Abstract
Host shifts–where a pathogen jumps between different host species–are an important source of emerging infectious disease. With on-going climate change there is an increasing need to understand the effect changes in temperature may have on emerging infectious disease. We investigated whether species’ susceptibilities change with temperature and ask if susceptibility is greatest at different temperatures in different species. We infected 45 species of Drosophilidae with an RNA virus and measured how viral load changes with temperature. We found the host phylogeny explained a large proportion of the variation in viral load at each temperature, with strong phylogenetic correlations between viral loads across temperature. The variance in viral load increased with temperature, while the mean viral load did not. This suggests that as temperature increases the most susceptible species become more susceptible, and the least susceptible less so. We found no significant relationship between a species’ susceptibility across temperatures, and proxies for thermal optima (critical thermal maximum and minimum or basal metabolic rate). These results suggest that whilst the rank order of species susceptibilities may remain the same with changes in temperature, some species may become more susceptible to a novel pathogen, and others less so.
Author summary
Emerging infectious diseases are often the result of a host shift, where a pathogen jumps from one host species into another. Understanding the factors underlying host shifts is a major goal for infectious disease research. This effort has been further complicated by the fact that host-parasite interactions are now taking place in a period of unprecedented global climatic warming. Here, we ask how host shifts are affected by temperature by carrying out experimental infections using an RNA virus across a wide range of related species, at three different temperatures. We find that as temperature increases the most susceptible species become more susceptible, and the least susceptible less so. This has important consequences for our understanding of host shift events in a changing climate as it suggests that temperature changes may affect the likelihood of a host shift into certain species.
Introduction
Temperature is arguably the most important abiotic factor that affects all organisms, having both indirect and direct effects on physiology and life history traits [1–3]. There is much to be learned about the impact of climate change on infectious diseases [1,4,5]. Changes in temperature can impact both host and parasite biology, leading to complex and difficult to predict outcomes [2,6].
Host shifts, where a parasite from one host species invades and establishes in a novel host species, are an important source of emerging infectious disease [7]. A successful host shift relies on a number of stages occurring [8]. Firstly, exposure of the host to the new pathogen species must occur in such a way that transmission is successful. Secondly, the pathogen must be able to replicate sufficiently to infect the novel host. Finally, there must be sufficient onwards transmission for the pathogen to become established in the new host species [7,9,10]. Some of the most deadly outbreaks of infectious diseases in humans including Ebola virus, HIV and SARS coronavirus have been linked to a host switch event [11–14] and many others have direct animal vectors or reservoirs (e.g. Dengue and Chikungunya viruses) [15,16]. The potential for novel host shifts may increase with changing temperatures due to, fluctuations in host and/or parasite fitness, or changes in species distributions and abundances [17,18]. Distribution changes may lead to new species assemblages, causing novel contacts between parasites and potential hosts [19–21].
Susceptibility to infection is known to vary with temperature, due to within individual physiological changes in factors such as the host immune response, metabolic rate or behavioural adaptations [22–25]. Thermally stressed hosts may face a trade-off between the resource investment needed to launch an immune response versus that needed for thermoregulation, or behavioural adaptations to withstand sub-optimal temperatures [26–29]. Temperature shifts could also cause asymmetrical or divergent effects on host and parasite traits [30]. For example, changes in temperature may allow differential production and survival of parasite transmission stages, and changes in replication rates, generation times, infectivity and virulence [31–33]. Temperature is also known to impact vector-borne disease transmission through multiple effects on both vector life cycles and transmission behaviours [20,34–37].
Host shifts have been shown to be more likely to occur between closely related species [38–40], but independently of this distance effect, clades of closely related hosts show similar levels of susceptibility [9,41]. Thermal tolerances − like virus susceptibility − are known to vary across species, with groups of closely related species having similar thermal limits, with a large proportion of the variation in these traits being explained by the phylogeny [42–45]. Previous studies on host shifts have assayed the susceptibility of species at a single temperature [9,39,41,46]. However, if the host phylogeny also explains much of the variation in thermal tolerance, then phylogenetic patterns in virus susceptibility could be due to differences between species’ natural thermal optima and the chosen assay temperatures. Therefore, for experiments carried out at a single temperature, phylogenetic signal in thermal tolerance may translate into phylogenetic signal in thermal stress. Any apparent phylogenetic signal in susceptibility could potentially be due to the effects of thermal stress, and may not hold true if each species was to be assayed at its optimal temperature. If this was indeed the case this would have implications for species distribution models that aim to use estimates of environmental conditions to predict host and pathogen ranges [5,47,48].
Here, we have asked how species’ susceptibilities change at different temperatures and whether susceptibility is greatest at different temperatures in different species. We infected 45 species of Drosophilidae with Drosophila C Virus (DCV; Dicistroviridae) at three different temperatures and measured how viral load changes with temperature. Viral load is used here as a measure of DCV’s ability to persist and replicate in a host, which has previously been shown to be tightly correlated to host mortality [41]. We are therefore examining one of the steps (“ability to infect a novel host”) needed for a host shift to successfully occur [7,9,10]. We also examine how proxies for thermal optima and cellular function (thermal tolerances and basal metabolic rate) relate to virus susceptibility across temperatures, as increasing temperatures may have broad effects on both host and parasite [43–45]. DCV is a positive sense RNA virus in the family Discistroviridae that was originally isolated from Drosophila melanogaster and in the wild has been found in D. melanogaster and D. simulans [49–51]. DCV infected flies show reduced metabolic rate and activity levels, develop an intestinal obstruction, reduced hemolymph pH and decreased survival [52–55]. This work examines how temperature can influence the probability of host shifts, and looks at some of the potential underlying causes.
Methods
Experimental infections
We used Drosophila C virus (DCV) clone B6A, which is derived from an isolate collected from D. melanogaster in Charolles, France [56]. The virus was prepared as described previously [57]; briefly DCV was grown in Schneider’s Drosophila line 2 cells and the Tissue Culture Infective Dose 50 (TCID50) per ml was calculated using the Reed-Muench end-point method [58].
Flies were obtained from laboratory stocks of 45 different species. All stocks were maintained in multi generation populations, in Drosophila stock bottles (Dutscher Scientific) on 50ml of their respective food medium at 22°C and 70% relative humidity with a 12 hour light-dark cycle (Table A in S1 Text). Each day, two vials of 0–1 day old male flies were randomly assigned to one of three potential temperature regimes; low, medium or high (17°C, 22°C and 27 °C respectively) at 70% relative humidity. Flies were tipped onto fresh vials of food after 3 days, and after 5 days of acclimatisation at the experimental temperature were infected with DCV. Flies were anesthetized on CO2 and inoculated using a 0.0125 mm diameter stainless steel needle that was bent to a right angle ~0.25mm from the end (Fine Science Tools, CA, USA)[9,41,57]. The bent tip of the needle was dipped into the DCV solution (TCID50 = 6.32×109) and pricked into the pleural suture on the thorax of the flies. We selected this route of infection as oral inoculation has been shown to lead to stochastic infection outcomes in D. melanogaster [55]. However, once the virus passes through the gut barrier, both oral and pin-pricked infections follow a similar course, with both resulting in the same tissues becoming infected with DCV [55]. One vial of inoculated flies was immediately snap frozen in liquid nitrogen to provide a time point zero sample as a reference to control for relative viral dose. The second vial of flies were placed onto a new vial of fresh cornmeal food and returned to their experimental temperature. After 2 days (+/- 1 hour) flies were snap frozen in liquid nitrogen. This time point was chosen based on pilot data as infected flies showed little mortality at 2 days post infection, and viral load plateaus from day 2 at 22°C. Temperatures were rotated across incubators in each block to control for incubator effects. All frozen flies were homogenised in a bead homogeniser for 30 seconds (Bead Ruptor 24; Omni international, Georgia, USA) in Trizol reagent (Invitrogen) and stored at -80°C for later RNA extractions.
These collections and inoculations were carried out over three replicate blocks, with each block being completed over consecutive days. The order that the fly species were infected was randomized each day. We aimed for each block to contain a day 0 and day 2 replicate for each species, at each temperature treatment (45 species × 3 temperatures × 3 experimental blocks). In total we quantified viral load in 12,827 flies over 396 biological replicates (a biological replicate = change in viral load from day 0 to day 2 post-infection), with a mean of 17.1 flies per replicate (range across species = 4–27). Of the 45 species, 42 had 3 biological replicates and three species had 2 biological replicates.
Measuring the change in viral load
The change in RNA viral load was measured using quantitative Reverse Transcription PCR (qRT-PCR). Total RNA was extracted from the Trizol homogenised flies, reverse-transcribed with Promega GoScript reverse transcriptase (Promega) and random hexamer primers. Viral RNA load was expressed relative to the endogenous control housekeeping gene RpL32 (RP49). RpL32 primers were designed to match the homologous sequence in each species and crossed an intron-exon boundary so will only amplify mRNA [9]. The primers in D. melanogaster were RpL32 qRT-PCR F (5’-TGCTAAGCTGTCGCACAAATGG -3’) and RpL32 qRT-PCR R (5’- TGCGCTTGTTCGATCCGTAAC -3’). DCV primers were 599F (5’-GACACTGCCTTTGATTAG-3’) and 733R (5’CCCTCTGGGAACTAAATG-3’) as previously described [41]. Two qRT-PCR reactions (technical replicates) were carried out per sample with both the viral and endogenous control primers, with replicates distributed across plates in a randomised block design.
qRT-PCR was performed on an Applied Biosystems StepOnePlus system using Sensifast Hi-Rox Sybr kit (Bioline) with the following PCR cycle: 95°C for 2min followed by 40 cycles of: 95°C for 5 sec followed by 60°C for 30 sec. Each qRT-PCR plate contained four standard samples. A linear model was used to correct the cycle threshold (Ct) values for differences between qRT-PCR plates. Any samples where the two technical replicates had cycle threshold (Ct) values more than 2 cycles apart after the plate correction were repeated. To estimate the change in viral load, we first calculated ΔCt as the difference between the cycle thresholds of the DCV qRT-PCR and the RpL32 endogenous control. For each species the viral load of day 2 flies relative to day 0 flies was calculated as 2-ΔΔCt; where ΔΔCt = ΔCtday0 –ΔCtday2. The ΔCtday0 and ΔCtday2 are a pair of ΔCt values from a day 0 biological replicate and a day 2 biological replicate. Calculating the change in viral load without the use of the endogenous control gene (RpL32) gave equivalent results (Spearman’s correlation between viral load calculated with and without endogenous control: ρ = 0.97, P< 0.005)
Critical thermal maximum and minimum assays
We carried out two assays to measure the thermal tolerances of species; a cold resistance measure to determine critical thermal minimum (CTmin) under gradual cooling, and a heat resistance measure through gradual heating to determine critical thermal maximum (CTmax). 0–1 day old males were collected and placed onto fresh un-yeasted cornmeal food vials. Flies were kept for 5 days at 22°C and 70% relative humidity and tipped onto fresh food every 2 days. In both assays individual flies were placed in 4 ml glass vials (ST5012, Ampulla, UK) and exposed to temperature change through submersion in a liquid filled glass tank (see Fig A in S1 Text). For CTmax the tank was filled with water and for CTmin a mixture of water and ethylene glycol (50:50 by volume) was used to prevent freezing and maintain a constant cooling gradient. Five biological replicates were carried out for each species for both CTmax and CTmin. Temperature was controlled using a heated/cooled circulator (TXF200, Grant Instruments, Cambridgeshire, UK) submerged in the tank and set to change temperatures at a rate of 0.1 °C/min, always starting from 22°C (the rearing temperature for stock populations). Flies were monitored continually throughout the assay and the temperature of knock down was ascertained by a disturbance method, whereby a fly was scored as completely paralysed if on gentle tapping of the vial wall the fly did not move any of its body parts.
Measuring metabolic rate
To examine how cellular function changes with temperature, we estimated the resting metabolic rate of each species at 17°C, 22°C and 27 °C to examine if changes in general cellular processes were related to changes in viral load. Following the same methods as the viral inoculation assay, groups of 10, 0–1 day old male flies from 44 species were acclimatised at the three experimental temperatures for 5 days (D. pseudoobscura was excluded as not enough individuals could be obtained from stocks for sufficient replication). Every 2 days flies were tipped onto fresh vials of cornmeal food. This was repeated in three blocks in order to get three repeat measures of metabolic rate for each of the species, at each of the three experimental temperatures. Flies were collected in a randomly assigned order across the three blocks.
Closed system respirometry was used to measure the rate of CO2 production (VCO2) as a proxy for metabolic rate [59]. Flies were held in 10ml-3 airtight plastic chambers constructed from Bev-A-Line V Tubing (Cole-Parmer Instrument Company, UK). All measures were carried out during the day inside a temperature controlled incubator, with constant light, that was set to each of the experimental temperatures that the flies had been acclimatised to. The set up followed that of Okada et al. (2011)[60]. Compressed air of a known concentration of oxygen and nitrogen (21% O2:79% N2) was scrubbed of any CO2 and water (with Ascarite II & Magnesium Perchlorate respectively) and pumped through a Sable Systems RM8 eight-channel multiplexer (Las Vegas, NV, USA) at 100 ml/min-1 (±1%) into the metabolic chambers housing the groups of 10 flies. The first chamber was left empty as a reference cell, to acquire a baseline reading for all subsequent chambers at the start and end of each set of runs, therefore seven groups of flies were assayed in each run. Air was flushed into each chamber for 2 minutes, before reading the previous chamber. Readings were taken every second for 10 minutes by feeding the exiting air through a LiCor LI-7000 infrared gas analyser (Lincoln, NE, USA). Carbon dioxide production was measured using a Sable Systems UI2 analog–digital interface for acquisition, connected to a computer running Sable Systems Expedata software (v1.8.2) [61]. The metabolic rate was calculated from the entire 10-minute recording period by taking the CO2 reading of the ex-current gas from the chamber containing the flies and subtracting the CO2 measure of the incurrent gas entering the chamber. These values were also corrected for drift away from the baseline reading of the empty chamber. Volume of CO2 was calculated as VCO2 = FR (Fe CO2 –Fi CO2) / (1-Fi CO2). Where FR is the flow rate into the system (100ml/min-1), Fe CO2 is the concentration of CO2 exiting and Fi CO2 is the concentration CO2 entering the respirometer. Species were randomly assigned across the respiration chambers and the order in which flies were assayed (chamber order) was corrected for statistically (see below).
Body size
To check for any potential effect of body size differences between species on viral load, wing length was measured as a proxy for body size [62]. A mean of 26 (range 20–30) males of each species were collected and immediately stored in ethanol during the collections for the viral load assay. Subsequently, wings were removed and photographed under a dissecting microscope. Using ImageJ software (version 1.48) the length of the IV longitudinal vein from the tip of the proximal segment to where the distal segment joins vein V was recorded, and the mean taken for each species.
Host phylogeny
The host phylogeny was inferred as described in Longdon et al (2015) [41], using the 28S, Adh, Amyrel, COI, COII, RpL32 and SOD genes. Briefly, any publicly available sequences were downloaded from Genbank, and any not available we attempted to Sanger sequence [9]. In total we had RpL32 sequences for all 45 species, 28s from 41 species, Adh from 43 species, Amyrel from 29 species, COI from 38 species, COII from 43 species and SOD from 25 species (see www.doi.org/10.6084/m9.figshare.6653192 full details). The sequences of each gene were aligned in Geneious (version 9.1.8, [63]) using the global alignment setting, with free end gaps and a cost matrix of 70% similarity. The phylogeny was constructed using the BEAST program (version 1.8.4,[64]). Genes were partitioned into three groups each with their own molecular clock models. The three partitions were: mitochondrial (COI, COII); ribosomal (28S); and nuclear (Adh, SOD, Amyrel, RpL32). A random starting tree was used, with a relaxed uncorrelated lognormal molecular clock. Each of the partitions used a HKY substitution model with a gamma distribution of rate variation with 4 categories and estimated base frequencies. Additionally, the mitochondrial and nuclear data sets were partitioned into codon positions 1+2 and 3, with unlinked substitution rates and base frequencies across codon positions. The tree-shape prior was set to a birth-death process. The BEAST analysis was run twice to ensure convergence for 1000 million MCMC generations sampled every 10000 steps. The MCMC process was examined using the program Tracer (version 1.6, [65]) to ensure convergence and adequate sampling, and the constructed tree was then visualised using FigTree (version 1.4.3, [66]).
Statistical analysis
All data were analysed using phylogenetic mixed models to look at the effects of host relatedness on viral load across temperature. We fitted all models using a Bayesian approach in the R package MCMCglmm [67,68]. We ran trivariate models with viral load at each of the three temperatures as the response variable similar to that outlined in Longdon et al. (2011) [9]. The models took the form:
Where y is the change in viral load of the ith biological replicate of host species h, for temperature t (high, medium or low). β are the fixed effects, with β1 being the intercepts for each temperature, β2 being the effect of basal metabolic rate, β3 the effect of wing size, and β4 and β5 the effects of the critical thermal maximum (CTmax) and minimum (CTmin) respectively. up are the random phylogenetic species effects and e the model residuals. We also ran models that included a non-phylogenetic random species effect (unp:ht) to allow us to estimate the proportion of variation explained by the host phylogeny [9,41,69]. We do not use this term in the main model as we struggled to separate the phylogenetic and non-phylogenetic terms. Our main model therefore assumes a Brownian motion model of evolution [70]. The random effects and the residuals are assumed to be multivariate normal with a zero mean and a covariance structure Vp ⊗ A for the phylogenetic affects and Ve ⊗ I for the residuals (⊗ here is the Kronecker product). A is the phylogenetic relatedness matrix, I is an identity matrix and the V are 3×3 (co)variance matrices describing the (co)variances between viral titre at different temperatures. The phylogenetic covariance matrix, Vp, describes the inter-specific variances in each trait and the inter-specific covariances between them. The residual covariance matrix, Ve, describes the within-species variance that can be both due to real within-species effects and measurement or experimental errors. The off-diagonal elements of Ve (the covariances) can not be estimated because no vial has been subject to multiple temperatures and so were set to zero. We excluded D. pseudoobscura from the full model as data for BMR was not collected, but included it in models that did not include any fixed effects, which gave equivalent results.
Diffuse independent normal priors were placed on the fixed effects (means of zero and variances of 108). Parameter expanded priors were placed on the covariance matrices resulting in scaled multivariate F distributions, which have the property that the marginal distributions for the variances are scaled (by 1000) F 1,1. The exceptions were the residual variances for which an inverse-gamma prior was used with shape and scale equal to 0.001. The MCMC chain was run for 130 million iterations with a burn-in of 30 million iterations and a thinning interval of 100,000. We confirmed the results were not sensitive to the choice of prior by also fitting models with inverse-Wishart and flat priors for the variance covariance matrices (described in [9]), which gave qualitatively similar results (10.6084/m9.figshare.6177191). All confidence intervals (CI’s) reported are 95% highest posterior density intervals.
Using similar model structures we also ran a univariate model with BMR and a bivariate model with CTmin and CTmax as the response variables to calculate how much of the variation in these traits was explained by the host phylogeny. Both of these models were also run with wing length as a proxy for body size as this is known to influence thermal measures [59]. We observed significant levels of measurement error in the metabolic rate data; this was partially caused by respiratory chamber order during the assay. We corrected for this in two different ways. First, we fitted a linear model to the data to control for the effect of respiratory chamber number and then used this corrected data in all further models. We also used a measurement error model that controls for both respiratory chamber number effects and random error. Both of these models gave similar results although the measurement error model showed broad CIs suggesting the BMR data should be interpreted with caution. All datasets and R scripts with the model parameterisation are provided as supporting information (S1 Text).
Results
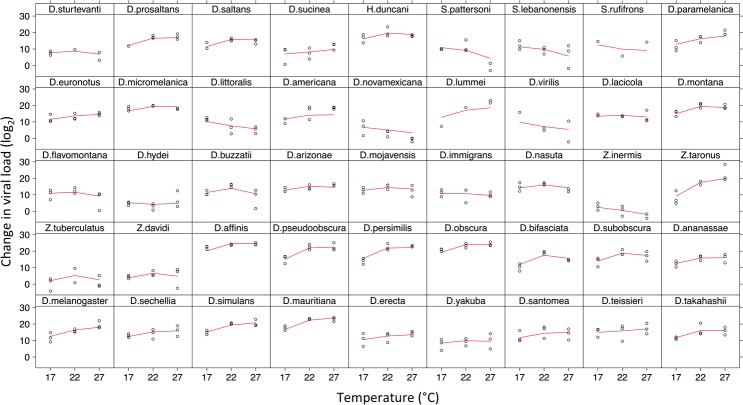
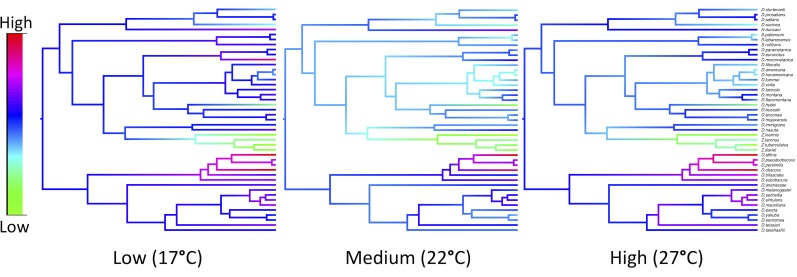
To investigate the effect of temperature on virus host shifts we quantified viral load in 12,827 flies over 396 biological replicates, from 45 species of Drosophilidae at three temperatures (Fig 1). DCV replicated in all host species, but viral load differed between species and temperatures (Fig 1). Species with similar viral loads cluster together on the phylogeny (Fig 2). Measurements were highly repeatable (Table 1), with a large proportion of the variance being explained by the inter-specific phylogenetic component (vp), with little within species or measurement error (vr) (Repeatability = vp/(vp + vr): Low = 0.90 (95% CI: 0.84, 0.95), Medium = 0.96 (95% CI: 0.93, 0.98), and High = 0.95, (95% CI: 0.89, 0.98)). We also calculated the proportion of between species variance that can be explained by the phylogeny as vp/(vp+ vs) [71], which is equivalent to Pagel’s lambda or phylogenetic heritability [69,72]. We found the host phylogeny explains a large proportion of the inter-specific variation in viral load across all three temperatures, although these estimates have broad confidence intervals due to the model struggling to separate the phylogenetic and non-phylogenetic components (Low = 0.77, 95% CI: 0.28, 0.99; Medium = 0.53, 95% CI: 0.31×10−5, 0.85; High = 0.40, 95% CI: 0.99×10−5, 0.74)
Fig 1. Change in viral load (log2) for 45 Drosophilidae species across three temperatures (17°C = Low, 22°C = Medium and 27°C = High).
Individual points are for each replicate (change in viral load between day 0 and day 2 post infection), the red line is the predicted values from the phylogenetic mixed model. Panels are ordered as on the tips of the phylogeny as in Fig 2.
Fig 2. Ancestral state reconstructions to visualise the change in viral load across the host phylogeny at three temperatures.
Ancestral states are plotted as colour gradients across the tree. The colour gradient represents the change in RNA viral load; red represents the highest and green the lowest viral load at that temperature. Ancestral states were estimated using a phylogenetic mixed model that partitioned the inter-specific variance into that explained by the host phylogeny under a Brownian model of evolution (vp), and a species-specific variance component that is not explained by the phylogeny (vs).
Table 1. Change in viral load with temperature.
| Temperature | Intercepts | Between-species Variance (vp) | Within-species Variance (vr) | |||
|---|---|---|---|---|---|---|
| Mean | 95% CIs | Mean | 95% CIs | Mean | 95% CIs | |
| Low | 11.9 | 9.5, 14.6 | 65.3 | 32.3, 110.3 | 6.9 | 4.8, 9.3 |
| Medium | 14.3 | 11.7, 17.1 | 172.2 | 90.2, 278.8 | 7.0 | 4.8, 9.2 |
| High | 13.5 | 10.8, 16.7 | 260.6 | 119.7, 413.7 | 12.8 | 8.9, 17.5 |
Intercepts are the temperature-specific intercepts when the other covariates (e.g. wing size) are set to their temperature specific means. They can be interpreted as the expected viral loads at the root of the phylogeny at each temperature. vp is the variance in between-species effects, which are structured by the phylogeny, and vr is the variance in within species effects attributable to between individual differences and measurement error.
To examine if species responded in the same or different way to changes in temperature we examined the relationships between susceptibilities across the different temperatures. We found strong positive phylogenetic correlations between viral loads across the three temperatures (Table 2). Our models showed that the variance in viral load increased with temperature, however the mean viral load showed no such upward trend (Table 1). This suggests that the changes in variance are not simply occurring due to an increase in the means, that is then driving an increase in variance.
Table 2. Interspecific correlations between viral loads at each temperature.
| Temperatures | Interspecific Correlation | 95% CIs |
|---|---|---|
| High-Low | 0.89 | 0.77, 0.98 |
| Medium-Low | 0.92 | 0.90, 0.99 |
| Medium-High | 0.97 | 0.93, 0.99 |
The high correlations suggest the rank order of susceptibility of the species is not changing with increasing temperature. However, the change in variance suggests that although the reaction norms are not crossing they are diverging from each other as temperature increases i.e. the most susceptible species are becoming more susceptible with increasing temperature, and the least susceptible less so [73]. For example, D. obscura and D. affinis are the most susceptible species at all three temperatures. The responses of individual species show that some species have increasing viral load as temperature increases (Fig 1, e.g. Z. taronus, D. lummei), while others decease (e.g. D. littoralis, D. novamexicana).
The changes we observe could be explained by the increase in temperature effectively increasing the rate at which successful infection is progressing (i.e. altering where in the course of infection we have sampled). However, this seems unlikely as at 2 days post infection at the medium temperature (22°C), viral load peaks and then plateaus [41]. Therefore, in those species where viral load increases at higher temperatures the peak viral load itself must be increasing, rather than us effectively sampling the same growth curve but at a later time point. Likewise, in those species where viral load decreased at higher temperatures, viral load would need to first increase and then decrease, which we do not observe in a time course at 22°C [41]. To check whether this also holds at higher temperatures we carried out a time course of infection in a subset of six of the 45 original experimental species at 27°C, where we would expect the fastest transition between the rapid viral growth and the plateau phase of infection to occur (Fig B in S1 Text). This allowed us to confirm that the decreasing viral loads observed in some species at higher temperatures are not due to general trend for viral loads to decline over longer periods of (metabolic) time.
We quantified the lower and upper thermal tolerances (CTmin and CTmax) across all 45 species with 3 replicates per species. Neither CTmax nor CTmin were found to be significant predictors of viral load (CTmin -0.21, 95% CI: -0.79, 0.93, pMCMC = 0.95 and CTmax 0.31, 95% CI: -0.11, 0.74, pMCMC = 0.152). When treated as a response in models we found the host phylogeny explained a large proportion of the variation in thermal maximum (CTmax: 0.95, 95% CI: 0.84, 1) and thermal minima (CTmin: 0.98, 95% CI: 0.92, 0.99, see S1 Text Fig C).
We also measured the basal metabolic rate of 1320 flies from 44 species, across the three experimental temperatures, to examine how cellular function changes with temperature. BMR was not found to be a significant predictor of viral load when included as a fixed effect in our model (slope = 9.09, 95% CI = -10.13, 20.2689, pMCMC = 0.548).
BMR increased with temperature across all species (mean BMR and SE: Low 0.64 ± 0.02, Medium 1.00 ± 0.04, High 1.2 ± 0.04 CO2ml/min-1, see S1 Text Fig D).
When BMR was analysed as the response in models, the phylogeny explained a small amount of the between species variation (Low 0.19, 95% CI: 2 × 10−8, 0.55, Medium 0.10, 95% CI: 5 × 10−7, 0.27, High 0.03, 95% CI: 8 × 10−9–0.13, S1 Text Fig E) indicating high within species variation or large measurement error. Consequently the mean BMRs for each species, at each temperature, were used in the analysis of viral load will be poorly estimated and so the effects of BMR will be underestimated with too narrow credible intervals. To rectify this we ran a series of measurement error models, the most conservative of which gave a slope of -9.8 but with very wide credible intervals (-62.5, 42.6). Full details of these models are given in the Supporting Information (S1 Text).
Discussion
We found that susceptibilities of different species responded in different ways to changes in temperature. The susceptibilities of different species showed differing responses as temperatures increased (Fig 1). There was a strong phylogenetic correlation in viral load across the three experimental temperatures (Table 2). However, the variance in viral load increased with temperature, whereas the mean viral load did not show the same trend. This suggests that the rank order of susceptibility of the species remains relatively constant across temperatures, but as temperature increases the most susceptible species become more susceptible, and the least susceptible less so.
Changes in global temperatures are widely predicted to alter host-parasite interactions and therefore the likelihood of host shifts occurring [5,21,47,74,75]. The outcome of these interactions may be difficult to predict if temperature causes a different effect in the host and pathogen species [18,37,76–78]. Our results show that changes in temperature may change the likelihood of pathogens successfully infecting certain species, although they suggest that it may not alter which species are the most susceptible to a novel pathogen.
The increase in phylogenetic variance with temperature is effectively a form of genotype-by-environment interaction [28,79–81]. However, it varies from the classically considered ecological crossing of reaction norms, as we do not see a change in the rank order of species susceptibly across the range of experimental temperatures. Instead, we find the species means diverge with increasing temperatures and so the between species differences increase [73,82]. It is also important to note that temperature may not simply be causing a change in effect size when considering the biological processes occurring during host-parasite interactions [22,83]. For example, virus replication may plateau at higher temperatures due to resource limitation. The observed level of susceptibility may be the combined outcome of both host and parasite traits, which may interact nonlinearly with temperature. We also note that by using a limited range of temperatures for practical reasons we may have not captured all unimodal relationships between viral load and temperature.
As temperature is an important abiotic factor in many cellular and physiological processes, we went on to examine the underlying basis of why viral load might change with temperature. Previous studies that found phylogenetic signal in host susceptibility were carried out at a single experimental temperature [9,41]. Therefore, the patterns observed could potentially be explained by some host clades being assayed at sub-optimal thermal conditions. We used CTmax and CTmin as proxies for thermal optima which, due to its multifaceted nature, is problematic to measure directly [84–86]. We also measured basal metabolic rate across three temperatures to see if the changes in viral load could be explained by general increases in enzymatic processes. We found that these measures were not significant predictors of the change in viral load with temperature. This may be driven by the fact that all temperature related traits are likely to be more complex than what any single measure can explore. Traits such as host susceptibility are a function of both the host and parasite thermal optima, as well as the shape of any temperature-trait relationship [37,78].
The host immune response and cellular components utilised by the virus are likely to function most efficiently at the thermal optima of a species, and several studies have demonstrated the outcomes of host-pathogen interactions can depend on temperature [26,28,76,81]. However, the mechanisms underlying the changes in susceptibility with temperature seen in this study are uncertain and a matter for speculation. Our results show that in the most susceptible species, viral load increases with temperature; this may be due to the virus being able to successfully infect and then freely proliferate, utilizing the host cells whist avoiding host immune defences. In less susceptible species viral load does not increase with temperature, and in some cases it actually appears to decreases. Here, temperature may be driving an increase in biological processes such as enhanced host immunity, or simply increasing the rate of degradation or clearance of virus particles that have failed to establish an infection of host cells.
We have investigated how an environmental variable can alter infection success following a novel viral challenge. However, temperature is just one of the potential environmental factors that will influence the different stages of a host shift event [8]. Using a controlled method of viral inoculation allows us to standardize inoculation dose so we can ask, given equal exposure, how does temperature affect the ability of a pathogen to persist and replicate in a given host? However, in nature hosts will be faced with variable levels of pathogen exposure, infected through various modes of transmission and often by multiple strains or genotypes [87]. Such variables may have consequences for the establishment and subsequent infection success of any potential host shift event. It is known that oral infection by DCV is stochastic and immune barriers such as the gut are important [55,88,89], therefore establishing the relevance of infection in the wild in this system would require further study using different potential routes of infection. The geographical distribution of a host will also influence factors such as diet and resource availability [28,90–93], and so further work on the role of nutrient and resource availability would therefore be needed to further explore the impact of these on potential host shifts.
In conclusion, we have found changes in temperature can both increase or decrease the likelihood of a host shift. Our results show the rank order of species’ susceptibilities remain the same across temperatures, suggesting that studies of host shifts at a single temperature can be informative in predicting which species are the most vulnerable to a novel pathogen. Changing global temperatures may influence pathogen host shifts; for example changes in distributions of both host and pathogen species may generate novel transmission opportunities. Our findings suggest that increases in global temperature could increase the likelihood of host shifts into the most susceptible species, and reduce it in others. Climate change may therefore lead to changing distributions of both host and pathogens, with pathogens potentially expanding or contracting their host range. Understanding how environmental factors might affect broader taxonomic groups of hosts and pathogens requires further study if we are to better understand host shifts in relation to climate change in nature.
Supporting information
(DOCX)
Acknowledgments
Many thanks to Darren Obbard and Frank Jiggins for useful discussion and Vanessa Kellerman and Johannes Overgaard for discussion about thermal assay methods. Thanks to Dave Hosken for use of BMR chambers and to the Drosophila Species Stock Centre for supplying flies. Thanks to two anonymous reviewers for constructive comments.
Data Availability
Data are available from Figshare, and listed in the supplementary materials with links. Sequences have been submitted to Genbank and all accession numbers of sequences used to infer the host phylogeny are also available in a data on Figshare. Full dataset for the project and scripts is available: https://figshare.com/account/home#/projects/30692. This includes all data and all scripts to run the analysis.
Funding Statement
KER and BL are supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (grant no. 109356/Z/15/Z). https://wellcome.ac.uk/funding/sir-henry-dale-fellowships. JDH is supported by a Royal Society Research Fellowship (UF150696). https://royalsociety.org/news/2017/06/royal-society-announces-university-research-fellowships-for-2017/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, et al. Climate warming and disease risks for terrestrial and marine biota. Science. American Association for the Advancement of Science; 2002;296: 2158–62. 10.1126/science.1063699 [DOI] [PubMed] [Google Scholar]
- 2.Poulin R. Global warming and temperature-mediated increases in cercarial emergence in trematode parasites. Parasitology. Cambridge University Press; 2006;132: 143–151. 10.1017/S0031182005008693 [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. Nature Publishing Group; 2011;470: 479–485. 10.1038/nature09670 [DOI] [PubMed] [Google Scholar]
- 4.Altizer S, Ostfeld RS, Johnson PTJ, Kutz S, Harvell CD. Climate change and infectious diseases: from evidence to a predictive framework. Science. 2013;341: 514–9. 10.1126/science.1239401 [DOI] [PubMed] [Google Scholar]
- 5.Redding DW, Moses LM, Cunningham AA, Wood J, Jones KE. Environmental-mechanistic modelling of the impact of global change on human zoonotic disease emergence: a case study of Lassa fever. Freckleton R, editor. Methods Ecol Evol. 2016;7: 646–655. 10.1111/2041-210X.12549 [DOI] [Google Scholar]
- 6.Mouritsen KN, Poulin R. Parasitism, climate oscillations and the structure of natural communities Oikos: Munksgaard International Publishers; 2002;97: 462–468. 10.1034/j.1600-0706.2002.970318.x [DOI] [Google Scholar]
- 7.Woolhouse MEJ, Haydon DT, Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol. Elsevier Current Trends; 2005;20: 238–244. 10.1016/j.tree.2005.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longdon B, Brockhurst MA, Russell CA, Welch JJ, Jiggins FM. The Evolution and Genetics of Virus Host Shifts. PLoS Pathog. 2014;10 10.1371/journal.ppat.1004395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longdon B, Hadfield JD, Webster CL, Obbard DJ, Jiggins FM. Host phylogeny determines viral persistence and replication in novel hosts Schneider DS, editor. PLoS Pathog. Elsevier Academic Press; 2011;7: e1002260 10.1371/journal.ppat.1002260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woolhouse MEJ. Population biology of emerging and re-emerging pathogens. Trends Microbiol. 2002;10: s3–s7. 10.1016/S0966-842X(02)02428-9 [DOI] [PubMed] [Google Scholar]
- 11.Villordo SM, Filomatori C V, Sánchez-Vargas I, Blair CD, Gamarnik A V. Dengue Virus RNA Structure Specialization Facilitates Host Adaptation. Nagy PD, editor. PLoS Pathog. 2015;11: 1–22. 10.1371/journal.ppat.1004604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musso D, Nilles EJ, Cao-Lormeau V-M. Rapid spread of emerging Zika virus in the Pacific area. Clin Microbiol Infect. 2014;20: O595–O596. 10.1111/1469-0691.12707 [DOI] [PubMed] [Google Scholar]
- 13.Leroy EM, Labouba I, Maganga GD, Berthet N. Ebola in West Africa: the outbreak able to change many things. Clin Microbiol Infect. 2014;20: O597–O599. 10.1111/1469-0691.12781 [DOI] [PubMed] [Google Scholar]
- 14.Caminade C, Kovats S, Rocklov J, Tompkins AM, Morse AP, Colón-González FJ, et al. Impact of climate change on global malaria distribution. Proc Natl Acad Sci U S A. National Academy of Sciences; 2014;111: 3286–91. 10.1073/pnas.1302089111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, et al. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci U S A. 2011;108: 7872–7877. 10.1073/pnas.1018344108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Messina JP, Brady OJ, Pigott DM, Golding N, Kraemer MUG, Scott TW, et al. The many projected futures of dengue. Nat Rev Microbiol. Nature Publishing Group; 2015;13: 230–239. 10.1038/nrmicro3430 [DOI] [PubMed] [Google Scholar]
- 17.Chen, Hill JK, Ohlemüller R, Roy DB, Thomas CD. Rapid range shifts of species associated with high levels of climate warming. Science. American Association for the Advancement of Science; 2011;333: 1024–6. 10.1126/science.1206432 [DOI] [PubMed] [Google Scholar]
- 18.Gehman A-LM, Hall RJ, Byers JE. Host and parasite thermal ecology jointly determine the effect of climate warming on epidemic dynamics. Proc Natl Acad Sci. 2018; 201705067. 10.1073/pnas.1705067115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks DR, Hoberg EP. How will global climate change affect parasite-host assemblages? Trends Parasitol. 2007;23: 571–574. 10.1016/j.pt.2007.08.016 [DOI] [PubMed] [Google Scholar]
- 20.Brady OJ, Golding N, Pigott DM, Kraemer MUGG, Messina JP, Reiner Jr RC, et al. Global temperature constraints on Aedes aegypti and Ae. albopictus persistence and competence for dengue virus transmission. Parasit Vectors. BioMed Central; 2014;7: 338 10.1186/1756-3305-7-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoberg EP, Brooks DR. Evolution in action: climate change, biodiversity dynamics and emerging infectious disease. Philos Trans R Soc Lond B Biol Sci. The Royal Society; 2015;370: 20130553–20130553. 10.1098/rstb.2013.0553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas MB, Blanford S. Thermal biology in insect-parasite interactions. Trends Ecol Evol. 2003;18: 344–350. 10.1016/S0169-5347(03)00069-7 [DOI] [Google Scholar]
- 23.Blanford S, Thomas MB. Host thermal biology: the key to understanding host-pathogen interactions and microbial pest control? Agric For Entomol. Blackwell Science Ltd; 1999;1: 195–202. 10.1046/j.1461-9563.1999.00027.x [DOI] [Google Scholar]
- 24.Labaude S, Moret Y, Cézilly F, Reuland C, Rigaud T, Ezilly FC, et al. Variation in the immune state of Gammarus pulex (Crustacea, Amphipoda) according to temperature: Are extreme temperatures a stress? Dev Comp Immunol. 2017;76: 25–33. 10.1016/j.dci.2017.05.013 [DOI] [PubMed] [Google Scholar]
- 25.Franke F , Armitage SAO , Kutzer MAM , Kurtz J , Scharsack JP. Environmental temperature variation influences fitness trade-offs and tolerance in a fish-tapeworm association. Parasit Vectors. BioMed Central; 2017;10: 252 10.1186/s13071-017-2192-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall ID. The influence of ambient temperature on the course of myxomatosis in rabbits. J Hyg (Lond). 1959;57: 484–497. 10.1017/S0022172400020325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamo SA, Lovett MME. Some like it hot: the effects of climate change on reproduction, immune function and disease resistance in the cricket Gryllus texensis. J Exp Biol. 2011;214: 1997–2004. 10.1242/jeb.056531 [DOI] [PubMed] [Google Scholar]
- 28.Mitchell SE, Rogers ES, Little TJ, Read AF. Host-Parasite and Genotype-By-Environment Interactions: Temperature Modifies Potential For Selection By a Sterilizing Pathogen. Evolution (N Y). 2005;59: 70–80. 10.1554/04-526 [DOI] [PubMed] [Google Scholar]
- 29.Killen SS, Marras S, Metcalfe NB, McKenzie DJ, Domenici P. Environmental stressors alter relationships between physiology and behaviour. Trends Ecol Evol. Elsevier Ltd; 2013;28: 651–658. 10.1016/j.tree.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 30.Kirk D, Jones N, Peacock S, Phillips J, Molnár PK, Krkošek M, et al. Empirical evidence that metabolic theory describes the temperature dependency of within-host parasite dynamics. Pawar S, editor. PLOS Biol. 2018;16: e2004608 10.1371/journal.pbio.2004608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Studer A, Thieltges DW, Poulin R. Parasites and global warming: Net effects of temperature on an intertidal host-parasite system. Mar Ecol Prog Ser. 2010;415: 11–22. 10.3354/meps08742 [DOI] [Google Scholar]
- 32.Macnab V, Barber I. Some (worms) like it hot: fish parasites grow faster in warmer water, and alter host thermal preferences. Glob Chang Biol. 2012;18: 1540–1548. 10.1111/j.1365-2486.2011.02595.x [DOI] [Google Scholar]
- 33.Cable J, Barber I, Boag B, Ellison AR, Morgan ER, Murray K, et al. Global change, parasite transmission and disease control: lessons from ecology. Philos Trans R Soc B Biol Sci. The Royal Society; 2017;372: 20160088 10.1098/rstb.2016.0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson LR, Ben-Horin T, Lafferty KD, McNally A, Mordecai E, Paaijmans KP, et al. Understanding uncertainty in temperature effects on vector-borne disease: a Bayesian approach. Ecology. 2015;96: 203–213. 10.1890/13-1964.1 [DOI] [PubMed] [Google Scholar]
- 35.Mordecai EA, Cohen JM, Evans M V, Gudapati P, Johnson R, Lippi CA, et al. Detecting the impact of temperature on transmission of Zika, dengue and chikungunya using mechanistic models Short title: Temperature predicts Zika, dengue, and chikungunya transmission 4. 2017; 1–18. 10.1101/063735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wesolowski A, Qureshi T, Boni MF, Sundsøy PR, Johansson MA, Rasheed SB, et al. Impact of human mobility on the emergence of dengue epidemics in Pakistan. Proc Natl Acad Sci U S A. National Academy of Sciences; 2015;112: 11887–92. 10.1073/pnas.1504964112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murdock CC, Paaijmans KP, Cox-Foster D, Read AF, Thomas MB. Rethinking vector immunology: the role of environmental temperature in shaping resistance. Nat Rev Microbiol. Nature Publishing Group; 2012;10: 869–876. 10.1038/nrmicro2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert GS, Webb CO. Phylogenetic signal in plant pathogen–host range. Proc Natl Acad Sci U S A. 2007;104: 4979–4983. Available: http://www.pnas.org/content/pnas/104/12/4979.full.pdf 10.1073/pnas.0607968104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Vienne DM, Hood ME, Giraud T. Phylogenetic determinants of potential host shifts in fungal pathogens. J Evol Biol. Blackwell Publishing Ltd; 2009;22: 2532–2541. 10.1111/j.1420-9101.2009.01878.x [DOI] [PubMed] [Google Scholar]
- 40.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin I V, McCracken GF, Rupprecht CE. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329: 676–9. 10.1126/science.1188836 [DOI] [PubMed] [Google Scholar]
- 41.Longdon B, Hadfield JD, Day JP, Smith SCL, McGonigle JE, Cogni R, et al. The causes and consequences of changes in virulence following pathogen host shifts. Schneider DS, editor. PLoS Pathog. Public Library of Science; 2015;11: e1004728 10.1371/journal.ppat.1004728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huey RB, Bennett AF. Phylogenetic studies of coadaptation:preferred temperatures versus optimal performance temperature of lizards. Evolution (N Y). 1987;41: 1098–1115. [DOI] [PubMed] [Google Scholar]
- 43.Kellermann V, Loeschcke V, Hoffmann AA, Kristensen TN, Fløjgaard C, David JR, et al. Phylogenetic Constraints In Key Functional Traits Behind Species’ Climate Niches: Patterns Of Desiccation And Cold Resistance Across 95 Drosophila Species. Evolution (N Y). 2012;66: 3377–89. 10.1111/j.1558-5646.2012.01685.x [DOI] [PubMed] [Google Scholar]
- 44.Kellermann V, Overgaard J, Hoffmann AA, Flojgaard C, Svenning J-C, Loeschcke V. Upper thermal limits of Drosophila are linked to species distributions and strongly constrained phylogenetically. Proc Natl Acad Sci. National Academy of Sciences; 2012;109: 16228–16233. 10.1073/pnas.1207553109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann AA, Chown SL, Clusella-Trullas S. Upper thermal limits in terrestrial ectotherms: how constrained are they? Fox C, editor. Funct Ecol. 2013;27: 934–949. 10.1111/j.1365-2435.2012.02036.x [DOI] [Google Scholar]
- 46.Perlman SJ, Jaenike J. Infection Success in Novel Hosts: an Experimental and Phylogenetic Study of Drosophila -Parasitic Nematodes. Evolution (N Y). 2003;57: 544–557. 10.1554/0014-3820(2003)057[0544:ISINHA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 47.Redding DW, Tiedt S, Lo Iacono G, Bett B, Jones KE. Spatial, seasonal and climatic predictive models of Rift Valley fever disease across Africa. Philos Trans R Soc B. 2017;372: 20160165 10.1098/rstb.2016.0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451: 990–993. 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christian PD. Studies on Drosophila C and A viruses in Australian populations of Drosophila melanogaster [Internet]. Australian National University; 1987. Available: http://hdl.handle.net/1885/142665 [Google Scholar]
- 50.Webster CL, Waldron FM, Robertson S, Crowson D, Ferrari G, Quintana JF, et al. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 2015;13: 1–33. 10.1371/journal.pbio.1002210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webster CL, Longdon B, Lewis SH, Obbard DJ. Twenty-five new viruses associated with the drosophilidae (Diptera). Evol Bioinforma. 2016;12: 13–25. 10.4137/EBO.S39454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnold PA, Johnson KN, White CR. Physiological and metabolic consequences of viral infection in Drosophila melanogaster. J Exp Biol. 2013;216: 3350–3357. 10.1242/jeb.088138 [DOI] [PubMed] [Google Scholar]
- 53.Chtarbanova S, Lamiable O, Lee K-Z, Galiana D, Troxler L, Meignin C, et al. Drosophila C virus systemic infection leads to intestinal obstruction. J Virol. 2014;88: 14057–69. 10.1128/JVI.02320-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson KN, Christian PD. The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J Gen Virol. 1998;79: 191–203. 10.1099/0022-1317-79-1-191 [DOI] [PubMed] [Google Scholar]
- 55.Ferreira ÁG, Naylor H, Esteves SS, Pais IS, Martins NE, Teixeira L. The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in Drosophila. PLoS Pathog. 2014;10 10.1371/journal.ppat.1004507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jousset FX, Plus N, Croizier G, Thomas M. [Existence in Drosophila of 2 groups of picornavirus with different biological and serological properties]. C R Acad Sci Hebd Seances Acad Sci D. 1972;275: 3043–6. Available: http://www.ncbi.nlm.nih.gov/pubmed/4631976 [PubMed] [Google Scholar]
- 57.Longdon B, Day JP, Alves JM, Smith SCL, Houslay TM, McGonigle JE, et al. Host shifts result in parallel genetic changes when viruses evolve in closely related species. PLoS Pathog. 2018;14: e1006951 10.1371/journal.ppat.1006951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reed L, Muench H. A Simple Method of Estimating Fifty Per Cent Endpoints. Am J Hyg. 1938;27: 231–239. [Google Scholar]
- 59.Lighton JRB. Measuring Metabolic Rates [Internet]. Measuring Metabolic Rates: A Manual For Scientists. Oxford University Press; 2008. 10.1093/acprof:oso/9780195310610.001.0001 [DOI] [Google Scholar]
- 60.Okada K, Pitchers WR, Sharma MD, Hunt J, Hosken DJ. Longevity, calling effort, and metabolic rate in two populations of cricket. Behav Ecol Sociobiol. 2011;65: 1773–1778. 10.1007/s00265-011-1185-3 [DOI] [Google Scholar]
- 61.Arnqvist G, Dowling DK, Eady P, Gay L, Tregenza T, Tuda M, et al. Genetic architecture of metabolic rate: environment specific epistasis between mitochondrial and nuclear genes in an insect. Evolution. United States; 2010;64: 3354–3363. 10.1111/j.1558-5646.2010.01135.x [DOI] [PubMed] [Google Scholar]
- 62.Huey RB, Moreteau B, Moreteau J-C, Gibert P, Gilchrist GW, Ives AR, et al. Sexual size dimorphism in a Drosophila clade, the D. obscura group. Zoology. 2006;109: 318–330. 10.1016/j.zool.2006.04.003 [DOI] [PubMed] [Google Scholar]
- 63.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drummond A, Suchard M, Xi ED, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology And Evolution 29; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rambaut A, Suchard M, Xie D, Drummond A. Tracer v1.6 [Internet]. 2014. Available: http://tree.bio.ed.ac.uk/software/tracer/
- 66.Rambaut A. FigTree [Internet]. 2006. Available: http://tree.bio.ed.ac.uk/software/figtree/.
- 67.Hadfield JD. MCMC methods for multi-respoinse generalized linear mixed models: The MCMCglmm R package. J Stat Softw. 2010;33: 1–22. 10.1002/ana.2263520808728 [DOI] [Google Scholar]
- 68.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- 69.Housworth EA, Martins EP, Lynch M. The phylogenetic mixed model. Am Nat. The University of Chicago Press; 2004;163: 84–96. 10.1086/380570 [DOI] [PubMed] [Google Scholar]
- 70.Felsenstein J. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am J Hum Genet. 1973;25: 471–492. 10.1038/ncomms15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freckleton RP, Harvey PH, Pagel M. Phylogenetic analysis and comparative data: a test and review of evidence. Am Nat. 2002;160: 712–26. 10.1086/343873 [DOI] [PubMed] [Google Scholar]
- 72.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401: 877–884. 10.1038/44766 [DOI] [PubMed] [Google Scholar]
- 73.Ingleby FC, Hunt J, Hosken DJ. The role of genotype-by-environment interactions in sexual selection. J Evol Biol. 2010;23: 2031–45. 10.1111/j.1420-9101.2010.02080.x [DOI] [PubMed] [Google Scholar]
- 74.Brooks DR, Hoberg EP. How will global climate change affect parasite–host assemblages? Trends Parasitol. Elsevier Current Trends; 2007;23: 571–574. 10.1016/J.PT.2007.08.016 [DOI] [PubMed] [Google Scholar]
- 75.Metcalf CJE, Walter KS, Wesolowski A, Buckee CO, Shevliakova E, Tatem AJ, et al. Identifying climate drivers of infectious disease dynamics: recent advances and challenges ahead. Proceedings Biol Sci. 2017;284: 20170901 10.1098/rspb.2017.0901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murdock CC, Paaijmans KP, Bell AS, King JG, Hillyer JF, Read AF, et al. Complex effects of temperature on mosquito immune function. Proceedings Biol Sci. The Royal Society; 2012;279: 3357–66. 10.1098/rspb.2012.0638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nowakowski AJ, Whitfield SM, Eskew EA, Thompson ME, Rose JP, Caraballo BL, et al. Infection risk decreases with increasing mismatch in host and pathogen environmental tolerances. Ecol Lett. 2016;19: 1051–1061. 10.1111/ele.12641 [DOI] [PubMed] [Google Scholar]
- 78.Cohen JM, Venesky MD, Sauer EL, Civitello DJ, McMahon TA, Roznik EA, et al. The thermal mismatch hypothesis explains host susceptibility to an emerging infectious disease. Ostfeld R editor. Ecol Lett. 2017;20: 184–193. 10.1111/ele.12720 [DOI] [PubMed] [Google Scholar]
- 79.Via S, Lande R. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution (N Y). 1985;39: 505–522. [DOI] [PubMed] [Google Scholar]
- 80.Gomulkiewicz R, Kirkpatrick M. Quantitative Genetics and the Evolution of Reaction Norms. Evolution (N Y). 1992;46: 390–411. 10.2307/2409860 [DOI] [PubMed] [Google Scholar]
- 81.Lazzaro BP, Flores HA, Lorigan JG, Yourth CP. Genotype-by-environment interactions and adaptation to local temperature affect immunity and fecundity in Drosophila melanogaster. PLoS Pathog. 2008;4: e1000025 10.1371/journal.ppat.1000025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mitchell SE, Halves J, Lampert W. Coexistence of Similar Genotypes of Daphnia magna in Intermittent Populations: Response to Thermal Stress. Oikos. 2004;106: 469–478. 10.1111/j.0030-1299.2004.13113.x [DOI] [Google Scholar]
- 83.Blanford S, Thomas MB, Pugh C, Pell JK. Temperature checks the Red Queen? Resistance and virulence in a fluctuating environment [Internet]. Ecology Letters. 2003. pp. 2–5. 10.1046/j.1461-0248.2003.00387.x [DOI] [Google Scholar]
- 84.Chown SL, Jumbam KR, Sørensen JG, Terblanche JS. Phenotypic variance, plasticity and heritability estimates of critical thermal limits depend on methodological context. Funct Ecol. 2009;23: 133–140. 10.1111/j.1365-2435.2008.01481.x [DOI] [Google Scholar]
- 85.Santos M, Castañeda LE, Rezende EL. Making sense of heat tolerance estimates in ectotherms: lessons from Drosophila. Funct Ecol. 2011;25: 1169–1180. 10.1111/j.1365-2435.2011.01908.x [DOI] [Google Scholar]
- 86.Overgaard J, Kristensen TN, Sørensen JG. Validity of thermal ramping assays used to assess thermal tolerance in arthropods. PLoS One. 2012;7: 1–7. 10.1371/journal.pone.0032758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Antonovics J, Wilson AJ, Forbes MR, Hauffe HC, Kallio ER, Leggett HC, et al. The evolution of transmission mode. Philos Trans R Soc B Biol Sci. 2017;372: 20160083 10.1098/rstb.2016.0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta V, Stewart CO, Rund SSC, Monteith K, Vale PF. Costs and benefits of sublethal Drosophila C virus infection. J Evol Biol. 2017;30: 1325–1335. 10.1111/jeb.13096 [DOI] [PubMed] [Google Scholar]
- 89.Cherry S, Perrimon N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol. 2004;5: 81–87. 10.1038/ni1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shikano I, Cory JS. Impact Of Environmental Variation On Host Performance Differs With Pathogen Identity: Implications For Host-Pathogen Interactions In A Changing Climate. Nat Publ Gr. 2015; 10.1038/srep15351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vale PF, Choisy M, Little TJ. Host nutrition alters the variance in parasite transmission potential. Biol Lett. 2013;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vale PF, Little TJ. Measuring parasite fitness under genetic and thermal variation. Heredity (Edinb). Nature Publishing Group; 2009;103: 102–9. 10.1038/hdy.2009.54 [DOI] [PubMed] [Google Scholar]
- 93.Lazzaro BP, Little TJ. Immunity in a variable world. Philos Trans R Soc Lond B Biol Sci. 2009;364: 15–26. 10.1098/rstb.2008.0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
Data are available from Figshare, and listed in the supplementary materials with links. Sequences have been submitted to Genbank and all accession numbers of sequences used to infer the host phylogeny are also available in a data on Figshare. Full dataset for the project and scripts is available: https://figshare.com/account/home#/projects/30692. This includes all data and all scripts to run the analysis.