia at 32°C in the dark. After three days of fungal growth, its white hyphae became attached to the Fuzi tissue surface, and the tissue blocks began to rot and give off a bad smell. All the symptoms of the tested Fuzi tissues were extremely similar to those of diseased Da-Hua Fuzi plants in the wild. These results verify previous inferences that A. rolfsii is the main causative pathogen of Fuzi root rot disease.
Supporting information
(TIF)
(TIF)
(TIF)
(XLSX)
Acknowledgments
This work was supported by the Key Research Project of the National Natural Science Foundation of China (81630101), the National Development and Reform Commission standardization project (ZYBZH-C-SC-51), the Comprehensive Development and Regional Development of Herbal Medicine in Sichuan Provincial Administration of Traditional Chinese Medicine (2016ZY008), and Scientific Research Funds of Chengdu University of Traditional Chinese Medicine (ZRYY1612 and 030029050).
Abbreviations
- TCM
typical traditional Chinese medicine
- ITS
internal transcribed spacer
- OTUs
operational taxonomic units
- GAP
Good Agricultural Practice of Medicinal Plants and Animals
- UPGMA
Unweighted Pair-group Method with Arithmetic Means
- NJ
Neighbour-Joining
Data Availability
Data underlying the study are available as follows. 17 soil replicates are frozen in the State Bank of Chinese Drug Germplam Resources, which is located in Chengdu university of Traditional Chinese Medicine, No. 1166 Liu Tai Road, Wenjiang District, Chengdu (http://grb.cdutcm.edu.cn/). High-throughput sequencing data is uploaded to the NCBI dababase with the number SRP142412. In addition, in our study, a new strain of A. rolfsii is named as FZ0816, and is also submitted to NCBI database with the number MH256035.
Funding Statement
This work was supported by Key Research Project of National Natural Science Foundation of China (81630101), National Development and Reform Commission standardization project (ZYBZH-C-SC-51), Comprehensive development and regional development of herbal medicine in Sichuan Provincial Administration of traditional Chinese Medicine (2016ZY008), Scientific Research Funds of Chengdu University of Traditional Chinese Medicine (ZRYY1612 and 030029050). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Singhuber J, Zhu M, Prinz S, Kopp B. Aconitum in traditional Chinese medicine: a valuable drug or an unpredictable risk? J Ethnopharmacol. 2009;126(1):18–30. 10.1016/j.jep.2009.07.031 [DOI] [PubMed] [Google Scholar]
- 2.Tai CJ, El-Shazly M, Wu TY, Lee KT, Csupor D, Hohmann J, et al. Clinical Aspects of Aconitum Preparations. Planta Med. 2015;81(12–13):1017–28. 10.1055/s-0035-1546183 [DOI] [PubMed] [Google Scholar]
- 3.Zhou G, Tang L, Zhou X, Wang T, Kou Z, Wang Z. A review on phytochemistry and pharmacological activities of the processed lateral root of Aconitum carmichaelii Debeaux. J Ethnopharmacol. 2015;160:173–93. 10.1016/j.jep.2014.11.043 [DOI] [PubMed] [Google Scholar]
- 4.Ghosh R, Tarafdar A, Sharma M. Rapid and sensitive diagnoses of dry root rot pathogen of chickpea (Rhizoctonia bataticola (Taub.) Butler) using loop-mediated isothermal amplification assay. Sci Rep. 2017;7:42737 10.1038/srep42737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaya J, Marhuenda FC, Pascual JA, Ros M. Microbiota Characterization of Compost Using Omics Approaches Opens New Perspectives for Phytophthora Root Rot Control. PLoS One. 2016;11(8):e0158048 10.1371/journal.pone.0158048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song JY, Seo MW, Kim SI, Nam MH, Lim HS, Kim HG. Genetic Diversity and Pathogenicity of Cylindrocarpon destructans Isolates Obtained from Korean Panax ginseng. Mycobiology. 2014;42(2):174–80. 10.5941/MYCO.2014.42.2.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A, Pandey V, Singh M, Pandey D, Saharan MS, Marla SS. Draft genome sequence of Karnal bunt pathogen (Tilletia indica) of wheat provides insights into the pathogenic mechanisms of quarantined fungus. PLoS One. 2017;12(2):e0171323 10.1371/journal.pone.0171323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaya J, Lacasa C, Lacasa A, Martinez V, Santisima-Trinidad AB, Pascual JA, et al. Characterization of Phytophthora nicotianae isolates in southeast Spain and their detection and quantification through a real-time TaqMan PCR. J Sci Food Agric. 2015;95(11):2354 10.1002/jsfa.7300 [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, He X, Wu B, Long Q, Shao T, Wang Z, et al. Time-Course Transcriptome Analysis Reveals Resistance Genes of Panax ginseng Induced by Cylindrocarpon destructans Infection Using RNA-Seq. PLoS One. 2016;11(2):e0149408 10.1371/journal.pone.0149408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halleen F, Schroers HJ, Groenewald JZ, Rego C, Oliveira H, Crous PW. Neonectria liriodendri sp. nov., the main causal agent of black foot disease of grapevines. Stud Mycol. 2006;55:227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao CP, Mi QL, Qiao XG, Zheng YK, Chen YW, Xu LH, et al. Rhizospheric fungi of Panax notoginseng: diversity and antagonism to host phytopathogens. J Ginseng Res. 2016;40(2):127–34. Epub 2016/05/10. 10.1016/j.jgr.2015.06.004 ; PubMed Central PMCID: PMC4845048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeon CS, Kim GH, Son KI, Hur JS, Jeon KS, Yoon JH, et al. Root Rot of Balloon Flower (Platycodon grandiflorum) Caused by Fusarium solani and Fusarium oxysporum. Plant Pathol J. 2013;29(4):440–5. 10.5423/PPJ.NT.07.2013.0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrego-Benjumea A, Basallote-Ureba MJ, Melero-Vara JM, Abbasi PA. Characterization of Fusarium isolates from asparagus fields in southwestern Ontario and influence of soil organic amendments on Fusarium crown and root rot. Phytopathology. 2014;104(4):403–15. 10.1094/PHYTO-08-13-0231-R [DOI] [PubMed] [Google Scholar]
- 14.Wunsch MJ, Bergstrom GC. Genetic and morphological evidence that Phoma sclerotioides, causal agent of brown root rot of alfalfa, is composed of a species complex. Phytopathology. 2011;101(5):594–610. 10.1094/PHYTO-04-10-0107 [DOI] [PubMed] [Google Scholar]
- 15.Pastor N, Carlier E, Andres J, Rosas SB, Rovera M. Characterization of rhizosphere bacteria for control of phytopathogenic fungi of tomato. J Environ Manage. 2012;95 Suppl:S332–7. [DOI] [PubMed] [Google Scholar]
- 16.Adhikari BN, Hamilton JP, Zerillo MM, Tisserat N, Levesque CA, Buell CR. Comparative genomics reveals insight into virulence strategies of plant pathogenic oomycetes. PLoS One. 2013;8(10):e75072 Epub 2013/10/15. 10.1371/journal.pone.0075072 ; PubMed Central PMCID: PMC3790786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon JH, Hong SB. Soft Rot of Tomato Caused by Mucor racemosus in Korea. Mycobiology. 2005;33(4):240–2. 10.4489/MYCO.2005.33.4.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hossain MM, Sultana F, Kubota M, Koyama H, Hyakumachi M. The plant growth-promoting fungus Penicillium simplicissimum GP17-2 induces resistance in Arabidopsis thaliana by activation of multiple defense signals. Plant Cell Physiol. 2007;48(12):1724–36. 10.1093/pcp/pcm144 [DOI] [PubMed] [Google Scholar]
- 19.Larena I, De Cal A, Melgarejo P. Solid substrate production of Epicoccum nigrum conidia for biological control of brown rot on stone fruits. Int J Food Microbiol. 2004;94(2):161–7. 10.1016/j.ijfoodmicro.2004.01.007 [DOI] [PubMed] [Google Scholar]
- 20.Koutb M, Ali EH. Potential of Epicoccum purpurascens Strain 5615 AUMC as a Biocontrol Agent of Pythium irregulare Root Rot in Three Leguminous Plants. Mycobiology. 2010;38(4):286–94. 10.4489/MYCO.2010.38.4.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Tarabily KA. Suppression of Rhizoctonia solani diseases of sugar beet by antagonistic and plant growth-promoting yeasts. J Appl Microbiol. 2004;96(1):69–75. [DOI] [PubMed] [Google Scholar]
- 22.Hilber-Bodmer M, Schmid M, Ahrens CH, Freimoser FM. Competition assays and physiological experiments of soil and phyllosphere yeasts identify Candida subhashii as a novel antagonist of filamentous fungi. BMC Microbiol. 2017;17(1):4 10.1186/s12866-016-0908-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maseko B, Burgess TI, Coutinho TA, Wingfield MJ. Two new Phytophthora species from South African Eucalyptus plantations. Mycol Res. 2007;111(Pt 11):1321–38. 10.1016/j.mycres.2007.08.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(XLSX)
Data Availability Statement
Data underlying the study are available as follows. 17 soil replicates are frozen in the State Bank of Chinese Drug Germplam Resources, which is located in Chengdu university of Traditional Chinese Medicine, No. 1166 Liu Tai Road, Wenjiang District, Chengdu (http://grb.cdutcm.edu.cn/). High-throughput sequencing data is uploaded to the NCBI dababase with the number SRP142412. In addition, in our study, a new strain of A. rolfsii is named as FZ0816, and is also submitted to NCBI database with the number MH256035.