Abstract
Macrophages represent one of the most numerous and diverse leukocyte types in the body. Furthermore, they are important regulators and promoters of many cardiovascular disease programs. Their functions range from sensing pathogens, digesting cell debris, modulating inflammation and producing key cytokines and other regulatory factors throughout the body. Macrophage research has undergone a renaissance in recent years, which has propelled a newfound interest in their heterogeneity as well as a new understanding of ontological differences in their development. In addition, recent technological advances such as single-cell mass-cytometry by time-of-flight (CyTOF) have enabled phenotype and functional analyses of individual immune myeloid cells, including macrophages, at unprecedented resolution. In this part 1 of a 4-part review series covering the macrophage in cardiovascular disease, we focus on the basic principles of macrophage development, heterogeneity, phenotype, tissue-specific differentiation and functionality as a basis to understand their role in cardiovascular disease.
Keywords: Macrophage, Cardiovascular, Atherosclerosis
Condensed Abstract:
Macrophages represent one of the most numerous and diverse leukocyte types in the body. Furthermore, they are important regulators and promoters of many cardiovascular disease programs. Macrophage research has undergone a renaissance in recent years, which has propelled a newfound interest in their biology and development. In addition, technological advances have enabled phenotype and functional analyses of immune myeloid cells, including macrophages, at unprecedented resolution. In this first part of a 4-part review series we focus on the basic principles of macrophage development, heterogeneity, phenotype, tissue-specific differentiation and functionality as a basis to understand their role in cardiovascular disease.
Introduction
Macrophages are among the most numerous and diverse leukocytes in the body. Their functions range from sensing pathogens, digesting cell debris, and being major producers of key cytokines and other regulatory factors throughout the body. Furthermore, macrophages are critical players in homeostasis and disease, and a vast amount of research has gone into unravelling their biology and contributions to various cardiovascular disease processes.
In this 4-part review series titled “The Macrophage in Cardiovascular Disease”, we cover and review the full breadth of macrophage biology and the cardiovascular system. Each of the 4 articles was written by a dedicated authorship team with acknowledged expertise in this field. This review series should serve as a benchmark and resource for the field moving forwards. A brief outline of the complete 4-part review series is provided in Table 1. Here, in part 1 of this series we cover basic macrophage biology, classification and emerging insights into the macrophage phenotype using advanced profiling techniques.
Table 1.
Overview of 4-part review series The Macrophage in Cardiovascular Disease
| Part 1: Macrophage Biology, Classification and Phenotype |
| Part 2: The Macrophage in Atherosclerosis - Trafficking, Inflammatory Resolution and Genomics |
| Part 3: Macrophage and Monocyte Dynamics in the Cardiovascular System - From Biology to Imaging |
| Part 4: The Macrophage and Cardiac Disease |
Macrophage Biology, Origins and Classification
Macrophage lineage development
Scientific background and historic perspective
Since their discovery in 1882, macrophages have been a favorite cell for biologists, likely a result of their ease of isolation, culturing, and diverse functional repertoire. An early pioneer of immunity, Eli Mechnikov, discovered and named macrophages, and did many of his seminal studies on these cells, which fostered the dawn of innate immunity and co-awarding of the 1908 Nobel Prize in Physiology or Medicine with Paul Ehrlich. Even with over a century of study, however, macrophage functions in homeostasis and disease remain to be fully understood. This is largely due to their immense diversity, plasticity, and presence in almost all tissues and disease systems. Furthermore, they possess an almost paradoxical behavior as an anti-inflammatory mediator of tissue repair but yet being pro-inflammatory cells in models of infection or inflammation. In many scenarios these phenotypes can be observed overlapping within the same tissue during processes of inflammation, leading to resolution.
Many cardiovascular diseases, including in particular atherosclerosis, have been recognized as inflammatory conditions characterized by the infiltration of monocytes and macrophage differentiation to promote localized inflammation (1). In atherosclerosis, scavenger receptor expressing foamy macrophages are particularly relevant hallmarks of overall disease burden and functionally connect to disease pathogenesis (2). Beginning with these key historical observations and moving into current literature, it is clear that the basic biology of macrophages must be fully understood in order to appreciate the roles these cells play in cardiovascular disease settings. The goal of this section in this macrophage review series is to highlight that background for the reader.
Yolk-sac, fetal liver, and bone marrow tracking of macrophage development
Modern myeloid lineage cell nomenclature derives from a proposal set forth in 1972 by Van Furth and colleagues, termed the mononuclear phagocyte system (3). This model proposed a classification system and basic developmental pathway for monocytes and macrophages. However, many key adjustments to this model have since been made with regard to the developmental origins of macrophages (4,5). Until the last decade, the concept that macrophages in adult organisms were replenished solely by monocytes was dominant. That is, macrophage replenishment was thought to occur by monocytes exiting from the circulation and undergoing a differentiation pathway in the tissues. While this key function of monocytes remains relevant (review of monocytes (6)), new technologies and in-depth analyses of macrophage development in the embryo have refined the historical paradigm that adult hematopoietic stem cell (HSC)derived monocytes are the primary or even sole source for macrophage differentiation.
Early observations dating back almost 30 years identified that macrophages exist in the embryonic yolk-sac prior to the formation of HSCs or monocyte detection (7–10). These observations led to the proposal, unpopular at the time, that embryonic derived macrophages were likely to contribute to the adult pool of macrophages (11). Unique embryonic progenitor cells are commonly described in ‘developmental waves’ that transiently populate tissues during embryogenesis. Importantly, progenitor cells from these distinct stages cannot be detected in the adult animal outside of the small windows in which they seed tissues with macrophages and other cell types. As depicted in the Central Illustration, the earliest wave leading to macrophage differentiation in the mouse begins at approximately embryonic day 7.0 (E7.0) observed in blood islands present in the yolk sac (12–14). At this time, these cells are derived from mesoderm and the progenitor cells contribute to a pool of macrophages, as well as erythroblasts and megakaryocytes (12,15). Unlike traditional bone marrow HSCs, these early macrophages are Myb-independent (16). The next wave from the yolk sac can be detected by E8.0–8.5 where the so-called erythro-myeloid precursor cells (EMP), can be identified and display myeloid cell potential and differentiation capabilities (17,18). Through a poorly understood mechanism, E9 EMPs can traffic through the newly developing blood vasculature to populate additional embryonic tissues (19) and transition to the fetal liver, where they subsequently are able to differentiate into monocytes, closely resembling monocytes from adult bone marrow HSCs. The fetal liver contains monocyte-progenitor cells until the development of bone marrow and these are considered the major source for seeding many tissue macrophage populations approximately within E11–17.5 (20). Current consensus is that these embryonic waves of progenitor cells contribute to origins of resident macrophages. However, there remains limited molecular understanding of early macrophage developmental pathways, and controversy remains with regard to the identification of key progenitor cells for specific macrophage tissue differentiation at early embryonic stages.
Central Illustration. Macrophage developmental origin and maintenance.
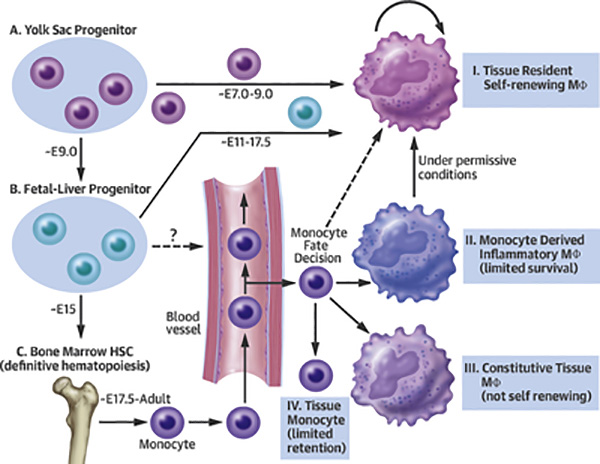
Macrophages are seeded within tissues during discrete time periods of embryogenesis. These stages can been separated into three distinct origins, (A) yolk sac, (B) fetal liver, and (C) bone marrow. Each stage is capable of seeding tissue-resident macrophage populations, over the developmental time points shown. Over embryonic day E7.0–9.0 macrophages are seeded from yolk sac precursors, from ~E11–17.5 monocyte-like cells are capable of seeding macrophages from the fetal liver, and from ~E17.5 through adulthood mature monocytes from bone marrow are able to seed a variety of macrophage lineages in the tissues. Displayed are four types of macrophage populations that can be found in tissues during steady or diseased states; I. tissue resident macrophage, which are the primary macrophage population found in most tissues, II. monocyte-derived inflammatory macrophage, which are typically expanded during injury, III. constitutive tissue macrophages deriving exclusively from monocytes, without possessing the ability to proliferate in tissues, and IV. monocytes migrating through tissues. Mȹ = macrophage
Macrophages and lineage tracking in the adult
The use of parabiosis - a surgical technique that links the microvasculature between two adult animals - to track the contributions and fate of circulating cells revealed that blood-borne cells make only marginal contributions to macrophage populations during steady state homeostasis (21–25). This approach, coupled with fate-mapping studies that have utilized inducible gene expression systems at discrete developmental time points to later detect cells in adult mice that derive from embryonically labeled cells, shows that adult monocytes are dispensable for the maintenance of many resident tissue macrophages. Given the diverse use of different genetic labeling approaches, including the promoters of Tie2, Runx, Cx3cr1, cKit, Cd115, and Flt3 as genetic drivers of the labeling, it is not surprising that there remain some minor discrepancies over detail that have been difficult to resolve (16,20,22,26–28). In-depth analysis of fate-mapping or lineage-tracking studies and discussion of gaps in our understanding of early macrophage differentiation were recently expertly reviewed (29,30). Although this work in mice is exciting and transformative, approaches to characterize human macrophage ontogeny have yet to be developed. Analogous human yolk sac progenitor populations were recently observed in fetal material collected from week 9 gestation samples, suggesting potential degrees of similarity between mouse and human (19). However, as anticipated, more studies will need to be completed to determine whether the features of murine macrophage development translate to human macrophage populations.
In the adult mouse, HSCs primarily reside in the bone marrow where they can differentiate into monocytes as the primary external source of macrophages for tissues. In some tissues, like the gut lamina propria, a subset of postpartum macrophages are continuously replenished from blood monocyte precursors (31,32), while others are long-lived and embryonically derived (33). There are two primary types of monocytes: classical (Ly6c+) monocytes, which derive from bone marrow precursors that use CCR2 to gain access to blood (5,28,34,35), and nonclassical (Ly6c−) monocytes that arise from classical monocytes (28,34). In humans, classical monocytes represent the major population in the circulating blood (~95%), whereas in mice they are more evenly mixed with classical monocytes representing 50–60% of the monocyte pool in normal blood. Using a heavy water labeling approach, human classical monocytes were found to have a half-life of approximately 1 day in the circulation; these cells enter tissues, die, or mature into nonclassical monocytes. Human nonclassical monocytes were found to have an extended half-life of approximately 7 days (36). Functionally, classical and nonclassical monocytes possess unique qualities. Classical monocytes are typically associated with recruitment into tissues in response to insults, although they do circulate through tissues in the steady state (25), whereas nonclassical monocytes patrol the endothelium to promote vascular health (37–41). Even with the known roles of monocytes expanding, monocytes likely remain an important contributor of macrophage homeostasis in tissues. The Central Illustration summarizes some of the known aspects of the origins of tissue macrophages and routes of differentiation that bone marrow derived monocytes may contribute to under different steady state or inflammatory conditions.
Tissue resident macrophage phenotypes
What defines and constitutes macrophage residency in tissue?
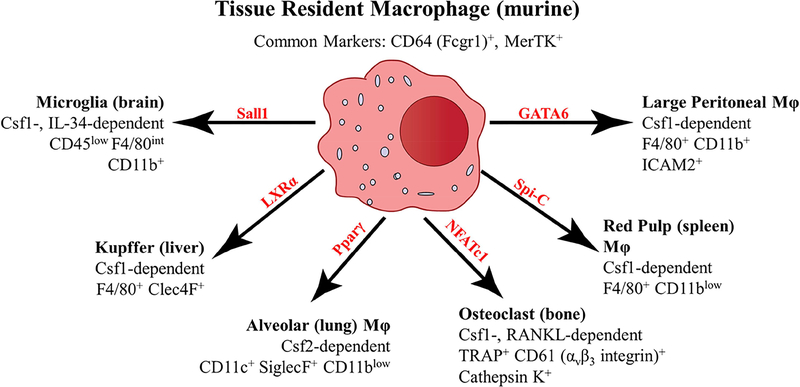
While our understanding of the ontological origins of resident macrophages has undergone a transformation in recent years, understanding what defines and constitutes macrophage residency in tissue has also progressed enormously and may be more informative for understanding their influence on disease. Macrophages have long been appreciated to have tissue-specific heterogeneity, suggesting that in each tissue macrophages take up unique functional tasks, while maintaining a general macrophage phenotype (42,43). Generating and using data from the Immgen Consortium project (44,45), Gautier et al. analyzed tissue resident macrophages from different organs to identify conserved and unique gene expression data associated with tissue macrophages (46). Through this approach, a set of shared macrophage genes across all organs was identified (including Fcgr1, Mertk, and Cd14), as well as the identification of tissue-specific gene signatures uniquely associated with each mature resident macrophage subset analyzed (46). Surprisingly, transcription factors identified prior to or as a result of this gene expression analysis led to the identification of tissue-specific regulators for maintenance of resident macrophages in specific organs, without affecting tissue macrophages in other organs. Figure 1 illustrates murine tissue resident macrophage populations from selected tissues with known transcriptional regulators unique to each given tissue, as well as unique cytokines needed for tissue resident macrophage survival. In addition, Figure 1 indicates some specific molecules for the identification of each tissue resident macrophage population.
Figure 1. Tissue resident macrophage heterogeneity and identity.
Macrophages share expression of core genes between all tissue resident populations, such as CD64 and MerTK (46). However, tissue macrophages possess niche-specific dependence on transcriptional and survival factors for their given microenvironment. Microglia (102,103), Kupffer (104), alveolar (105–107), osteoclast (108,109), red pulp (110), and large peritoneal macrophages (111–113) are displayed as representative illustrations of tissue resident macrophages that are dependent on unique transcription factors (displayed in red) for differentiation, and growth factors (Csf1 (MCSF), Csf2 (GM-CSF), IL-34, or RANKL (TNFSF11)) for survival. In addition to the shared CD64 and MerTK macrophage markers, specific tissue resident markers are displayed for identification of these unique macrophage populations.
Thus, while in some studies, ‘tissue resident macrophage’ refers to macrophages deriving from embryonic origins, it is increasingly clear that the true definition of the tissue resident macrophage is one that acquires the expression of genes that are unique to macrophages in the given organ. During inflammation, monocytes are able to differentiate to macrophages (cells expressing Mertk and other canonical macrophage markers), without necessarily turning on resident macrophage genes (47). In addition, to be called a resident macrophage, typically there is an intrinsic ability for self-maintenance through proliferation. Shortly following the recent discovery that many resident macrophage pools were derived from embryonic precursors, the idea developed that the adult bone marrow monocyte could not repopulate the resident macrophage that was derived from embryonic precursors. However, recent studies reveal that blood monocytes, in contrast to already mature macrophages from other tissues, can indeed fill open niches to become full resident macrophages (48).
Indeed, Guilliams now proposes that monocytes are perfectly capable of becoming resident macrophages but that doing so requires that there is an open niche available (49). In an extremely interesting progression of concept and data, Perlman et al. showed that monocytes recruited in inflammation indeed have the capacity to become lung resident alveolar macrophages (50). However, the process of full differentiation is rather slow, and remarkably, intermediate stages of differentiation lead to macrophages that drive disease pathology (eg., lung fibrosis in a bleomycin model) (50). This very new concept, which states that macrophage differentiation from monocytes is a slow, progressive event that involves pro-inflammatory (and pro-pathological) intermediate steps, allows for a re-examination of past studies in a new light. It may still be the case that proliferating resident macrophages indeed have a greater ability to replenish the resident macrophage niche than recruited cells when both are available (24), but under circumstances in which the original resident pool is partially or totally eliminated there is a reliance on the blood monocyte for replenishing resident macrophages (47). That reliance may come at a cost involving increased inflammation.
It has been thought that tissue resident macrophages are often complemented by a minor non-resident monocyte-derived macrophage population that requires constant replenishment for maintenance (37). However, while there are often multiple populations of resident macrophages in a single organ, there is confusion about their origins, and there may be differences between organs. For instance, lung interstitial or small peritoneal macrophages, and those expressing CD11b in particular, are often not considered tissue resident even though they can be defined by a unique gene expression signature. In fact, their turnover is as slow as alveolar macrophages and all evidence points to the fact that they are not replenished by monocytes on an ongoing basis (25,51). However, in the peritoneal cavity, the quantitatively minor resident macrophages (socalled ‘small macrophages’) are monocyte-derived and require constant replacement (52).
Additional resident macrophage populations that derive from embryonic origins show a slow but steady replacement over time by monocyte-derived macrophages. For example, peritoneal macrophages show replacement within weeks of birth and slowly expand the “monocyte-derived” pool of resident cells with aging (21). In another model, macrophages found in the aortic adventitia were shown to display embryonic origins, but required a large influx of monocyte-derived cells following birth (53). Whether this low-grade replacement occurs as a result of insults or even microbe exposure has yet to be exhaustively approached. Nevertheless, it appears that these tissue-resident gene signatures and functions are products of the microenvironments in which these cells reside (54,55).
Human tissue resident macrophages have also been shown to possess intrinsic abilities to maintain themselves independent of circulating precursor cells. These studies have largely been performed through allogenic transplantation. Specifically, in models of HSC transplantation where graft versus host disease (GVHD) was evident, prolonged survival of recipient dermal macrophage populations occurred in GVHD lesions, whereas other myeloid populations such as DCs were completely replaced by donor cells (56). Additionally, human alveolar macrophages were identified and even showed signs of proliferation in the lungs at time points greater than one year post transplantation (57,58). In hand allograft transplantation, donor Langerhans cells in the epidermis persisted even 4 years post transplantation (59). Less is known about human macrophage heterogeneity, and whole-body wide gene expression analysis is needed. However, elegant in vitro studies have been performed to identify gene signatures to help define macrophages. Xue et al. used genetic approaches in combination with a multitude of activation scenarios to develop a gene signature to identify and distinguish macrophages from monocytes and dendritic cells in humans. Human macrophages were defined by high level expression of CD14, MerTK, CD64 (FCGR1), CD32 (FCGR2A), and CD13 (ANPEP) (60). Some of these genes were coexpressed by dendritic cells and monocytes. However macrophages in all examined conditions maintained high expression of all these markers. Overall, even with limited studies, these human data support the concepts developed from murine macrophage research of tissue residency and shared gene expression signatures across multiple macrophage lineages.
Tissue resident macrophages and their microenvironment
Education by the microenvironment can be observed by chromatin remodeling that occurs to help specialize macrophages to their tissue of residence (54,55). In one experiment, following transfer into the lungs of mice, resident peritoneal macrophages were shown to adopt a gene expression signature resembling alveolar macrophages, leading to the argument that even after differentiation to a mature tissue resident macrophage population, a high degree of plasticity remains (55). However, this argument was largely debunked by Guilliams and colleagues, who repeated the experiment and showed that such adoptive transfer could not reverse lethal proteinosis, whereas adoptive transfer of monocytes was able to generate lung macrophages sufficiently well to reverse proteinosis (48). Thus, the plasticity of fully mature resident macrophages remains elusive.
Functional properties associated with macrophage origins, and how this interacts with microenvironmental influences, remains a topic of great controversy and waits to be thoroughly vetted. Thus far, it appears as though only some resident macrophages of embryonic-origin possess intrinsic functional abilities, while others can be repopulated by monocytes and replace the embryonic derived population without detrimental outcomes. Studies of liver Kupffer cells and lung alveolar macrophages showed minimal changes in gene expression associated with embryonic and monocyte origin cells (48,61,62). However, it is plausible that epigenetic changes, as a result of origins, could influence their ability to respond to microenvironmental cues or to react to an insult. Recruited monocytes that differentiated into alveolar macrophages were hyperinflammatory following challenge compared to their embryonic origin counterparts (63). Importantly, it was found that differing origins of pancreatic macrophages contributed to the degree of protection from tumors, such that while monocyte-derived macrophages were involved with antigen presentation, embryonically derived macrophages were pro-fibrotic (64). However, one problem with these studies using RNA-Seq (RNA sequencing) analyses that were performed between recruited and resident populations is that temporal resolution is limited, such that the recruited population is likely observed at multiple states of differentiation and, unless examined by single-cell RNA-Seq (scRNA-seq), the reported gene expression differences may have more to do with differentiation status than with authentic end-stage differences. It remains to be seen whether resident macrophages in cardiovascular tissues contribute to pathogenesis or have primarily protective roles in models of cardiovascular disease.
Macrophage activation states and innate immune memory
Macrophages are the first line of immune defense in the body, and they are noted for often initiating an appropriate and measured immune response through a diverse array of bacterial and viral sensing receptors. As such, macrophages maintain a high degree of functional flexibility to effectively respond to the diverse array of agents they may encounter. In response to infection or environmental cues through surface receptors, macrophages are able to adjust gene expression profiles and function to address new challenges (reviewed (42,65)). Historically, this feature of macrophage-acquired cellular resistance, including persistence of such resistance, was identified as early as the 1960’s (66,67). Over time, it has become clear that major patterns of macrophage activation exist to give rise to distinct functional states, including states frequently referred to as M1 and M2 modes of activation, which are readily observed after Interferongamma or IL-4 exposure, respectively (68), but which can be overlaid with a wide variety of other stimuli, including antibody engagement via Fc Receptors that can affect the duration and character of the response. Indeed, these stimuli and the macrophage response to them can be of such a significant magnitude and duration that it is reasonable to consider the activation states to possess ‘memory-like’ or ‘trained’ phenotypes that allow the macrophage to be better prepared for future insults (69).
This concept of “trained” innate immunity refers to the ability of the innate immune compartment to adjust its responsiveness to previous exposures (reviewed (70)). Unlike the adaptive immune system where T cells with a TCR-specificity for a given target are preferentially selected and develop into memory cells, the innate compartment utilizes changes in chromatin structure and polarity of tissue macrophages to become better sensors and responders to previously experienced insults. The class of RNAs that post-transcriptionally control gene expression, microRNAs, also regulates trained immunity in macrophages (69).
Trained immunity in macrophages can be seen in a variety of models. For example, in vitro cultured macrophages that were previously activated through granulocyte-macrophage colony-stimulating factor (GM-CSF) show elevated Dectin-1 expression compared to macrophages previously challenged with lipopolysaccharide (71). Similar observations in shifts of epigenetic and transcriptional landscapes were found in human monocyte-derived macrophages in response to activation with lipopolysaccharide or Beta-glucan (72). Finally, protection derived from trained immunity was demonstrated in vivo using models of lethal Candida albicans infection (72). Further studies are needed to better understand the long-term consequences that previous stimulus exposure may have on macrophages within tissues, and these studies may prove valuable in the development of therapeutic interventions.
Deep Profiling Approaches to Study Macrophage Biology
Emerging insights into macrophage heterogeneity using single-cell approaches
As discussed in the preceding section, the myeloid cell system is highly heterogeneous. Even small differences between individual monocyte and macrophage cells may be important in orchestrating both physiologic and pathologic immune responses, and some differences may be regulated post-transcriptionally by microRNAs, protein turnover, and other processes like autophagy or phagocytosis. Therefore, there is the urgent need to dissect this diversity according to ontogeny, phenotype, and distribution of macrophages across tissues. Recent technological advances have been highly successful and enabled single-cell analyses of phenotype and function of immune myeloid cells, including macrophages, at unprecedented resolution, revealing a significant variation of the myeloid system both in health and disease (73–76). Before moving into recent advances in our understanding of the myeloid compartment, we will provide essential information on single-cell mass cytometry and scRNA-seq to guide a comprehensive understanding of these studies.
Single-cell mass-cytometry by time-of-flight (CyTOF) and single-cell RNA sequencing
Since its introduction in the late 1960s, flow-cytometry has been widely used and traditionally represented the “gold standard” technology to study the immune system with singlecell resolution. Despite technical advances that allow the use of 18 markers per single-cell, most flow-cytometry experiments are generally limited to 6–10 single-cell parameters to avoid erroneous interpretation of data and ensure efficiency (77).
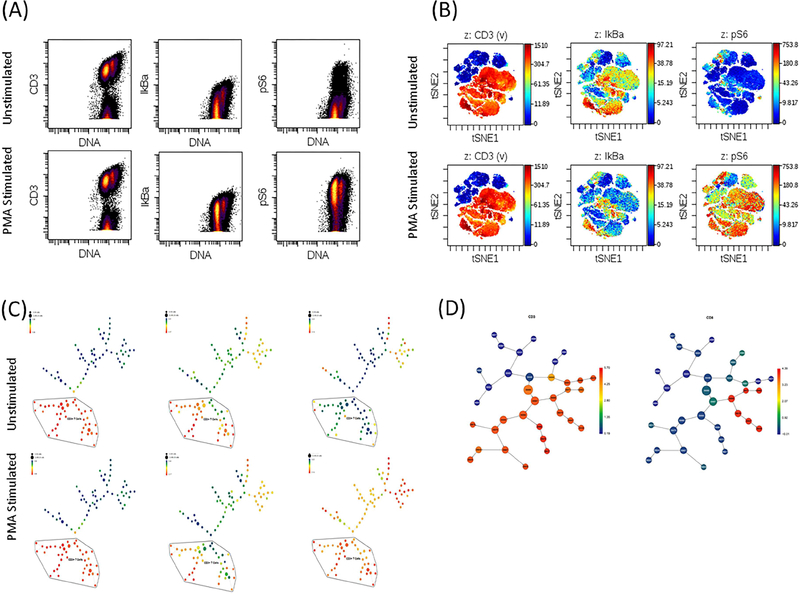
The recent introduction of innovative high-dimensional technologies like single-cell mass-cytometry by time-of-flight (CyTOF) has overcome many of these obstacles because it uses transition element isotopes to tag antibodies, which allows the routine measurement of up-to 40 single cell parameters with minimal overlap between channels (78). This high-dimensional approach has enabled a much deeper and accurate analysis of cell diversity, based on an objective segregation of cells according to the global expression patterns of selected markers. The resulting high-dimensional data offer other advantages, including the ability to perform unbiased analysis with no prior knowledge that can capture both expected and unexpected cell populations and subsets (78). Due to the large amount of data obtained for each analyzed sample by CyTOF, bioinformatics analyses are typically performed using dimensionality reduction approaches combined with unsupervised clustering algorithms like viSNE (79) or Phenograph (80). For example, viSNE is as a novel dimensionality-reducing visualization tool that projects cells onto a two dimensional map such that the distances between cells reflects their distance or similarity in high-dimensional space. Other algorithms like CITRUS (Cluster Identification, Characterization and Regression) offer other features that are important in immune monitoring population studies (81). SPADE (Spanning-tree Progression Analysis of Density-normalized Events) (82) is another clustering algorithm that allows extracting a cellular hierarchy from highdimensional data. Additional computational approaches and tools have been developed to optimize the analysis of CyTOF data (83–85). However, a detailed description of these tools is beyond the scope of this review and readers are referred to the above articles for further details regarding these approaches. Figure 2 shows a workflow summary of a typical mass cytometry analysis.
Figure 2. CyTOF analysis of immune responses for unstimulated and PMA-stimulated human peripheral blood mononuclear cells.
(A) Traditional flow-cytometry dot plots showing expression of CD3, IκBα and pS6. (B) ViSNE analysis of the same cell populations. (C) SPADE analysis of the same cell populations. (D) CITRUS analysis of the same cell populations. This figure was prepared using original data provided by Dr. C Giannarelli.
As a different but complementary approach, the ability to perform scRNA-seq is another recent technological advance that allows for transcriptome-wide genomic analyses of immune cells, revealing exciting biological insights. Although a systematic analysis of different scRNA-seq methods is beyond the scope of this review, several approaches are available with different sensitivity, read coverage across transcripts, and single cell capture methods (86,87). scRNA-seq has allowed single-cell transcriptional profiling of immune cells, revealing unanticipated immune heterogeneity in different diseases (74,86,88–90). These recent new data add to our understanding of the myeloid immune system, but also highlight the need to fully exploit CyTOF, scRNA-seq and other single-cell technological advances in future studies.
Murine myeloid cell system characterization
Using CyTOF and unbiased computational pipelines described above, recent studies have made new inroads on dissecting the complexity of the murine and human myeloid immune systems. In a seminal work, Becher and colleagues (73) used a holistic approach to perform an in-depth analysis of the myeloid immune system using a 38 parameter CyTOF panel across eight tissues (lung, spleen, mesenteric lymph nodes, liver, thymus, brain, kidney, and bone marrow) from C57BL/6 mice. After identifying major myeloid cell populations across tissues using traditional gating approaches, unbiased approaches were applied. While a biased gating identified only 55% of lung myeloid cells, using unbiased automatic detection algorithms nearly 100% of all myeloid cells were detected; indicating that non-linear dimensionality reduction (in this case using viSNE), followed by automatic clustering, can identify both expected and unexpected myeloid populations in heterogeneous samples. Furthermore, an unbiased analysis of a composite dataset of all cellular phenotypes spanning all tissues identified a total of 28 myeloid cell clusters, and provided a comprehensive view of myeloid cells across all eight tissues. Interestingly, while dendritic cells, plasmacytoid dendritic cells, monocytes and granulocytes clustered largely together, macrophages showed more distant features reflecting specialized phenotypes in distinct tissues. In particular, among all analyzed myeloid cells, some clusters of macrophages were specialized for some tissues, like brain microglia, lung alveolar macrophages, or red pulp macrophages; while for other tissues cells overlapped, such as spleen and kidney macrophages (73). Applying the same unbiased approach to analyze the same eight tissues from Csf2rb−/− mice, in which the genesis and maturation of lung macrophages is affected due to deficient GM-CSF signaling, revealed the absence of lung alveolar macrophages. Among the 28 myeloid clusters, other alterations were evident including a reduction in the detection of non-alveolar macrophages. Similarly, 3 clusters of monocytes showed highly variable phenotypes and frequencies across tissues, likely reflecting different stages of monocyte-macrophage differentiation. Above all else, this important study clearly highlighted that CyTOF is a powerful tool for unambiguous and unbiased characterization of the myeloid system (73).
While the macrophage in atherosclerosis is the subject of the 2nd article in this review series, it is notable that 3 recent simultaneous publications used these advanced technologies to provide significant insights about the composition of the atherosclerotic immune compartment (89–91). Using a combined scRNA-seq and CyTOF strategy, Winkels et al. (89) performed a deep analysis of the immune landscape of the aortas from Ldlr−/− and ApoE−/− atherosclerosis-prone mice (20 weeks of age) fed a Western diet (from 8 weeks of age) or regular chow diet (throughout the study). They identified 11 principal leukocyte clusters in the scRNA-seq merged dataset obtained from atherosclerotic aortas of Western diet and chow few mice. In contrast, only 5 clusters of leukocytes were detectable in aortas from younger (8 week old) ApoE−/− mice fed regular chow, indicating a less diverse immune composition in younger aortas with minimal or no atherosclerosis. While two subsets of macrophages that co-expressed the tissue-resident marker CX3CR1 were found in the relatively normal aortas of young ApoE−/− mice, only one subset of macrophages co-expressing CX3CR1 and Lyve1 was found in atherosclerotic aortas (from mice that received Western diet), possibly indicating that the aortic macrophage population is sustained by in situ proliferation. Furthermore, the fraction of monocytes (comprised mainly of Ly6c+ inflammatory monocytes) in the aorta was relatively higher in 8 week old ApoE−/− mice, possibly indicating an innate immune response that precedes overt atherosclerosis, which is consistent with prior observations (92). In contrast, the monocyte population was decreased in the atherosclerotic aorta and largely dominated by Ly6clow monocytes (89). Simultaneous with the above study, Cochain et al. (90) published an in-depth scRNA-seq analysis of aortic macrophages, which represented the largest cell population in the atherosclerotic aorta (28.9% of total CD45+ leukocytes). In this analysis, macrophages were divided into three clusters: inflammatory (47.0%), resident-like (34.4%) and a new type of macrophage expressing high levels of TREM2 (18.6%). Interestingly, inflammatory macrophages were enriched in both M1 (pro-atherogenic phenotype)-associated genes (Interleukin (IL)-1β, tumor necrosis factor, Cxcl10, Cxcl2, Ccl2) and in the gene encoding the Mox-associated transcription factor NRF2 (encoded by Nfe2l2), while resident-like macrophages expressed M2 (anti-atherogenic phenotype) genes (Mrc1, Folr2, F13a1). Of note, Mrc1 (encoding the mannose receptor, CD206 which is typically used to define M2 anti-atherogenic macrophages) was also expressed in a subset of inflammatory macrophages, suggesting that the traditional classification of macrophages into M1 and M2 phenotypes does not fully capture the diversity of the population in vivo. This possibility is supported by the observation that TREM2hi macrophages showed no clear signature of either M1 or M2.
In a systematic CyTOF study of the myeloid immune compartment, Cole et al. (91) identified 20 myeloid clusters (13 monocyte, 5 macrophage and 2 undefined) in the atherosclerotic aorta, as opposed to 10 clusters identified by Winkels et al. (89) using the same unsupervised clustering analysis, likely due to differences in the antibody panels. Cole et al. (91) also showed that Western (high fat) diet feeding, used to accelerate the formation of atherosclerotic plaques in Ldlr−/− and ApoE−/− mice, affects the distribution of the monocyte and macrophage subsets in the aorta via an increase of Ly6c+ and Ly6c− monocytes and CD11c+ macrophages, combined with a decrease of CD206+CD196+CD209b+ and CD206+CD169+CD209b− macrophages and conventional type 2 dendritic cells (cDC2).
While a great deal remains to be understood, these murine studies highlight the enormous promise of these advanced profiling techniques, and the importance of systematic approaches to resolve the diversity of macrophage and other cell populations.
Human myeloid cell system characterization
Until now, while murine monocyte and macrophage studies have allowed investigators to trace origins, plasticity and adaptability to specialized tissue microenvironments both in health and disease (i.e. inflammation), similar studies have been particularly challenging in humans. However, CyTOF is ideal for analyzing limited clinical samples using low input single cell suspensions from blood (typically peripheral blood mononuclear cells), skin and other tissues (93). Thus, using CyTOF, it has become possible to perform translational studies into the phenotype and function of monocytes and macrophages in humans. Among other things, CyTOF has already been successfully translated from basic to clinical studies allowing deep phenotype and functional analyses of the human immune system, and it is increasingly used for clinical studies and trials involving immune monitoring (i.e. NCT01882569, NCT02476084, NCT02718573, NCT03335605, NCT02680652, NCT03207854, NCT03003728). Furthermore, by applying new barcoding approaches (93,94) that improve accuracy and allow the analysis of multiple samples at the same time, it has become possible to simultaneously process multiple samples from different patients and tissues, thereby reducing technical variability. While certain limitations remain to be addressed such as a lack of temporal resolution and standardized protocols in these clinical studies, collectively, CyTOF has opened new opportunities to study and understand human immune phenotypes and their functional plasticity.
What has CyTOF informed us about the human immune system? Already, prior classification systems have been brought into question. In a recent study (76), using a panel of 36 markers to analyze human monocytes, the traditional classification of classical, non-classical and transitional intermediate monocytes based on CD14 and CD16 expression alone was challenged by evidence of contamination across subsets. The authors proposed a new gating and classification strategy based on additional markers identified by CyTOF, including CCR2, CD36, HLA-DR and CD11c to better discriminate among the three populations. In another study (75), using a panel of 38 phenotype, activation and polarization markers, 4 subsets of monocytes aligned with canonical monocyte populations were identified in the peripheral blood of healthy donors. Monocytes were distinguished by high expression of CD33, CD36, and CCR2 and low expression of CD163 and CD274. Following a workflow analysis that excluded B (CD19+), T (CD3+) and NK (CD3−CD16+CD45RA+) lymphocytes, SPADE analysis revealed that monocytes were 85% classical (CD14+CD16−), 3% SLANlow non-classical (CD14lowCD16hiSLANlow), 3% SLANhi non-classical (CD14lowCD16hiSLANhi) and 9% intermediate (CD14+CD16+). As expected, non-classical SLANhi and SLANlow monocytes expressed low levels of CD36, CD64, CCR2 and CD14.
New insights have also been gained on the differentiation of human monocytes to macrophages. Villani et al. (95) studied the differentiation of peripheral monocytes into macrophages using macrophage colony-stimulating factor (M-CSF) followed by stimulation using conventional differentiation protocols and various standard stimuli (i.e. IL-4, IL-10, lipopolysaccharide, interferon-γ, IL-6, tristetraprolin). Each stimulation condition resulted in a specific polarized macrophage phenotype that was also in agreement with segregation based on transcriptional data from previous studies (95). Using CyTOF, new phenotype patterns reflecting each different polarization state compared to baseline macrophage (M-CSF only) were identified. Lipopolysaccharide-induced macrophages were characterized by high levels of CD13 and CD86 and low levels of CD163 and CD206. Macrophages differentiated by IL-4 were CD274hi and CD64low. Tristetraprolin-induced macrophages were CD14hi and HL-DRlow. Interferon-γ-induced macrophages were CD64hi and CD86hi, while IL-10-induced macrophages showed high expression of CD14, CCR2, and CD163. Finally, macrophages differentiated by IL-6 were CD11chi and CD33hi.
While important, a limitation of the above studies is that these novel monocyte and macrophage populations were only described based on surface markers (75,76) and gene expression analysis (95). Future studies will be needed to characterize the functional state of these cell populations. The importance of integrating functional characteristics of immune cells in human studies is highlighted by the observation that specific immune responses in patients can predict clinical outcomes. For example, where CyTOF was implemented at the bedside of surgical patients, immune correlates pertaining to the functional (i.e., signaling) state of CD14+ monocytes were associated with patient recovery (96). In a subsequent study, CyTOF analysis of the preoperative immune state to predict surgical recovery identified a critical role of the TLR4 signaling pathway in monocytes (97). Finally, CyTOF has been used to demonstrate a precise tuning of immune events in human term pregnancy (98), and distinct immune features that can discriminate between patients with a history of term and preterm birth (96). Considering that monocytes are precursors of macrophages in tissue, these data indicate the importance of the myeloid system in determining clinical outcomes and suggest that specific macrophage immune responses may be critical in driving the disease at sites of tissue injury.
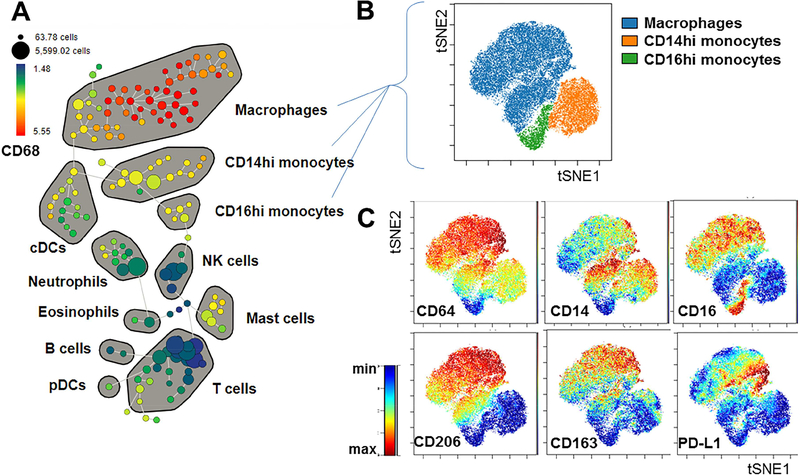
As already mentioned, macrophage characterization in humans is hampered by the limited availability of tissue samples. In recent work, highlighting the capabilities of this approach with small samples, Lavin et al. (74) used a paired CyTOF approach and scRNA-seq to perform an in-depth analysis of the immune landscape of early lung carcinoma by comparing immune cells infiltrating tumor tissue, normal lung and in blood from the same patients. Among all identified clusters of macrophages, one was unique to tumor tissue showing a distinct transcriptional signature from macrophages in normal lung tissue. CyTOF analysis confirmed a higher expression of PPAR-γ, CD64, CD14 and lower CD86, CD206 and IL-6 in tumor macrophages, defined as PPARγhiCD64hiCD14hiIL-6hi, compared to macrophages in normal lung tissue. Functionally, tumor macrophages were characterized by increased production of IL-6 indicating a pro-tumorigenic phenotype. Figure 3 shows the characterization of human tissue monocyte and macrophage heterogeneity in lung tissue by mass-cytometry.
Figure 3. Characterization of human tissue monocyte and macrophage subsets by mass cytometry.
Lung tissue from a surgical resection specimen was dissociated and analyzed by CyTOF. (A) SPADE analysis of all viable CD45+ cells, showing several major immune subsets as identified by canonical marker expression patterns. The SPADE tree is colored to show the relative expression of CD68 across all populations. (B) ViSNE analysis of gated macrophage and monocyte populations. (C) ViSNE plots showing expression patterns of specific markers, highlighting the phenotypic heterogeneity between and within these subsets. This figure was generated using data of “CyTOF analysis of paired blood, tumor and non-involved lung from NSCLC patients”, a shared public dataset on flow repository.com.
In the cardiovascular system, our knowledge of immune cell composition is still largely based on traditional immunohistochemistry approaches that fail to provide a comprehensive immune atlas of the disease. For example, estimates of the overall immune composition of human atherosclerotic lesions in large cohorts of patients have been reported as largely dominated by macrophages (89). However, these data should be interpreted cautiously when considering that cell frequencies were derived from bulk gene expression data using deconvolution methods (99), which therefore did not provide single-cell resolution of the samples. In fact, to infer cell frequency estimates from bulk RNA-seq data, deconvolution methods rely on the underlying assumption that gene expression signatures derived from peripheral mononuclear cells are equivalent to other tissues (99), which may not hold true in cardiovascular system (100). Future single-cell CyTOF and scRNA-seq studies will assuredly shed light on these issues.
Conclusion
Advances in defining the origins of macrophages have dramatically shaped our understanding of macrophage development and influenced our current understanding of progenitor cell differentiation by microenvironment factors to mature resident macrophages. In the current era, analyses of macrophages in any given organ should begin with a detailed assessment of tissue macrophage phenotype and identity. As a timely technological advance that will greatly facilitate this assessment, single-cell CyTOF is an exciting new tool that is now being used to characterize and expand our understanding of the immune myeloid system in patients. Although, until now, most analyses of the myeloid system using CyTOF focused on circulating immune cells like monocytes, emerging data show that this technology can be used to analyze clinical tissue samples. The use of this unbiased approach to characterize tissue macrophages, particularly when combined with scRNA-seq, offers the potential of identifying previously unknown macrophage clusters implicated in driving disease pathology in humans, and of identifying phenotypic or molecular markers that relate to specific clinical cardiovascular functions or disease traits.
Looking ahead even further, technology and innovation is moving at such a rapid pace that within a few years even scRNA-seq and CyTOF may be superseded by highly advanced methods that combine elements of these approaches into a unified technique (101). By building on our strong foundational knowledge of monocytes/macrophages, the methodical application of these tools will serve to propel this field to exciting new levels of understanding that should, optimistically, see macrophage-specific biomarkers and therapeutic interventions become a clinical reality in the fight against cardiovascular disease.
Acknowledgements:
Jesse Williams was supported by American Heart Association grant 17POST33410473 and NIH K99HL138163. Gwendalyn Randolph was supported by NIH R01 HL118206, R37 AI049653, and DP1 DK109668. Chiara Giannarelli is funded by NIH K23HL111339, R03HL135289, R21TR001739 and UH2TR002067; and acknowledges support from the American Heart Association (16SDG27250090; A14SFRN20840000). Jason Kovacic acknowledges research support from the National Institutes of Health (R01HL130423), the American Heart Association (14SFRN20490315; 14SFRN20840000) and The Leducq Foundation (Transatlantic Network of Excellence Award). The data used for generating Figures 2 and 3 were obtained using mass cytometry instrumentation supported by NIH S10OD023547. We also thank Dr. Dawn Fernandez for her assistance in the preparation of Figure 2.
Abbreviations
- CyTOF
mass cytometry by time-of-flight
- EMPs
erythro-myeloid precursor cells
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- GVHD
graft versus host disease
- HSC
hematopoietic stem cell
- IL
interleukin
- M-CSF
macrophage colony-stimulating factor
- RNA-seq
RNA sequencing
- scRNA-seq
single-cell RNA sequencing
Footnotes
Disclosures: The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ross R Atherosclerosis--an inflammatory disease. N Engl J Med 1999;340:115–26. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. Lipoprotein metabolism in the macrophage: implications for cholesterol deposition in atherosclerosis. Annu Rev Biochem 1983;52:223–61. [DOI] [PubMed] [Google Scholar]
- 3.van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull World Health Organ 1972;46:845–52. [PMC free article] [PubMed] [Google Scholar]
- 4.Guilliams M, Ginhoux F, Jakubzick C et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol 2014;14:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziegler-Heitbrock L, Ancuta P, Crowe S et al. Nomenclature of monocytes and dendritic cells in blood. Blood 2010;116:e74–80. [DOI] [PubMed] [Google Scholar]
- 6.Jakubzick CV, Randolph GJ, Henson PM. Monocyte differentiation and antigenpresenting functions. Nat Rev Immunol 2017;17:349–362. [DOI] [PubMed] [Google Scholar]
- 7.Naito M, Takahashi K, Nishikawa S. Development, differentiation, and maturation of macrophages in the fetal mouse liver. J Leukoc Biol 1990;48:27–37. [DOI] [PubMed] [Google Scholar]
- 8.Naito M, Yamamura F, Nishikawa S, Takahashi K. Development, differentiation, and maturation of fetal mouse yolk sac macrophages in cultures. J Leukoc Biol 1989;46:1–10. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamamura F, Naito M. Differentiation, maturation, and proliferation of macrophages in the mouse yolk sac: a light-microscopic, enzyme-cytochemical, immunohistochemical, and ultrastructural study. J Leukoc Biol 1989;45:87–96. [DOI] [PubMed] [Google Scholar]
- 10.Morris L, Graham CF, Gordon S. Macrophages in haemopoietic and other tissues of the developing mouse detected by the monoclonal antibody F4/80. Development 1991;112:517–26. [DOI] [PubMed] [Google Scholar]
- 11.Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res 1999;117:145–52. [DOI] [PubMed] [Google Scholar]
- 12.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 1999;126:5073–84. [DOI] [PubMed] [Google Scholar]
- 13.Palis J, Yoder MC. Yolk-sac hematopoiesis: the first blood cells of mouse and man. Exp Hematol 2001;29:927–36. [DOI] [PubMed] [Google Scholar]
- 14.Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 2007;446:1056–61. [DOI] [PubMed] [Google Scholar]
- 15.Tober J, Koniski A, McGrath KE et al. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood 2007;109:1433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz C, Gomez Perdiguero E, Chorro L et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012;336:86–90. [DOI] [PubMed] [Google Scholar]
- 17.Frame JM, McGrath KE, Palis J. Erythro-myeloid progenitors: “definitive” hematopoiesis in the conceptus prior to the emergence of hematopoietic stem cells. Blood Cells Mol Dis 2013;51:220–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mass E, Ballesteros I, Farlik M et al. Specification of tissue-resident macrophages during organogenesis. Science 2016;353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stremmel C, Schuchert R, Wagner F et al. Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat Commun 2018;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeffel G, Chen J, Lavin Y et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 2015;42:665–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epelman S, Lavine KJ, Beaudin AE et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 2014;40:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ginhoux F, Greter M, Leboeuf M et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010;330:841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guilliams M, De Kleer I, Henri S et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med 2013;210:1977–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto D, Chow A, Noizat C et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 2013;38:792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakubzick C, Gautier EL, Gibbings SL et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity 2013;39:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoeffel G, Wang Y, Greter M et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med 2012;209:1167–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng J, Ruedl C, Karjalainen K. Most Tissue-Resident Macrophages Except Microglia Are Derived from Fetal Hematopoietic Stem Cells. Immunity 2015;43:382–93. [DOI] [PubMed] [Google Scholar]
- 28.Yona S, Kim KW, Wolf Y et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013;38:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ginhoux F, Guilliams M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 2016;44:439–449. [DOI] [PubMed] [Google Scholar]
- 30.Perdiguero EG, Geissmann F. The development and maintenance of resident macrophages. Nat Immunol 2016;17:2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bain CC, Bravo-Blas A, Scott CL et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 2014;15:929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zigmond E, Jung S. Intestinal macrophages: well educated exceptions from the rule. Trends Immunol 2013;34:162–8. [DOI] [PubMed] [Google Scholar]
- 33.Shaw TN, Houston SA, Wemyss K et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J Exp Med 2018;215:1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gautier EL, Jakubzick C, Randolph GJ. Regulation of the migration and survival of monocyte subsets by chemokine receptors and its relevance to atherosclerosis. Arterioscler Thromb Vasc Biol 2009;29:1412–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol 2006;7:311–7. [DOI] [PubMed] [Google Scholar]
- 36.Patel AA, Zhang Y, Fullerton JN et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med 2017;214:1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auffray C, Fogg D, Garfa M et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 2007;317:666–70. [DOI] [PubMed] [Google Scholar]
- 38.Carlin LM, Stamatiades EG, Auffray C et al. Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell 2013;153:362–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcovecchio PM, Thomas GD, Mikulski Z et al. Scavenger Receptor CD36 Directs Nonclassical Monocyte Patrolling Along the Endothelium During Early Atherogenesis. Arterioscler Thromb Vasc Biol 2017;37:2043–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quintar A, McArdle S, Wolf D et al. Endothelial Protective Monocyte Patrolling in Large Arteries Intensified by Western Diet and Atherosclerosis. Circ Res 2017;120:1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams JW, Randolph GJ, Zinselmeyer BH. A Polecat’s View of Patrolling Monocytes. Circ Res 2017;120:1699–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol 2005;5:953–64. [DOI] [PubMed] [Google Scholar]
- 43.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol 2005;23:901–44. [DOI] [PubMed] [Google Scholar]
- 44.Tufano RP, Randolph GW. Arguments for and against attempting to perform a true total thyroidectomy for differentiated thyroid cancer. JAMA Otolaryngol Head Neck Surg 2014;140:415–6. [DOI] [PubMed] [Google Scholar]
- 45.http://www.immgen.org/. Accessed January 2nd, 2018.
- 46.Gautier EL, Shay T, Miller J et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 2012;13:1118–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gautier EL, Ivanov S, Lesnik P, Randolph GJ. Local apoptosis mediates clearance of macrophages from resolving inflammation in mice. Blood 2013;122:2714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Laar L, Saelens W, De Prijck S et al. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity 2016;44:755–68. [DOI] [PubMed] [Google Scholar]
- 49.Guilliams M, Scott CL. Does niche competition determine the origin of tissue-resident macrophages? Nat Rev Immunol 2017;17:451–460. [DOI] [PubMed] [Google Scholar]
- 50.Misharin AV, Morales-Nebreda L, Reyfman PA et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med 2017;214:2387–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibbings SL, Thomas SM, Atif SM et al. Three Unique Interstitial Macrophages in the Murine Lung at Steady State. Am J Respir Cell Mol Biol 2017;57:66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim KW, Williams JW, Wang YT et al. MHC II+ resident peritoneal and pleural macrophages rely on IRF4 for development from circulating monocytes. J Exp Med 2016;213:1951–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ensan S, Li A, Besla R et al. Self-renewing resident arterial macrophages arise from embryonic CX3CR1(+) precursors and circulating monocytes immediately after birth. Nat Immunol 2016;17:159–68. [DOI] [PubMed] [Google Scholar]
- 54.Gosselin D, Link VM, Romanoski CE et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 2014;159:1327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lavin Y, Winter D, Blecher-Gonen R et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014;159:1312–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haniffa M, Ginhoux F, Wang XN et al. Differential rates of replacement of human dermal dendritic cells and macrophages during hematopoietic stem cell transplantation. J Exp Med 2009;206:371–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eguiluz-Gracia I, Schultz HH, Sikkeland LI et al. Long-term persistence of human donor alveolar macrophages in lung transplant recipients. Thorax 2016;71:1006–1011. [DOI] [PubMed] [Google Scholar]
- 58.Nayak DK, Zhou F, Xu M et al. Long-Term Persistence of Donor Alveolar Macrophages in Human Lung Transplant Recipients That Influences Donor-Specific Immune Responses. Am J Transplant 2016;16:2300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanitakis J, Petruzzo P, Dubernard JM. Turnover of epidermal Langerhans’ cells. N Engl J Med 2004;351:2661–2. [DOI] [PubMed] [Google Scholar]
- 60.Xue J, Schmidt SV, Sander J et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014;40:274–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibbings SL, Goyal R, Desch AN et al. Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood 2015;126:1357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott CL, Zheng F, De Baetselier P et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun 2016;7:10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mould KJ, Barthel L, Mohning MP et al. Cell Origin Dictates Programming of Resident versus Recruited Macrophages during Acute Lung Injury. Am J Respir Cell Mol Biol 2017;57:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu Y, Herndon JM, Sojka DK et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity 2017;47:323–338 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity 2010;32:593–604. [DOI] [PubMed] [Google Scholar]
- 66.Mackaness GB. Cellular resistance to infection. J Exp Med 1962;116:381–406. [PubMed] [Google Scholar]
- 67.Mackaness GB. The Immunological Basis of Acquired Cellular Resistance. J Exp Med 1964;120:105–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gordon S Alternative activation of macrophages. Nat Rev Immunol 2003;3:23–35. [DOI] [PubMed] [Google Scholar]
- 69.Locati M, Mantovani A, Sica A. Macrophage activation and polarization as an adaptive component of innate immunity. Adv Immunol 2013;120:163–84. [DOI] [PubMed] [Google Scholar]
- 70.Netea MG, Joosten LA, Latz E et al. Trained immunity: A program of innate immune memory in health and disease. Science 2016;352:aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willment JA, Lin HH, Reid DM et al. Dectin-1 expression and function are enhanced on alternatively activated and GM-CSF-treated macrophages and are negatively regulated by IL-10, dexamethasone, and lipopolysaccharide. J Immunol 2003;171:4569–73. [DOI] [PubMed] [Google Scholar]
- 72.Saeed S, Quintin J, Kerstens HH et al. Epigenetic programming of monocyte-tomacrophage differentiation and trained innate immunity. Science 2014;345:1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Becher B, Schlitzer A, Chen J et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol 2014;15:1181–9. [DOI] [PubMed] [Google Scholar]
- 74.Lavin Y, Kobayashi S, Leader A et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 2017;169:750–765 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roussel M, Ferrell PB Jr, Greenplate AR et al. Mass cytometry deep phenotyping of human mononuclear phagocytes and myeloid-derived suppressor cells from human blood and bone marrow. J Leukoc Biol 2017;102:437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomas GD, Hamers AAJ, Nakao C et al. Human Blood Monocyte Subsets: A New Gating Strategy Defined Using Cell Surface Markers Identified by Mass Cytometry. Arterioscler Thromb Vasc Biol 2017;37:1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol 2004;4:648–55. [DOI] [PubMed] [Google Scholar]
- 78.Bendall SC, Simonds EF, Qiu P et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011;332:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amir el AD, Davis KL, Tadmor MD et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 2013;31:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levine JH, Simonds EF, Bendall SC et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 2015;162:184–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci U S A 2014;111:E2770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qiu P, Simonds EF, Bendall SC et al. Extracting a cellular hierarchy from highdimensional cytometry data with SPADE. Nat Biotechnol 2011;29:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bendall SC, Davis KL, Amir el AD et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 2014;157:714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krishnaswamy S, Spitzer MH, Mingueneau M et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science 2014;346:1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Setty M, Tadmor MD, Reich-Zeliger S et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol 2016;34:637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buenrostro JD, Corces MR, Lareau CA et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 2018;173:1535–1548 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ziegenhain C, Vieth B, Parekh S, Hellmann I, Enard W. Quantitative single-cell transcriptomics. Brief Funct Genomics 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gorman MJ, Caine EA, Zaitsev K et al. An Immunocompetent Mouse Model of Zika Virus Infection. Cell Host Microbe 2018;23:672–685 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winkels H, Ehinger E, Vassallo M et al. Atlas of the Immune Cell Repertoire in Mouse Atherosclerosis Defined by Single-Cell RNA-Sequencing and Mass Cytometry. Circ Res 2018;122:1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cochain C, Vafadarnejad E, Arampatzi P et al. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res 2018;122:1661–1674. [DOI] [PubMed] [Google Scholar]
- 91.Cole JE, Park I, Ahern D et al. Immune cell census in murine atherosclerosis: cytometry by time of flight illuminates vascular myeloid cell diversity. Cardiovasc Res 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Erbilgin A, Siemers N, Kayne P, Yang WP, Berliner J, Lusis AJ. Gene expression analyses of mouse aortic endothelium in response to atherogenic stimuli. Arterioscler Thromb Vasc Biol 2013;33:2509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yao Y, Liu R, Shin MS et al. CyTOF supports efficient detection of immune cell subsets from small samples. J Immunol Methods 2014;415:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lai L, Ong R, Li J, Albani S. A CD45-based barcoding approach to multiplex masscytometry (CyTOF). Cytometry A 2015;87:369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Villani AC, Satija R, Reynolds G et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017;356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gaudilliere B, Ganio EA, Tingle M et al. Implementing Mass Cytometry at the Bedside to Study the Immunological Basis of Human Diseases: Distinctive Immune Features in Patients with a History of Term or Preterm Birth. Cytometry A 2015;87:817–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fragiadakis GK, Gaudilliere B, Ganio EA et al. Patient-specific Immune States before Surgery Are Strong Correlates of Surgical Recovery. Anesthesiology 2015;123:1241–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aghaeepour N, Ganio EA, McIlwain D et al. An immune clock of human pregnancy. Sci Immunol 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Newman AM, Liu CL, Green MR et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schelker M, Feau S, Du J et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat Commun 2017;8:2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stoeckius M, Hafemeister C, Stephenson W et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buttgereit A, Lelios I, Yu X et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol 2016;17:1397–1406. [DOI] [PubMed] [Google Scholar]
- 103.Wang Y, Szretter KJ, Vermi W et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol 2012;13:753–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Joseph SB, Bradley MN, Castrillo A et al. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell 2004;119:299–309. [DOI] [PubMed] [Google Scholar]
- 105.Gautier EL, Chow A, Spanbroek R et al. Systemic analysis of PPARgamma in mouse macrophage populations reveals marked diversity in expression with critical roles in resolution of inflammation and airway immunity. J Immunol 2012;189:2614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol 2014;15:1026–37. [DOI] [PubMed] [Google Scholar]
- 107.Shibata Y, Berclaz PY, Chroneos ZC, Yoshida M, Whitsett JA, Trapnell BC. GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU.1. Immunity 2001;15:557–67. [DOI] [PubMed] [Google Scholar]
- 108.Takayanagi H, Kim S, Koga T et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell 2002;3:889–901. [DOI] [PubMed] [Google Scholar]
- 109.Takayanagi H, Kim S, Matsuo K et al. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature 2002;416:744–9. [DOI] [PubMed] [Google Scholar]
- 110.Kohyama M, Ise W, Edelson BT et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature 2009;457:318–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gautier EL, Ivanov S, Williams JW et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J Exp Med 2014;211:1525–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 2014;157:832–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rosas M, Davies LC, Giles PJ et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science 2014;344:645–648. [DOI] [PMC free article] [PubMed] [Google Scholar]