Abstract
Rabbit hemorrhagic disease virus (RHDV) is an important member of the Caliciviridae family and a highly lethal pathogen in rabbits. Although the cell receptor of RHDV has been identified, the mechanism underlying RHDV internalization remains unknown. In this study, the entry and post-internalization of RHDV into host cells were investigated using several biochemical inhibitors and RNA interference. Our data demonstrate that rabbit nucleolin (NCL) plays a key role in RHDV internalization. Further study revealed that NCL specifically interacts with the RHDV capsid protein (VP60) through its N-terminal residues (aa 285–318), and the exact position of the VP60 protein for the interaction with NCL is located in a highly conserved region (472Asp-Val-Asn474; DVN motif). Following competitive blocking of the interaction between NCL and VP60 with an artificial DVN peptide (RRTGDVNAAAGSTNGTQ), the internalization efficiency of the virus was markedly reduced. Moreover, NCL also interacts with the C-terminal residues of clathrin light chain A, which is an important component in clathrin-dependent endocytosis. In addition, the results of animal experiments also demonstrated that artificial DVN peptides protected most rabbits from RHDV infection. These findings demonstrate that NCL is involved in RHDV internalization through clathrin-dependent endocytosis.
Author summary
Rabbit hemorrhagic disease virus (RHDV) is the causative agent of a highly contagious and lethal disease in rabbits. Since, at present, there is no robust cell culture system for the propagation of the virus, the molecular mechanism of RHDV internalization remains poorly understood. Here, we demonstrated that rabbit nucleolin (NCL) efficiently mediates RHDV internalization by interacting with the RHDV capsid protein (VP60). The exact function domains of the interaction between NCL and VP60 were also determined. Notably, these functional domains are highly conserved in all RHDV genotypes. Further study results revealed that NCL was involved in clathrin-dependent endocytosis through interactions with the C-terminal residues of clathrin light chain A. In addition, the artificial peptide (RRTGDVNAAAGSTNGTQ; DVN peptide) based on the functional domain, which is responsible for RHDV VP60 binding to NCL, is able to inhibit RHDV infection, indicating that the DVN peptide might be a candidate target for the design of antiviral drugs against RHDV infection.
Introduction
Rabbit hemorrhagic disease virus (RHDV) is a non-enveloped, single-stranded, positive sense RNA virus, belonging to the Caliciviridae family [1], and it is the causative agent of a highly contagious and lethal disease in rabbits which is strongly associated with liver degeneration and diffuse hemorrhage [2,3]. RHDV was first isolated in China in 1984 [4] and it has been subsequently detected in rabbit populations throughout Asia, Europe, Australia, and the Americas, resulting in the death of millions of wild and domestic adult rabbits [3].
It is well known that the first step in viral infection is viral entry. However, a suitable cell culture capable of supporting authentic RHDV has not yet been established, thereby greatly impeding the progress of investigations into the mechanisms underlying the pathogenesis, translation, and replication of RHDV. Consequently, studies on the viral entry of RHDV have relied on the self-assembly of the capsid protein into virus-like particles when expressed in Escherichia coli or insect cells. RHDV has been shown to bind to oligosaccharides H type 2 and A type 2, which are histo-blood group antigens (HBGAs) expressed on the surfaces of cells lining the duodenal surface and trachea of rabbits, thereby presenting two possible viral entry points [5]. RHDV isolates from six different genetic groups bind specifically to different HBGAs, which act as attachment factors that facilitate infection [6,7].
Following attachment of RHDV to the cell surface, internalization by an unknown mechanism and desencapsidation occur, leading to the release of the viral genome into the cytoplasm of the host cell. In 2017, we successfully constructed and cultured a mutant RHDV (mRHDV) in RK-13 cells in vitro, which has a specific receptor-recognition motif (Arg-Gly-Asp) on the surface of the capsid protein that is characterized by two amino acid (aa) substitutions. mRHDV is recognized by the intrinsic membrane receptor (integrin α3β1) of RK-13 cells and then gains entry, replicates, and imparts apparent cytopathic effects [8].
In previous studies, we found that the RHDV capsid protein (VP60) was associated with many host proteins, including NCL, through affinity purification of infected cells [8]. NCL is a phosphoprotein that is ubiquitously and abundantly expressed in many growing eukaryotic cells and highly conserved during evolution, as it is involved in a remarkably large number of cellular activities [9]. Generally, NCL is mainly distributed in the nucleolus, but also exists in the nucleoplasm and cytoplasm, and on the cell surface, where its specific functions vary [10,11]. NCL controls a wide range of fundamental cellular processes, such as ribosome biogenesis, proliferation, and cell cycle regulation, and also plays important roles in the replication and intracellular trafficking of multiple viruses [12–16]. For example, the interaction between NCL and the 3′ untranslated region (UTR) of tombusvirus RNA has been shown to inhibit replication by interfering with the recruitment of viral RNA [17]. Similarly, the interaction between NCL and the UTRs of feline calicivirus [18] and poliovirus [19,20] stimulate translation of viral proteins. In addition, NCL binds to a protein of herpes simplex virus type 1 to facilitate exportation of US11 from the cell nucleus to the cytoplasm [21]. Notably, cell surface NCL is involved in viral infection by promoting viral attachment and internalization. For example, NCL acts as a receptor of human respiratory syncytial virus [22] and as a low-affinity receptor of human immunodeficiency virus [23]. NCL also mediates cellular attachment and internalization of enterovirus 71 [24]. Moreover, the internalization of multiple influenza A viruses, including H1N1, H3N2, H5N1, and H7N9, is reduced by suppression of NCL function and expression on the cell surface [25–29]. A recent study showed that NCL interacts with the capsid protein of dengue virus, suggesting a role in viral morphogenesis [30]. In addition, NCL is important for lyssavirus infection [31].
Here, we demonstrated that RHDV VP60 protein directly interacts with NCL. Of note, this interaction was found to play a key role in RHDV internalization. Furthermore, our data showed that NCL is involved in clathrin-dependent endocytosis, which is an important pathway of RHDV infection, by interacting with the C-terminal residues of clathrin light chain A (CLTA). Collectively, these findings further illustrate the molecular mechanism underlying RHDV infection.
Results
NCL is involved in RHDV internalization
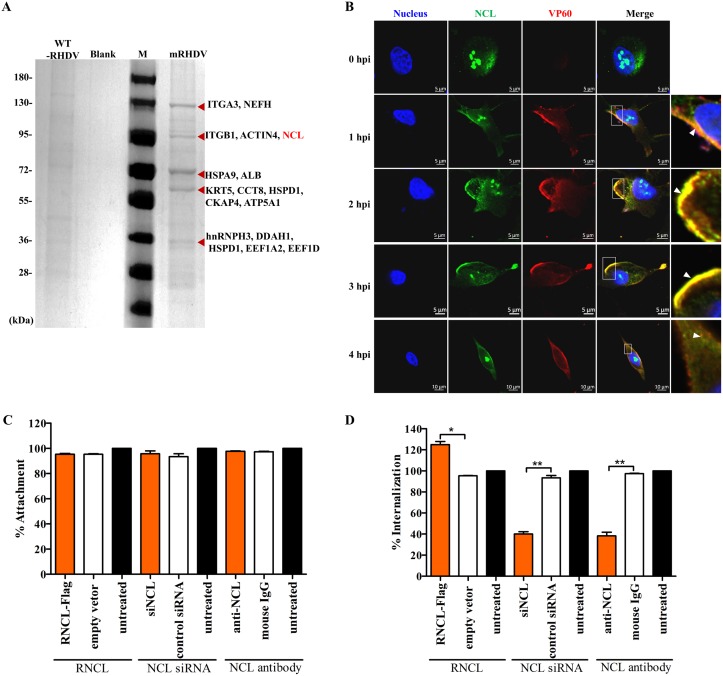
We previously reported a novel strategy for the construction of an mRHDV that could be recognized by host cells and grows well in RK-13 cells [8]. Furthermore, we performed affinity purification with RHDV capsid protein VP60 mAb from RK-13 cells, which were infected with mRHDV for 48 h, followed by mass spectrometry analysis. As shown in Fig 1A, many host proteins were associated with VP60, including NCL [8]. In this study, we found that RHDV and NCL were co-localized on the cell membrane surface at the early stage of mRHDV infection. This phenomenon was most apparent at 2–3 hours post-infection (hpi) (Fig 1B). Therefore, we hypothesized that NCL is important for RHDV entry.
Fig 1. NCL is involved in RHDV internalization.
(A) Screening of host proteins that interacted with the RHDV capsid protein. SDS-PAGE analysis of immunoprecipitation-purified host factors of RK-13 cells that interact with the mRHDV capsid protein. The red arrows indicate the bands identified by mass spectrometry. Lane M is the protein marker and the lane titled “blank” is the negative control (RK-13 cells not infected with mRHDV) [8]. (B) Immunofluorescence co-localization analysis of NCL (green) and VP60 (red). mRHDV-infected cells incubated with mAbs against NCL and VP60 at 0, 1, 2, 3, and 4 h, respectively. The small white boxes represent amplified random co-localization spots within the merged image, and the co-localization spots are indicated with white arrowheads. (C) The effect of changing the expression level of NCL on RHDV attachment. RK-13 cells were pre-transfected with Flag-tagged NCL plasmids or NCL siRNA for 24 h, or pre-incubated with anti-NCL mAb for 1 h at 37°C. The cells were then chilled and mRHDV was added at a MOI of 1. After viral attachment at 4°C for 2 h, the RK-13 cells were washed and lysed. The numbers of attached viral RNA copies were determined by qRT-PCR. Percent changes in VP60 RNA copy numbers were derived by comparing the number of VP60 RNA copies in the treated samples to those of the untreated RK-13 cells. The p3×FLAG-CMV-14 vector, non-specific siRNA, and mouse IgG were used as negative controls. (D) The effect of NCL on RHDV internalization. The treatment of cells in each group was the same as that for the attachment assay. RK-13 cells were incubated with mRHDV (MOI = 1) for 2 h at 4°C and then washed with chilled PBS. The temperature was then shifted to 37°C to facilitate viral internalization. At 30 min post infection, when RHDV internalization was proportionally increasing, the cells were washed in pre-chilled acidic PBS (pH = 2.5) to remove non-internalized viruses. After cell lysis, the amount of internalized mRHDV VP60 RNA copies was determined by qRT-PCR. The percentage of internalized VP60 copies was calculated as a ratio to the value obtained from untreated RK-13 cells. The student’s t-tests and analysis of variance were used for statistical analyses. *p < 0.05 and **p < 0.01. All experiments were conducted in triplicate, which produced similar results.
To determine if NCL is required for RHDV infection, the expression level of NCL was changed by siRNA (S1 Fig) or transfection with plasmids (S2A Fig). For the attachment assay, RK-13 cells were pre-transfected with Flag-tagged NCL plasmids or NCL siRNA for 24 h, or pre-incubated with anti-NCL mAb for 1 h at 37°C. The cells were then chilled and mRHDV was added at a multiplicity of infection (MOI) of 1 (S3A Fig). After viral attachment at 4°C for 2 h, the RK-13 cells were washed and lysed. qRT-PCR determined the number of attached viral RNA copies. Percent changes in VP60 RNA copy numbers were derived by comparing the number of VP60 RNA copies in the treated samples to that of the untreated RK-13 cells. As shown in Fig 1C, mRHDV attachment to RK-13 cells was not obviously affected by changing or blocking NCL regulation.
Next, we investigated whether NCL was required for RHDV internalization. For the internalization assays, RK-13 cells were incubated with mRHDV (MOI = 1) for 2 h at 4°C and then washed with chilled PBS. The temperature was then shifted to 37°C to facilitate viral internalization. At 30 min post infection, when RHDV internalization was proportionally increasing (S3B Fig), the cells were washed in pre-chilled acidic PBS (pH = 2.5) to remove non-internalized viruses. After cell lysis, the amount of internalized mRHDV VP60 RNA copies was determined by qRT-PCR. As shown in Fig 1D, the percentage of mRHDV internalized in RK-13 cells was 40% or 38% after treatment with NCL siRNA or NCL Ab, respectively. In contrast, overexpression of NCL increased the percentage of internalized mRHDV by 25%.
Collectively, these data show that NCL is required during RHDV internalization, but not attachment.
RHDV VP60 binds to the N-terminal domain of NCL
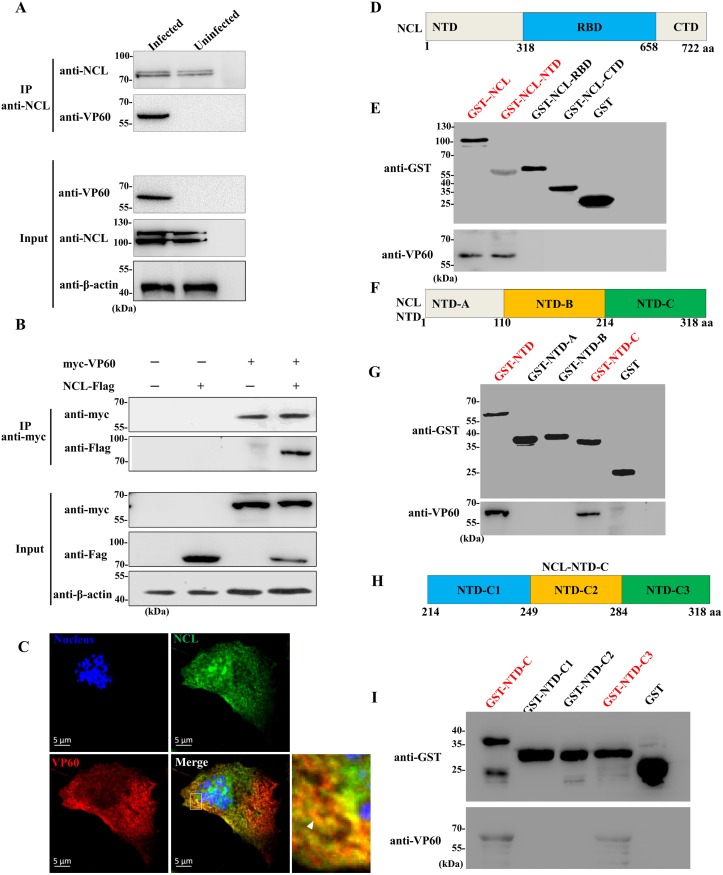
Previous studies have reported that NCL is involved in the entry of dengue virus and enterovirus 71 by interacting with the capsid proteins [24,30]. The only capsid protein present in RHDV, VP60, is responsible for antigenicity and binding to host proteins, and plays a key role in RHDV entry [7]. To determine if NCL binds to VP60 during RHDV internalization, we assessed the interaction between NCL and VP60 in RK-13 cells in the presence and absence of mRHDV infection for 2h at 37°C. The results of an immunoprecipitation (IP) assay performed on cell lysates using NCL mAb showed that NCL and VP60 associated in infected cells (Fig 2A). Moreover, to determine whether NCL directly interacts with VP60, co-IP assays were employed with a myc mAb in RK-13 cells, which were co-transfected with pVP60-myc and pNCL-Flag eukaryotic expression plasmids. Western blot analysis using a mAb against the Flag tag showed a band corresponding to NCL in the myc co-IP assay, indicating a direct interaction between RHDV VP60 and NCL (Fig 2B). In addition, an IFA was performed using mAbs against VP60 and NCL in RK-13 cells co-transfected with pVP60-myc and pNCL-Flag plasmids for 24 h. As shown in Fig 2C, NCL was co-localized with RHDV VP60 in the RK-13 cell cytoplasm.
Fig 2. RHDV VP60 interacted with the N-terminal domain of NCL.
(A) NCL binds to VP60 during RHDV internalization. An IP assay was performed on cell lysates using NCL mAb in RK-13 cells that were infected or uninfected with mRHDV, then immunoblotted with mAbs against NCL or VP60. β-actin was employed as an internal control. The cells uninfected with mRHDV served as negative control. (B) Validation of the interaction of VP60 with NCL by the Co-IP assay. RK-13 cells were co-transfected with plasmids expressing myc-VP60 and NCL-Flag. Cell lysates were prepared 48 h post-transfection and the proteins were immunoprecipitated using anti-myc mAb and then immunoblotted with mAbs against tag myc or Flag. β-actin was employed as an internal control. (C) Confocal microscopy analysis of NCL (green) and VP60 (red) in RK-13 cells co-transfected with myc-VP60 and NCL-Flag plasmids for 24 h with mAbs against NCL and VP60. The small white box represents amplified random co-localization spots within the merged image, and the co-localization spot is indicated with a white arrowhead. (D) Schematic representation of the strategy for constructing full-length GST-fusion proteins and the NTD, RBD, and CTD domains of NCL for the GST pull-down assay. The numbers indicate aa positions. (E) GST-fusion protein of NCL and its three domains were purified on glutathione-sepharose beads, and incubated in cell lysates obtained from RK13 cells transfected with the myc-VP60 plasimd for 48 h. After extensive washing, the binding of VP60 was determined by western blot analysis with an anti-VP60 mAb. The GST protein acted as a negative control. (F) Schematic illustration of the full-length and truncated forms of GST-NTD for the GST pull-down assay. The numbers indicate aa positions. (G) GST-NTD proteins were purified on glutathione-sepharose beads and incubated in RK-13 cells lysates prepared 48 h post-transfection with plasmids expressing myc-VP60. After extensive washing, bound VP60 was immunoblotted with anti-VP60 mAb. The GST protein acted as a negative control. (H) Schematic illustration of the full-length and truncated forms of GST-NTD-C for the GST pull-down assay. The numbers indicate aa positions. (I) Western blot analysis of glutathione affinity pull-down assays that were performed to map the locus of NCL binding. GST-fusions of various NCL-NTD fragments (NTD-C1, NTD-C2, and NTD-C3) were used as the bait and VP60 expressed in RK-13 cells as the prey. Binding of the VP60 protein was determined by western blot analysis with anti-VP60 mAb. The GST protein acted as a negative control.
To characterize the critical domain of NCL for NCL-RHDV VP60 interactions, GST-fusion proteins corresponding to NCL and sub-fragments of NCL (GST-NCL, GST-NCL-NTD, GST-NCL-RBD and GST-NCL-CTD, respectively) were prepared for use as bait proteins in glutathione pull-down assays to determine their abilities to interact with the VP60 protein expressed in RK-13 cells (Fig 2D). The results of these assays showed that both GST-NCL and GST-NCL-NTD bound to RHDV VP60, but did not to GST-NCL-RBD, GST-NCL-CTD, and GST (Fig 2E). Subsequently, sub-fragments of NCL-NTD GST-fusion proteins (i.e., GST-NTD-A, GST-NTD-B, and GST-NTD-C) were prepared for use as bait proteins in the GST pull-down assay (Fig 2F). As shown in Fig 2G, both GST-NTD-C and GST-NCL-NTD bound to RHDV VP60, while the other proteins were undetectable. In addition, NCL-NTD-C was split into the fragments NTD-C1 (aa 214–249), NTD-C2 (aa 250–284), and NTD-C3 (aa 285–318), which were fused with GST and expressed (Fig 2H). The results of further pull-down assays showed that NTD-C3 and NTD-C bound to VP60, while the other proteins did not (Fig 2I).
Together these results suggest that RHDV VP60 directly and specifically interacts with the N-terminal residues 285–318 of NCL.
A conserved sequence of RHDV VP60 is critical for binding of the NCL NTD domain
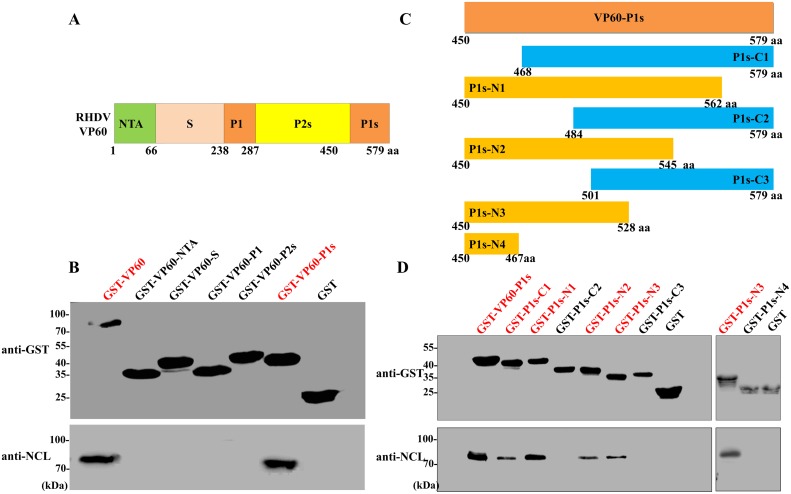
RHDV VP60 is composed of three domains: N-terminal arm (NTA), shell (S), and protrusion (P). The P domain is further divided into the P1 and P2 sub-domains [32]. To identify functional areas of VP60 that interacts with NCL, the GST-VP60 fusion protein and its sub-fragments (i.e., GST-VP60-NTA, GST-VP60-S, GST-VP60-P1, GST-VP60-P2s, and GST-VP60-P1s) (Fig 3A) were prepared for use as bait proteins in GST pull-down assays to determine their abilities to interact with the NCL protein. As shown in Fig 3B, only GST-VP60-P1s and GST-VP60 bound to NCL. These results confirm that binding to NCL requires the P1s domain of VP60.
Fig 3. RHDV VP60 interacts with NCL residues 468–484.
(A) Schematic diagram illustrating the construction of the full-length GST-fusion protein and the five domains of the VP60 protein for the GST pull-down assay. The numbers indicate the aa positions of each domain. (B) Western blotting analysis of glutathione affinity pull-down assays was performed to map the binding domain of the RHDV VP60 protein. GST-fusions of various VP60 domains (VP60-NTA, VP60-S, VP60-P1, VP60-P2s, and VP60-P1s) were used as the bait, while NCL expressed by RK-13 cells was used as the prey. Bound NCL was immunoblotted with anti-NCL mAb. The GST protein acted as a negative control. (C) Schematic illustration of the full-length and truncated forms of GST-VP60 P1s for the GST pull-down assay. The numbers indicate aa positions. (D) GST-P1s proteins were purified on glutathione-sepharose beads and incubated in RK-13 cell lysates containing the NCL protein. After extensive washing, NCL binding was determined by western blot analysis with anti-NCL mAb. The GST protein acted as a negative control.
To further map the VP60 P1s segments responsible for the VP60-NCL interactions, GST-fusion proteins corresponding to sub-fragments of the VP60-P1s domain were prepared by removing 50 aa residues from the N- and C- terminals of VP60-P1s (i.e., GST-P1s-N1, GST-P1s-N2, GST-P1s-N3, GST-P1s-N4, GST-P1s-C1, GST-P1s-C2, and GST-P1s-C3) (Fig 3C). The results of a set of pull-down assays showed that GST-P1s-N1, GST-P1s-N2, GST-P1s-N3, GST-P1s-C1, and GST-P1s interacted with NCL, while the other proteins did not (Fig 3D). These findings indicate that RHDV VP60 interacts with NCL via aa residues 468–484.
To pinpiont the key aa responsible for binding of the RHDV VP60 P1s domain with NCL, blocks of three or four aa substitutions were introduced within and beyond the conserved sequence motif. The following non-conservative substitutions to the GST-tagged RHDV VP60 protein were made: 468RRTG468DDPP, 472DVN472RSP, 475AAA475FPQ, and 482GTQ482KPA. Furthermore, the sequence GSGSGS was inserted after aa residue 483 (Fig 4A). It has been reported that GSGSGS is a flexible peptide for use in different expression systems to separate functional proteins [33]. The wild-type and mutant RHDV VP60 P1s proteins were used as bait proteins in the GST pull-down assays to determine their abilities to bind to NCL. As shown in Fig 4B, the 472DVN472RSP mutation was not able to bind to NCL, while the binding capacities of the other mutations to the residues at positions 468–484 were reduced to different extents. Moreover, as predicted with the SWISS-MODEL online tool (https://swissmodel.expasy.org/), wild-type DVN peptides formed a structure similar to a “claw,” which provides a structural basis for interactions with NCL. However, once the key aa were changed (DVN mutated to RSP), the “claw” structure is broken and loses the ability to bind to NCL (Fig 4C). These results indicate that this “claw” structure is the structural basis for the interaction between VP60 and NCL. In addition, analysis of the VP60 sequence of the G1–G6 genotypes of RHDV showed that the DVN motif was highly conserved (Fig 4D).
Fig 4. The P1s domain of the RHDV VP60 protein contains a conserved sequence motif that binds to the NCL NTD domain.
(A) Sequence alignment showing the location of aa substitutions introduced into the GST-tagged VP60-P1s-C1. (B) Western blot analysis of the glutathione affinity pull-down assays using GST-tagged VP60-P1s-C1 mutants as the bait and NCL expressed in RK-13 cells as the prey. Bound NCL was immunoblotted with anti-VP60 mAb. The GST-P1s-C1 protein served as a positive control and the GST protein as a negative control. (C) Modeling of the RHDV VP60 P1s domain (aa 468–484). For clarity, the structure of the RHDV VP60 protein (aa 468–484) was obtained from the Protein Data Bank under the identification number 4egt.1.B (https://www.rcsb.org/). The structure of the 472DVN472RSP mutation, as predicted by the SWISS-MODEL online tool and based on homology molecules found in the Protein Data Bank. The DVN peptide is shown in red. (D) Alignments of representative aa sequences of the VP60 protein of the G1–G6 genogroups of RHDV. The representative strains used in the alignments were G1 RHDV-SD (GenBank accession no. Z29514.1), G2 RHDV strain WX/China/1984 (GenBank accession no. AF402614.1), G3 RHDV isolate 13086-PT11 (GenBank accession no. KJ683906.1), G4 RHDV isolate 00–13 (GenBank accession no. AJ495856), G5 RHDV isolate Hagenow (GenBank accession no. Y15441), G6 RHDV isolate 29778-PT08 (GenBank accession no. KJ683905), and RHDV strain JX/CHA/97 (GenBank accession no. DQ205345.1). Sequence alignment was performed with the ClustalW algorithm (http://www.clustal.org/). The conserved NCL-binding motif is boxed. Selected aa from this motif are indicated with RHDV VP60 numbering.
These observations confirm that RHDV VP60 interacts with NCL via the DVN (Asp-Val-Asn) motif, which is a conserved sequence in RHDV.
The interaction between VP60 and NCL plays a key role in RHDV internalization
The above results indicate that NCL was required during RHDV internalization and interacted with the RHDV capsid protein (VP60). Therefore, we speculated that NCL is involved in RHDV internalization via binding to VP60. To verify this hypothesis, we blocked the binding site of NCL-VP60 with a synthesized DVN peptide (RRTGDVNAAAGSTNGTQ) and examined its effect on RHDV entry. The results of the attachment and internalization assays showed that mRHDV internalization was drastically reduced in RK-13 cells treated with the DVN peptide, with only 40% internalization, but there was no obvious effect on mRHDV attachment (Fig 5A).
Fig 5. Synthetic DVN peptides significantly inhibit RHDV internalization.
(A) RK-13 cells were pretreated with DVN or control peptides at a concentration of 100 μg/mL for 6 h. Then, the attachment and internalization assays were performed with mRHDV-infected cells (MOI = 1) and the cells harvested for RNA analysis. The percentage of RHDV VP60 copies was calculated as a ratio to the value obtained from untreated RK-13 cells. The control peptide was employed as a negative control. (B) Kinetics of mRHDV internalization into RK-13 cells. Internalization of mRHDV (MOI = 1) or mRHDV-FITC into RK-13 cells was assessed by mRHDV incubation for 10, 30, and 60 min at 37°C. The amount of internalized virus was determined by qRT-PCR and expressed as VP60 RNA copies per 100,000 of GADPH. (C) RK-13 cells were pretreated with the DVN and control peptides at concentrations of 5, 10, 20, 40, 80, and 100 μg/mL for 6 h, respectively. mRHDV-FITC-infected cells (MOI = 1) were then used for internalization assays and harvested for flow cytometry analyses. The control peptide was employed as a negative control. The data are representative of the means and standard deviations (error bars) of three samples per group (**p < 0.01). All experiments were repeated at least three times with consistent results. (D) RK-13 cells were pretreated with the DVN peptides (100 μg) or anti-NCL mAb (50 μg) for 6 h. Then, the cells were used for the internalization assay with mRHDV-FITC (MOI = 1) and subsequent immunofluorescence analysis. The RHDV virion is green and cell nucleus in blue. Mouse IgG and control peptides were employed as negative controls.
To image the internalization of single RHDV particles in live cells, the fluorescent dye Alexa Fluor 488 was conjugated to purified virions. This labeling process did not significantly reduce the viral titer, as measured by the qRT-PCR assay (Fig 5B). Alexa Fluor 488-conjugated mRHDV (RHDV-FITC) was used to evaluate the efficiency of viral internalization in response to different doses of the DVN peptide as well as a control peptide (RHDV VP60 residues 434–450: VTYTPQPDRIVTTPGTP). RK-13 cells were pretreated with DVN peptide or a control peptide at a concentration of 5, 10, 20, 40, 80, or 100 μg/mL, respectively for 6 h, and then incubated with RHDV-FITC (MOI = 1) for 2 h at 4°C and washed with chilled PBS. The temperature was then shifted to 37°C to facilitate viral internalization. At 30 min post infection, when RHDV internalization was proportionally increasing, the cells were washed in pre-chilled acidic PBS (pH = 2.5) to remove non-internalized viruses. The amount of internalized mRHDV was quantitated by flow cytometry. As shown in Fig 5C, with the increase in the DVN peptide, the amount of RHDV internalized in RK-13 cells was significantly reduced, and when the cells were treated with DVN peptide (80 μg/mL), the amount of virus in the treated cells was only about half of the control group. In addition, fluorescence microscopy also showed that the amount of internalized mRHDV was drastically reduced in RK-13 cells treated with either the DVN peptide or NCL mAb (Fig 5D).
These results suggest that blocking the binding site of the interaction between VP60 and NCL resulted in a significant reduction in RHDV internalization.
Clathrin-dependent endocytosis is involved in RHDV internalization
It is well known that viruses typically make use of various pinocytic mechanisms of endocytosis that serve the cell by promoting the uptake of fluid solutes, and small particles. The mechanisms include clathrin-dependent endocytosis, the most commonly used mechanism for virus, macropinocytosis, a transient, ligand-induced, actin-dependent mechanism, and lipid raft-dependent endocytosis, a lipid raft-dependent mechanism mainly used by polymaviruses [34]. A previous study reported that RHDV VLPs use clathrin-dependent endocytosis to enter mouse and human dendritic cells [35]. However, the mechanism by which RHDV enters rabbit cells remains unclear. In order to determine the type of endocytosis involved in RHDV internalization, a series of experiments were conducted to examine RHDV entry into RK-13 cells. First, the cells were treated with 30 mM CPZ, an inhibitor of clathrin-dependent endocytosis, which inhibits clathrin triskelions assembly, 100 mM Nys, an inhibitor of lipid raft-dependent endocytosis, which disrupts membrane lipid rafts, 50 mM 5-EIPA, an inhibitor of macropinocytosis, which is a selective blocker of Na+/H+ pump exchanger, or DMSO, respectively. The effect of inhibitors on RHDV-FITC internalization was quantified by qRT-PCR, flow cytometry, and fluorescence microscopy. The results showed that CPZ, as well as NCL siRNA, inhibited RHDV internalization, as the percentage of internalization was about 40%, but not Nys or EIPA (Fig 6A–6C), suggesting that clathrin-dependent endocytosis is involved in the process of RHDV entry into rabbit cells. Our results are consistent with those of previous studies, suggesting that clathrin-dependent endocytosis is involved in RHDV internalization.
Fig 6. Clathrin-dependent endocytosis is involved in RHDV internalization and is regulated by NCL.
(A) Clathrin-dependent endocytosis is involved in RHDV internalization. RK-13 cells of three groups were pretreated with CPZ (30 mM), EIPA (50 mM), or Nys (100 mM) for 2 h. Another group of cells was transfected with NCL siRNA (100 nM) for 24 h. Cells were then prompted to internalize mRHDV (MOI = 1) and harvested for RNA analysis. The percentage of RHDV VP60 copies was calculated as a ratio to the value obtained from untreated RK-13 cells. DMSO and non-specific siRNA were used as negative controls. The data are presented as the mean and standard deviation (error bars) of three samples per group (**p < 0.01). All experiments were repeated at least three times and showed consistent results. (B) mRHDV-FITC (MOI = 1) infected RK-13 cells, treated with procedures described above, were subjected to the internalization assay and harvested for flow cytometry analysis. (C) RK-13 cells of three groups were pretreated with CPZ (30 mM), EIPA (50 mM), or Nys (100 mM) for 2 h. Another group cells was transfected with NCL siRNA (100 nM) for 24 h. Then, the cells were used for the internalization assay with mRHDV-FITC (MOI = 1) and subsequent immunofluorescence analysis. The RHDV virion is green, cell nucleus is blue. DMSO and control siRNA were employed as negative controls.
NCL plays a key role in clathrin-dependent endocytosis by interacting with the C-terminal domain of CLTA
Previous studies have shown that NCL is required for clathrin-dependent endocytosis in human and murine cells [27,36]. Here, FITC-conjugated ligands were used to evaluate the efficiency of cargo uptake by siRNA-treated and untreated RK-13 cells. The effect of NCL siRNA on all endocytic pathways was quantified by flow cytometry. The results showed that NCL siRNA inhibited the uptake of transferrin and EGF, which were used as representative markers of clathrin-dependent endocytosis [37], but had no effect on the uptake of the representative markers of the lipid raft-dependent endocytosis (CD4 [38]) and macropinocytosis (dextran [39]) (S4A–S4D Fig). These results showed that NCL also plays a key role in clathrin-dependent endocytosis in RK-13 cells.
However, the molecular mechanism underlying NCL involvement in clathrin-dependent endocytosis remains unclear. To elucidate the molecular mechanism, RK-13 cell lysates were used for affinity purification with NCL mAb. Briefly, the eluted protein complexes were resolved by SDS-PAGE and the protein bands were visualized by silver staining and identified by mass spectrometry analysis. As shown in Fig 7A, many host proteins were bound to NCL, including adaptor protein 2 (AP2) and CLTA, which are necessary for clathrin-dependent endocytosis. CLTA participates in several membrane traffic pathways involving both clathrin and actin by binding with actin-organizing huntingtin-interacting proteins [40]. Moreover, IP experiments were performed with an NCL mAb in RK-13 cells. Western blot analysis using a mAb against AP2 or CLTA showed a band corresponding to CLTA, but not AP2, indicating an interaction between CLTA and NCL (Fig 7B). Furthermore, the crucial domain of the interactions between NCL and CLTA was investigated using the glutathione pull-down assays. The GST-fusion proteins corresponding to CLTA and sub-fragments of CLTA (GST-CLTA, GST-CLTA-N, GST-CLTA (aa 81–120), GST-CLTA (aa 121–160), and GST-CLTA-C, respectively) were prepared for use as bait proteins to determine their abilities to interact with the NCL protein expressed in RK-13 cells (Fig 7C). These results showed that CLTA-GST and GST-CLTA-C bound to NCL, but no GST-CLTA-N, GST-CLTA(aa 81–120), GST-CLTA(aa 121–160) or GST (Fig 7D).
Fig 7. NCL binds to the C-terminal domain of CLTA.
(A) Screening of host proteins that interacted with the NCL protein. SDS-PAGE analysis of immunoprecipitation-purified host factors of RK-13 cells that interact with the NCL protein was performed with NCL mAb. The bands were identified by mass spectrometry. In addition, M is a protein marker and the negative control was cell lysates immunoprecipitation-purified with mouse IgG. (B) Validation of the interaction between CLTA and NCL with the IP assay. Proteins in the lysates of RK-13 cells were immunoprecipitated with anti-CLTA mAb, anti-AP2 mAb, or mouse IgG, and then immunoblotted with anti-NCL mAb. β-actin was employed as an internal control. (C) Schematic illustration of the full-length and truncated forms of GST-CLTA for the GST pull-down assay. The numbers indicate aa positions. (D) GST-CLTA proteins were purified on glutathione-sepharose beads and incubated in RK-13 cells lysates containing the NCL protein. After extensive washing, the binding of NCL was determined by western blot analysis with anti-NCL mAb. The GST protein acted as a negative control.
Taken together, these results suggest that NCL plays a key role in clathrin-dependent endocytosis by directly and specifically interacting with CLTA via the C-terminal aa residues 161–219.
The interaction between CLTA and NCL plays an important role in RHDV internalization
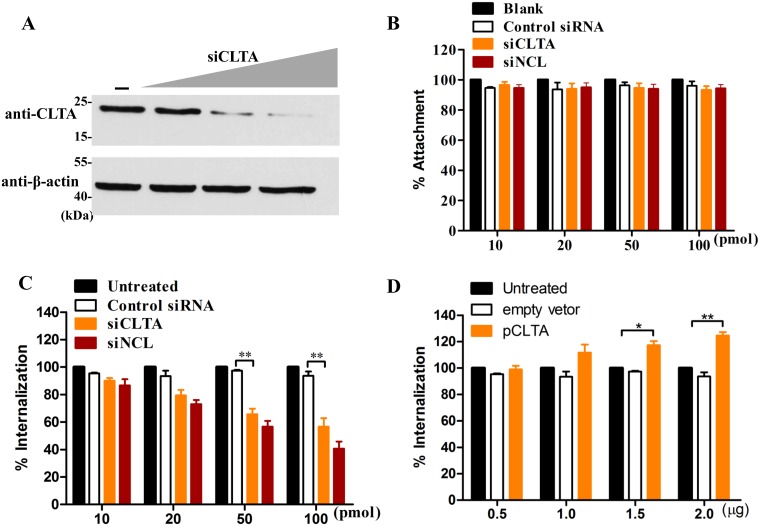
The results of the above studies showed that NCL is involved in RHDV internalization by binding to VP60. In addition, clathrin-dependent endocytosis is involved in RHDV internalization and NCL participates in clathrin-dependent endocytosis by interacting with CLTA. To investigate the role of the interaction between CLTA and NCL in RHDV internalization, the expression level of CLTA was changed by siRNA (Fig 8A) or myc-CLTA plasmid (S2B Fig) transfection. The results of the attachment and internalization assays showed that mRHDV internalization was reduced in RK-13 cells treated with CLTA or NCL siRNA (Fig 8C), and increased by overexpression of CLTA (Fig 8D), athough there was no obvious influence on mRHDV attachment (Fig 8B). Of note, the effect on mRHDV internalization is dependent on the dose of CLTA siRNA, NCL siRNA or the myc-CLTA plasmid. The percentage of mRHDV internalization in RK-13 cells was about 41%, 56% and 125%, following treatment with CLTA siRNA (100 pmol), NCL siRNA (100 pmol) or myc-CLTA (2 μg), respectively (Fig 8C and 8D). These data showed that CLTA is involved in RHDV internalization, confirming that the interaction between CLTA and NCL plays an important role in RHDV internalization.
Fig 8. The interaction between CLTA and NCL plays an important role in RHDV internalization.
(A) The expression level of CLTA was determined by western blot analysis with anti-CLTA mAb. Cell lysates were obtained from RK-13 cells transfected with CLTA siRNA (10, 20, 50, or 100 nM) for 24 h. β-actin was employed as an internal control. (B) The effect of CLTA siRNA on RHDV attachment. Before the addition of RHDV (MOI = 1) for 2 h at 4°C, RK-13 cells were transfected with CLTA siRNA. Non-specific siRNA was used as a control. siNCL acted as a positive control. The percentage of attached RHDV VP60 copies was calculated as a ratio to the value obtained from untreated RK-13 cells. (C-D) The effect of CLTA on RHDV internalization. Before the addition of RHDV (MOI = 1) for 2 h at 4°C, RK-13 cells were transfected with CLTA siRNA (C) or the pCLTA plasmid (D). Non-specific siRNA or pCMV-Myc was used as negative control. siNCL acted as a positive control. The percentage of internalized VP60 copies was calculated as a ratio to the value obtained from untreated RK-13 cells. The student’s t-tests and analyses of variance were used for statistical analyses, *p < 0.05 and **p < 0.01. The experiments were conducted in triplicate and produced similar results.
The DVN peptide provides some protection against RHDV infection in rabbits
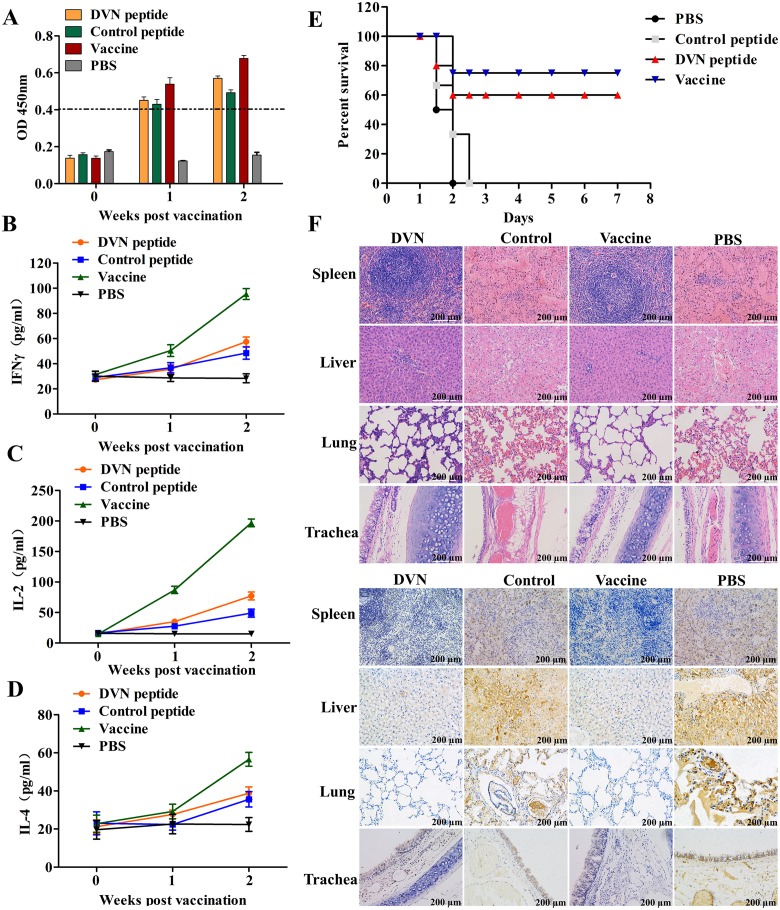
The above experimental results were obtained with RK-13 cells infected with mRHDV. In order to assess the role of NCL-VP60 interaction during infection with wild-type RHDV, rabbits were immunized with the DVN peptide, control peptide, commercial inactivated RHDV vaccine, or PBS. The VP60-specific Ab responses of immunized rabbits were determined using an indirect ELISA. As shown by the results presented in Fig 9A, the Ab titers of VP60 from the DVN peptide, control peptide, and commercial vaccine groups gradually increased. Statistically, the Ab titers were significantly higher (p < 0.05) in the DVN peptide, control peptide, and commercial vaccine groups than in the PBS group at 0, 1 and 2 weeks post-immunization. Simultaneously, in order to investigate cell-mediated immune responses, the amounts of IFN-γ, IL-2, and IL-4 were measured by ELISA. As shown in Fig 9B–9D, there was no significant difference in the levels of IFN-γ, IL-2, and IL-4 over time between the DVN peptide and control peptide groups. However, the levels of IFN-γ, IL-2, and IL-4 were significantly higher (p < 0.05) in the commercial vaccine group than in the PBS group at 1 and 2 weeks post-immunization. These results indicated that the DVN peptide was unable to activate the cellular immune response in rabbits. At 21 days post immunization, all rabbits were challenged intramuscularly with wild-type RHDV. As shown in Fig 9E, the survival rates of rabbits against virulent RHDV in the DVN peptide and commercial vaccine groups were 60% and 80%, respectively. However, all rabbits in the control peptide group and PBS group (negative control) died within 48–72 hpi with virulent RHDV. These negative control animals exhibited clinical symptoms of RHDV infection. Moreover, histopathological analysis indicated that typical pathological changes, such as diffuse bleeding of tissues and organs, were observed in the control peptide and PBS groups, but the tissues and organs of rabbits in the DVN peptide and commercial vaccine groups showed no pathological changes. Similarly, immunohistochemical analysis indicated large amounts of RHDV virions in the tissues of rabbits in the control and PBS groups, but not in the DVN peptide and commercial vaccine groups (Fig 9F).
Fig 9. DVN peptides provide some protection to rabbits against RHDV infection.
(A) RHDV-specific Ab responses of rabbits to the DVN peptide. Twenty 8-week-old rabbits that were seronegative for RHDV were randomly distributed into four groups (n = 5/group) and immunized with the DVN peptide (1 mg), control peptide (1 mg), commercial inactivated RHDV vaccine, or PBS, respectively. Rabbit sera from different groups were collected at 0, 1, and 2 weeks post-immunization, and subjected to an indirect ELISA. Optical density (OD) values at a wavelength of 450 nm were measured in quadruplicate. (B) IFN-γ production by lymphocytes in immunized rabbits. (C) IL-2 production by lymphocytes in immunized rabbits. (D) IL-4 production by lymphocytes in immunized rabbits. Serum levels of IL-2, IL-4, and IFN-γ were measured by an ELISA. Samples were tested in quadruplicate and data are presented as the mean ± standard deviation. (E) The survival of rabbits infected with RHDV JX/CHA/97. All rabbits were challenged intramuscularly with 100 LD50 of RHDV at 21 days post-immunization and subsequently clinically examined daily for 7 days. (F) The results of postmortem histopathological and immunohistochemical analyses. All animals were necropsied and organs were collected. For the histopathological analysis, samples were fixed in 10% neutral buffered formalin solution, sectioned, and stained with hematoxylin and eosin. Alternatively, immunohistochemical analysis was conducted with anti-VP60 mAb.
These data show that the DVN peptide provides partial protection against RHDV in rabbits, suggesting that the interaction between VP60 and NCL plays a key role in RHDV internalization.
Discussion
NCL is well known as a multifunctional protein that is mainly localized in the nucleolus, but is also found in the nucleoplasm and cytoplasm, as well as on the cell membrane [10]. The multi-functionality of NCL mainly results from its multi-domain structure. Sequence comparison of different species revealed a high degree of evolutionary conservation of this large protein, and biophysical and biochemical studies have shown that NCL is composed of three main structural domains: the N-terminal domain, the central domain, and the C-terminal domain [41]. The N-terminal domain of NCL contains acidic regions, rich in glutamic acid and aspartic acid, which are the sites of phosphorylation, and participate in the transcription of rRNA and interact with components of the pre-rRNA processing complex [16]. The central domain of NCL contains four RNA-binding domains (RBDs), also known as RNA-recognition motifs, which are involved in a variety of biological processes, including RNA packaging, pre-mRNA splicing, poly-A tail synthesis and maturation, translational control, and mRNA stability [11]. The C-terminal region of NCL contains a glycine- and arginine-rich domain, through which NCL interacts with target mRNAs and proteins, including ribosomal proteins [9].
NCL is involved in several cellular functions, including ribosome biogenesis, DNA and RNA metabolism, cell proliferation, apoptosis regulation, and stress responses [42]. Moreover, increasing evidence suggests that NCL is also involved in the pathogenesis of many viruses. For viral infection, cell surface NCL has been shown to be involved by promoting either initial attachment of the virus to the cell surface or entry into host cells. For instance, NCL was found to serve as a cellular receptor during infection with human respiratory syncytial virus [22]. Cell surface NCL also mediates the binding and internalization of enterovirus 71 [43]. By targeting NCL on the cell surface, HB-19 pseudo-peptides inhibit attachment of human immunodeficiency virus to the cell surface as well as subsequent viral entry into the host cell [44]. Furthermore, cell surface NCL ligands, such as midkine and lactoferrin, have similar effects during viral infection [45]. In addition, NCL serves as a conserved cellular factor that is required for cell entry of multiple influenza A viruses, including H1N1, H3N2, H5N1, and H7N9. Suppression of the expression or function of cell surface NCL by siRNA or blocking Abs substantially reduced internalization of the influenza virus [9,25,26,29].
In this study, we found that NCL is also involved in RHDV infection and plays a key role in RHDV internalization. It has been reported that HBGAs are functional receptors of RHDV and play key roles in viral attachment to the cell surface [5]. Data derived from the three-dimensional structure of RHDV suggest that HBGAs mainly bind to the P region of the RHDV capsid protein, thereby triggering RHDV infection of host cells [7,9]. However, the molecular mechanism underlying RHDV invasion of host cells remains unknown. The results of the present study showed that NCL was involved in RHDV internalization. In other words, following RHDV binding to the cell receptor (HBGA), the viral internalization process is completed with the help of NCL, which acts as a bridge that connects RHDV VP60 and CLTA. In this study, the NTD domain of NCL was found to bind to the RHDV VP60 protein via a conserved sequence at the P1s domain of VP60, and the internalization efficiency of RHDV was significantly inhibited by blocking these interactions. This finding was also confirmed by the animal experiments. Rabbits were partially protected from RHDV assault (~ 60%) after inoculation with synthetic DVN peptides. In the designed experiment, activation of the humoral immune response of rabbits and resulted in the production of specific Abs against DVN. However the cellular immune response was very low. We speculated that the specific Abs induced by DVN peptides blocked the internalization of RHDV, and provided protection to the rabbits.
However, the role of NCL in RHDV infection differs from that of other viruses. The results of this study showed that NCL does not directly mediate viral entry into cells as a functional receptor for RHDV, but still plays a key role in the RHDV internalization stage, as NCL was found to mediate the entry of RHDV into the cells by clathrin-dependent endocytosis via binding to the C-terminus of CLTA. It is well known that clathrin-dependent endocytosis is a classical endocytic pathway that transports extracellular substances into the cell, and most enveloped and non-enveloped viruses enter the host cell by this endocytic pathway [34]. Clathrin consists of light and heavy chains, and forms a triangular complex structure. When an extracellular substance binds to the receptor, the clathrin complex is recruited to the cell membrane surface in response to endocytic signaling and pulls the substance down to the cell membrane, where it becomes trapped and forms clathrin-invaded pits. Under the action of dynein, the invaginated pits leave the cell membrane and form clathrin-coated vesicles that eventually pass the endocytosed material to endosomes [40]. Previous studies have shown that NCL is involved in the internalization of the EGF receptor, which is mediated by clathrin-dependent endocytosis [46]. Also, NCL is involved in clathrin-dependent endocytosis through interactions with the C-terminus of CLTA. This discovery should prove helpful to further understand clathrin-dependent endocytosis. Inhibition of clathrin-dependent endocytosis by drugs inhibited the internalization of RHDV, which further demonstrated that clathrin-dependent endocytosis indeed plays an important role in RHDV infection. Together, these results demonstrated that RHDV is internalized by the host cell via clathrin-dependent endocytosis, which is mediated by NCL.
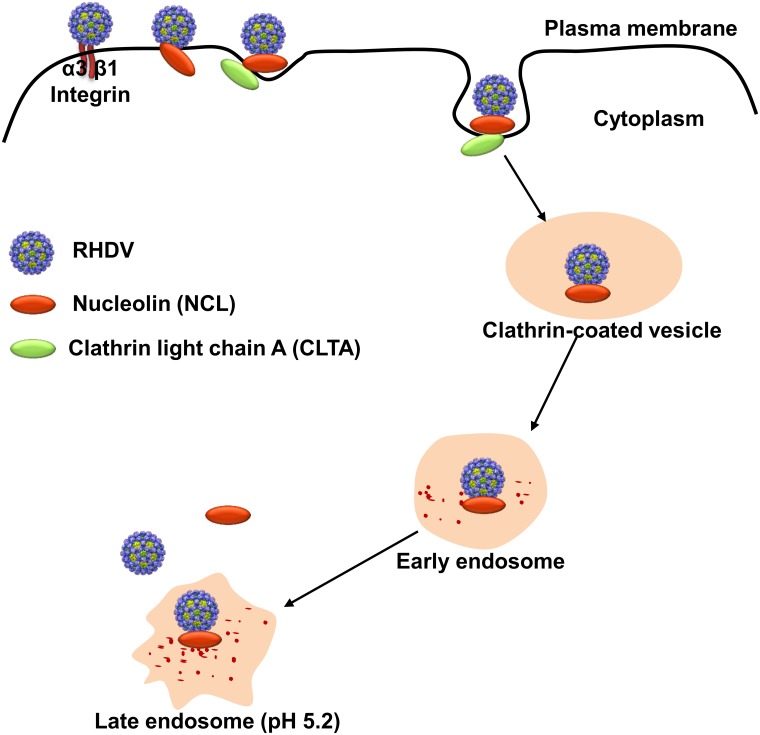
In conclusion, this study is the first to report that NCL plays a key role in the infection mechanism of RHDV. As shown in Fig 10, the capsid protein of RHDV and the host clathrin are connected through NCL and form a triprotein complex, thereby completing the intrusion process of RHDV through clathrin-dependent endocytosis. To our knowledge, this is the first report of the infection mechanism of RHDV at the molecular level. These results enrich current knowledge of the pathogenic mechanism of RHDV and provide important clues for the development of novel vaccines against RHDV infection.
Fig 10. RHDV internalization model.
First, RHDV attaches to the RK-13 cell surface by binding to specific cell receptors. Then, the capsid protein of RHDV and the host clathrin are bridged by NCL, and form a protein complex. Finally, RHDV is internalized by the host cell via clathrin-dependent endocytosis.
Materials and methods
Ethics statement
All experiments were performed in a secondary biosecurity laboratory. All experiments involving rabbits and mice were carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China, and all efforts were made to minimize suffering. All animal procedures were approved by the Institutional Animal Care and Use Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (permit number: SHVRIAU-7-0103).
Cell lines and viruses
Rabbit kidney cells (RK-13, ATCC CCL37) and 293T cells (ATCC, CRL-3216) were grown in minimal essential medium (Life Technologies, Carlsbad, CA, USA) or Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum under an atmosphere of 5% CO2 at 37°C. RHDV strain JX/CHA/97 was isolated in 1997 during an outbreak of RHDV in China and stored in our laboratory. mRHDV was constructed in 2017 with a reverse genetics technique and stored in our laboratory [8].
Plasmid constructs
The p3×FLAG-CMV-14 vector (Sigma-Aldrich Corporation, St. Louis, MO, USA) and pCMV-Myc (Clontech Laboratories, Inc., Mountain View, CA, USA) were used to create mammalian expression constructs. The pGEX-4T-1 vector (GE Healthcare Life Sciences, Chicago, IL, USA) was expressed in competent E. coli BL21-CodonPlus (DE3) cells. NCL (GenBank accession number XM_017343189.1) and CLTA (GenBank accession number XM_002707987.3) sequences were amplified by reverse transcription polymerase chain reaction (RT-PCR) from a RK-13 cell cDNA library. Total RNA was isolated from RK-13 cells using TRIzol reagent (Invitrogen Corporation, Carlsbad, CA, USA) according to the manufacturer’s instructions. DNA was removed from the isolated RNA using DNaseI (Takara Bio, Inc., Shiga, Japan) and cDNA was produced with Moloney murine leukemia virus reverse transcriptase (Promega Corporation, Madison, WI, USA) and random hexamers. The RHDV VP60 gene was amplified by RT-PCR from RHDV cDNA. The genomic sequence of RHDV CHA/JX/97 was retrieved from the GenBank database (accession number DQ205345). Viral cDNA was generated as described in our previous report [47]. All plasmids were created with In-Fusion HD Cloning Kits (Clontech Laboratories, Inc.) according to the manufacturer’s instructions. All RT-PCR amplifications for cloning were performed with TransStart FastPfu Fly DNA Polymerase (TransGen Biotech Co., Ltd., Beijing, China) according to the manufacturer’s instructions. RT-PCR products were separated by agarose gel electrophoresis and purified with the SanPrep Column DNA Gel Extraction Kit (Sangon Biotech Co., Ltd., Shanghai, China). Restriction digests were performed using commercial kits (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s instructions. All plasmid sequences were amplified by RT-PCR and analyzed by Sanger sequencing to verify the sequence fidelity and reading frames (Sangon Biotech Co., Ltd.). Details of all constructs used in the study, including residue numbers, expression vectors, and tags, are summarized in S1 Table. In addition, the primers used in this research are listed in S2 Table.
Bacterial expression of recombinant proteins and purification
All proteins were expressed in competent E. coli BL21-CodonPlus (DE3) cells (TransGen Biotech Co., Ltd.) that were seeded in 1 mL of an overnight starter culture and then grown in 100 mL of lysogeny broth at 220 rpm and 37°C until the mid-log phase (OD600 = ~0.6–0.8). Then, the cells were typically induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside and incubated for approximately 16 h at 16°C and 220 rpm. Cells were pelleted by centrifugation at ~5,000 g and stored at -80°C. Bacterial pellets were resuspended in lysis buffer (20 mM Tris/HCl pH 7.4, 60 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1 mg/mL lysozyme, 1 mM dithiothreitol, and 0.1% Triton X-100) supplemented with complete protease inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA, USA) for 1 h on ice. Nuclease was then added and the lysate was incubated for 1 h at ambient temperature under rotation. The lysates were centrifuged at 4°C for 10 min at 12,000 × g. Glutathione-Sepharose 4B beads (Pierce Biotechnology, Waltham, MA, USA) were added to the clarified supernatants and the mixtures were incubated overnight at 4°C under rotation. The beads were washed with lysis buffer, followed by three washes with phosphate-buffered saline (PBS), and then stored at 4°C in an equal volume of PBS.
Glutathione-S-transferase (GST) pulldown assay
For the in vitro binding assay, Flag- or myc-tagged NCL, CLTA, and VP60 proteins were expressed in RK-13 cells. In accordance with the manufacturer’s instructions, the GST pull-down assay was performed by incubating 50 μL of a 50% slurry of glutathione-sepharose beads containing 25 μM GST fusion protein in lysis buffer with a three-fold molar excess of prey protein (Pierce Biotechnology). The bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then subjected to western blot analysis.
Co-immunoprecipitation (Co-IP) analysis
RK-13 cells were co-transfected with the bait and prey plasmids. At 48 h after transfection, total protein was isolated from RK-13 cells using IP lysis buffer. Co-IP analysis was conducted using a commercial Co-IP kit (Pierce Biotechnology) according to the manufacturer’s instructions. AminoLink Plus Coupling Resin was incubated with anti-NCL monoclonal antibody (mAb) (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or anti-VP60 mAb, which was prepared in our laboratory, and then subjected to SDS-PAGE. Immunoblot analysis of the proteins was subsequently conducted using mAbs against NCL, VP60, CLTA, and AP2 (Abcam, Cambridge, UK).
Western blot analysis
Protein samples were separated on 12% gels and then transferred to nitrocellulose membranes (Hybond-C; Amersham Life Sciences, Little Chalfont, UK) using a semi-dry transfer apparatus (Bio-Rad Laboratories, Hercules, CA, USA). The membranes were blocked with 5% (w/v) nonfat milk in TBST buffer (150 mM NaCl, 20 mM Tris, and 0.1% Tween-20, pH 7.6) for 3 h at 4°C and then stained overnight at 4°C with a primary antibody (Ab). After washing three times for 10 min each time, the membranes were incubated with a secondary Ab against immunoglobulin G (IgG) conjugated to horseradish peroxidase (Sigma-Aldrich Corporation) in PBST buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, and 0.1% Tween-20 pH 7.4) for 1 h at room temperature (RT). Finally, after washing three times for 10 min each time, the proteins were detected using an automatic chemiluminescence imaging analysis system (Tanon Science & Technology Co., Ltd., Shanghai, China).
Immunofluorescence assay (IFA)
Cells were fixed in 3.7% paraformaldehyde in PBS (pH 7.5) at RT for 30 min and subsequently permeabilized by incubation in methanol at -20°C for 30 min. The fixed cells were blocked with 5% (w/v) nonfat milk in PBST buffer for 3 h at 4°C and then stained with a primary Ab for 2 h at 37°C. After washing three times for 10 min each time, the cells were incubated with a secondary Ab against IgG conjugated to fluorescein isothiocyanate (FITC) (Sigma-Aldrich Corporation) in PBST buffer for 1 h at RT. Finally, after washing three times for 10 min each time the samples were observed under a fluorescence microscope equipped with a video documentation system (Nikon Corporation, Tokyo, Japan).
Attachment and internalization of RHDV
The kinetics of RHDV attachment was studied by adding the virus at a MOI of 0.25–2 to chilled RK-13 cells. After incubation for 2 h at 4°C, the cells were washed extensively with chilled PBS and then lysed. For the internalization analysis, the cells were washed extensively with chilled PBS following attachment for 2 h and the temperature was shifted to 37°C to allow internalization of the attached viruses. At specific time points post-internalization, the cells were washed extensively with acidic PBS (pH 2.5) and PBS to remove viruses attached to the cell surface. The washed cells were lysed and the total RNA of each lysate sample was extracted with the Spin Column RNA Cleanup & Concentration Kit (Sangon Biotech Co., Ltd.).
Fluorescent ligand uptake and measurement
NCL siRNA-treated (24 h post-transfection) RK-13 cells were serum-starved in DMEM for 3 h, rinsed extensively with chilled PBS, and cold-bound with Alexa Fluor 488-conjugated epidermal growth factor (EGF), Alexa Fluor 488-conjugated transferrin, Alexa Fluor 488-conjugated dextran, and Alexa Fluor 488-conjugated CD4 at concentrations of 2 μg/mL for 1 h at 4°C, as recommended by the manufacturer (Life Technologies). Unbound ligands were removed by extensive washing with cold PBS and the cells were incubated in warm DMEM at 37°C for the designated times. At the indicated time-points, non-internalized ligands were removed by washing with acidic PBS (pH 2.5). Internalized fluorescent signals in the cells were measured by flow cytometry.
To image the internalization of single RHDV particles in live cells, purified virions were conjugated to the fluorescent dye Alexa Fluor 488 (Molecular Probes, Invitrogen Corporation), solubilized in dimethyl sulfoxide (DMSO) at 10 mg/mL, and incubated at a final concentration of 50 μg/mL with purified RHDV (1 mg/mL) in 0.1 M NaHCO3 (pH 8.3) for 90 min at RT. The viruses were separated from free dye by differential centrifugation, solubilized in PBS containing 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; pH 7.4), and stored at -80°C. To measure the effect of labeling on viral internalization in RK-13 cells, equivalent amounts of total viral protein from labeled and unlabeled preparations were titrated by quantitative real-time polymerase chain reaction (qRT-PCR).
Effects of inhibitors on RHDV internalization
RK-13 cells were pretreated for 30 min at 37°C with each of the inhibitors followed by cold-synchronized infection. Where indicated, inhibitors were added to previously infected cells at 2 or 3 hpi. Stock solutions of 50 mM nystatin (Nys) (Sigma-Aldrich Corporation) and 50 mM 5-ethylisopropyl amiloride (EIPA) (Sigma-Aldrich Corporation) were dissolved in DMSO. Water was used as a solvent for 10 mM chlorpromazine (CPZ) (Sigma-Aldrich Corporation). All inhibitors were present throughout the experiments. This study was required given that we used the highest doses reported in the literature to ensure the efficacy of the inhibitors. We also assessed the cytotoxic activity of the organic solvent DMSO. Based on the results of these experiments, optimal non-toxic working concentrations were selected for the internalization assay. For the blocking experiments, RK-13 cells were pretreated in serum-free minimal essential medium containing DMSO as a control, 100 mM Nys, 50 mM EIPA, or 30 mM CPZ. These drug-treated cells were subsequently infected with RHDV at a MOI of 1, incubated in growth medium for 1 h at 37°C, and harvested for flow cytometry or qRT-PCR analysis.
qRT-PCR analysis
The vRNA-VP60 copy numbers of attached and internalized mRHDV were estimated by qRT-PCR. The amount of mRNA in each sample was normalized to that of rabbit glyceraldehyde 3-phosphate dehydrogenase (GADPH). Total RNA was extracted from cell lysate using TRIzol reagent (Invitrogen Corporation) according to the manufacturer’s instructions. All qRT-PCR reactions were performed with the SYBR PreMix Ex Taq II kit (Takara Bio, Inc.) with an ABI 7500 Fast Real Time PCR system (Applied Biosystems, Carlsbad, CA, USA). For VP60, the qRT-PCR reaction was performed as follows: one cycle at 95°C for 30 s, then 40 cycles at 95°C for 5 s, 58°C for 5 s and 72°C for 30 s, and one cycle to generate a melting curve. The primers are listed in S2 Table.
Immunization and viral challenge
Twenty 8-week-old New Zealand male rabbits seronegative for RHDV were randomly distributed into four groups (n = 5/group) and housed in individual ventilated cages. All experimental protocols were reviewed by a state ethics commission and have been approved by the competent authority. The details of the protection assay are shown in S4 Table. Rabbits were immunized with DVN peptide (1 mg), a control peptide (1 mg), a commercial inactivated RHDV vaccine (Nanjing Tianbang Bio-industry Co., Ltd., Nanjing, China), or PBS, respectively. All of the groups were immunized on day 0. Blood samples were collected from the marginal ear vein before each immunization and 2 weeks after the last injection to analyze the level and specificity of the Ab response against RHDV. Ab titers were assessed using an enzyme-linked immunosorbent assay (ELISA). Briefly, RHDV strain JX/CHA/97 (100 μL, 1 μg/mL, incubated overnight at 4°C) was used to capture Abs in the sera (incubated for 1 h at 37 °C) and then detected with 100 μL of horseradish peroxidase-conjugated mouse anti-rabbit IgG (Sigma-Aldrich Corporation) per well (diluted 1:5000 in PBS containing 0.5% Tween 20 and 10% fetal bovine serum), followed by 100 μL of tetramethylbenzidine liquid substrate (Sigma-Aldrich Corporation) per well for 30 min at RT in the dark. End-point titers were defined as the highest plasma dilution that resulted in an absorbance value (OD450). Data are presented as log10 values. In addition, all rabbits were challenged intramuscularly with 100 × the median lethal dose (LD50) of RHDV at 21 days after immunization. The rabbits were clinically examined daily for 7 days post-challenge. All animals were necropsied and serum and organs were collected and stored at -30°C. For histopathological analysis, samples were fixed in 10% neutral buffered formalin solution, sectioned, and stained with hematoxylin and eosin. Alternatively, immunohistochemical analysis was conducted with a VP60-specific mAb. To evaluate the efficiency of the cellular responses, the serum levels of interferon (IFN)-γ, interleukin (IL)-2 and IL-4 at weeks 0, 1, and 2 post-immunization were measured using commercially available ELISA kits (R&D Systems, Inc., Minneapolis, MN, USA) according to the manufacturer’s instructions.
Mass spectrometry
Jingjie PTM BioLab Co., Ltd. (Hangzhou, China) performed all mass spectrometry analyses.
Statistical analyses
Statistical analyses were conducted with the Student’s t-test and one-way analysis of variance using SAS 9.1 software (SAS Institute, Cary, NC, USA). Probability (p) values of < 0.05 and < 0.01 were considered as significant and extremely significant, respectively.
Supporting information
The expression level of NCL was determined by western blot analysis with anti-NCL mAb. RK-13 cells were transfected with NCL siRNA (100 nM) or non-specific siRNA (100 nM) for 24 h and then lysed. β-actin was employed as an internal control.
(TIF)
The expression levels of NCL (A) or CLTA (B) were determined by western blot analysis with anti-Flag mAb or anti-Myc mAb. RK-13 cells were transfected with pNCL-Flag (2 μg) or pMyc-CLTA-Flag (2 μg) for 48 h and then lysed. The pCMV-Myc vector and p3×FLAG-CMV-14 vector acted as negative controls. β-actin was employed as an internal control.
(TIF)
(A) Kinetics of mRHDV attachment to RK-13 cells. Increasing amounts of mRHDV were added (0.25–2 MOI) to chilled RK-13 cells. After 2 h of attachment, the cells were washed and lysed. The amount of attached virus was determined by qRT-PCR and expressed as VP60 RNA copies per 100,000 copies of GADPH. (B) Kinetics of mRHDV internalization into RK-13 cells. Internalization of mRHDV (MOI = 1) into RK-13 cells was assessed by mRHDV incubation for 10, 30, and 60 min at 37°C. The amount of internalized virus was determined by qRT-PCR and expressed as VP60 RNA copies per 100,000 of GADPH.
(TIF)
Uptake of EGF-Alexa 488 (A), transferrin-Alexa 488 (B), dextran-Alexa 488 (C) or CD4-Alexa 488 (D) by RK-13 cells treated with NCL siRNA or non-specific siRNA, as quantified by flow cytometry.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
The authors wish to thank Dr. Lens H. Kuhn and the reviewers for their valuable suggestions pertaining to this manuscript. In addition, we thank International Science Editing for linguistic assistance during the preparation of this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by grants from the National Natural Science Foundation of China (31672572, http://www.nsfc.gov.cn/), the Key Project of Agriculture Science and Technology of Shanghai (2016043, https://www.xmgl.org/), the Foundation of Shanghai Key Laboratory of Veterinary Biotechnology (klab201712, http://www.agri.sjtu.edu.cn/) and the National Key Research and Development Program of China (2016YFD0500108, http://www.nmp.gov.cn/). The funders had no role in the study design, data collection and interpretation, or the decision to submit this work for publication.
References
- 1.Meyers G, Wirblich C, Thiel HJ (1991) Rabbit hemorrhagic disease virus—molecular cloning and nucleotide sequencing of a calicivirus genome. Virology 184: 664–676. [DOI] [PubMed] [Google Scholar]
- 2.Gregg DA, House C, Meyer R, Berninger M (1991) Viral haemorrhagic disease of rabbits in Mexico: epidemiology and viral characterization. Rev Sci Tech 10: 435–451. [DOI] [PubMed] [Google Scholar]
- 3.Abrantes J, van der Loo W, Le Pendu J, Esteves PJ (2012) Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet Res 43: 12 10.1186/1297-9716-43-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu SJ, Xue H. P., Pu B. Q., Qian SH (1984) A new viral disease in rabbits. Anim Hubs Vet Med 16: 253–255. [Google Scholar]
- 5.Ruvoen-Clouet N, Ganiere JP, Andre-Fontaine G, Blanchard D, Le Pendu J (2000) Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J Virol 74: 11950–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nystrom K, Le Gall-Recule G, Grassi P, Abrantes J, Ruvoen-Clouet N, et al. (2011) Histo-blood group antigens act as attachment factors of rabbit hemorrhagic disease virus infection in a virus strain-dependent manner. PLoS Pathog 7: e1002188 10.1371/journal.ppat.1002188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leuthold MM, Dalton KP, Hansman GS (2015) Structural analysis of a rabbit hemorrhagic disease virus binding to histo-blood group antigens. J Virol 89: 2378–2387. 10.1128/JVI.02832-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J, Miao Q, Tan Y, Guo H, Liu T, et al. (2017) Inclusion of an Arg-Gly-Asp receptor-recognition motif into the capsid protein of rabbit hemorrhagic disease virus enables culture of the virus in vitro. J Biol Chem 292: 8605–8615. 10.1074/jbc.M117.780924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia W, Yao Z, Zhao J, Guan Q, Gao L (2017) New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sci 186: 1–10. 10.1016/j.lfs.2017.07.025 [DOI] [PubMed] [Google Scholar]
- 10.Tuteja R, Tuteja N (1998) Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit Rev Biochem Mol Biol 33: 407–436. 10.1080/10409239891204260 [DOI] [PubMed] [Google Scholar]
- 11.Abdelmohsen K, Gorospe M (2012) RNA-binding protein nucleolin in disease. RNA Biol 9: 799–808. 10.4161/rna.19718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bicknell K, Brooks G, Kaiser P, Chen H, Dove BK, et al. (2005) Nucleolin is regulated both at the level of transcription and translation. Biochem Biophys Res Commun 332: 817–822. 10.1016/j.bbrc.2005.05.022 [DOI] [PubMed] [Google Scholar]
- 13.Hirano K, Miki Y, Hirai Y, Sato R, Itoh T, et al. (2005) A multifunctional shuttling protein nucleolin is a macrophage receptor for apoptotic cells. J Biol Chem 280: 39284–39293. 10.1074/jbc.M505275200 [DOI] [PubMed] [Google Scholar]
- 14.Becherel OJ, Gueven N, Birrell GW, Schreiber V, Suraweera A, et al. (2006) Nucleolar localization of aprataxin is dependent on interaction with nucleolin and on active ribosomal DNA transcription. Hum Mol Genet 15: 2239–2249. 10.1093/hmg/ddl149 [DOI] [PubMed] [Google Scholar]
- 15.Mongelard F, Bouvet P (2007) Nucleolin: a multiFACeTed protein. Trends Cell Biol 17: 80–86. 10.1016/j.tcb.2006.11.010 [DOI] [PubMed] [Google Scholar]
- 16.Durut N, Saez-Vasquez J (2015) Nucleolin: dual roles in rDNA chromatin transcription. Gene 556: 7–12. 10.1016/j.gene.2014.09.023 [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y, Li Z, Nagy PD (2010) Nucleolin/Nsr1p binds to the 3’ noncoding region of the tombusvirus RNA and inhibits replication. Virology 396: 10–20. 10.1016/j.virol.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancio-Lonches C, Yocupicio-Monroy M, Sandoval-Jaime C, Galvan-Mendoza I, Urena L, et al. (2011) Nucleolin interacts with the feline calicivirus 3’ untranslated region and the protease-polymerase NS6 and NS7 proteins, playing a role in virus replication. J Virol 85: 8056–8068. 10.1128/JVI.01878-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumi RE, Valdez B, Banerjee R, Srivastava M, Dasgupta A (2001) Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res 76: 17–29. [DOI] [PubMed] [Google Scholar]
- 20.Waggoner S, Sarnow P (1998) Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J Virol 72: 6699–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greco A, Arata L, Soler E, Gaume X, Coute Y, et al. (2012) Nucleolin interacts with US11 protein of herpes simplex virus 1 and is involved in its trafficking. J Virol 86: 1449–1457. 10.1128/JVI.06194-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, et al. (2011) Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med 17: 1132–1135. 10.1038/nm.2444 [DOI] [PubMed] [Google Scholar]
- 23.Said EA, Krust B, Nisole S, Svab J, Briand JP, et al. (2002) The anti-HIV cytokine midkine binds the cell surface-expressed nucleolin as a low affinity receptor. J Biol Chem 277: 37492–37502. 10.1074/jbc.M201194200 [DOI] [PubMed] [Google Scholar]
- 24.Su PY, Wang YF, Huang SW, Lo YC, Wang YH, et al. (2015) Cell surface nucleolin facilitates enterovirus 71 binding and infection. J Virol 89: 4527–4538. 10.1128/JVI.03498-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan Y, Du Y, Wang G, Li K (2017) Non-structural protein 1 of H3N2 influenza A virus induces nucleolar stress via interaction with nucleolin. Sci Rep 7: 17761 10.1038/s41598-017-18087-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar D, Broor S, Rajala MS (2016) Interaction of Host Nucleolin with Influenza A Virus Nucleoprotein in the Early Phase of Infection Limits the Late Viral Gene Expression. PLoS One 11: e0164146 10.1371/journal.pone.0164146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan CM, Chu H, Zhang AJ, Leung LH, Sze KH, et al. (2016) Hemagglutinin of influenza A virus binds specifically to cell surface nucleolin and plays a role in virus internalization. Virology 494: 78–88. 10.1016/j.virol.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 28.Melen K, Tynell J, Fagerlund R, Roussel P, Hernandez-Verdun D, et al. (2012) Influenza A H3N2 subtype virus NS1 protein targets into the nucleus and binds primarily via its C-terminal NLS2/NoLS to nucleolin and fibrillarin. Virol J 9: 167 10.1186/1743-422X-9-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murayama R, Harada Y, Shibata T, Kuroda K, Hayakawa S, et al. (2007) Influenza A virus non-structural protein 1 (NS1) interacts with cellular multifunctional protein nucleolin during infection. Biochem Biophys Res Commun 362: 880–885. 10.1016/j.bbrc.2007.08.091 [DOI] [PubMed] [Google Scholar]
- 30.Balinsky CA, Schmeisser H, Ganesan S, Singh K, Pierson TC, et al. (2013) Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J Virol 87: 13094–13106. 10.1128/JVI.00704-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oksayan S, Nikolic J, David CT, Blondel D, Jans DA, et al. (2015) Identification of a role for nucleolin in rabies virus infection. J Virol 89: 1939–1943. 10.1128/JVI.03320-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Xu F, Liu J, Gao B, Liu Y, et al. (2013) Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS Pathog 9: e1003132 10.1371/journal.ppat.1003132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leen EN, Sorgeloos F, Correia S, Chaudhry Y, Cannac F, et al. (2016) A Conserved Interaction between a C-Terminal Motif in Norovirus VPg and the HEAT-1 Domain of eIF4G Is Essential for Translation Initiation. PLoS Pathog 12: e1005379 10.1371/journal.ppat.1005379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercer J, Schelhaas M, Helenius A (2010) Virus entry by endocytosis. Annu Rev Biochem 79: 803–833. 10.1146/annurev-biochem-060208-104626 [DOI] [PubMed] [Google Scholar]
- 35.Win SJ, Ward VK, Dunbar PR, Young SL, Baird MA (2011) Cross-presentation of epitopes on virus-like particles via the MHC I receptor recycling pathway. Immunol Cell Biol 89: 681–688. 10.1038/icb.2010.161 [DOI] [PubMed] [Google Scholar]
- 36.Song N, Ding Y, Zhuo W, He T, Fu Z, et al. (2012) The nuclear translocation of endostatin is mediated by its receptor nucleolin in endothelial cells. Angiogenesis 15: 697–711. 10.1007/s10456-012-9284-y [DOI] [PubMed] [Google Scholar]
- 37.Hanover JA, Willingham MC, Pastan I (1984) Kinetics of transit of transferrin and epidermal growth factor through clathrin-coated membranes. Cell 39: 283–293. [DOI] [PubMed] [Google Scholar]
- 38.Fragoso R, Ren D, Zhang X, Su MW, Burakoff SJ, et al. (2003) Lipid raft distribution of CD4 depends on its palmitoylation and association with Lck, and evidence for CD4-induced lipid raft aggregation as an additional mechanism to enhance CD3 signaling. J Immunol 170: 913–921. [DOI] [PubMed] [Google Scholar]
- 39.Al Soraj M, He L, Peynshaert K, Cousaert J, Vercauteren D, et al. (2012) siRNA and pharmacological inhibition of endocytic pathways to characterize the differential role of macropinocytosis and the actin cytoskeleton on cellular uptake of dextran and cationic cell penetrating peptides octaarginine (R8) and HIV-Tat. J Control Release 161: 132–141. 10.1016/j.jconrel.2012.03.015 [DOI] [PubMed] [Google Scholar]
- 40.Robinson MS (2015) Forty Years of Clathrin-coated Vesicles. Traffic 16: 1210–1238. 10.1111/tra.12335 [DOI] [PubMed] [Google Scholar]
- 41.Ginisty H, Sicard H, Roger B, Bouvet P (1999) Structure and functions of nucleolin. J Cell Sci 112 (Pt 6): 761–772. [DOI] [PubMed] [Google Scholar]
- 42.Tajrishi MM, Tuteja R, Tuteja N (2011) Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol 4: 267–275. 10.4161/cib.4.3.14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakarai C, Osawa K, Akiyama M, Matsubara N, Ikeuchi H, et al. (2015) Expression of AKR1C3 and CNN3 as markers for detection of lymph node metastases in colorectal cancer. Clin Exp Med 15: 333–341. 10.1007/s10238-014-0298-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nisole S, Said EA, Mische C, Prevost MC, Krust B, et al. (2002) The anti-HIV pentameric pseudopeptide HB-19 binds the C-terminal end of nucleolin and prevents anchorage of virus particles in the plasma membrane of target cells. J Biol Chem 277: 20877–20886. 10.1074/jbc.M110024200 [DOI] [PubMed] [Google Scholar]
- 45.Alete DE, Weeks ME, Hovanession AG, Hawadle M, Stoker AW (2006) Cell surface nucleolin on developing muscle is a potential ligand for the axonal receptor protein tyrosine phosphatase-sigma. FEBS J 273: 4668–4681. 10.1111/j.1742-4658.2006.05471.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, et al. (2008) Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell 15: 209–219. 10.1016/j.devcel.2008.06.012 [DOI] [PubMed] [Google Scholar]
- 47.Liu G, Zhang Y, Ni Z, Yun T, Sheng Z, et al. (2006) Recovery of infectious rabbit hemorrhagic disease virus from rabbits after direct inoculation with in vitro-transcribed RNA. J Virol 80: 6597–6602. 10.1128/JVI.02078-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression level of NCL was determined by western blot analysis with anti-NCL mAb. RK-13 cells were transfected with NCL siRNA (100 nM) or non-specific siRNA (100 nM) for 24 h and then lysed. β-actin was employed as an internal control.
(TIF)
The expression levels of NCL (A) or CLTA (B) were determined by western blot analysis with anti-Flag mAb or anti-Myc mAb. RK-13 cells were transfected with pNCL-Flag (2 μg) or pMyc-CLTA-Flag (2 μg) for 48 h and then lysed. The pCMV-Myc vector and p3×FLAG-CMV-14 vector acted as negative controls. β-actin was employed as an internal control.
(TIF)
(A) Kinetics of mRHDV attachment to RK-13 cells. Increasing amounts of mRHDV were added (0.25–2 MOI) to chilled RK-13 cells. After 2 h of attachment, the cells were washed and lysed. The amount of attached virus was determined by qRT-PCR and expressed as VP60 RNA copies per 100,000 copies of GADPH. (B) Kinetics of mRHDV internalization into RK-13 cells. Internalization of mRHDV (MOI = 1) into RK-13 cells was assessed by mRHDV incubation for 10, 30, and 60 min at 37°C. The amount of internalized virus was determined by qRT-PCR and expressed as VP60 RNA copies per 100,000 of GADPH.
(TIF)
Uptake of EGF-Alexa 488 (A), transferrin-Alexa 488 (B), dextran-Alexa 488 (C) or CD4-Alexa 488 (D) by RK-13 cells treated with NCL siRNA or non-specific siRNA, as quantified by flow cytometry.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.