Abstract
Introduction
Pancreatic cancer is a highly lethal malignancy with high invasion metastasis, which is difficult to diagnose and treat. MicroRNA-216b (miR-216b) plays an important role in many types of tumors. In this study, we explore how miR-216b affected human pancreatic cancer cell development by targeting KRAS.
Material and methods
Expression level of miR-216b and KRAS in tissue samples and cells were detected by RT-PCR and western blot. Immunohistochemical assay analysed the expressions of KRAS protein in tumor and adjacent tissues. The target relationship between miR-216b and KRAS was validated by dual-luciferase reporter assay. Pancreatic cancer cell proliferation, migration, invasion and apoptosis abilities of cells transfected with miR-216b mimics and KRAS-siRNA, Panc-1 were detected by MTT assay, transwell assay and flow cytometry assay respectively. Prognosis of patients with different expression levels of miR-216b and KRAS were analyzed by Kaplan-Meier survival analysis and Cox proportional hazards regression model.
Results
The expression of miR-216b in pancreatic cancer tissue and cell line was down-regulated (p < 0.01), while KRAS expression was up-regulated (p < 0.01) compared with adjacent normal tissues. Both the expressions of miR-216b and KRAS have a strong influence on prognosis of the pancreatic cancer patients (p = 0.024 and p = 0.017). The dual-luciferase reporter assay verified that miR-216b directly targeted KRAS in pancreatic cancer cells. Overexpression of miR-216b reduced the expression of mRNA and protein of KRAS (p = 0.013 and p = 0.003), but silencing KRAS had no effect on miR-216b expression (p = 0.706). By silencing KRAS or up-regulation of miR-216b could suppress cell proliferation, migration and invasion of pancreatic cancer cells and promote apoptosis.
Conclusions
MiR-216b might inhibit pancreatic cancer cell progression and stimulate apoptosis by silencing KRAS.
Keywords: pancreatic cancer, miR-216b, KRAS
Introduction
Pancreatic cancer is a devastating cancer with a high morbidity and poor prognosis. It is reported that only 20% of patients will be alive 5 years after pancreatic resection and the survival rate of pancreatic cancer patients is less than 30% [1, 2]. Radiotherapy and chemotherapy are widely used for the treatment of pancreatic cancer. However, as pancreatic cancer is highly resistant to chemotherapy and radiotherapy, the efficacy of current treatment for pancreatic cancer still cannot reach expectations [3]. Therefore, it is urgent for the treatment of pancreatic cancer to develop an effective therapeutic strategy.
MicroRNAs (miRNAs), as endogenous small and non-coding RNAs, play a crucial role in regulating gene expression within the target messenger RNA (mRNA) 3′-untranslated region (UTR) [4]. There is considerable evidence suggesting that miRNAs present aberrant expressions in multiple cancer tissue and cells [5]. Yuan et al. demonstrated that miR-216b inhibited the multi-drug resistance of hepatocellular cancer cells through down-regulating autophagy [6]. Zheng et al. revealed that miR-216b may serve as a potential therapeutic agent for hepatocellular carcinoma by targeting an oncogene [7]. Zhang et al. inferred that miR-216b suppressed pancreatic cancer by inducing apoptosis [8]. Nonetheless, the mechanism of miR-216b in pancreatic cancer cells needs to be further explored.
In normal tissue signaling, the normal KRAS protein has an essential function, while a mutation of the KRAS gene may lead to many unpredictable cancers. One study showed that KRAS mutation is a critical predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer [9]. Another study revealed that PanINs and PDAC can develop from pancreatic acinar cells when mutant KRAS is restricted using an acinar-specific promoter [10]. Moreover, a recent study revealed that the establishment of a highly metastatic KRAS mutant in lung cancer is fatal [11]. All the above studies suggested that KRAS plays a significant role in cancer development. However, the relationship between KRAS and pancreatic cancer remains incompletely elucidated.
In the current study, we investigated how miR-216b took effect in human pancreatic cancer by interacting with KRAS. Our study provided clues for the role of miR-216b-KRAS interaction in regulating the cancer cell biology of pancreatic cancer, which distinguished it from previous studies, and indicated the therapeutic potential for miR-216b in pancreatic cancer treatment.
Material and methods
Clinical samples
Twenty-seven pairs of pancreatic cancer tumor tissues and adjacent normal tissues were obtained from pancreatic cancer patients undergoing surgery at the Third Affiliated Hospital of Soochow University from May 2013 to May 2015. Clinical and pathologic characteristics of pancreatic cancer patients are shown in Table I. The adjacent tissues were taken at a distance of 1.5 cm from the tumor. All the tissues were snap-frozen with liquid nitrogen at –80°C. No treatment such as radiotherapy and chemotherapy had been conducted on any patients before surgery. The study was sanctioned by the Ethics Committee of the Third Affiliated Hospital of Soochow University. All patients were informed of the study and gave informed consent.
Table I.
Clinical and pathological characteristics of 27 patients with pancreatic cancers
| Variables | Numbers |
|---|---|
| Age [years]: | |
| ≤ 60 | 9 |
| > 60 | 18 |
| Gender: | |
| Male | 17 |
| Female | 10 |
| Maximum tumor diameter [cm]: | |
| < 5.0 | 19 |
| ≥ 5.0 | 8 |
| Clinical stage: | |
| I and II | 14 |
| III | 13 |
| Tumor localization: | |
| Pancreatic head | 21 |
| Periampullary | 6 |
Cell culture and transfection
BxPC-3, Panc-1, and CFPAC-1 (cell lines of pancreatic cancer) and HPDE6-C7 (a normal pancreatic cell line) were obtained from Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences (Shanghai, China). Cultured in High Glucose Dulbecco’s modified Eagle medium (DMEM) (Gibco, Paisley, UK), all cells were supplemented with 10% fetal bovine serum (FBS) and maintained in a humidified atmosphere at 37°C. Cell transfection was not performed until cell growth reached about 80% confluence. MiR-216b mimics and KRAS-siRNA were synthesized by Shanghai Bioengineering Engineering Inc. (Shanghai, China) and transfected into Panc-1 cells by Lipofectamine 2000 reagent (Life Technologies, Grand Island, NY, USA) according to the manufacturer’s manual. The siRNA sequences used for KRAS siRNA experiments are shown in Table II. Four groups of cells were constructed: control group (non-transfection), NC group (transfection with negative control mimics or siRNA), miR-216b group (transfection with miR-216b mimics), and KRAS-siRNA group (transfection with KRAS siRNA).
Table II.
Sequences for KRAS siRNA
| KRAS siRNA | Sequence (5’-3’) |
|---|---|
| Sense1 | 5′-CCAACAAUAGAGGAUUCCUACAGGA-3′ |
| Antisense1 | 5′-UCCUGUAGGAAUCCUCUAUUGUUGG-3′ |
| Sense2 | 5′-CAAGACAGAGAGUGGAGGAUGCUUU-3′ |
| Antisense2 | 5′-AAAGCAUCCUCCACUCUCUGUCUUG-3′ |
| Sense3 | 5′-CAUUGGUGAGGGAGAUCCGACAAUA-3′ |
| Antisense3 | 5′-UAUUGUCGGAUCUCCCUCACCAAUG-3′ |
qRT-PCR
Trizol reagent (Invitrogen) was first used to lyse cells and extract total RNA. Next, ReverTra Ace qPCR RT Kit (Toyobo, Japan) was used to perform reverse transcription of RNA according to the instructions. Real-time quantitative PCR was then carried out on the MiniOpticon real-time PCR system (Bio-Rad, Hercules, CA, USA), using SYBR-Green RealMasterMix (Bio-Rad). U6 acted as an internal reference for miR-216b, while KRAS mRNA expression was normalized relative to GAPDH by the 2–∆∆Ct method. Primers were designed and provided by Sangon Biotech (Shanghai, China); their sequences are displayed in Table III.
Table III.
Primer sequences in qRT-PCR
| Variables | Forward primer | Reverse primer |
|---|---|---|
| miR-216b | 5’-GCCGCGCTAAAGTGCTTA-3’ | 5’-CACCAGGGTCCGAGGT-3’ |
| U6 | 5’-TGCGGGTGCTCGCTTCGGC-3’ | 5’-CCAGTGCAGGGTCCGAGGT-3’ |
| KRAS | 5’-TCTCCTTCTCAGGATTCCTACAG-3’ | 5’-ACAAAGAAAGCCCTCCCCAGT-3’ |
| GADPH | 5’-ACAACTTTGGTATCGTGGAAGG-3’ | 5’-GCCATCACGCCACAGTTTC-3’ |
Western blot
Panc-1 cells in the logarithmic growth phase were first collected and digested with 0.25% trypsin. Next, radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, Shanghai, China) was applied to extract total protein. A bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL, USA) was employed for the quantification of protein concentrations and 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Bio-Rad) was utilized for protein separation. After that, polyvinylidene fluoride (PVDF) membranes (Invitrogen) were employed for protein transference following the producer’s guidelines and sealed in 5% skim milk at 37°C for 2 h. The membranes were placed into a plastic bag in the next step and incubated overnight with primary antibodies against KRAS (ab180772, 1 : 500, Abcam, Cambridge, MA, USA) and GAPDH antibody (ab9485, 1 : 2000, Abcam) at 4°C. After 3 times 5-min washing with phosphate buffered saline (PBS), at room temperature, 1.5-h incubation of membranes with secondary antibody horseradish peroxidase-conjugated goat anti-rabbit IgG H&L (1 : 2000) was conducted, followed by again washing 3 times for 5 min with PBS. An electrochemiluminescent (ECL) detection system (Thermo Scientific, MA, USA) was employed for signal detection. The density of protein bands was quantified or analyzed using Quantity One v4.6.2 software (Bio-Rad).
Immunohistochemistry (IHC)
Immunohistochemical staining was carried out on 4 μm-thick slides. Briefly, the tissues were embedded in paraffin, then the slides were deparaffinized and rehydrated through graded alcohols and washed in phosphate buffered saline (PBS) 2 times for 10 min. Next the sections were incubated on the slides overnight with rabbit polyclonal primary antibody of KRAS (ab55391, Abcam) at a 5 µg/ml concentration. Then the sections were incubated with 45 μl of secondary antibody horseradish peroxidase-conjugated goat polyclonal anti-mouse IgG H&L (HRP) (1 : 500, ab6789, Abcam) at 37°C for 30 min. Slides were stained with 3,3′-diaminobenzidine (DAB) working solution for 3 min, then washed in water for 10 min. Slides were counterstained with hematoxylin. After rewashing the slides in water for 10 min, we finally dehydrated and cleared them. The slides were then ready for microscopic observation.
MTT assay
After 24-h transfection, cells were inoculated into 96-well plates at a density of 1 × 104 cells/well and stored in DMEM with FBS in it. At 37°C, cells were incubated for 4 h with 50 µl of MTT (0.5 mg/ml, Sigma-Aldrich) added into each well and had their growth condition observed at 0, 24, 48, and 72 h. To solubilize the crystals, 150 µl of dimethylsulfoxide (DMSO) was added into each well after the supernatant was subsequently removed. The optical density (OD) was measured at 570 nm by a microplate reader (Bio-Rad).
Transwell assay
Cells were first placed on a transwell plate. After being diluted with 100 μl of serum-free medium, Matrigel (200 mg/ml, BD Biosciences, CA, USA) was added to the upper chamber of the bottom membranes. Cells were lysed by trypsin and collected by centrifugation 48 h after transfection. Afterwards, the cells were washed with PBS twice, re-suspended in the serum-free medium with bovine serum albumin (BSA) in it and then inoculated into the upper chamber. At 37°C atmosphere, 250 μl DMEM with 10% FBS in it was added into the lower chamber for 24-h incubation. Non-invading cells were gently removed using a cotton swab. Migratory or invasive cells were fixed with 95% alcohol, stained with hematoxylin for 15 min and counted under an Olympus inverted microscope (200×) (Olympus, Tokyo, Japan).
Flow cytometry assay
After 48-h transfection, cells were digested by 0.25% trypsin and harvested by centrifugation at 800 rpm after 5 min. Next, at 4°C, the cells were washed with 0.01 mol/l ice-cold PBS twice, 5 min each time, and resuspended in binding buffer for 15 min. Annexin-V FITC (5 μl) and 5 μl of propidium iodide (PI; BD Bioscience) were added to the cells. After incubation for 20–30 min in the dark, the apoptotic cells were detected by flow cytometry (EPICS XL; Beckman-Coulter, Fullerton, CA).
Dual-luciferase reporter assay
The sequences of KRAS 3′UTR wild-type (wt) and KRAS 3′UTR mutated type (mut) were synthesized by Sangon Biotech (Shanghai, China). After amplification, the KRAS 3′UTR wt and KRAS 3′UTR mut were subcloned into the pmirGLO vector (Promega, Madison, USA). When 80% confluence was reached, using Lipofectamine 2000 reagent (Invitrogen), Panc-1 cells were co-transfected with miR-216b mimic or negative control mimic and pmirGLO-KRAS 3′UTR-wt or pmirGLO-KRAS 3′UTR-mut. Luciferase activity was detected at 48 h after transfection using a dual-luciferase reporter gene assay kit (Promega, USA).
Statistical analysis
All data were processed and analyzed by SPSS 21.0 statistical software (SPSS, Chicago, IL, USA). Data were presented as the mean ± standard deviation (SD). All in vitro experiments were carried out in triplicate. Comparison of data from two groups was performed using Student’s t-test when the data had a normal distribution; otherwise the Mann-Whitney U-test was used. Comparison between three groups or more was performed by one-way ANOVA. The association between miR-216b expression and KRAS expression levels was analyzed by Spearman’s method. Categorical variables of prognosis were contrasted by the χ2 test. Kaplan-Meier survival analysis and Cox proportional hazards models were applied to determine the link between the expression levels of miR-216b and KRAS and prognosis of patients. A p-value less than 0.05 was defined as statistically significant.
Results
Abnormal expression of miR-216b and KRAS in pancreatic cancer tissues and cells
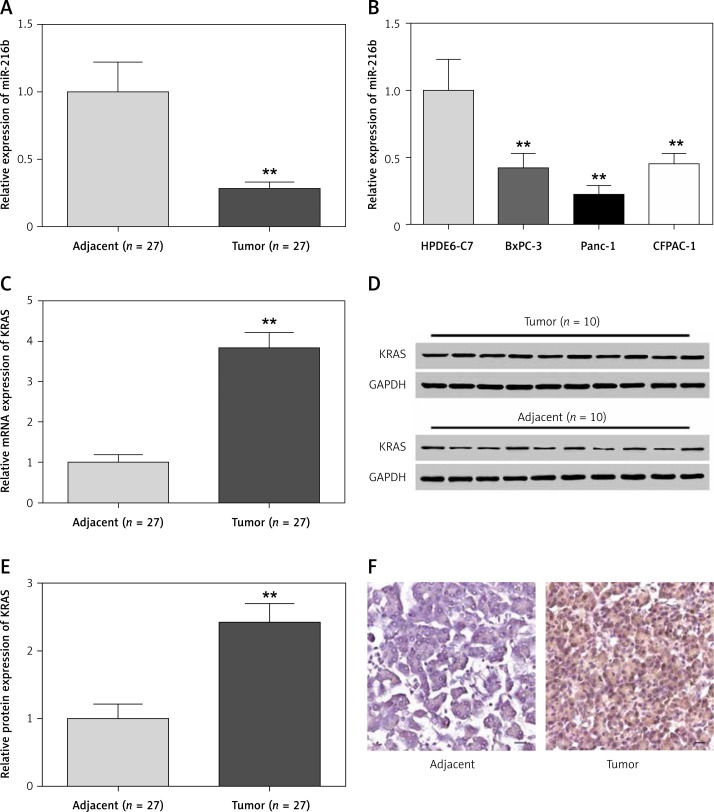
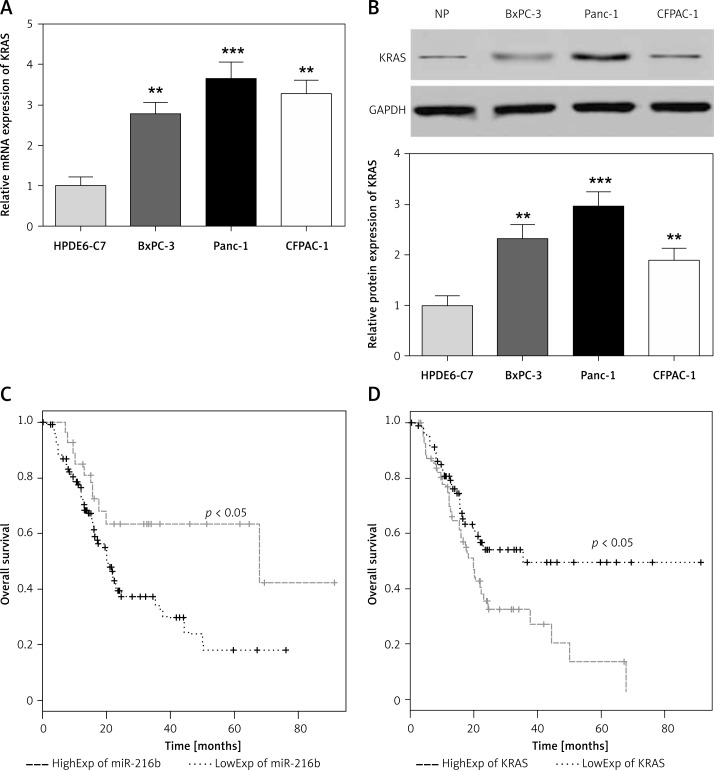
The results of qRT-PCR showed that compared with that of normal pancreatic tissues and cells, miR-216b mRNA expression in pancreatic cancer tissues and cells was obviously lower (p < 0.01, Figures 1 A, B). Conversely, the expression level of KRAS mRNA and protein were considerably higher in pancreatic tumor tissues (all, p < 0.01, Figures 1 C–E). Also, the expression of KRAS protein was higher in pancreatic tumor tissues than normal pancreatic tissues according to IHC assay (Figure 1 F). Moreover, pancreatic cancer cells presented remarkably higher expression of KRAS than normal pancreatic cells (p < 0.01, Figures 2 A, B). As the Panc-1 cell line presented a more significant difference than the other two cancer cell lines, it was selected for further experiments.
Figure 1.
The expression levels of miR-216b were lower and the expression levels of KRAS were higher in pancreatic tissues and cell samples. A, B – qRT-PCR indicated that the expression levels of miR-216b were significantly lower in pancreatic cancer tissues and cells than in adjacent tissues and normal pancreatic cells HPDE6-C7. **P < 0.01, compared with adjacent tissues and HPDE6-C7 cells. C–E – qRT-PCR and western blot results indicated that the expression levels of KRAS mRNA and protein in tumor tissues were significantly higher than those in adjacent tissues. **P < 0.01, compared with adjacent tissues. F – IHC assay results suggested that the expression levels of KRAS protein in tumor tissues were higher than those in adjacent tissues
Figure 2.
High expression of miR-216b and low expression of KRAS was beneficial for the prognosis of pancreatic cancer patients. A, B – qRT-PCR and western blot results showed that the expression levels of KRAS mRNA and protein in pancreatic cancer cell lines (BxPC-3, Panc-1, and CFPAC-1) were remarkably higher than those of normal pancreatic cell line HPDE6-C7. **P < 0.01, ***p < 0.001, compared with HPDE6-C7 cells. C, D – Kaplan-Meier survival analysis confirmed that the high expression of miR-216b and low expression of KRAS were related to the better prognosis of pancreatic cancer patients (log-rank test, all p < 0.05)
The TCGA database was analyzed to compare the clinicopathological characteristics with the expression of miR-216b and KRAS in pancreatic cancer patients. Patients were divided into two groups according to the mean expression of miR-216b or KRAS. The χ2 method was used and the results are shown in Tables IV and V. The low miR-216b expression group shows higher lymphatic metastasis and TNM staging compared to the high miR-216b group (p < 0.05). Meanwhile, patients with a high KRAS level suffered from a high probability of lymphatic metastasis, a larger tumor size and an advanced TNM staging (p < 0.05). In addition, the overall survival (OS) analysis of patients with low-expressed miR-216b was significantly poorer than those with high-expressed miR-216b (Figure 2 C, p < 0.05), while the prognosis of patients with low-expressed KRAS was observably better than those with high-expressed KRAS (Figure 2 D, p < 0.05). To further study the factors and their contribution to the prognosis of pancreatic cancers, we performed both univariable and multivariable analysis of overall survival. The results confirmed KRAS as an independent indicator for the prognosis of patients with pancreatic cancers (Table VI).
Table IV.
Relationship between miR-216b expression and clinicopathological characteristics in pancreatic cancer patients
| Characteristic | Number | High expression of miR-216b (n = 89) | Low expression of miR-216b (n = 89) | P-value |
|---|---|---|---|---|
| Age [years]: | ||||
| > 60 | 101 | 52 | 47 | 0.546 |
| ≤ 60 | 77 | 37 | 42 | |
| Gender: | ||||
| Male | 96 | 49 | 46 | 0.764 |
| Female | 82 | 40 | 43 | |
| Tumor location: | ||||
| Pancreatic head | 110 | 59 | 55 | 0.640 |
| Periampullary | 68 | 30 | 34 | |
| Tumor size [cm]: | ||||
| < 5 | 109 | 55 | 47 | 0.289 |
| ≥ 5 | 69 | 34 | 42 | |
| Lymphatic metastasis: | ||||
| Yes | 107 | 35 | 57 | 0.002* |
| No | 71 | 54 | 32 | |
| Neural invasion: | ||||
| Present | 91 | 40 | 52 | 0.099 |
| Absent | 87 | 49 | 37 | |
| TNM staging: | ||||
| Early stage (I–II) | 82 | 49 | 33 | 0.012* |
| Advanced (III–IV) | 96 | 40 | 56 | |
Categorical variables were compared by the χ2 test. *P < 0.05 was recognized as a significant difference.
Table V.
Relationship between KRAS expression and clinicopathological characteristics in pancreatic cancer patients
| Characteristic | Number | High expression of KRAS (n = 89) | Low expression of KRAS (n = 89) | P-value |
|---|---|---|---|---|
| Age [years]: | 0.0961 | |||
| > 60 | 101 | 56 | 45 | |
| ≤ 60 | 77 | 33 | 44 | |
| Gender: | 0.1327 | |||
| Male | 96 | 53 | 43 | |
| Female | 82 | 36 | 46 | |
| Tumor location: | 0.6437 | |||
| Pancreatic head | 110 | 53 | 57 | |
| Periampullary | 68 | 36 | 32 | |
| Tumor size [cm]: | 0.0089* | |||
| < 5 | 109 | 63 | 46 | |
| ≥ 5 | 69 | 26 | 43 | |
| Lymphatic metastasis: | < 0.0001* | |||
| Yes | 107 | 68 | 39 | |
| No | 71 | 21 | 50 | |
| Neural invasion: | 0.1335 | |||
| Present | 91 | 51 | 40 | |
| Absent | 87 | 38 | 49 | |
| TNM staging: | 0.0238* | |||
| Early stage (I–II) | 82 | 33 | 49 | |
| Advanced (III–IV) | 96 | 56 | 40 |
Categorical variables were compared by the χ2 test. P < 0.05 was recognized as a significant difference.
Table VI.
Univariate and multivariate analysis for overall survival of patients with pancreatic cancer
| Factors | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Relative risk | Pa | Relative risks | Pb | |
| Age | 1.83 | 0.233 | ||
| Gender | 0.61 | 0.674 | ||
| Tumor location | 2.83 | 0.081 | ||
| Tumor size | 6.22 | 0.002* | 4.11 | 0.284 |
| Lymphatic metastasis | 5.31 | 0.005* | 5.17 | 0.023* |
| Neural invasion | 4.88 | 0.061 | ||
| TNM staging | 6.29 | 0.001* | 7.12 | 0.002* |
| miR-216b expression | 3.97 | 0.004* | 2.74 | 0.325 |
| KRAS expression | 4.69 | 0.004* | 6.17 | 0.007* |
Analysis was performed using Kaplan-Meier method (a) and Cox proportional hazards regression model (b).
P < 0.05 represents a significant difference.
MiR-216b directly targeted at KRAS
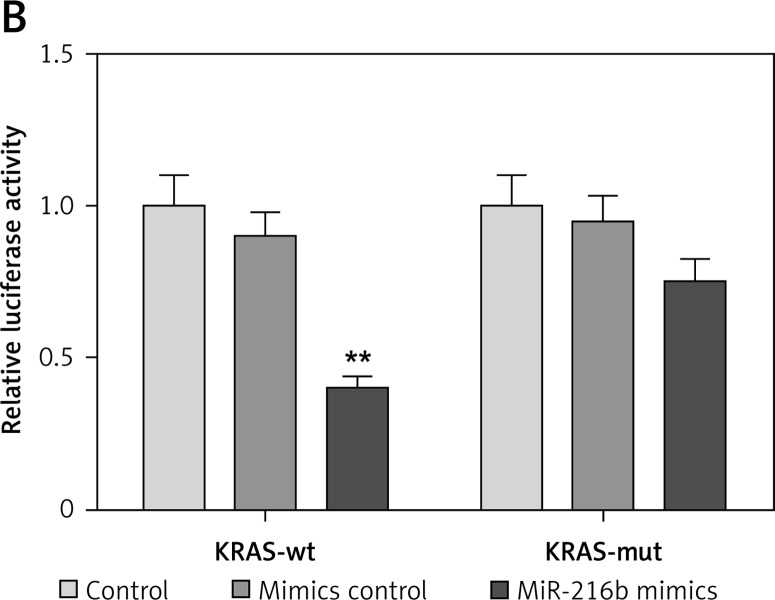
We first predicted the potential binding sites of miR-216b in the 3′UTR of KRAS through the TargetScan database (Figure 3 A). In the meantime, as Figure 3 B shows, the luciferase activity of KRAS 3′UTR-wt in Panc-1 cells transfected with miR-216b mimics was remarkably weaker than cells transfected with negative control mimics (p < 0.01). Nonetheless, no significant difference of luciferase activity of KRAS 3′UTR-mut was observed between the miR-216b group and NC group (p > 0.05). The above results indicated that miR-216b directly targeted at KRAS and overexpression of miR-216b significantly repressed the luciferase activity of the wild-type 3′UTR of KRAS in Panc-1 cells.
Figure 3.
MiR-216b directly targeted KRAS. A – TargetScan algorithm was used to predict the binding site of miR-216b in KRAS 3′UTR. B – The luciferase activity of KRAS 3′UTR-wt in Panc-1 cells transfected with miR-216b mimics was significantly weaker compared with the cells transfected with negative control mimics. However, there was no significant difference in the luciferase activity of KRAS 3′UTRmut between the miR-216b group and NC group
**P < 0.05, compared with NC group
MiR-216b overexpression inhibited proliferation, migration and invasion, and induced apoptosis of pancreatic cancer cells by down-regulating KRAS.
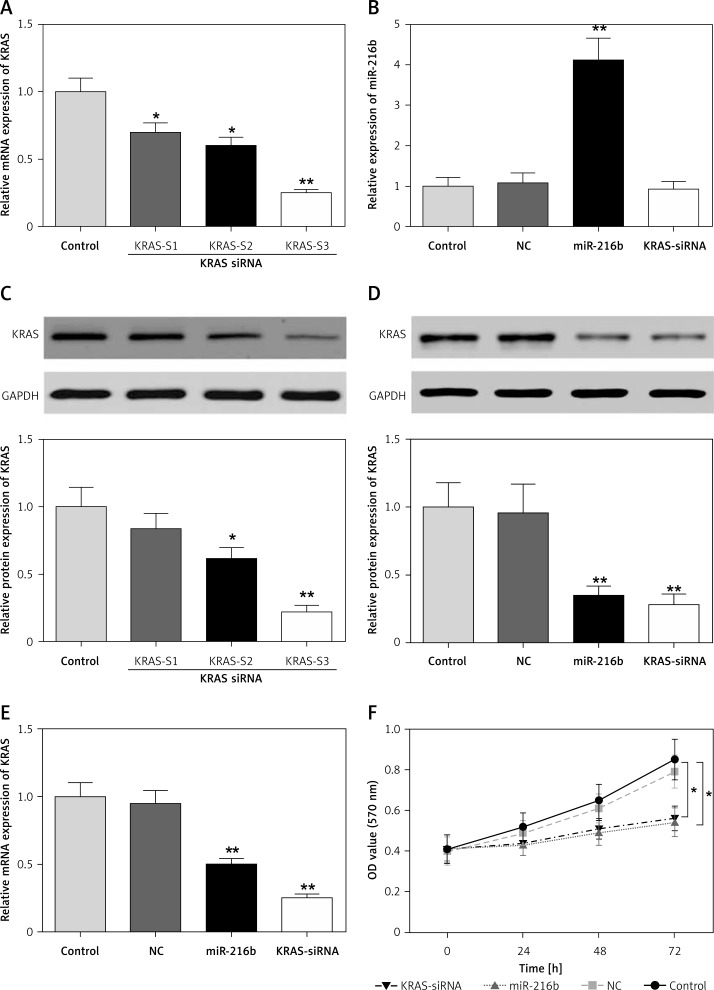
We determined the transfection efficiency of three KRAS siRNA transfected groups by qRT-PCR (Figures 4 A, C) and found that the third KRAS siRNA transfection group had the lowest KRAS mRNA and protein expression. Hence, we used the third KRAS siRNA transfection group for subsequent experiments. Western blot and qRT-PCR attested that after transfection with miR-216b mimics, miR-216b expression was remarkably up-regulated, while that of KRAS dramatically decreased in comparison with the control group (Figures 4 B, D, E). At the same time, as Figure 4 F shows, the cell viabilities in the miR-216b overexpression group and KRAS-siRNA group were significantly attenuated (p < 0.05), suggesting that miR-216b could prevent the proliferation of pancreatic cancer cells by down-regulating KRAS.
Figure 4.
MiR-216b suppressed pancreatic cancer cell proliferation through targeting KRAS. A, C – The transfection efficiency of three KRAS siRNA transfected groups was determined. *P < 0.05, **p < 0.01, compared with control group. B, D, E – The expression of miR-216b in Panc-1 cells was significantly up-regulated, while the expression of KRAS was remarkably down-regulated after being transfected with KRAS-siRNA detected by qRT-PCR and western blot. **P < 0.01 compared with control group. F – The proliferation ability of Panc-1 cells after transfection with miR-216b mimics and KRAS-siRNA was significantly repressed, as observed by MTT assay. *P < 0.05, compared with control group
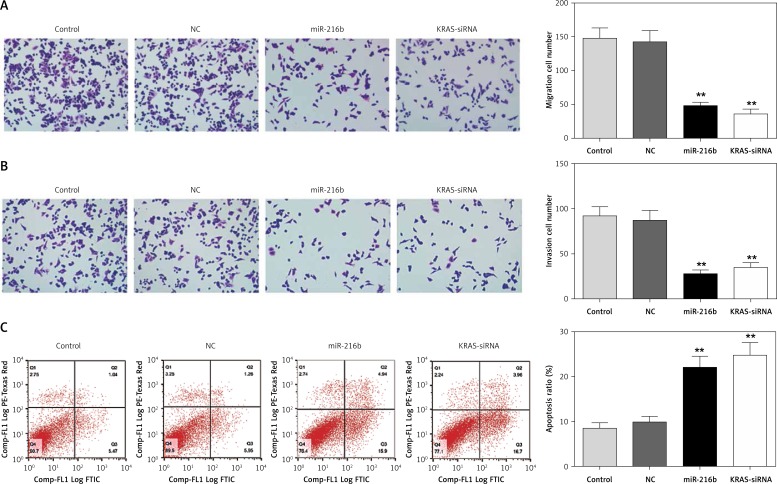
In addition, results from transwell assay suggested that the number of migratory and invasive cells in the miR-216b group and KRAS-siRNA group was lower than that of the control group (p < 0.05, Figures 5 A, B), indicating that miR-216b overexpression and under-expression of KRAS could hinder cell migration and invasion abilities. Furthermore, flow cytometry assay results indicated that the apoptosis rates of Panc-1 cells in the miR-216b group and KRAS-siRNA group were observably higher than the control group (both p < 0.05, Figure 5 C). Taken together, miR-216b could inhibit pancreatic cancer cell migration and invasion, and promote cell apoptosis via down-regulation of KRAS expression.
Figure 5.
MiR-216b inhibited pancreatic cancer cell migration and invasion and promoted apoptosis by targeting KRAS. A, B – The migration and invasion abilities of Panc-1 cells significantly decreased after transfection with the miR-216b mimics and KRAS-siRNA observed by transwell assay. C – The apoptosis ability of Panc-1 cells significantly increased after transfection with the miR-216b mimics and KRAS-siRNA detected by flow cytometry assay. **P < 0.01, compared with control group
Discussion
There was a previous study proving the targeting relationship between miR-216 and KRAS in the ELa-KRASG12D mouse model [10]. In our study, we laid more emphasis on the human tissues in an in vitro experiment. We came to the conclusion that by silencing KRAS, miR-216b could slow down the progression of human pancreatic cancer cells.
It is well known that KRAS is a critical gene leading to the deterioration of pancreatic cancer. One previous study has already reported that KRAS gene mutation was a major cancer initiating event and may incur pancreatic carcinogenesis [12]. According to previous studies, KRAS had high expression when endometriosis occurred [13]. It was also revealed that KRAS had high expression in lung adenocarcinoma [14]. Another study revealed that KRAS mutations led to poor prognosis in patients with microsatellite-stable stage III colon cancer [15]. Similarly, KRAS gene mutations were verified to lead to a poorer prognosis of patients with pancreatic cancer [16]. Intriguingly, in this research, we substantiated that KRAS was highly expressed in pancreatic cancer and the prognosis with low KRAS expression was better, which are also consistent with some previous study results.
MiR-216 has been found to modulate gene expression and presented down-regulated expression in various cancers. For instance, one typical study revealed that miR-216b expression was down-regulated in pancreatic ductal adenocarcinoma (PDAC) and that miR-216b expression was reduced 2.7-fold in the cases that did not benefit from therapy [17]. Another study revealed that there was significantly decreased expression of miR-216b in human gastric adenocarcinoma [18]. What is more, researchers also found out that the high expression of miR-216 was related to higher overall survival [19]. In this study, we have already verified the low expression of miR-216b and the better prognosis with high miR-216b expression in pancreatic cancer.
There is considerable evidence suggesting that miR-216b functions as a key inhibitor in regulating gene expression through base pairing within the target messenger RNA (mRNA) 3′-untranslated region (UTR) [4]. Our study results demonstrated that miR-216b targeted KRAS in pancreatic cancer cells. One previous study suggested that miR-216b directly targeted KRAS in RInk-1 cells in pancreatic cancer [20]. Another study indicated that miR-216b targeted KRAS in nasopharyngeal carcinoma [21]. A similar article also revealed that miR-216a could actually target KRAS in PDAC [10]. These lines of evidence all support our conclusion.
In addition, some researchers hypothesized that miR-216a induced apoptosis in pancreatic cancer cells by silencing MALAT1 expression [8], indicating that up-regulation of miR-216a may inhibit the oncogene and suppress the cancer. In the current study, we mainly investigated the role miR-216b played in pancreatic cells through regulating KRAS and validated that miR-216b down-regulated the expression of KRAS, inhibiting pancreatic cancer cell proliferation, migration and invasion and promoting apoptosis. Moreover, some previous studies have demonstrated that KRAS could induce invasion and proliferation of pancreatic cancer cells [22]. The results in a similar study showed that kinase Pim-1 could regulate the oncogene KRAS, and hence suppressed human PDAC cell growth [23], while other results indicated that miR-216a could also significantly inhibit cell growth and promote cell apoptosis in pancreatic cancer [24]. Furthermore, some researchers also found out that miR-216a exerted its tumor suppressor function through inhibiting the KRAS-related pathway [21]. Our conclusions were strongly supported.
However, some limitations need to be overcome in future studies. For one thing, the signaling pathway is still under investigation. For another, the number of samples in our study was also limited due to the shortage of funds.
In conclusion, our results revealed a correlation between miR-216b and KRAS in human pancreatic cancer cells. All these data suggested that miR-216b was functional in inhibiting oncogenic KRAS and could act as a novel therapeutic target for the treatment of pancreatic cancer.
Our study demonstrated that miR-216b suppressed pancreatic cancer cell proliferation, migration and invasion and promoted apoptosis via down-regulation of the oncogene KRAS, providing a novel approach for pancreatic cancer treatment.
Acknowledgments
Xinquan Wu and Weibo Chen contributed equally to this work.
This study was supported by a Youth Project of the National Natural Science Foundation of China (no. 81502002).
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers (Basel) 2017;9 doi: 10.3390/cancers9050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreyer SB, Chang DK, Bailey P, Biankin AV. Pancreatic cancer genomes: implications for clinical management and therapeutic development. Clin Cancer Res. 2017;23:1638–46. doi: 10.1158/1078-0432.CCR-16-2411. [DOI] [PubMed] [Google Scholar]
- 3.Keklikoglou I, Hosaka K, Bender C, et al. MicroRNA-206 functions as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene. 2015;34:4867–78. doi: 10.1038/onc.2014.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–33. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 5.Hou LJ, Zhai JJ. Aberrant expression profile of translationally controlled tumor protein and tumor-suppressive microRNAs in cervical cancer. J BUON. 2015;20:1504–9. [PubMed] [Google Scholar]
- 6.Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2alpha-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478:1067–73. doi: 10.1016/j.bbrc.2016.08.065. [DOI] [PubMed] [Google Scholar]
- 7.Zheng WW, Zhou J, Zhang CH, Liu XS. MicroRNA-216b is downregulated in hepatocellular carcinoma and inhibits HepG2 cell growth by targeting Forkhead box protein M1. Eur Rev Med Pharmacol Sci. 2016;20:2541–50. [PubMed] [Google Scholar]
- 8.Zhang Y, Tang X, Shi M, Wen C, Shen B. MiR-216a decreases MALAT1 expression, induces G2/M arrest and apoptosis in pancreatic cancer cells. Biochem Biophys Res Commun. 2017;483:816–22. doi: 10.1016/j.bbrc.2016.12.167. [DOI] [PubMed] [Google Scholar]
- 9.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–6. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 10.Azevedo-Pouly AC, Sutaria DS, Jiang J, et al. miR-216 and miR-217 expression is reduced in transgenic mouse models of pancreatic adenocarcinoma, knockout of miR-216/miR-217 host gene is embryonic lethal. Funct Integr Genom. 2017;17:203–12. doi: 10.1007/s10142-016-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano T, Kanai Y, Amano Y, et al. Establishment of highly metastatic KRAS mutant lung cancer cell sublines in long-term three-dimensional low attachment cultures. PLoS One. 2017;12:e0181342. doi: 10.1371/journal.pone.0181342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins MA, Pasca di Magliano M. Kras as a key oncogene and therapeutic target in pancreatic cancer. Front Physiol. 2013;4:407. doi: 10.3389/fphys.2013.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo JY, Kim TH, Fazleabas AT, et al. KRAS activation and over-expression of SIRT1/BCL6 contributes to the pathogenesis of endometriosis and progesterone resistance. Sci Rep. 2017;7:6765. doi: 10.1038/s41598-017-04577-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Justilien V, Ali SA, Jamieson L, et al. Ect2-dependent rRNA synthesis is required for KRAS-TRP53-driven lung adenocarcinoma. Cancer Cell. 2017;31:256–69. doi: 10.1016/j.ccell.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taieb J, Le Malicot K, Shi Q, et al. Prognostic value of BRAF and KRAS mutations in MSI and MSS stage III colon cancer. J Natl Cancer Inst. 2016:109. doi: 10.1093/jnci/djw272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao LY, Zhang LF, Xiu DR, Yuan CH, Ma ZL, Jiang B. Prognostic significance of K-ras mutations in pancreatic cancer: a meta-analysis. World J Surg Oncol. 2016;14:146. doi: 10.1186/s12957-016-0888-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egeli U, Tezcan G, Cecener G, et al. miR-216b targets FGFR1 and confers sensitivity to radiotherapy in pancreatic ductal adenocarcinoma patients without EGFR or KRAS mutation. Pancreas. 2016;45:1294–302. doi: 10.1097/MPA.0000000000000640. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Xu P, Yao J, et al. MicroRNA-216b is down-regulated in human gastric adenocarcinoma and inhibits proliferation and cell cycle progression by targeting oncogene HDAC8. Target Oncol. 2016;11:197–207. doi: 10.1007/s11523-015-0390-9. [DOI] [PubMed] [Google Scholar]
- 19.Zhan MX, Li Y, Hu BS, et al. [Expression of serum microRNAs (miR-222, miR-181, miR-216) in human hepatocellular carcinoma and its clinical significance] Zhonghua Yi Xue Za Zhi. 2013;93:1830–2. [PubMed] [Google Scholar]
- 20.Ali S, Banerjee S, Logna F, et al. Inactivation of Ink4a/Arf leads to deregulated expression of miRNAs in K-Ras transgenic mouse model of pancreatic cancer. J Cell Physiol. 2012;227:3373–80. doi: 10.1002/jcp.24036. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Deng M, Tang H, Zhou Y, et al. miR-216b suppresses tumor growth and invasion by targeting KRAS in nasopharyngeal carcinoma. J Cell Sci. 2011;124:2997–3005. doi: 10.1242/jcs.085050. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Xu XF, Zhao Y, Tang MC, Zhou YQ, Gao FH. NS-398 promotes pancreatic cancer cell invasion by CD147 and MMP-2 via the activation of P38. Mol Med Rep. 2016;13:2208–14. doi: 10.3892/mmr.2016.4783. [DOI] [PubMed] [Google Scholar]
- 23.Xu D, Allsop SA, Witherspoon SM, et al. The oncogenic kinase Pim-1 is modulated by K-Ras signaling and mediates transformed growth and radioresistance in human pancreatic ductal adenocarcinoma cells. Carcinogenesis. 2011;32:488–95. doi: 10.1093/carcin/bgr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Chen X, Tang M. MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2. Oncol Rep. 2014;32:2824–30. doi: 10.3892/or.2014.3478. [DOI] [PubMed] [Google Scholar]