Abstract
Hutchinson-Gilford progeria syndrome (HGPS) is a very rare fatal disease characterized for accelerated aging. Although the causal agent, a point mutation in LMNA gene, was identified more than a decade ago, the molecular mechanisms underlying HGPS are still not fully understood and, currently, there is no cure for the patients, which die at a mean age of thirteen. With the aim of unraveling non-previously altered molecular pathways in the premature aging process, human cell lines from HGPS patients and from healthy parental controls were studied in parallel using Next-Generation Sequencing (RNAseq) and High-Resolution Quantitative Proteomics (iTRAQ) techniques. After selection of significant proteins and transcripts and crosschecking of the results a small set of protein/transcript pairs were chosen for validation. One of those proteins, ribose-phosphate pyrophosphokinase 1 (PRPS1), is essential for nucleotide synthesis. PRPS1 loss-of-function mutants present lower levels of purine. PRPS1 protein and transcript levels are detected as significantly decreased in HGPS cell lines vs. healthy parental controls. This modulation was orthogonally confirmed by targeted techniques in cell lines and also in an animal model of Progeria, the ZMPSTE24 knock-out mouse. In addition, functional experiments through supplementation with S-adenosyl-methionine (SAMe), a metabolite that is an alternative source of purine, were done. Results indicate that SAMe has a positive effect in the proliferative capacity and reduces senescence-associated Beta-galactosidase staining of the HPGS cell lines. Altogether, our data suggests that nucleotide and, specifically, purine-metabolism, are altered in premature aging, opening a new window for the therapeutic treatment of the disease.
Introduction
Mutations in LMNA gene are the causal agent of subset of genetic diseases affecting mesoderm tissues called laminopathies [1]. This term refers to a highly heterogeneous group of disorders affecting the integrity of the nuclear lamina [2]. Among them, Hutchinson Guilford progeria syndrome (HGPS) or progeria is a fatal disease with a very low incidence characterized by a typical clinical picture of elderly pathologies [3]. HPGS-affected patients begin to show symptoms of accelerated aging at age of two, and usually die in the second decade of life due to cardiovascular deficiencies. HGPS is due, in most cases, to a point mutation (G608G) in LMNA gene encoding lamin A and C, major structural components of the nuclear lamina [4]. The mutation causes the occurrence of a cryptic alternative processing site in lamin A, generating a truncated isoform called progerin -PG- or Δ50 lamin A. This aberrant isoform remains farnesylated since the deletion includes the specific recognition sequence for the metalloprotease FACE1 that, in normal conditions removes the farnesylated C-terminal end of the Prelamin A to form the mature lamin A [5]. Accumulation of PG promotes defects on nuclear structure, replication, chromatin organization and stem cell differentiation, causing a senescence phenotype at the cellular level, and a premature aging of the organism [6,7]. A mouse model of HGPS, the knock-out deficiency for the metalloproteinase ZMPSTE24, the FACE1 homolog, fails to form mature lamin A, accumulates the permanently farnesylated precursor and recapitulates most of the symptoms of the disease [8].
So far, most of the efforts put on the reversion of the harmful effects of the nuclear accumulation of PG have focused on the use of farnesyl-transferase inhibitors (FTI) [9].
Aging is characterized by physiological alterations that compromise the health of the organism and represents one of the major risk factors for the development of a plethora of pathologies, including cardiovascular disease, cancer, musculoskeletal degeneration and neurodegenerative disorders [10]. At the molecular level, aging is accompanied by a set of hallmarks [11]. Among them, cellular senescence [12] mitochondrial dysfunction (13) [13], stem cell exhaustion [14], loss of proteostasis [15] and genomic instability [16] are the most characteristic. Aging is now considered a multi-factorial process. For this reason, the combination of complementary scientific approaches is necessary to fully understand the molecular aspects of aging and to delay or even reverse their detrimental aspects.
Proteomics provides tools to globally analyze cellular activity at protein level. Besides, this proteomic profiling will allow the elucidation of connections between broad cellular pathways and molecules that were previously impossible to predict using only traditional biochemical analysis. However, so far, the results obtained must be orthogonally validated with other approaches. Next-Generation Sequencing (NGS) technology, together with novel methods of pattern recognition and network analyses, has revolutionized cellular pathways [17]. Our intention for this study was to combine both shotgun proteomics and NGS to unravel new molecular pathways modulated in HGPS.
Materials and methods
Cell culture
Human HGPS-derived fibroblast cell lines (AG3513, AG3199 and AG8467) and their respective parental healthy controls (AG3512, AG3257 and AG8468) were acquired from Coriell Cell Repositories (CCR, Camdem, NJ, USA). Cells were cultured according to CCR instructions in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and expanded when they reached 70% confluence to 100mm culture dishes. For SAMe supplementation SAMe was periodically added to the medium at a concentration of 10 μg/mL.
RNA isolation and preparation for NGS
Cells expanded in 100 mm culture dishes were harvested following standard procedures for RNA isolation using the TRIzol® extraction method. The quality of 1 μL of each RNA sample was checked using the Bioanalyzer 2100 (Agilent Techologies, St. Clara, USA) to determine the RIN (RNA Integrity) score using the 6000 Nanochip and reagents (Agilent). Samples with a RIN score > 7 were retained and converted to cDNA by SureSelect Strand Specific RNA library Prep for Illumina multiplexed sequencing method.
Protein preparation for quantitative proteomics
Cells expanded in 100 mm culture dishes were gently scrapped with 200 μL of RIPA buffer with protease inhibitors at 4°C. Cells were transferred to 1.5 mL tubes and incubated at 100°C for 10 minutes. After centrifugation protein supernatants were collected for determination of the concentration by Bradford assay and checking of the protein integrity by SDS-PAGE and silver-staining.
RNA sequencing
Sample preparation was carried out as recommended by Agilent SureSelect Strand- Specific RNA Library Prep (Agilent, St.Clara, United States) for Illumina multiplex sequencing method209. The study was designed to screen the complete transcriptome sequence per normal (AG03257 and AG03512) and HGPS (AG03199 and AG03513) human skin fibroblasts. RNA from AG08647 failed to be retro-transcribed and this cell line and its counter partner AG08648 were not included in the study. The experiment could not be repeated due to our tight budget and the lack of extra funding. 1 μg of total RNA per sample was used. Fragmented DNA was end-repaired and the sequencing data was generated on Hiseq 1500 (Illumina, San Diego, United States) on a rapid mode flow-cell (Illumina, San Diego, United States). All samples were sequenced twice and they were prepared in duplicate.
Bioinformatics analysis of NGS data
An average of 25 million paired-end 100 bases pairs (bp) reads was obtained per sample in transcriptome analysis. The raw RNA-sequencing reads for each sample were aligned to the reference Homo sapiens (human) genome browser (GRCh38.p12) assembly using Bowtie2 (bowtie-bio.sourceforge.net/index.shtml/) and Tophat2 (http://tophat.cbcb.umd.edu/). After alignment, raw sequence read depths was converted to estimate transcript abundance measures as fragments per kilo base of exons per million (FPKM) values with Cufflinks (http://cufflinks.cbcb.umd.edu/). Identified transcripts were considered statistically significant when q-value ≤ 0.05.
iTRAQ labeling
Total protein extracts (100 μg per condition) were processed for iTRAQ labeling following manufacturers instruction (Sciex). Two biological replicates were done in parallel (iTRAQ 1 and 2, Fig 1). Briefly, the total proteins were precipitated with overnight incubation with six volumes of cold acetone at −20°C, and denatured with 2% sodium dodecyl sulphate in 1 M triethylammonium bicarbonate buffer (ABSciex, Foster City, CA, USA). After incubation for 1 hour at 60°C using 50 mM tris-(2-carboxyethy) phosphine (Sciex), cysteines were blocked with 84 mM iodoacetamide (Sigma-Aldrich) at room temperature for 30 minutes in absence of ligth. Digestion was done with spectrometry-grade trypsin (Promega, Madison, WI, USA) at a concentration of 1:50 trypsin/protein for 16 hours at 37°C. The next morning each peptide solution was labeled for 1.5 hours at room temperature using the iTRAQ reagents (Sciex). Samples were labeled with iTRAQ reagents as follows: AG3513: tag 113; AG3199: tag 114; AG8467: tag 115; AG3512: tag 116; AG3257: tag 117; and AG8468: tag 118. As an internal control of the labeling a duplicate of AG3513 was labeled with tag 119. The reaction was stopped by reduction of the percentage of the organic phase with the addition of de-ionized water, and all the labeled samples were combined in one tube. Homemade stage tips were used for desalting prior LC analysis.
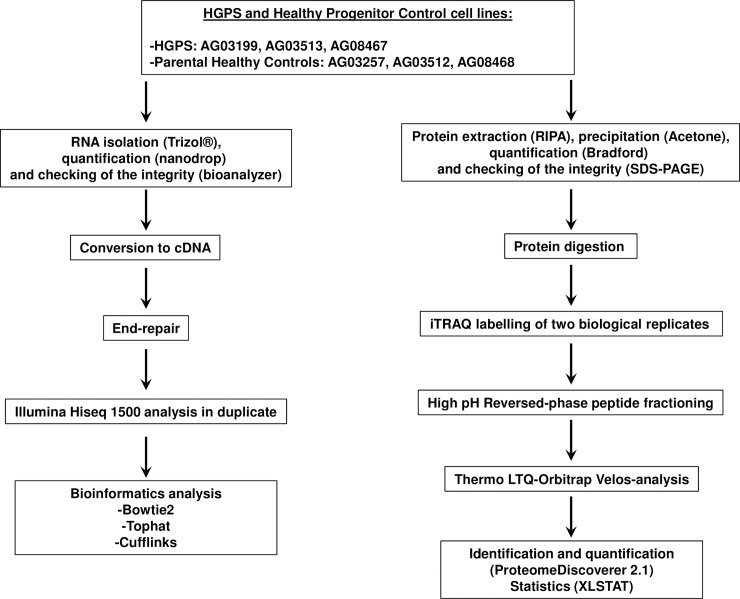
Fig 1. Parallel genomic and proteomic workflows.
Schematic representation of the workflow followed for the genomic (left) and proteomic (right) large-scale approaches. HGPS and Healthy Progenitor cell lines were processed for RNA and total protein isolation. RNA was converted to cDNA for analysis in a Illumina Hi seq 1500 platform. Two technical replicates were analyzed for each sample. Raw data was processed and statistical analysis was done using Bowtie2, TopHat and Cufflinks programs. Total protein extracts were labeled using iTRAQ 8-plex. Two biological replicates were processed in parallel (iTRAQ1 and iTRAQ2). Labelled peptides were fractionated by Basic Reversed-phase chromatography and fractions were analyzed in a LTQ-Orbitrap Velos platform. Protein quantification was done using Proteome Discoverer 2.1 software and XLSTAT software was used for statistical analysis.
High pH reversed phase fractionation
Each of the iTRAQ labeled tryptic digests was subjected to high pH fractionation with High pH reversed phase peptide fractionation kit (Pierce) following the manufacturer’s instructions. Briefly the samples were loaded onto a spin column in 0.1% trifluoroacetic acid (TFA), washed and buffer exchanged with high pH buffer and then eluted in 9 fractions of increasing acetonitrile (ACN) concentration (f1 = 10%ACN; f2 = 12.5%ACN; f3 = 15%ACN; f4 = 17.5%ACN; f5 = 20%ACN; f6 = 22.5%ACN; f7 = 25%ACN; f8 = 50%ACN; f9 = 75%ACN). Flow through and wash fractions were pooled and also analyzed as FT-wash fraction. The fractions were dried down in a speed-vacuum centrifuge and kept at -80°C until LC-MSMS analysis.
LC-MSMS analysis
The 10 dried-down fractions were analyzed in a nano-Acquity liquid chromatograph (Waters) coupled to a LTQ-Orbitrap Velos (Thermo Scientific) mass spectrometer. Tryptic labeled peptides were resuspended in 2% ACN/1% formic acid (FA) solution and an aliquot (corresponding to 500 ng) was injected for their chromatographic separation. Peptides were trapped on a Symmetry C18TM trap column (5μm 180μm x 20mm; Waters), and were separated using a C18 reverse phase capillary column (75 μm Øi, 25 cm, nano Acquity, 1.7μm BEH column; Waters). The gradient used for the elution of the peptides was 2 to 35% B in 155 minutes, followed by a gradient from 35% to 45% in 20 min (A: 0.1% FA; B: 100% ACN, 0.1%FA), with a 250 nL/min flow rate.
Eluted peptides were subjected to electrospray ionization in an emitter needle (PicoTipTM, New Objective) with an applied voltage of 2000V. Peptide masses (m/z 300–1800) were analyzed in data dependent mode where a full Scan MS in the Orbitrap with a resolution of 30,000 FWHM at 400m/z was obtained. Up to the 10 most abundant peptides (minimum intensity of 2000 counts) were selected from each MS scan and then fragmented using HCD (Higher Energy Collision Dissociation) in C-trap using nitrogen as collision gas, with 40% normalized collision energy and analyzed in the Orbitrap with a resolution of 7,500 FWHM at 400m/z. The scan time settings were: Full MS: 250 ms (1 micro-scan) and MSn: 300 ms (2 micro-scans).Generated .raw data files were collected with Thermo Xcalibur (v.2.2).
Raw data was processed using Proteome Discoverer 2.1 software (Thermo Scientific) with SequestHT as search engine against the latest SwissProt/Uniprot Human database. 10 .raw files corresponding to the 10 injections from the MS analyses were used to perform as a single search against this database. Both a target and a decoy database were searched to obtain a false discovery rate (FDR; <1%), and thus estimate the number of incorrect peptide-spectrum matches that exceed a given threshold. The following search parameters were applied: Database/Taxonomy: SwissProt/Uniprot Human plus Contaminants (March 2017); Enzyme: Trypsin; Missed cleavage: 2; Fixed modifications: Carbamidomethyl of cystein, iTRAQ8plex (N-term), iTRAQ8plex (K); Variable modifications: oxidation of methionine, iTRAQ8plex (Y); Peptide tolerance: 10 ppm and 0.1 Da (respectively for MS and MS/MS spectra). Thermo software Proteome Discover (v.2.1) was also used for quantification, using an isobarically labeled reporter ion quantification method (iTRAQ 8-plex).
Statistical analysis of iTRAQ data
XLSTAT software was used to analyze the individual ratios obtained after Proteome Discoverer 2.1 processing. Briefly, the ratios for all the proteins quantified in both replicates were extracted and classified as “equal” (twelve ratios HGPS/HGPS and Control/Control) or “opposite” (eighteen ratios Control/HGPS). After checking that the ratios follow a normal distribution using the Kolmogorov-Smirnov test we applied a Student T-test to identify those proteins that present significant (p-value ≤ 0.001) differences when comparing HGPS/HGPS and Control/Control (equal) versus Control/HGPS (opposite) ratios.
Real-time PCR of PRPS1
RNA was transformed to complementary DNA (cDNA) using NZY First-Strand cDNA synthesis kit (NZYTECH, Lisbon, Portugal) according to manufacturer's instructions. cDNA was amplified using specific primers for the PRPS1 gene. The design of primers was carried out using the software Primer3 (http://biotools.umassmed.edu/bioapps/primer3). The sequences of the primers were as follows; Forward: tcagactgcctgctgacttc; Reverse: tacctcaacgtgctcagtgg. Quantitative RT-PCR was carried out in a LightCycler 480 Instrument (Roche Applied Science) using Light Cycler 4800 SYBR Green I Master kit (Roche, Basel, Switzerland). The amplification program consisted on initial denaturation of 92 oC for 2 min followed by 40 cycles at 92 oC for 15 s, annealing at 55–62 oC, depending on the gene, for 30 s and extension at 72 oC for 15 s. qRT-PCR were done in triplicate, with each set of assays repeated three times. For control experiments no reverse transcriptase was used.
Immunohistochemistry of PRPS1 in liver sections
Full-depth sections (4 μm) of paraffin-embedded livers from ZMPSTE24 null, heterozygous and wild-type mouse strains (blocks kindly donated by B. Caramés, INIBIC, A Coruña, Spain) were cut with a microtome and fixed in 4% (w/v) paraformaldehyde (Sigma-Aldrich) in PBS at pH 7.6. All sections were immuno-stained with rabbit polyclonal antibody for PRPS1 (1:100) (Abcam). Biotinylated secondary antibodies were detected using a peroxidase-labeled biotin-streptavidin complex with diaminobenzidine substrate Ennvision Kit (Dako) following manufacturer instructions.
Cell proliferation assay
Cell lines were cultured at a density of 10.000 cells/well in 96-well plates, with or without periodic addition of SAMe (Sigma-Aldrich) at a final concentration of 10 μg/mL. At different times of culture (3, 5, 7 and 10 days), 10 μl of Thiazolyl Blue Tetrazolium Bromide (MTT, from Sigma-Aldrich) was added per well to achieve a final concentration of 0.45 mg/ml and incubated by 4 hours at 37°C. The medium was removed and 100 μl of Dimethyl sulfoxide (Sigma-Aldrich) was added per well to dissolve formazan crystals for 10 min and absorbance was recorded at 570 nm in a Nanoquant platform (Tecam).
Senescence-associated Beta-galactosidase staining
X-Gal cytochemical staining for SA-β-gal was performed as previously described [18]. The cells were fixed and then stained with freshly prepare SA-β-gal staining solution overnight at 37°C according to the manufacturer's protocols (Cell Signaling Technology, Beverly, MA, USA). The number of SA-β-gal-positive cells in randomly selected fields was expressed as a percentage of all cells counted.
Results
RNAseq analysis of HGPS and healthy control cell lines
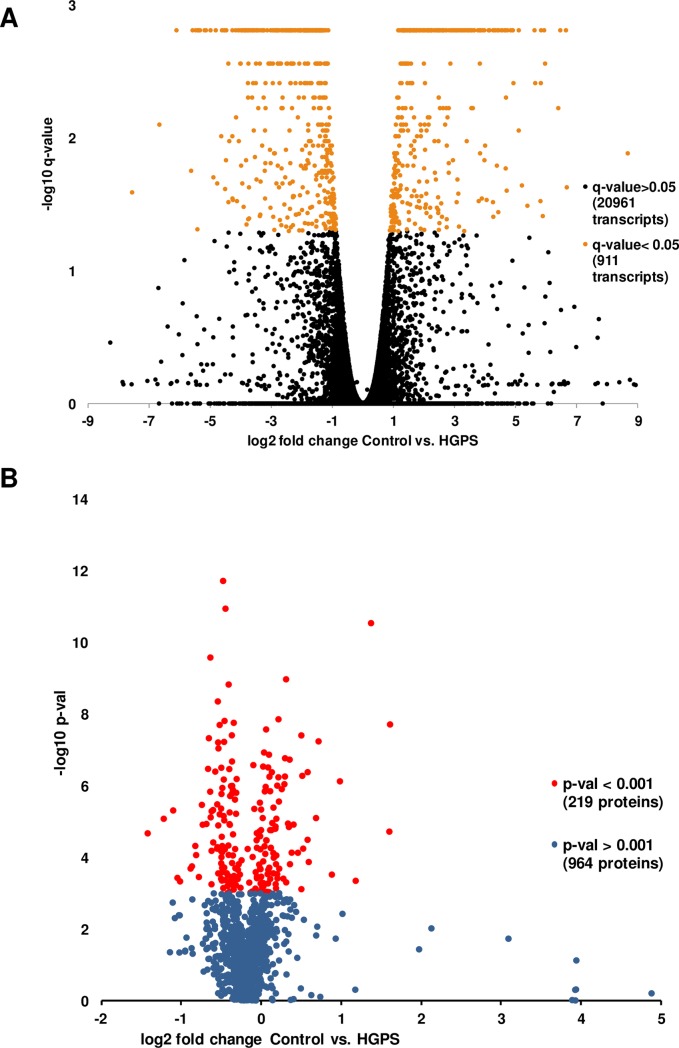
A schematic workflow of the analysis is represented in Fig 1. Genomic datasets are deposited at the Gene Expression Omnibus (GEO-NCBI) repository (http://www.ncbi.nlm.nih.gov/geo/). Raw files are public and freely accessible (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113648). A total of 21872 transcripts were detected and quantified in both conditions (Control and HGPS) by NGS following our approach (S1 File and Fig 2A) Among them, 911 were differentially expressed with a q-value ≤ 0.05 representing roughly a 4 per cent of the total sequenced transcriptome.
Fig 2. Transcripts and proteins significantly modulated in HGPS-derived cells versus healthy parental control cell lines.
Volcano plot representations of the RNAseq (A) and shotgun proteomics (B) analysis of the HGPS and Control cell lines. For RNAseq, a threshold of q-value = 0.05 was set for significance according the statistical package used for analysis. A total of 911 transcripts present a q-value ≤ 0.05. For shotgun proteomics a threshold of p-value = 0.001 was set for significance according to the statistical package used. In this case 219 proteins were detected as significant with a p-value ≤ 0.001.
iTRAQ analysis of HGPS and healthy control cell lines
Proteomic datasets are deposited at MassIVE repository (www.massive.ucsd.edu). Raw and processed files (Quantitative Proteomics Study (iTRAQ) of Progeria Cell Lines; #MSV000081576) are public and freely accessible. A summary of the quantitative proteomics analysis is represented in Fig 2B and S1 Fig, which show the quantification results in both experiments (S1A Fig), showing a nice correlation between the results (S1B Fig). Statistical analysis of the quantification results (S2 File and Fig 2B) showed that 219 proteins present significant (p-value ≤ 0.001) differences in the Control/HGPS versus Control/Control and HGPS/HGPS abundance ratios.
Comparison of the proteomics and transcriptomics results
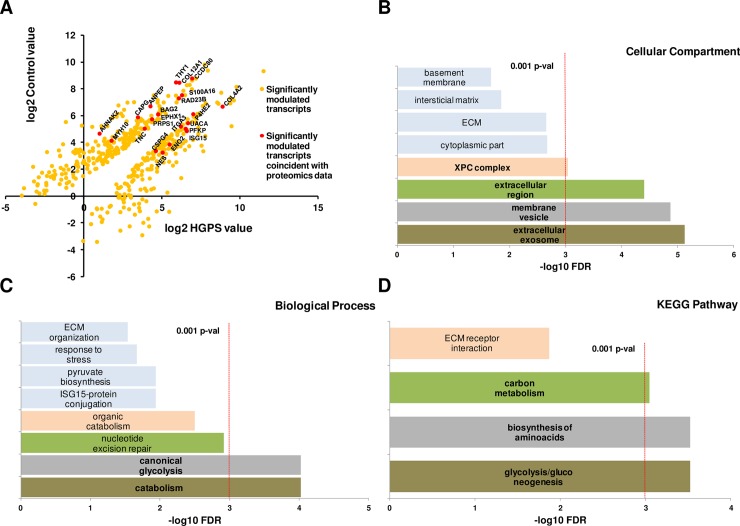
The difference in the amount of identified transcripts and proteins is inherent to the differences in the sensitivity of the techniques used. The list of significant proteins was crosschecked against the list of significant transcripts (Fig 3). From the 219 proteins we found that 24 of their corresponding transcripts were detected as differentially expressed with a q-value ≤ 0.05. As showed in Table 1, the correlation of those 24 protein/transcript pairs is very high. Most of those proteins appear to play important roles in cell shape and adhesion (THY1, TNC and COL12A1), proliferation and apoptosis (CSPG4) and metabolism and generation of energy (ENO2, PFKP, ANPEP and PRPS1). PRPS1 codifies for Ribose-phosphate pyrophosphokinase 1, a key protein responsible for de novo purine synthesis in the nucleotide metabolism. Our data showed a decrease of the amount of PRPS1 in HGPS cell lines at both transcript and protein levels when compared to the parental healthy controls. Due to the novelty of this link with HGPS, and the importance of de-regulation of the metabolism and imbalance of energy generation in this disease, we decided to focus our validation in this protein.
Fig 3. Crosschecking of the genomic and proteomic results shows that HGPS present features of a metabolic disorder.
Representation of the modulation in HGPS versus Control cell lines of the transcripts detected as significant and those correlating with the proteomic approach (red dots in A). String 10.4 gene ontology statistical study of the 22 transcript/protein coincident pairs (B, C, D) demonstrate that most of the proteins are membrane-bounded or secreted and have a role on glycolysis, energy generation and synthesis of bio-molecules.
Table 1. Crosschecking of the RNAsec and ITRAQ data.
| Gene name | Protein name | RNAsec data: Up in | iTRAQ data: Up in | Function |
|---|---|---|---|---|
| AHNAK2 | Protein AHNAK2 | Control | Control | Regulation of calcium channels. |
| ANPEP | Aminopeptidase N | Control | Control | Digestion of proteins. |
| BAG2 | BAG family molecular chaperone regulator 2 | Control | Control | Protein folding. |
| CAPG | Macrophage-capping protein | Control | Control | Cytoplasmic and nuclear structure. |
| CCDC80 | Coiled-coil domain-containing protein 80 | Control | Control | Extracellular matrix organization. |
| COL12A1 | Collagen alpha-1(XII) chain | Control | Control | Extracellular matrix organization. |
| COL4A2 | Collagen alpha-2(IV) chain | HGPS | HGPS | Extracellular matrix organization. |
| CSPG4 | Chondroitin sulfate proteoglycan 4 | HGPS | HGPS | Regulation of cell proliferation and migration |
| ENO2 | Gamma-enolase | HGPS | HGPS | Glycolysis. |
| EPHX1 | Epoxide hydrolase 1 | Control | Control | Catabolism of aromatic compounds. |
| GPNMB | Transmembrane glycoprotein NMB | HGPS | Control | Cell migration and adhesion. |
| ISG15 | Ubiquitin-like protein ISG15 | HGPS | HGPS | Defense against bacteria. |
| ITGA3 | Integrin alpha-3 | HGPS | HGPS | Cell adhesion and cell-cell interaction. |
| MYH10 | Myosin-10 | Control | Control | Cell shape and cytokynesis. |
| NES | Nestin | HGPS | HGPS | Intermediate fillament binding. |
| P4HA2 | Prolyl 4-hydroxylase subunit alpha-2 | Control | HGPS | Proline hydroxilation in collagen. |
| PFKP | ATP-dependent 6-phosphofructokinase | HGPS | HGPS | Glycolysis. |
| PRPS1 | Ribose-phosphate pyrophosphokinase 1 | Control | Control | Nucleotide biosynthesis. |
| PSME2 | Proteasome activator complex subunit 2 | HGPS | HGPS | Proteosomal degradation. |
| RAD23B | UV excision repair protein RAD23 homolog B | Control | Control | Proteosomal degradation, part of the XPC complex. |
| S100A16 | Protein S100-A16 | Control | Control | Calcium binding protein, adipocyte differentiation. |
| THY1 | Thy-1 membrane glycoprotein | Control | Control | Cell adhesion and cell-cell interaction. |
| TNC | Tenascin | Control | Control | Extracellular matrix organization. |
| UACA | Uveal autoantigen with coiled-coil domains and ankyrin repeats | HGPS | HGPS | Regulation of stress-induced apoptosis. |
List of the 24 protein/transcript pairs selected after the crosschecking of the proteomics and transcriptomics data.
Orthogonal validation of PRPS1 levels in HGPS and healthy control cell lines and in the ZMPSTE 24 KO mouse
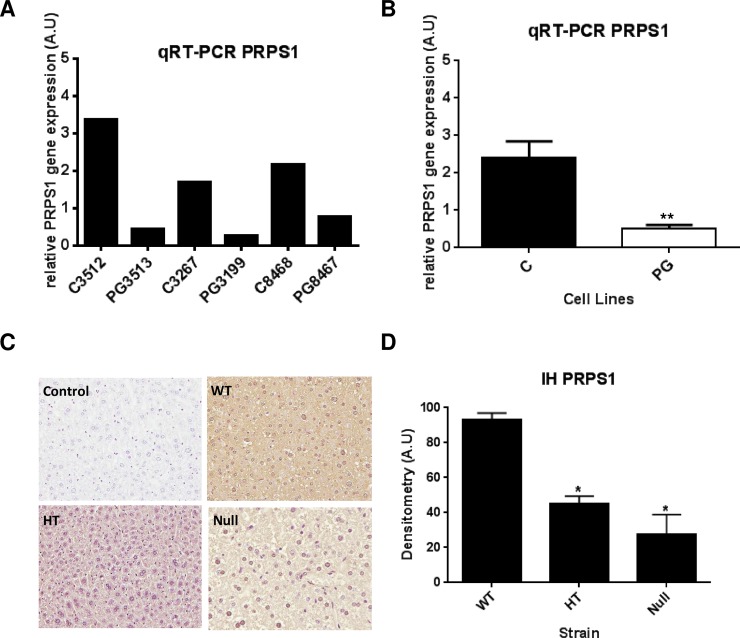
Quantitative Real Time PCR (qt-RT-PCR) analysis of PRPS1 in the cell lines used for the omics-study confirms the reduced levels of PRPS1 transcripts in the three HGPS cell lines (Fig 4A and 4B). Statistical analysis showed a significant difference (Fig 4B). We then decided to verify if this difference in expression occurs also in vivo. ZMPSTE24 knock-out mouse is the most used mammal model of HGPS. Since the liver is the main organ where purine de novo synthesis occurs, we checked PRPS1 levels by immunohistochemistry in liver sections of ZMPSTE24-null mice comparing to the heterozygous (HT) and the wild-type (WT) mice (Fig 4C). PRPS1 levels are significantly higher in WT versus both HT and null mice, thus corroborating the in vitro results.
Fig 4. Targeted validation of PRPS1 shows down-regulation of transcript and protein levels in HGPS-cells and the animal model of HPGS, respectively.
Orthogonal validation of PRPS1 modulation by targeted techniques in cell lines (A, B) and in the mouse model of HGPS (C, D). Real-Time PCR (RT-PCR) of HGPS and control cell lines (A) showing that PRPS1 transcript is down-regulated in the three HGPS cell lines when compare to their respective controls. Statistical analysis (B) demonstrates that the difference between control and HGPS is significant (p-value ≤ 0.01). Representative images (C) of the immunohystochemistry of the PRPS1 protein in liver sections of the ZMPSTE24-null mice strain (magnification: 20x). Statistical analysis shows that wild-type mice (WT) present significant (p-value ≤ 0.05) higher levels of the protein than heterozygous and null mice.
Study of the effect of the incubation with an alternative purine source in the proliferation capacity and senescence phenotype in HGPS cell lines
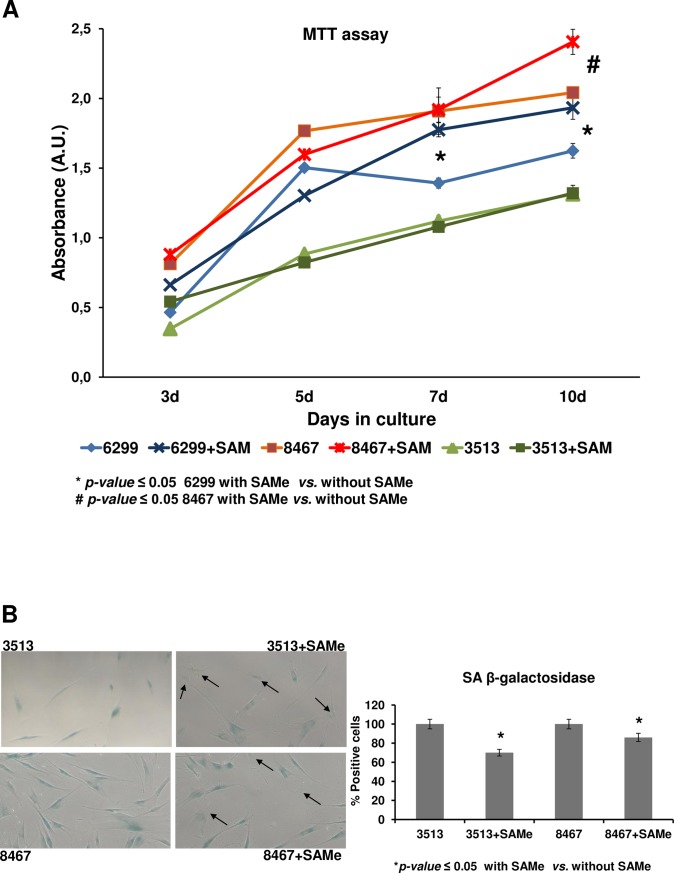
To further investigate the link between cellular senescence in premature aging and defective purine synthesis we incubate HGPS cell lines with S-adenosyl methionine (SAMe), a compound that provides an alternative source of adenosine, the final metabolite in the purine synthesis pathway. The periodic addition of SAMe to the culture medium promotes a significant increase in the proliferative capacity of two out of the three HGPS cell lines studied (Fig 5A). This effect is accompanied by a decrease of senescence-associated β-galactosidase staining (Fig 5B).
Fig 5. An alternative source of purine partially reverts the premature phenotype aging in HGPS-derived cells.
Effect of incubation with SAMe on the premature-aging phenotype of HGPS cell lines. MTT-based proliferation assay (A) showing that periodic addition of SAMe to the culture media at a final concentration of 10 μg/mL, have a significant (p-value≤ 0.05) positive effect in the proliferation capacity of two out of the three cell lines after 10 days in culture. Representative images (magnification: 20x) of the staining of Senescence Associated Beta-galactosidase (SA-β-gal) staining in HGPS cells with and without SAMe addition to the culture media (B), showing a higher number of negative cells (arrows) in SAMe-treated cells. Densitometry analysis of the staining signal shows a decrease in two of the HGPS cell lines after incubation with SAMe (C).
Discussion
Aging is a complex and multi-factorial process that, to the date, is poorly understood at the mechanistic level [19]. Genetic, epigenetic, metabolic and environmental factors contribute to the accumulation of molecular alterations that promote a negative imbalance in the regeneration/degeneration ratio of the tissues increasing the risk of disease and death at the organism level. Metabolism is a key factor influencing aging, since caloric restriction and down-regulation of the insulin/insulin-like growth factor 1 (IGF-1) pathway drive to an extension of the lifespan in nematodes, insects and mammals [20–22]. Premature aging syndromes share many molecular and phenotype features with aging-associated diseases [10]. The implication of de-regulation of the metabolism in accelerated aging has been also demonstrated. HGPS is characterized by a dramatic lipodystrophy [23] and has been recently associated to increased protein synthesis [15, 24].
Large-scale omic-techniques have experienced an exponential development in the last 5 years in terms of sensitivity, reproducibility and throughput capacity. These tools are now of great help to deeply investigate the molecular mechanisms underlying cell homeostasis and organism healthiness but also the altered molecular pathways in disease [25].
In the present work we have found a good correlation between the shotgun quantitative proteomics and the Next-Generation Sequencing after crosschecking of the results. The corresponding transcripts of 24 out of the 219 modulated proteins were detected as significantly modulated (Fig 3). As Table 1 shows, 22 out of the 24 protein/transcript pairs present the same kind of modulation, (i.e. increased in control or, alternatively, in HGPS in both approaches). Our intention for this study was to focus on this group of candidates. The analysis of the functional protein association networks using String 10.5 demonstrates that the crosschecked modulated proteins are mainly membrane-bounded or secreted and participate in glycolysis and metabolism (Fig 3), suggesting that HGPS presents features typical of a metabolic disorder.
Among them, one protein took specially our attention due to the novelty of its link with premature aging. PRPS1 gene codifies for Phosphoribosyl pyrophosphate synthetase-1, a key enzyme in nucleotide metabolism [26]. PRPS1 catalyzes the first step of the de novo Purine synthesis pathway, the formation of phosphoribosyl-pyrophosphate (PRPP) from Ribose-5-phosphate plus a molecule of adenosine triphosphate (ATP). PRPS1 loss-of-function mutations have been previously related to disease phenotypes like Arts Syndrome, diabetes insipidus or hearing loss [27].
De novo nucleotide synthesis is tightly regulated in the cells by a multi-enzymatic complex, the purinosome, integrated by six different enzymes that catalyze ten different reactions [28]. The resulting metabolites are essential for key biological processes such as cell signaling, energy generation and enzymatic activity, besides being the structural constituents of DNA and RNA [29]. The purinosome is a transient structure, that is subjected to periodic assembly/disassembly cycles in proliferating cells [30]. Purinosome activity is up-regulated during G1/S-phases, accompanied by an increase in PRPP intracellular levels, and decreases dramatically by G2/M-phases [31].
In the present study, PRPS1 levels are detected as significantly lower in HGPS vs. healthy progenitor cell lines in both massive approaches. This is, to our knowledge, the first scientific evidence linking premature aging and de novo purine synthesis. Further orthogonal verification by targeted techniques is in accordance with the results. Real-Time PCR demonstrated diminished levels of PRPS1 in the three HGPS cell lines versus the correspondent control cell lines. Then we decided to investigate if this down-regulation is observed in vivo. ZMPSTE24-null mouse is the most used animal model for the study of HGPS [32]. ZMPSTE24 is the mouse-homolog of human FACE1, the metalloprotease that converts pre-lamin A to mature lamin A. These mice accumulate permanently-farnesylated pre-lamin A, which exerts similar harmful effects than progerin, and display most of the phenotypic features of the disease [33]. Immunohistochemical analysis in ZMPSTE24-null mice liver sections showed diminished levels of PRPS1 when compared to both heterozygous and wild-type strains.
S-adenosyl-methionine (SAMe) is the central methyl donor in many cellular methylation reactions, catalyzed by cellular methyltransferases and hydrolases, and is also an alternative source of purine in living cells [34]. This metabolite was first discovered more than sixty years ago [35] and several studies have demonstrated its therapeutic value for the treatment of osteoarthritis [36], depression [37] and liver disease [38]. In vitro, SAMe has a positive effect on chondrocyte proliferation at a dose of 10 μg/mL [39]. In our hands, incubation of HGPS derived cells with SAMe ameliorates the senescence phenotype and increase the proliferation rate of the cells, suggesting that purine deprivation could be in part responsible for premature aging phenotype.
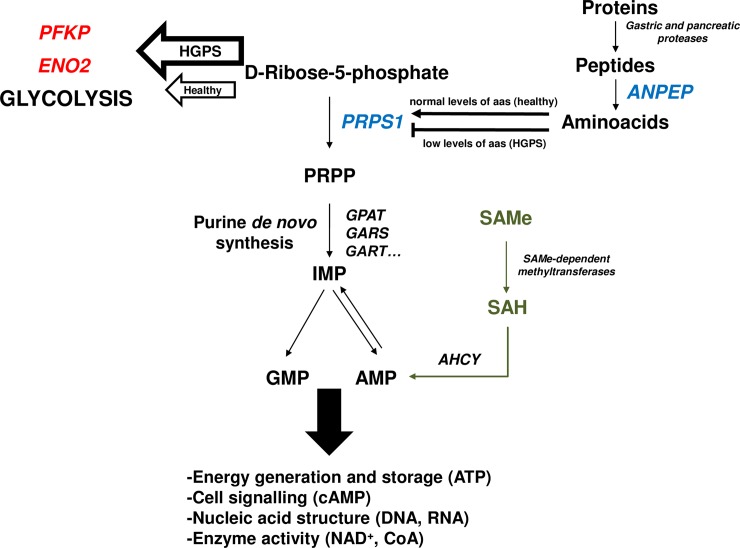
PRPS1 activity has been previously reported to be negatively modulated by both amino-acid starvation and a decrease of available D-Ribose-5-phosphate [40]. D-Ribose-5-phosphate is an intermediate of the pentose phosphate pathway, specifically the first product of the non-oxidative set of reactions. Interestingly, glycolysis has been also described as enhanced in ZMPSTE24-null mice using proteomics approaches [41]. This is consistent with our results, since we have found in the present work two of the key glycolytic enzymes, ATP-dependent 6-phosphofructokinase (PFKP) and gamma-enolase (ENO2), as up-regulated in HGPS cell lines. In this scenario (Fig 6), the increase in the glycolysis due to lamin A de-regulation, would produce a decrease in the amount of available D-Ribose-5-phosphate necessary for purine biosynthesis, thus promoting a reduction in the activity of PRPS1 affecting the entire de novo purine pathway. On the other hand, regarding the effect of amino-acid starvation on the PRPS1 activity, it is noteworthy the reduction of the levels of aminopeptidase N (ANPEP) that we have detected by both large-scale approaches. ANPEP, also known as CD13, is one of the enzymes that participate in the final digestion of peptides coming from the gastric hydrolysis of proteins [42] and participates in the rennin-angiotensin system that regulates vasopressin release, thus regulating arterial blood pressure and the pathogenesis of hypertension [43]. A reduction of the levels of ANPEP in HGPS would be responsible for the inhibition of PRPS1 through amino-acid starvation and, even more importantly, would contribute to the hypertension and high blood pressure levels that, long term, derive in the cardiovascular malfunction characteristic of the disease.
Fig 6. Proposed model to explain the down-regulation of PRPS1 in HGPS.
Enzymes in blue are detected as down-regulated and those in red as up-regulated in the present study. High glycolitic rate in HGPS drives to a decrease in the levels of D-Ribose-5-phosphate available for de novo purine synthesis. This reduction compromises PRPS1 activity. Furthermore, this inhibition is complemented by free amino-acid starvation resulting from down-regulation of ANPEP. As a result, de novo nucleotide synthesis is affected thus promoting defects in key biological processes that contribute to the premature aging phenotype. Supplementation of SAMe helps to restore the levels of AMP which, in turn, could be transformed to GMP by the cellular machinery, partially ameliorating the premature aging phenotype. Abbreviations: PFKP: ATP-dependent 6-phosphofructokinase; ENO2: gamma-enolase; PRPS1: ribose-phosphate pyrophosphokinase 1; ANPEP: aminopeptidase N; GPAT: glutamine-phosphorybosil aminotransferase; GARS: glycinamide-ribonucleotide synthetase; GART: glycinamide-ribonucleotide transformylase; IMP: inosine monophosphate; GMP: guanosine monophosphate; AMP: adenosine monophosphate; SAMe: S-adenosylmethionine; SAH: S-adenosylhomocysteine; AHCY: S-adenosylhomocysteine hydrolase.
Altogether, our results indicate a non-previously described alteration of the de novo purine synthesis cellular machinery in HGPS. An attractive possibility is that LMNA de-regulation promotes down-regulation of the PRPS1 gene, resulting in a decrease of PRPS1 levels in HGPS. In this case, purine synthesis deprivation would compromise energy generation and cell signaling. Further investigation is needed for a deeper understanding of this novel link between premature aging and de-regulation of purine synthesis and to explore new therapeutic windows for the treatment of the disease.
Supporting information
749 proteins in common were quantificated in both iTRAQ replicates (A). Graphical representation of the Control/HGPS ratios shows a good correlation between the two replicates (B).
(TIF)
RNAseq analysis of Control vs. HGPS cell lines.
(XLSX)
iTRAQ analysis (replicates 1 and 2) of Control cells vs. HGPS cell lines.
(XLSX)
Acknowledgments
We are very grateful to Maria José Sánchez-Dopico, and Noa Goyanes for excellent technical help and Beatriz Caramés for donation of ZMPSTE24 paraffin-embedded liver blocks. The authors declare no competing financial interests.
Abbreviations
- HGPS
Hutchinson-Gilford progeria syndrome
- PG
Progerin
- NGS
Next-Generation Sequencing
- iTRAQ
isobaric tags for absolute and relative quantitation
- RIN
RNA integrity
- RIPA
radioimmunoprecipitation assay
- FDR
false discovery rate
- PBS
phosphate buffered saline
- qt-RT-PCR
quantitative Real-Time Polimerase Chain Reaction
- PFKP
ATP-dependent 6-phosphofructokinase
- ENO2
gamma-enolase
- PRPS1
ribose-phosphate pyrophosphokinase 1
- ANPEP
aminopeptidase N
- GPAT
glutamine-phosphorybosil aminotransferase
- GARS
glycinamide-ribonucleotide synthetase
- GART
glycinamide-ribonucleotide transformylase
- IMP
inosine monophosphate
- GMP
guanosine monophosphate
- AMP
adenosine monophosphate
- SAMe
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- AHCY
S-adenosylhomocysteine hydrolase
- FTI
farnesyltransferase inhibitors
Data Availability
Genomic datasets are deposited at the Gene Expression Omnibus (GEO-NCBI) repository (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113648). Proteomic datasets are deposited at MassIVE repository (https://massive.ucsd.edu/). Raw and processed files (Quantitative Proteomics Study (iTRAQ) of Progeria Cell Lines; #MSV000081576) are public and freely accessible.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2(2):a000760 10.1101/cshperspect.a000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guénantin AC, Briand N, Bidault G, Afonso P, Béréziat V, Vatier C, et al. Nuclear envelope-related lipodystrophies. Semin Cell Dev Biol. 2013. [DOI] [PubMed] [Google Scholar]
- 3.Hennekam RC. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am J Med Genet A. 2006;140(23):2603–24. 10.1002/ajmg.a.31346 [DOI] [PubMed] [Google Scholar]
- 4.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 423 England2003. p. 293–8. 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy S, Comai L. Lamin A, farnesylation and aging. Exp Cell Res. 2012;318(1):1–7. 10.1016/j.yexcr.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viteri G, Chung YW, Stadtman ER. Effect of progerin on the accumulation of oxidized proteins in fibroblasts from Hutchinson Gilford progeria patients. Mech Ageing Dev. 2010;131(1):2–8. 10.1016/j.mad.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidak S, Foisner R. Molecular insights into the premature aging disease progeria. Histochem Cell Biol. 2016;145(4):401–17. 10.1007/s00418-016-1411-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osorio FG, Varela I, Lara E, Puente XS, Espada J, Santoro R, et al. Nuclear envelope alterations generate an aging-like epigenetic pattern in mice deficient in Zmpste24 metalloprotease. Aging Cell. 2010;9(6):947–57. 10.1111/j.1474-9726.2010.00621.x [DOI] [PubMed] [Google Scholar]
- 9.Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2012;109(41):16666–71. 10.1073/pnas.1202529109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubben N, Misteli T. Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat Rev Mol Cell Biol. 2017;18(10):595–609. 10.1038/nrm.2017.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernadotte A, Mikhelson VM, Spivak IM. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging (Albany NY). 2016;8(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birch-Machin MA, Bowman A. Oxidative stress and ageing. Br J Dermatol. 2016;175 Suppl 2:26–9. [DOI] [PubMed] [Google Scholar]
- 14.Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 2009;5(3):279–89. 10.1016/j.stem.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mateos J, Landeira-Abia A, Fafián-Labora JA, Fernández-Pernas P, Lesende-Rodríguez I, Fernández-Puente P, et al. iTRAQ-based analysis of progerin expression reveals mitochondrial dysfunction, reactive oxygen species accumulation and altered proteostasis. Stem Cell Res Ther. 2015;6(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16(10):593–610. 10.1038/nrm4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Zhao K, Kirberger M, Liao W, Yan Y. Next generation sequencing technologies in cancer diagnostics and therapeutics: A mini review. Cell Mol Biol (Noisy-le-grand). 2015;61(5):91–102. [PubMed] [Google Scholar]
- 18.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4(12):1798–806. 10.1038/nprot.2009.191 [DOI] [PubMed] [Google Scholar]
- 19.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277(5328):942–6. [DOI] [PubMed] [Google Scholar]
- 21.Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat Rev Mol Cell Biol. 2007;8(9):681–91. 10.1038/nrm2234 [DOI] [PubMed] [Google Scholar]
- 22.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–5. 10.1038/nature08221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hennekam RC. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am J Med Genet A. 2006;140(23):2603–24. 10.1002/ajmg.a.31346 [DOI] [PubMed] [Google Scholar]
- 24.Buchwalter A, Hetzer MW. Nucleolar expansion and elevated protein translation in premature aging. Nat Commun. 2017;8(1):328 10.1038/s41467-017-00322-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jalili M, Gebhardt T, Wolkenhauer O, Salehzadeh-Yazdi A. Unveiling network-based functional features through integration of gene expression into protein networks. Biochim Biophys Acta. 2018. [DOI] [PubMed] [Google Scholar]
- 26.Kim HJ, Sohn KM, Shy ME, Krajewski KM, Hwang M, Park JH, et al. Mutations in PRPS1, which encodes the phosphoribosyl pyrophosphate synthetase enzyme critical for nucleotide biosynthesis, cause hereditary peripheral neuropathy with hearing loss and optic neuropathy (cmtx5). Am J Hum Genet. 2007;81(3):552–8. 10.1086/519529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mittal R, Patel K, Mittal J, Chan B, Yan D, Grati M, et al. Association of PRPS1 Mutations with Disease Phenotypes. Dis Markers. 2015;2015:127013 10.1155/2015/127013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320(5872):103–6. 10.1126/science.1152241 [DOI] [PubMed] [Google Scholar]
- 29.Pedley AM, Benkovic SJ. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem Sci. 2017;42(2):141–54. 10.1016/j.tibs.2016.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fridman A, Saha A, Chan A, Casteel DE, Pilz RB, Boss GR. Cell cycle regulation of purine synthesis by phosphoribosyl pyrophosphate and inorganic phosphate. Biochem J. 2013;454(1):91–9. 10.1042/BJ20130153 [DOI] [PubMed] [Google Scholar]
- 31.Chan CY, Zhao H, Pugh RJ, Pedley AM, French J, Jones SA, et al. Purinosome formation as a function of the cell cycle. Proc Natl Acad Sci U S A. 2015;112(5):1368–73. 10.1073/pnas.1423009112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet. 2002;31(1):94–9. 10.1038/ng871 [DOI] [PubMed] [Google Scholar]
- 33.Peinado JR, Quirós PM, Pulido MR, Mariño G, Martínez-Chantar ML, Vázquez-Martínez R, et al. Proteomic profiling of adipose tissue from Zmpste24-/- mice, a model of lipodystrophy and premature aging, reveals major changes in mitochondrial function and vimentin processing. Mol Cell Proteomics. 2011;10(11):M111.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, et al. S-Adenosylmethionine and methylation. FASEB J. 1996;10(4):471–80. [PubMed] [Google Scholar]
- 35.Catoni GL. S-Adenosylmethionine; a new intermediate formed enzymatically from L-methionine and adenosinetriphosphate. J Biol Chem. 1953;204(1):403–16. [PubMed] [Google Scholar]
- 36.Bradley JD, Flusser D, Katz BP, Schumacher HR Jr., Brandt KD, Chambers MA, et al. A randomized, double blind, placebo controlled trial of intravenous loading with S-adenosylmethionine (SAM) followed by oral SAM therapy in patients with knee osteoarthritis. J Rheumatol. 1994;21(5):905–11. [PubMed] [Google Scholar]
- 37.Bressa GM. S-adenosyl-l-methionine (SAMe) as antidepressant: meta-analysis of clinical studies. Acta Neurol Scand Suppl. 1994;154:7–14. [DOI] [PubMed] [Google Scholar]
- 38.Lieber CS. Role of S-adenosyl-L-methionine in the treatment of liver diseases. J Hepatol. 1999;30(6):1155–9. [DOI] [PubMed] [Google Scholar]
- 39.Harmand MF, Vilamitjana J, Maloche E, Duphil R, Ducassou D. Effects of S-adenosylmethionine on human articular chondrocyte differentiation. An in vitro study. Am J Med. 1987;83(5A):48–54. [DOI] [PubMed] [Google Scholar]
- 40.Boss GR. Decreased phosphoribosylpyrophosphate as the basis for decreased purine synthesis during amino acid starvation of human lymphoblasts. J Biol Chem. 1984;259(5):2936–41. [PubMed] [Google Scholar]
- 41.Peinado JR, Quiros PM, Pulido MR, Marino G, Martinez-Chantar ML, Vazquez-Martinez R, et al. Proteomic profiling of adipose tissue from Zmpste24-/- mice, a model of lipodystrophy and premature ageing, reveals major changes in mitochondrial function and vimentin processing. Mol Cell Proteomics. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong AH, Zhou D, Rini JM. The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J Biol Chem. 2012;287(44):36804–13. 10.1074/jbc.M112.398842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danziger RS. Aminopeptidase N in arterial hypertension. Heart Fail Rev. 2008;13(3):293–8. 10.1007/s10741-007-9061-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
749 proteins in common were quantificated in both iTRAQ replicates (A). Graphical representation of the Control/HGPS ratios shows a good correlation between the two replicates (B).
(TIF)
RNAseq analysis of Control vs. HGPS cell lines.
(XLSX)
iTRAQ analysis (replicates 1 and 2) of Control cells vs. HGPS cell lines.
(XLSX)
Data Availability Statement
Genomic datasets are deposited at the Gene Expression Omnibus (GEO-NCBI) repository (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113648). Proteomic datasets are deposited at MassIVE repository (https://massive.ucsd.edu/). Raw and processed files (Quantitative Proteomics Study (iTRAQ) of Progeria Cell Lines; #MSV000081576) are public and freely accessible.