Abstract
Osteoarthritis (OA), a common joint disease in elderly, causes serious social and economic burdens worldwide. Previous studies indicated that some differentially expressed circular RNAs (circRNAs) participated in the initiation and progression of OA. These findings suggested that circRNAs may act as promising diagnostic biomarkers and therapeutic targets for OA. In this review, we summarize the biogenesis and biological functions of circRNAs and explore the underlying roles of circRNAs in OA, which may enlighten further studies and contribute to the early diagnosis and intervention of OA.
Keywords: biogenesis, CircRNA, Osteoarthritis
Introduction
Circular RNAs (circRNAs), a new type of RNAs originating from skipping of exons during alternative splicing of pre-mRNAs, extensively exist in organisms ranging from prokaryotes and eukaryotes to mammalians [1–3]. Unlike their linear counterparts, such as mRNAs and lncRNAs, circRNAs are characterized as covalently closed loop structure without 5′ caps and 3′ poly-A tails [4]. Also, the structure feature gives them the ability to resist the digestion of RNase R and RNA exonuclease, which enables them to stably express in organisms [4,5]. CircRNAs were once regarded as splicing errors without additional biological functions [6]. However, recent studies indicated that circRNAs can sponge miRNAs or functional proteins to regulate relevant biological functions at the transcriptional or post-transcriptional level [7,8]. Moreover, further studies revealed that circRNAs, such as Circ-FBXW7 and circ-SHPRH, can be translated into functional proteins with potential prognostic implications in cancer [9,10]. Therefore, circRNAs are not ‘splicing rubbish’, but a class of regulatory molecules with some important biological functions. Studies also indicated that dysfunction of circRNAs was associated with initiation and progression of several diseases, such as malignant tumors, atherosclerosis, degenerative diseases, and nervous system diseases [11–16]. Moreover, accumulated evidence demonstrated that circRNAs were promising diagnostic biomarkers and therapeutic targets for many diseases [17,18].
Osteoarthritis (OA), a common joint disease in the elderly, is associated with increasing medical burden in these years [19]. A recent study indicated that OA would pose a threat to the health of some 3.1 million people with relevant medical costs exceeding 2.9 billion Australian dollars until 2030 [20]. Persistent pain and progressive disability of affected joints are the main clinical symptoms of OA [21]. Generally, risk factors for OA are multi-factorial and complicated, which mainly include heredity, aging, gender, obesity, injury, and inflammation [22–25]. Considering the heterogeneity in etiology, symptoms, and prognosis among OA patients, some researchers suggested that more detailed-defined OA classification should be performed, which may contribute to individualized treatment for OA [26]. For instance, Herrero-Beaumont et al. [27] conducted an etiological classification of primary OA, including genetically determined OA, estrogen hormone-dependent OA, and aging-related OA. Also, Deveza et al. [28] classified OA patients into four phenotypes, which were mechanistic subgroups, pain subgroups, prognostic subgroups, and subgroups based on response to therapy. Some clinical guidelines recommended non-pharmacologic and pharmacologic therapies for early-stage OA, such as physical activities, oral and topical NSAIDs, and intra-articular injection treatments [29,30]. Regardless of the potential effectiveness in relieving symptoms, the pathological progress of OA can hardly be inhibited by following the aforementioned treatments and most of the OA patients still end up with joint replacement [31,32]. With the development of imaging techniques and biochemical markers, it is easier to diagnose OA than before. However, it is still hard to detect OA at an early stage [33]. The reason for this is that the key molecules and mechanisms switching on OA progression are still largely unclear. Therefore, it is essential for us to have a greater depth of understanding of OA pathogenesis. Recent studies found that numerous circRNAs were associated with OA, which suggested that they may play important roles in the initiation and progression of OA [34,35].
In this review, we will summarize the evidence on the biogenesis and biological functions of circRNAs and unveil potential mechanisms involved in OA. Furthermore, we will explore their possible clinical implications as early diagnostic biomarkers and therapeutic targets in OA.
The discovery journey of circRNAs
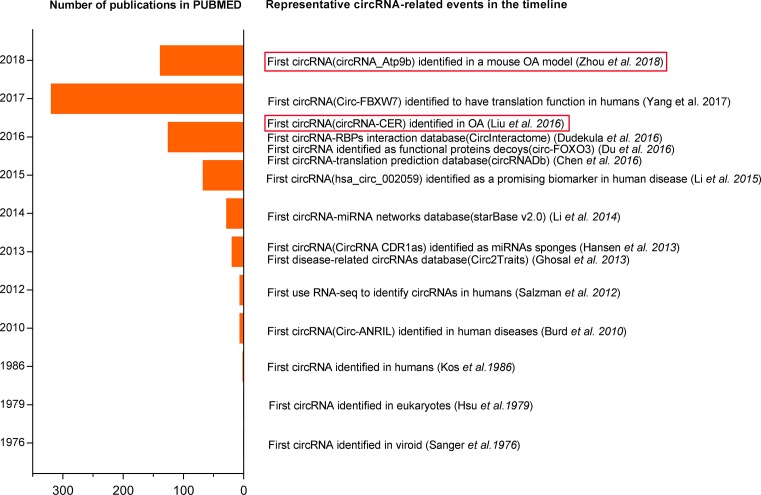
CircRNAs were first discovered in plant-based viroids and eukaryotes using electron microscopy in 1970s [36,37]. And then, PCR amplification and sequencing verified the expression of circRNAs in humans in 1986 [38]. With the development of RNA-seq and bioinformatics, thousands of circRNAs were found in various species from virus, saccharomyces cerevisiae, mammals, to humans [39]. Although numerous circRNAs were found in humans, they were regarded as the splicing ‘by-products’ without additional biological functions [6]. Accidentally, Brud et al. [40] found that a novel non-polyadenylated circular RNA (circ-ANRIL) in 2010, which can resist RNAse R digestion, was associated with atherosclerosis risk, but the underlying pathogenesis related to circ-ANRIL in atherosclerosis was unclear. Since Hansen et al. [41] first found the ‘miRNA sponge’ function of ciRS-7 in 2013, researchers attached more attention to ‘the ancient molecules’. They demonstrated that circRNAs (CDR1as and Sry), like lncRNAs and mRNAs, were a class of important competitive endogenous RNAs (ceRNAs) with miRNAs response elements (MREs) and they can regulate the expression of target gene through binding to paired miRNAs [7]. From then, increasing studies verified that circRNAs had important biological functions including miRNA ‘sponge’, functional proteins decoys and translation function, and that the dysfunction of circRNAs was associated with the initiation and progression of various human diseases [8,9,42]. Interestingly, Liu et al. [35] first reported that 71 differentially expressed circRNAs participated in initiation and progression of OA in 2016; and of these, CircRNA-CER can act as a sponge of miR-136 to regulate the expression of MMP13, thus inducing extracelluar matrix (ECM) degradation. Subsequently, Zhou et al. [34] found that 255 circRNAs were differentially expressed in a mouse OA model; and of these, circRNA_Atp9b can promote ECM catabolism and inflammation in OA. The discovery journey of circRNAs is summarized in Figure 1.
Figure 1. A timeline of representative events in circRNA research.
Biogenesis of circRNAs
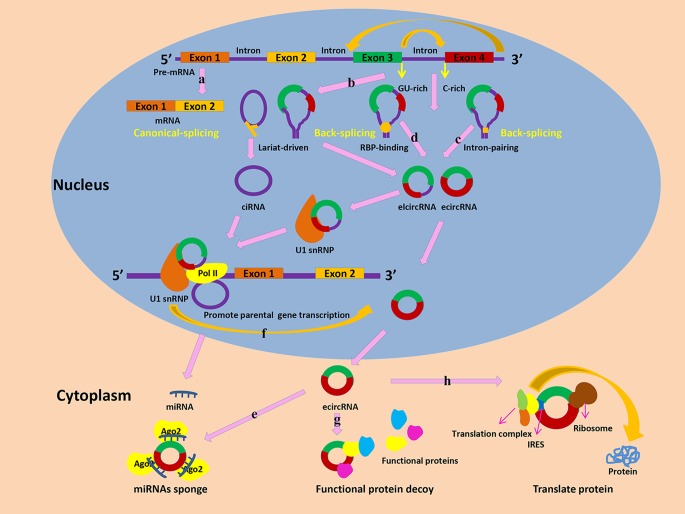
According to their genomic origin and structure feature, circRNAs are divided into three subclasses: exonic circRNAs (ecircRNAs), intronic circRNAs (ciRNAs), and Exon-Intron circRNAs (EIciRNAs) [2]. Both circRNAs and mRNA originate from the alternative splicing of pre-mRNAs (Figure 2A) [43]. However, unlike the canonical splicing of pre-mRNA into mRNA, the special splicing process of circRNAs is called ‘the back-splicing’ that a downstream 3′ splice site is joined with an upstream 5′ splice site [4,44]. Circularization of circRNAs mainly includes lariat-driven circularization, intron-pairing driven circularization, and RNA-binding protein (RBPs) driven circularization. Lariat-driven circularization or exon skipping is an important model for circRNAs formation [45]. In the process of exon skipping, lariats with intron(s) and exon(s) are produced as the by-products of the canonical linear alternative splicing. And then, ecircRNA or EIciRNA is synthesized by the combination of 3′ tail of a downstream exon and the 5′ head of an upstream exon following the internal splicing of the lariats to remove intron(s) [46]. Also, the base pairing of intronic reverse complementary is important for the function of lariat driven circularization, which involves a seven-nucleotide GU-rich element located in the 5′ splice site and an 11-nucleotide C-rich element close to the branch point site (Figure 2B) [47]. Intron-pairing driven circularization is another important formation model of circRNAs [48,49]. The special ALU reverse complementary flanking sequences of intronic regions play critical roles in catalyzing the circularization (Figure 3B) [50,51]. Also, studies indicated that the special ALU sequences can maintain the balance between circRNAs and their linear counterparts, although the potential mechanism was unclear [51]. Similar to reverse complementary sequences in introns, some RBPs, such as QKI and MBL, also play vital roles in intron-pairing driven circulation model [44,52]. These RBPs with intronic binding sites interact with corresponding targeted introns to promote circularization (Figure 2D). A recent study indicated that the formation of ecircRNAs was triggered by insertion of the QKI sequence into linear RNA [53]. Besides the positive roles, RBPs also negatively regulate circularization. For instance, ADARs, a RBP with RNA-editing function, destroys the stem RNA structure to restrain the formation of circRNA. Furthermore, knockdown of ADARs promote the expression of circRNAs [47,54].
Figure 2. The biogenesis and biological functions of circRNAs.
(A) The canonical-splicing of pre-mRNA into mRNA. (B) Lariat-driven circularization. (C) Intron-pairing driven circularization. (D) RBP-binding driven circularization. (E) CircRNAs can act as miRNA sponges to inhibit miRNA function by forming miRNA-Ago2 complexes. (F) EIciRNAs and ciRNAs can interact with transcription complexes, such as U1 snRNP, to promote the transcription of their host genes. (G) CircRNAs can interact with functional proteins to affect relevant functions. (H) Translation function of circRNAs.
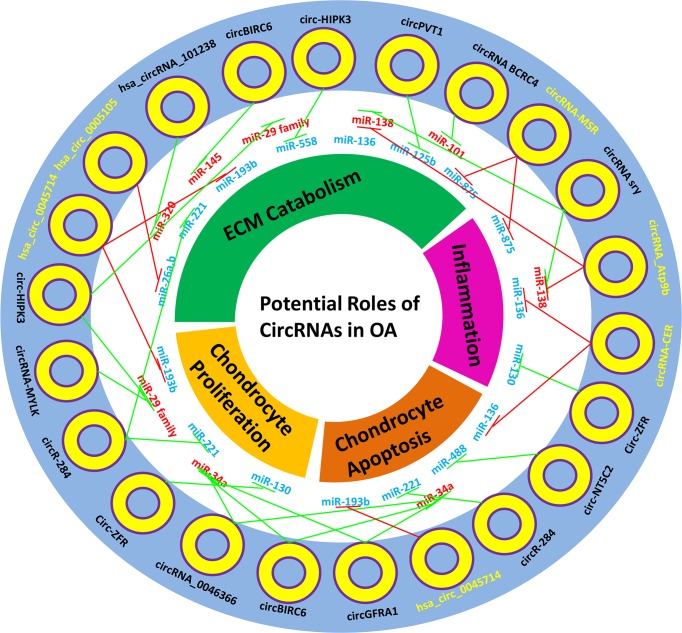
Figure 3. Potential ‘miRNAs sponges’ roles of circRNAs in OA.
Blue miRNAs denote down-regulated miRNAs in OA; Red miRNAs denote up-regulated miRNAs in OA; Yellow circRNAs denote experimentally-verified circRNAs in OA; Black circRNAs denote literature-supported circRNAs which may sponge OA-related miRNAs; Red edges denote experimentally-verified sponge relationships in OA; Green edges denote possible sponge relationships in OA.
Biological functions of circRNA
CircRNAs used to be regarded as splicing ‘rubbish’ without biological functions. However, increasing studies indicate that circRNAs extensively exist in eukaryotes and play important roles in the transcriptional, post-transcriptional, and translational levels. Generally, ecircRNAs are mainly located in cytoplasm and regulate post-transcriptional and translational levels, while ciRNAs or EIcircRNAs from nucleus have potential effects on transcription. Here, we summarize the biological functions of circRNAs.
As a ceRNA or miRNA sponge
MiRNAs, a class of non-coding RNAs with some 20 nucleotides, bind to 3′ UTR of mRNAs to inhibit their translation [55]. Actually, the expression of mRNAs function orderly in organisms, which suggests that there may exist some potential mechanisms to suppress the functions of miRNAs. Of these, the competing endogenous RNA mechanism is a critical model to regulate the functions of miRNAs and balance gene expression [56]. Two studies first found that circRNAs with miRNA response elements acted as ‘miRNA sponges’ to regulate the expression of targeted genes in 2013 [7,41]. They documented that Cirs-7 was a circRNA containing more than 70 putative binding sites for miRNA-7 and it acted as a powerful miR-7 sponge to counteract its inhibited roles for targeted mRNAs (Figure 2E). Circular SRY, another famous circRNA, functions as a sponge for miR-138 to regulate the expression of targeted gene [41]. With the development of RNA-seq and bioinformatics, many new circRNAs are identified to contain MREs and act as ‘miRNAs sponges’, which further enrich the complexity of ceRNAs [57].
Protein-binding functions
Beside miRNAs, circRNAs also bind to proteins to modulate the corresponding functions. The ciRNAs and EIcircRNAs, which are mainly located in the nucleus, do not function as ceRNAs, but regulate gene transcription through directly interacting with RBPs (Figure 2F) [58]. For instance, Ci-ankrd52 is found to richly distribute in transcription initiation site of the relevant gene and it can bind to RNA polymerase II to promote the transcription of the parental gene [59]. Knockdown of Ci-ankrd52 inhibits the expression of the linear mRNA ankrd52, but not its upstream or downstream genes. Two EIciRNAs, EIciEIF3J, and EIciPAIP2 can interact with U1 snRNP and Pol II to facilitate transcription of the host genes [58]. The circRNAs located in cytoplasm act as a decoy of functional proteins (Figure 2G). For example, circ-Foxo3 acts as a decoy of several cellular stress protein factors, such as the antisenescent protein ID-1, the transcription factor E2F1, the antistress proteins FAK, and HIF1alpha, to regulate senescence of mouse embryonic fibroblasts [60]. Also, circ-Foxo3 binds to CDK2 and P21 to develop a ternary complex, subsequently inhibiting cell cycle progression [8].
Translation function
CircRNAs were used to be regarded as a class of non-coding RNAs. However, some facts always remind us of their potential translation function. For instance, most of circRNAs originate from protein-coding genes and many circRNAs located in the cytoplasm contain the translation start codon and open reading frames according to the CSCD database [61]. Previous studies have found that functional proteins are produced following by transfecting engineered circRNAs with internal ribosome entry sites, which is necessary for circRNA translation [62]. A recent study by Legnini et al. [63] have showed that Circ-ZNF609 is translated into a functional protein to regulate myoblast proliferation. Also, Pamudurti et al. [64] found that CircMbl3, a special circRNA from fly heads, is translated into a novel protein in a cap-independent way. More interestingly, Zhang and colleagues have demonstrated that cricRNAs are translated into functional proteins in humans for the first time. They show that circ-FBXW7 is translated into a novel 21-kDa protein, FBXW7-185aa, which is positively linked with overall survival of glioblastoma patients [9]. Also, they verify that SHPRH-146aa, a 17-kDa protein from circ-SHPRH, acts as a tumor suppressor in human glioblastoma (Figure 2H) [10]. Just consistent with the above prediction, are those circRNAs with translation functions originated from protein-coding genes, and contain the translation start codon and open reading frame. All of them are translated into functional protein using a splicing-dependent and cap-independent model.
CircRNAs in OA: a coming journey to find a treasure
Potential roles of circRNAs in OA
Generally, some intracellular and extracellular stress from OA-associated risk factors activates relevant signaling pathways and transcriptional factors, thereby resulting in the dysfunction of chondrocytes and the imbalance of extracellular matrix homeostasis. Previous studies indicated that circRNAs were differentially expressed at different pathological status of OA. Liu et al. [35] found that 71 circRNAs were aberrantly expressed in articular cartilage of OA patients. Of these, circRNA-CER was obviously up-regulated and silencing of circRNA-CER inhibited MMP13 expression and promoted ECM formation. They also found that 104 differentially expressed circRNAs were associated with mechanical stress-induced OA and silencing of circRNA-MSR inhibited TNF-α expression and increased ECM formation [65]. Zhou et al. [34] demonstrated that a total of 255 circRNAs were differentially expressed in a IL-1β-treated OA mouse model and knockdown of circRNA_Atp9b increased the expression of ECM and inhibited the release of inflammatory factors, such as COX-2 and IL-6 [66]. Actually, the exact mechanisms of these circRNAs in OA cartilage are largely unclear, but it is speculated that they are associated with stress and play important regulatory roles in pathogenesis of OA. It is plausible that some circRNAs may be stimulated to prevent the pathological development or dysregulated circRNAs aggravate the progression of OA [67]. Increasing evidences have suggested that circRNAs take a part in human diseases via ‘miRNA sponge’. Until now, only sporadic studies have reported the special roles of circRNAs in OA, with the similar conclusion that some circRNAs, act as miRNAs ‘sponge’ to modulate the expression of targeted mRNA in OA (Table 1) [34,35,65,66,68,69]. Actually, previous studies have revealed that ectopically expressed miRNAs from chronic stress, such as miR-29 family (a,b,c), miR-558, and miR-221 are associated with the initiation and progression of OA [70–72]. Moreover, several studies have indicated that some circRNAs, such as circ-HIPK3 and circRNA-284 can act as ‘sponges’ of these stress-related miRNAs to regulate the expression of targeted genes [73,74]. Therefore, it is essential to explore the potential roles of these circRNAs in OA, which may provide novel evidence of the ‘miRNA sponge’ mechanism in OA (Figure 3). On the other hand, is ‘sponge’ the only way to regulate progression of OA for circRNAs? Obviously, the answer is ‘No!’. Judging from the discovery history of circRNA functions, we track back to 2013, when Hansen et al. first identified the ‘sponge’ function of ciRS-7. Subsequently, their biogenesis and other regulatory functions, such as decoys of functional proteins and translation function, are gradually recognized. Moreover, RNA-seq and bioinformatics analysis largely promotes the understanding of circRNAs in human diseases. Thus, we can forecast that future studies will unveil more regulatory roles of circRNAs in OA, considering that the first study about circRNAs in OA is merely published in 2016. Anyway, these previous studies provide advantageous implications to enrich our understanding of circRNAs’ roles in OA.
Table 1. The expression of circRNAs in OA.
| Author | Year | Design | Main findings | References (PMID) |
|---|---|---|---|---|
| Liu et al. [35] | 2016 | Microarray; Bioinformatics analysis; qRT-PCR | A total of 71 circRNAs were aberrantly expressed in articular cartilage of OA patients. Of these, circRNA-CER was obviously up-regulated and silencing of circRNA-CER can inhibit MMP13 expression and promote ECM formation. | 26931159 |
| Liu et al. [65] | 2017 | Microarray; Bioinformatics analysis; qRT-PCR | A total of 104 differentially expressed circRNAs were identified in damaged versus intact cartilage. CircRNAs-MSR participated in TNF-α expression and was associated with cartilage matrix degradation. | 28624198 |
| Li et al. [69] | 2017 | qRT-PCR | Hsa_circ_0045714-miR-193b-IGF1R axis played an important role in extracellular matrix synthesis and chondrocytes proliferation and apoptosis. | 28795385 |
| Wu et al. [68] | 2017 | qRT-PCR | Hsa_circ_0005105-miR-26a-NAMPT axis played an important role in cartilage matrix degradation and expression of inflammation factors. | 28276108 |
| Zhou et al. [34] | 2018 | CircRNA sequencing; Bioinformatics analysis; qRT-PCR | A total of 255 circRNAs were identified to be differentially expressed in IL-1β-treated chondrocytes. | 29247798 |
| Zhou et al. [66] | 2018 | qRT-PCR | Knockdown of circRNA_Atp9b increased the expression of ECM and inhibited the release of such inflammatory factors as COX-2 and IL-6. | 29305974 |
CircRNA: a promising biomarkers in OA
The covalently closed ring structure of circRNAs gives them the ability to endure the degradation of RNase, which makes them express stably in the body. A recent study has found that thousands of circRNAs exist in human peripheral whole blood and some circRNAs have higher abundance than their linear counterparts, which suggests the underlying roles of circRNAs as diagnostic and prognostic biomarkers in the easily accessible body fluid [75]. Li et al. [76] have suggested that hsa-circRNA11783-2 in peripheral blood acts as a useful diagnostic biomarker for T2DM patients with coronary artery disease. Furthermore, the study by Zhao et al. [77] indicates that 22 circRNAs are differentially expressed in the peripheral blood of coronary artery disease patients and of these, hsa_circ_0124644 with the largest AUC is a promising diagnostic biomarker for coronary artery disease. Additionally, many aberrantly-expressed circRNAs are identified as the sensitive prognosis biomarkers in cancers. For instance, Xia et al. [78] have found that hsa_circ_0067934, a significantly up-regulated circRNA, is associated with poor differentiation, I-II T stage, and I-II TNM stage in esophageal squamous cell carcinoma. Han et al. [42] have demonstrated that low circMTO1 expression is related to poor prognosis in HCC patients. A recent finding by Jiang et al. [79] suggested that Cdr1as was up-regulated in cholangiocarcinoma and the overexpression of Cdr1as was linked to advanced TNM stage, lymph node invasion, postoperative recurrence, and worst overall survival. The conventional diagnostic methods for OA are based on imageology examination (i.e. X-ray and MRI) and clinical symptoms. However, these available methods are merely effective for detection of advanced OA, while they hardly recognize OA in earlier stage. Given that dysfunction of circRNAs is associated with the onset and progression of OA, circRNAs may play potential roles as early diagnostic and prognostic biomarkers in OA. There are no available studies to verify differentially expressed circRNAs in the accessible body fluids of OA patients, so further studies should be warranted to explore the potential diagnostic and prognostic value of circRNAs in OA.
The therapeutic potentials of circRNAs in OA
Apart from potentials as biomarkers, circRNAs may act as promising therapeutic targets in OA. Zhang et al. [80] have suggested that circRNA_100269 is down-regulated and its targeted miRNA, miR-630, is up-regulated in gastric cancer. Furthermore, cell proliferation of gastric cancer cells is obviously inhibited following overexpression of circRNA_100269. Also, Li et al. [81] found that the expression of circ-104916 was down-regulated in gastric cancer and up-regulating circ-104916 inhibited the proliferation, migration, and invasion of gastric cancer cells. Additionally, a recent study has indicated that the treatment of hsa_circ_0045714 inhibits the progression of OA through promoting the expression of type II collagen and aggrecan, and chondrocyte proliferation [69]. On the other hand, numerous studies suggested that the dysfunction of miRNAs played vital roles in the onset and progression of OA [82]. The traditional methods to inhibit miRNA function contain gene knockout, antisense oligonucleotides, and miRNA sponges [83,84]. The construction of animal knockout models may spend much time and money, and the miRNA inhibitor function of antisense oligonucleotides may work well in short-term experiment, but not in long-term experiment [84,85]. Interestingly, miRNA sponges have similar miRNA-inhibited ability to antisense oligonucleotides in vitro [83,86]. More importantly, circRNA, a ‘super sponge’ with several MREs, may have more powerful ability to bind to targeted miRNAs when compared with its linear counterparts, such as mRNA and lncRNA. Jost et al. [87] investigated that artificial circRNAs sponges can down-regulate the level of miR-122, thus regulating the expression of corresponding proteins in a HCV model system. Therefore, artificial regulation of circRNAs may act as a promising gene therapy against functions of miRNAs to prevent the development of OA.
CircRNAs in other joint diseases
As with OA, dysfunction of circRNAs is associated with the pathogenesis of other joint diseases, such as rheumatoid arthritis (RA) and intervertebral disc degeneration (IDD). Ouyang et al. [88] found that five circRNAs (092516, 003524, 103047, 104871, and 101873) were significantly unregulated in peripheral blood mononuclear cells from RA patients. Of these, circRNA_104871 could function as a potential diagnostical biomarker for RA. Zheng et al. [89] performed microarray analysis for peripheral blood mononuclear cells of 10 RA patients, which indicated that 255 circRNAs were significantly up-regulated and 329 down-regulated in the RA samples. Recent studies also indicate that circRNAs are also involved in development of IDD [90]. Cheng et al. [91] reveal that the level of CircVMA21 is significantly down-regulated in cytokines-treated nucleus pulposus (NP) cells and degenerative NP tissues. Furthermore, overexpression of CircVMA21 can alleviate the progression of IDD through miR-200c-XIAP axis in vivo and in vitro. The study performed by Guo et al. [15] indicates that circ-GRB10 can activate ERBB2 signaling pathway via sequestering miR-328-5p, thus inhibiting NP cell apoptosis and promote cell proliferation in vitro. Therefore, circ-GRB10 may act as novel diagnostic biomarkers and therapeutic targets for IDD. Actually, circRNA-related studies in non-OA joint diseases are still scarce, so further studies are necessary to explore the regulatory roles of circRNAs in these diseases.
Conclusion and perspectives
Until now, only six studies explored the potential roles of circRNAs in OA. All of these studies suggested that the circRNAs–miRNAs–mRNAs axis played important roles in the pathogenesis of OA. However, we should be alert that the ‘sponge’ mechanism may be a potential pitfall in the pathogenesis of diseases. For instance, Li et al. [69] found that hsa_circ_0045714-miR-193b-IGF1R axis played an important role in extracellular matrix synthesis and chondrocytes proliferation and apoptosis. However, the expression level of hsa_circ_0045714 is down-regulated, while miR-193b up-regulated in OA. It is questionable to the ‘sponge’ role of hsa_circ_0045714 for miR-193b in vivo. Consequently, it is worthwhile to explore the potential roles of some low-abundance circRNAs in pathogenesis of diseases, when considering that they could hardly affect the functions of predicted miRNAs with higher abundance. In theory, at least seven binding sites are necessary for circRNA to ‘sponge’ miRNA. Therefore, it is questionable for some studies which verified the ‘sponge’ function in vitro, but with less than seven circRNA–miRNA binding sites from bioinformatics prediction. For example, Wu et al. [68] demonstrated that hsa_circ_0005105 acted as a sponge of miR-26a to promote extracellular matrix degradation in OA. However, only three consecutive binding sites were predicted between hsa_circ_0005105 and miR-26a in the study, so the ‘sponge’ relationship was needed for further experimental verification. The current available studies have merely unveiled the potential roles of OA-related circRNAs in ECM degradation, chondrocytes proliferation, apoptosis, and inflammation. Actually, previous studies indicate that differentially expressed miRNAs are associated with dysfunction of chondrocytes autophagy, oxidative stress and signaling pathways, which are correlated with pathogenesis of OA [92]. Therefore, it is essential to elucidate whether circRNAs also participated in these pathophysiological processes in OA. Apart from ‘sponge’ functions for downstream miRNAs, some circRNAs, such as circRNA FOXO3, are verified to act as decoys of several functional proteins to regulate relevant functions in human diseases [8,60]. Currently, there is no study focused on circRNA–protein interactions in OA. Some experimental methods, such as RNA pull-down assay, RNA immunoprecipitation, RNase protection assay, and fluorescence in situ hybridization techniques, are useful for exploring this novel mechanism in OA [93]. Moreover, recent studies indicated that some circRNAs, such as Circ-FBXW7 and circ-SHPRH, can be translated into functional protein, which participated in the development and progression of diseases [9,10]. The proteins translated from circRNA may be relatively non-functional under physiological conditions. However, intracellular and extracellular stress may promote the activation of the cap-independent translation model, so proteins encoded by circRNAs may play critical roles in pathological conditions [94]. Anyway, the special cap-independent translation mechanism of circRNAs is still largely unclear, which needs to be further investigated. Also, the underlying translation function of circRNAs in OA is worth exploring in future studies. One the other hand, the development of RNA-seq and bioinformatics promotes construction of such circRNA-related databases as circBase, circRNABase, RegRNA 2.0, Circ2Traits, circinteractome, circnet, deepbase version 2.0, circRNADb, CSCD, and circlncRNAnet, which contributes to further clarify the potential roles of circRNAs in OA (Supplementary Table S1) [61,95–103]. CircRNAs may be identified as promising diagnostic biomarkers and therapeutic targets in OA, but relevant studies are limited until now. Thus, further studies should be warranted to explore the roles of circRNAs in accessible body fluid of OA patients, such as peripheral blood and saliva, which may act as novel diagnostic biomarkers and therapeutic targets for OA.
In conclusion, although the circRNA-related studies in OA and other joint diseases are limited, these findings indicate that circRNAs participate in the pathogenesis of OA. Therefore, further studies should be performed to clarify the potential roles of circrRNAs in OA, which may contribute to the early diagnosis and intervention for OA.
Supporting information
Table S1. CircRNA-related databases and their useful functions.
Abbreviations
- ceRNA
competitive endogenous RNA
- circRNA
circular RNA
- ECM
extracelluar matrix
- IDD
intervertebral disc degeneration
- MRE
miRNAs response element
- NP
nucleus pulposus
- OA
osteoarthritis
- RA
rheumatoid arthritis
- RBP
RNA-binding protein
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant numbers 81772384; 81572174). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D.. et al. (1991) Scrambled exons. Cell 64, 607–613 10.1016/0092-8674(91)90244-S [DOI] [PubMed] [Google Scholar]
- 2.Chen L., Huang C., Wang X. and Shan G. (2015) Circular RNAs in eukaryotic cells. Curr. Genomics 16, 312–318 10.2174/1389202916666150707161554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N.. et al. (2014) Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 9, e90859 10.1371/journal.pone.0090859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lasda E. and Parker R. (2014) Circular RNAs: diversity of form and function. RNA 20, 1829–1842 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki H. and Tsukahara T. (2014) A view of pre-mRNA splicing from RNAse R resistant RNAs. Int. J. Mol. Sci. 15, 9331–9342 10.3390/ijms15069331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerelle C., Mascrez B., Hetuin D. and Bailleul B. (1993) Mis-splicing yields circular RNA molecules. FASEB J. 7, 155–160 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- 7.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A.. et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 8.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P. and Yang B.B. (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and cdk2. Nucleic Acids Res. 44, 2846–2858 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F.. et al. (2018) Novel role of fbxw7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst., 110, 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F.. et al. (2018) A novel protein encoded by the circular form of the shprh gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814 10.1038/s41388-017-0019-9 [DOI] [PubMed] [Google Scholar]
- 11.Yu J., Xu Q.G., Wang Z.G., Yang Y., Zhang L., Ma J.Z.. et al. (2018) Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 68, 1214–1227 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 12.Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T.. et al. (2018) Circhipk3 promotes colorectal cancer growth and metastasis by sponging mir-7. Cell Death Dis. 9, 417 10.1038/s41419-018-0454-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Song C.L., Wang J.P., Xue X., Liu N., Zhang X.H., Zhao Z.. et al. (2017) Effect of circular anril on the inflammatory response of vascular endothelial cells in a rat model of coronary atherosclerosis. Cell. Physiol. Biochem. 42, 1202–1212 10.1159/000478918 [DOI] [PubMed] [Google Scholar]
- 14.Shan K., Liu C., Liu B.H., Chen X., Dong R., Liu X.. et al. (2017) Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 136, 1629–1642 10.1161/CIRCULATIONAHA.117.029004 [DOI] [PubMed] [Google Scholar]
- 15.Guo W., Zhang B., Mu K., Feng S.Q., Dong Z.Y., Ning G.Z.. et al. (2018) Circular RNA GRB10 as a competitive endogenous RNA regulating nucleus pulposus cells death in degenerative intervertebral disk. Cell Death Dis. 9, 319 10.1038/s41419-017-0232-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J.L., Qin M.C., Zhou Y., Xu Z.H., Yang S.M., Zhang F.. et al. (2018) Comprehensive analysis of differentially expressed profiles of alzheimer’s disease associated circular RNAs in an alzheimer’s disease mouse model. Aging 10, 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M. and Xin Y. (2018) Circular RNAs: a new frontier for cancer diagnosis and therapy. J. Hematol. Oncol. 11, 21 10.1186/s13045-018-0569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F., Nazarali A.J. and Ji S. (2016) Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 6, 1167–1176 [PMC free article] [PubMed] [Google Scholar]
- 19.Cross M., Smith E., Hoy D., Nolte S., Ackerman I., Fransen M.. et al. (2014) The global burden of hip and knee osteoarthritis: estimates from the global burden of disease 2010 study. Ann. Rheum. Dis. 73, 1323–1330 10.1136/annrheumdis-2013-204763 [DOI] [PubMed] [Google Scholar]
- 20.Ackerman I.N., Pratt C., Gorelik A. and Liew D. (2018) Projected burden of osteoarthritis and rheumatoid arthritis in australia: a population-level analysis. Arthritis Care Res. 70, 877–883 10.1002/acr.23414 [DOI] [PubMed] [Google Scholar]
- 21.Glyn-Jones S., Palmer A.J., Agricola R., Price A.J., Vincent T.L., Weinans H.. et al. (2015) Osteoarthritis. Lancet 386, 376–387 10.1016/S0140-6736(14)60802-3 [DOI] [PubMed] [Google Scholar]
- 22.Loeser R.F. (2009) Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthr. Cartil. 17, 971–979 10.1016/j.joca.2009.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grotle M., Hagen K.B., Natvig B., Dahl F.A. and Kvien T.K. (2008) Obesity and osteoarthritis in knee, hip and/or hand: an epidemiological study in the general population with 10 years follow-up. BMC Musculoskelet. Disord. 9, 132 10.1186/1471-2474-9-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain S.M., Cicuttini F.M., Alyousef B. and Wang Y. (2018) Female hormonal factors and osteoarthritis of the knee, hip and hand: a narrative review. Climacteric 21, 132–139 10.1080/13697137.2017.1421926 [DOI] [PubMed] [Google Scholar]
- 25.Magnusson K., Scurrah K., Ystrom E., Orstavik R.E., Nilsen T., Steingrimsdottir O.A.. et al. (2017) Genetic factors contribute more to hip than knee surgery due to osteoarthritis - a population-based twin registry study of joint arthroplasty. Osteoarthr. Cartil. 25, 878–884 10.1016/j.joca.2016.12.015 [DOI] [PubMed] [Google Scholar]
- 26.Roman-Blas J.A., Bizzi E., Largo R., Migliore A. and Herrero-Beaumont G. (2016) An update on the up and coming therapies to treat osteoarthritis, a multifaceted disease. Expert Opin. Pharmacother. 17, 1745–1756 10.1080/14656566.2016.1201070 [DOI] [PubMed] [Google Scholar]
- 27.Herrero-Beaumont G., Roman-Blas J.A., Castaneda S. and Jimenez S.A. (2009) Primary osteoarthritis no longer primary: three subsets with distinct etiological, clinical, and therapeutic characteristics. Semin. Arthritis Rheum. 39, 71–80 10.1016/j.semarthrit.2009.03.006 [DOI] [PubMed] [Google Scholar]
- 28.Deveza L.A. and Loeser R.F. (2018) Is osteoarthritis one disease or a collection of many? Rheumatology 57, iv34–iv42 10.1093/rheumatology/kex417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochberg M.C., Altman R.D., April K.T., Benkhalti M., Guyatt G., McGowan J.. et al. (2012) American college of rheumatology 2012 recommendations for the use of nonpharmacologic and pharmacologic therapies in osteoarthritis of the hand, hip, and knee. Arthritis Care Res. 64, 465–474 10.1002/acr.21596 [DOI] [PubMed] [Google Scholar]
- 30.Rausch Osthoff A.K., Niedermann K., Braun J., Adams J., Brodin N., Dagfinrud H.. et al. (2018) 2018 eular recommendations for physical activity in people with inflammatory arthritis and osteoarthritis. Ann. Rheum. Dis. 77, 1251–1260 10.1136/annrheumdis-2018-213585 [DOI] [PubMed] [Google Scholar]
- 31.Hunt L.P., Ben-Shlomo Y., Clark E.M., Dieppe P., Judge A., MacGregor A.J.. et al. (2014) 45-day mortality after 467,779 knee replacements for osteoarthritis from the national joint registry for england and wales: an observational study. Lancet 384, 1429–1436 10.1016/S0140-6736(14)60540-7 [DOI] [PubMed] [Google Scholar]
- 32.Hunt L.P., Ben-Shlomo Y., Whitehouse M.R., Porter M.L. and Blom A.W. (2017) The main cause of death following primary total hip and knee replacement for osteoarthritis: a cohort study of 26,766 deaths following 332,734 hip replacements and 29,802 deaths following 384,291 knee replacements. J. Bone Joint Surg. Am. 99, 565–575 10.2106/JBJS.16.00586 [DOI] [PubMed] [Google Scholar]
- 33.Bijlsma J.W., Berenbaum F. and Lafeber F.P. (2011) Osteoarthritis: an update with relevance for clinical practice. Lancet 377, 2115–2126 10.1016/S0140-6736(11)60243-2 [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z., Du D., Chen A. and Zhu L. (2018) Circular RNA expression profile of articular chondrocytes in an il-1beta-induced mouse model of osteoarthritis. Gene 644, 20–26 10.1016/j.gene.2017.12.020 [DOI] [PubMed] [Google Scholar]
- 35.Liu Q., Zhang X., Hu X., Dai L., Fu X., Zhang J.. et al. (2016) Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a mir-136 ‘sponge’ in human cartilage degradation. Sci. Rep. 6, 22572 10.1038/srep22572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanger H.L., Klotz G., Riesner D., Gross H.J. and Kleinschmidt A.K. (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 73, 3852–3856 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu M.T. and Coca-Prados M. (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280, 339–340 10.1038/280339a0 [DOI] [PubMed] [Google Scholar]
- 38.Kos A., Dijkema R., Arnberg A.C., van der Meide P.H. and Schellekens H. (1986) The hepatitis delta (delta) virus possesses a circular RNA. Nature 323, 558–560 10.1038/323558a0 [DOI] [PubMed] [Google Scholar]
- 39.Salzman J., Gawad C., Wang P.L., Lacayo N. and Brown P.O. (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7, e30733 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z. and Sharpless N.E. (2010) Expression of linear and novel circular forms of an ink4/arf-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 6, e1001233 10.1371/journal.pgen.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K.. et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 42.Han D., Li J., Wang H., Su X., Hou J., Gu Y.. et al. (2017) Circular RNA circmto1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66, 1151–1164 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 43.Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.H.. et al. (2015) Exon circularization requires canonical splice signals. Cell Rep. 10, 103–111 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 44.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M.. et al. (2014) CircRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 45.Kelly S., Greenman C., Cook P.R. and Papantonis A. (2015) Exon skipping is correlated with exon circularization. J. Mol. Biol. 427, 2414–2417 10.1016/j.jmb.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 46.Barrett S.P., Wang P.L. and Salzman J. (2015) Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 4, e07540 10.7554/eLife.07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R.. et al. (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 48.Liang D. and Wilusz J.E. (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28, 2233–2247 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L. and Yang L. (2014) Complementary sequence-mediated exon circularization. Cell 159, 134–147 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 50.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J.. et al. (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daniel C., Behm M. and Ohman M. (2015) The role of ALU elements in the cis-regulation of RNA processing. Cell. Mol. Life Sci. 72, 4063–4076 10.1007/s00018-015-1990-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A.. et al. (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 53.Suiko T., Kobayashi K., Aono K., Kawashima T., Inoue K., Ku L.. et al. (2016) Expression of quaking RNA-binding protein in the adult and developing mouse retina. PLoS ONE 11, e0156033 10.1371/journal.pone.0156033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi L., Yan P., Liang Y., Sun Y., Shen J., Zhou S.. et al. (2017) Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 8, e3171 10.1038/cddis.2017.556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krol J., Loedige I. and Filipowicz W. (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- 56.Militello G., Weirick T., John D., Doring C., Dimmeler S. and Uchida S. (2017) Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief. Bioinform. 18, 780–788 [DOI] [PubMed] [Google Scholar]
- 57.Szabo L. and Salzman J. (2016) Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 17, 679–692 10.1038/nrg.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X.. et al. (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H.. et al. (2013) Circular intronic long noncoding rnas. Mol. Cell 51, 792–806 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 60.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z.. et al. (2017) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 38, 1402–1412 [DOI] [PubMed] [Google Scholar]
- 61.Xia S., Feng J., Chen K., Ma Y., Gong J., Cai F.. et al. (2018) Cscd: a database for cancer-specific circular RNAs. Nucleic Acids Res. 46, D925–D929 10.1093/nar/gkx863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y. and Wang Z. (2015) Efficient backsplicing produces translatable circular mRNAs. RNA 21, 172–179 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O.. et al. (2017) Circ-znf609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22.e29–37.e29 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L.. et al. (2017) Translation of circRNAs. Mol. Cell 66, 9.e27–21.e27 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Q., Zhang X., Hu X., Yuan L., Cheng J., Jiang Y.. et al. (2017) Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol. Ther. Nucleic Acids 7, 223–230 10.1016/j.omtn.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou Z.B., Du D., Huang G.X., Chen A. and Zhu L. (2018) Circular RNA atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting mir-138-5p. Gene 646, 203–209 10.1016/j.gene.2017.12.064 [DOI] [PubMed] [Google Scholar]
- 67.Fischer J.W. and Leung A.K. (2017) CircRNAs: a regulator of cellular stress. Crit. Rev. Biochem. Mol. Biol. 52, 220–233 10.1080/10409238.2016.1276882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y., Zhang Y., Zhang Y. and Wang J.J. (2017) CircRNA hsa_circ_0005105 upregulates nampt expression and promotes chondrocyte extracellular matrix degradation by sponging mir-26a. Cell Biol. Int. 41, 1283–1289 10.1002/cbin.10761 [DOI] [PubMed] [Google Scholar]
- 69.Li B.F., Zhang Y., Xiao J., Wang F., Li M., Guo X.Z.. et al. (2017) Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of mir-193b target gene igf1r. Hum. Cell 30, 311–318 10.1007/s13577-017-0177-7 [DOI] [PubMed] [Google Scholar]
- 70.Park S.J., Cheon E.J. and Kim H.A. (2013) MicroRNA-558 regulates the expression of cyclooxygenase-2 and il-1beta-induced catabolic effects in human articular chondrocytes. Osteoarthr. Cartil. 21, 981–989 10.1016/j.joca.2013.04.012 [DOI] [PubMed] [Google Scholar]
- 71.Zheng X., Zhao F.C., Pang Y., Li D.Y., Yao S.C., Sun S.S.. et al. (2017) Downregulation of mir-221-3p contributes to il-1beta-induced cartilage degradation by directly targeting the SDF1/CXCR4 signaling pathway. J. Mol. Med. 95, 615–627 10.1007/s00109-017-1516-6 [DOI] [PubMed] [Google Scholar]
- 72.Le L.T., Swingler T.E., Crowe N., Vincent T.L., Barter M.J., Donell S.T.. et al. (2016) The microRNA-29 family in cartilage homeostasis and osteoarthritis. J. Mol. Med. 94, 583–596 10.1007/s00109-015-1374-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C.. et al. (2017) Circhipk3 sponges mir-558 to suppress heparanase expression in bladder cancer cells. Hum. Cell 18, 1646–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B.. et al. (2016) Circular RNA profiling reveals an abundant circhipk3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7, 11215 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Memczak S., Papavasileiou P., Peters O. and Rajewsky N. (2015) Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 10, e0141214 10.1371/journal.pone.0141214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li X., Zhao Z., Jian D., Li W., Tang H. and Li M. (2017) Hsa-circRNA11783-2 in peripheral blood is correlated with coronary artery disease and type 2 diabetes mellitus. Diab. Vasc. Dis. Res. 14, 510–515 10.1177/1479164117722714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Z., Li X., Gao C., Jian D., Hao P., Rao L.. et al. (2017) Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 7, 39918 10.1038/srep39918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xia W., Qiu M., Chen R., Wang S., Leng X., Wang J.. et al. (2016) Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci. Rep. 6, 35576 10.1038/srep35576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang X.M., Li Z.L., Li J.L., Xu Y., Leng K.M., Cui Y.F.. et al. (2018) A novel prognostic biomarker for cholangiocarcinoma: circRNA cdr1as. Eur. Rev. Med. Pharmacol. Sci. 22, 365–371 [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y., Liu H., Li W., Yu J., Li J., Shen Z.. et al. (2017) CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting mir-630. Aging 9, 1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang P., Zuo Z., Shang W., Wu A., Bi R., Wu J.. et al. (2017) Identification of differentially expressed circular RNAs in human colorectal cancer. Tumour Biol. 39, 1010428317694546 [DOI] [PubMed] [Google Scholar]
- 82.Trachana V., Ntoumou E., Anastasopoulou L. and Tsezou A. (2018) Studying microRNAs in osteoarthritis: critical overview of different analytical approaches. Mech. Ageing Dev. 171, 15–23 10.1016/j.mad.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 83.Ruberti F., Barbato C. and Cogoni C. (2012) Targeting microRNAs in neurons: tools and perspectives. Exp. Neurol. 235, 419–426 10.1016/j.expneurol.2011.10.031 [DOI] [PubMed] [Google Scholar]
- 84.He J., Xie Q., Xu H., Li J. and Li Y. (2017) Circular RNAs and cancer. Cancer Lett. 396, 138–144 10.1016/j.canlet.2017.03.027 [DOI] [PubMed] [Google Scholar]
- 85.Baigude H. and Rana T.M. (2014) Strategies to antagonize miRNA functions in vitro and in vivo. Nanomedicine 9, 2545–2555 10.2217/nnm.14.162 [DOI] [PubMed] [Google Scholar]
- 86.Tay F.C., Lim J.K., Zhu H., Hin L.C. and Wang S. (2015) Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv. Drug. Deliv. Rev. 81, 117–127 10.1016/j.addr.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 87.Jost I., Shalamova L.A., Gerresheim G.K., Niepmann M. and Bindereif A. (2018) Functional sequestration of microRNA-122 from hepatitis c virus by circular RNA sponges. RNA Biol. 1–8 10.1080/15476286.2018.1435248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ouyang Q., Wu J., Jiang Z., Zhao J., Wang R., Lou A.. et al. (2017) Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell. Physiol. Biochem. 42, 651–659 10.1159/000477883 [DOI] [PubMed] [Google Scholar]
- 89.Zheng F., Yu X., Huang J. and Dai Y. (2017) Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol. Med. Rep. 16, 8029–8036 10.3892/mmr.2017.7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lan P.H., Liu Z.H., Pei Y.J., Wu Z.G., Yu Y., Yang Y.F.. et al. (2016) Landscape of RNAs in human lumbar disc degeneration. Oncotarget 7, 63166–63176 10.18632/oncotarget.11334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng X., Zhang L., Zhang K., Zhang G., Hu Y., Sun X.. et al. (2018) Circular RNA vma21 protects against intervertebral disc degeneration through targeting mir-200c and x linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. 10.1136/annrheumdis-2017-212056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Portal-Nunez S., Esbrit P., Alcaraz M.J. and Largo R. (2016) Oxidative stress, autophagy, epigenetic changes and regulation by miRNAs as potential therapeutic targets in osteoarthritis. Biochem. Pharmacol. 108, 1–10 10.1016/j.bcp.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 93.Du W.W., Zhang C., Yang W., Yong T., Awan F.M. and Yang B.B. (2017) Identifying and characterizing circRNA-protein interaction. Theranostics 7, 4183–4191 10.7150/thno.21299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li M., Ding W., Sun T., Tariq M.A., Xu T., Li P.. et al. (2018) Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 285, 220–232 10.1111/febs.14191 [DOI] [PubMed] [Google Scholar]
- 95.Glazar P., Papavasileiou P. and Rajewsky N. (2014) Circbase: a database for circular RNAs. RNA 20, 1666–1670 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li J.H., Liu S., Zhou H., Qu L.H. and Yang J.H. (2014) Starbase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale clip-seq data. Nucleic Acids Res. 42, D92–D97 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang H.Y., Chien C.H., Jen K.H. and Huang H.D. (2006) Regrna: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res. 34, W429–W434 10.1093/nar/gkl333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ghosal S., Das S., Sen R., Basak P. and Chakrabarti J. (2013) Circ2traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 4, 283 10.3389/fgene.2013.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K. and Gorospe M. (2016) Circinteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 13, 34–42 10.1080/15476286.2015.1128065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Y.C., Li J.R., Sun C.H., Andrews E., Chao R.F., Lin F.M.. et al. (2016) Circnet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 44, D209–D215 10.1093/nar/gkv940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen X., Han P., Zhou T., Guo X., Song X. and Li Y. (2016) CircRNAdb: a comprehensive database for human circular RNAs with protein-coding annotations. PLoS ONE 6, 34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng L.L., Li J.H., Wu J., Sun W.J., Liu S., Wang Z.L.. et al. (2016) Deepbase v2.0: identification, expression, evolution and function of small RNAs, lncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 44, D196–D202 10.1093/nar/gkv1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu S.M., Liu H., Huang P.J., Chang I.Y., Lee C.C., Yang C.Y.. et al. (2018) CirclncRNAnet: an integrated web-based resource for mapping functional networks of long or circular forms of noncoding RNAs. Gigascience 7, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]