Abstract
Introduction
Gliomas are the most frequent primary tumors in the human brain. Recent studies have identified a class of long noncoding RNAs, named lncRNAs, which were reported to participate in regulating the development of various diseases, including gliomas. In our previous studies, we found that lncRNA UBE2CP3-001 was overexpressed in gliomas but not in normal tissue. However, the molecular functions of UBE2CP3-001 in glioma are largely unknown.
Material and methods
The presence of UBE2CP3-001 in U87 cells, glioma tissues and normal brain tissues was detected by real-time RT-PCR. The ability of U87 cells to migrate was analyzed using a cellular wound healing assay after downregulation of UBE2CP3-001. The survival rate of U87 cells after UBE2CP3-001 knockdown was also analyzed using the CCK8 assay. In vivo tumor weights from xenograft tumors transfected with UBE2CP3-001 shRNA were further analyzed using in vivo animal experiments. The expression levels of MMP-9 and TRAF3IP2 were determined by Western blot.
Results
Our data showed that UBE2CP3-001 was overexpressed in most glioma tissues (p < 0.01). Downregulation of UBE2CP3-001 could inhibit cell migration (p < 0.01) and invasiveness (p < 0.01) of U87 cells. Downregulation of UBE2CP3-001 in U87 cells also suppressed the cell proliferation (p < 0.01) and promoted apoptosis (p < 0.01). Furthermore, in vivo studies confirmed that knockdown of UBE2CP3-001 could retard the growth of U87 xenograft tumors (p < 0.01). Western blot analysis showed that knockdown of UBE2CP3-001 could effectively inhibit the expression of MMP-9 (p < 0.01) and TRAF3IP2 (p < 0.01) in U87 glioma cells.
Conclusions
These data suggest an important role of UBE2CP3-001 in glioma and indicate its potential application in anti-glioma therapy.
Keywords: lncRNA UBE2CP3-001, TRAF3IP2, glioma, proliferation, apoptosis
Introduction
Gliomas are primary tumors occurring mostly in the central nervous system of adults [1]. Despite recent advances in the control of gliomas, their occurrence has not changed significantly [2]. Surgery, radiation, and chemotherapy are still the main therapy strategies; thus, patients with malignant gliomas have very few viable treatment alternatives [3]. The deadly nature of gliomas is due to its rapid diffuse infiltrative growth and the inherent complexity of the tumors. However, efforts to increase patient survival rates have been stagnant, due to the complex gene interactions and molecular modulations involved in the development of gliomas [4]. Therefore, understanding the molecular mechanisms involved in the development of gliomas is important to propose effective therapy [5].
Recently, it was discovered that long non-coding RNAs (lncRNAs) function in various aspects of cell biology and potentially contribute to tumor development [6]. LncRNAs are non-protein coding transcripts that contain more than 200 nucleotides [7] and are involved in a number of important events, such as epigenetic regulation, transcriptional regulation, and posttranscriptional regulation [8, 9]. With the progress in transcriptome profiling, lncRNAs detected by microarrays are characterized as natural antisense lncRNAs, intronic antisense lncRNAs, bidirectional lncRNAs, exon sense overlapping lncRNAs, intron-sense overlapping lncRNAs and intergenic lncRNAs. Some reports in the literature indicate that intronic antisense lncRNAs are enriched in the introns of genes and are involved in the regulation of transcription. Most of the intronic antisense lncRNAs have the same tissue expression patterns as the corresponding coding genes and may stabilize protein-coding transcripts or regulate their alternative splicing [10]. A study on prostate cancers suggested that the presence of intronic antisense lncRNAs was correlated with the degree of tumor differentiation [11]. Aberrantly expressed lncRNAs can be discovered through comparative transcriptome analysis. Accumulating evidence indicates that lncRNAs are associated with a diverse range of biological functions in cells, including those in glioma [12, 13]. Some researchers have shown that lncRNAs such as HOTAIR, MEG3, and H19 play an important role in the development of gliomas [14–16].
In our previous study, we used lncRNA expression profiling in microarrays to identify aberrant expression of lncRNAs in glioma tissues compared to benign tissues. LncRNA ENST0000502715, also referred to as UBE2CP3-001, is an intronic antisense that was found to be overexpressed in the microarray data. LncRNA UBE2CP3-001 is encoded by UBE2CP3 that is located on chromosome 4q12, but its function is unknown. To the best of our knowledge, this is the first study to investigate the expression of UBE2CP3-001 in gliomas and its function in gliomas.
Material and methods
Tumor tissue samples and cell lines
Forty specimens of glioma tissues and adjacent benign tissues were collected from the Neurosurgery Department of Nanjing Brain Hospital, Affiliated with Nanjing Medical University. Histopathological characteristics were evaluated by the hospital pathologist. Written informed consent was obtained from all patients who participated in the study. This study was approved by the Ethics Committee of the university. Following collections, specimens were preserved in liquid nitrogen and shipped. Detailed patient information is provided in Table I. Human U87 cell lines were purchased from the Cell Bank Type Culture Collection of Chinese Academy of Science (Shanghai, China). Cells were grown in DMEM (Hyclone, Thermo Fisher Scientific, Waltham, MA, USA) and supplemented with 10% FBS (Gibco, Invitrogen, Carlsbad, CA, USA).
Table I.
Detailed patient information
| Sample number | Gender | Age [years] | Pathological category | WHO grade |
|---|---|---|---|---|
| 1 | F | 57 | Anaplastic astrocytoma | WHO III |
| 2 | M | 37 | Anaplastic astrocytoma | WHO III |
| 3 | M | 44 | anaplastic astrocytoma | WHO III |
| 4 | F | 66 | Anaplastic astrocytoma | WHO III |
| 5 | M | 61 | Anaplastic astrocytoma | WHO III–IV |
| 6 | F | 56 | Anaplastic astrocytoma | WHO II–IV |
| 7 | M | 52 | Anaplastic oligoastrocytoma | WHO III |
| 8 | M | 34 | Anaplastic oligoastrocytoma | WHO III |
| 9 | F | 58 | Anaplastic oligoastrocytoma | WHO III |
| 10 | F | 48 | Anaplastic oligodendroglial | WHO III–IV |
| 11 | F | 59 | Anaplastic oligodendroglial | WHO III |
| 12 | M | 40 | Anaplastic oligodendroglial | WHO III |
| 13 | M | 50 | Anaplastic oligodendroglial | WHO II–III |
| 14 | M | 50 | Astrocyte | WHO II–III |
| 15 | F | 60 | Astrocyte | WHO III |
| 16 | M | 49 | Astrocyte | WHO II |
| 17 | F | 49 | Astrocyte | WHO II |
| 18 | M | 54 | Astrocyte | WHO II |
| 19 | M | 48 | Astrocyte | WHO III |
| 20 | F | 58 | Astrocytoma | WHO II |
| 21 | F | 62 | Gangliocytoma | WHO III |
| 22 | F | 53 | Gangliocytoma | WHO III |
| 23 | M | 63 | Glioblastoma | WHO IV |
| 24 | M | 56 | Glioblastoma | WHO IV |
| 25 | M | 47 | Glioblastoma | WHO IV |
| 26 | M | 60 | Glioblastoma | WHO IV |
| 27 | M | 72 | Glioblastoma | WHO IV |
| 28 | M | 62 | Glioblastoma | WHO IV |
| 29 | F | 13 | Glioblastoma | WHO IV |
| 30 | F | 70 | Glioblastoma | WHO IV |
| 31 | F | 46 | Glioblastoma | WHO IV |
| 32 | M | 39 | Oligoastrocytoma | WHO II |
| 33 | F | 61 | Oligoastrocytoma | WHO III–IV |
| 34 | F | 49 | Oligoastrocytoma | WHO II |
| 35 | F | 36 | Oligodendroglioma | WHO II |
| 36 | M | 49 | Oligodendroglioma | WHO II |
| 37 | F | 48 | Oligodendroglioma | WHO II |
| 38 | F | 48 | Oligodendroglioma | WHO II |
| 39 | F | 70 | Oligodendroglioma | WHO III |
| 40 | M | 40 | Pilocytic astrocytoma | WHO I |
RNA extraction
Total RNA was isolated using TRIzol reagent (Invitrogen, CA, USA) as follows: 1) Glioma and adjacent normal tissues in liquid nitrogen were pulverized with a mortar and pestle. Tissue samples comprising 50 mg tissue/1 ml Trizol were homogenized until all the liquid nitrogen had evaporated; 2) The homogenate was pipetted up and down several times and transferred to an Eppendorf tube; 3) Homogenized samples were incubated for 5 min at room temperature and then centrifuged at 12,000 rpm for 5 min according to the protocol. Next, 200 μl of chloroform was added per 1 ml of TRIzol. Samples were incubated for 15 min at room temperature and centrifuged at 12,000 rpm for 15 min at 4°C. The upper soluble layer was isolated and mixed with isopropanol and TRIzol. Samples were incubated for 10 min at room temperature and centrifuged at 12,000 rpm for 10 min at 4°C. The supernatant was removed and washed with 1 ml 75% ethanol. Samples were then centrifuged at 8,000 rpm for 5 min at 4°C. This was followed by brief incubation at room temperature to allow the samples to dry. Finally, RNA samples were dissolved in 50 μl of DEPC water and stored at –80°C until use. The concentration of each sample was determined using the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Inc., USA). RNA integrity was evaluated by electrophoresis using denaturing agarose gel.
Quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol (Invitrogen, CA, USA). Reverse transcription was performed using the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Dalian, China) according to the manufacturer’s protocol. Expression of lncRNA UBE2CP3-001 was determined using the SYBR Green detection method (TaKaRa, Dalian, China). Samples were run on an ABI7500 System (Applied Biosystems, Foster City, CA). Primer sequences for lncRNA detection are listed in Table II. β-Actin was used as a control for normalization. The qPCR reaction was carried out in a total volume of 20 μl. The reaction consisted of 10 μl 2× SYBR Green Master mix, 1 μl PCR Forward Primer (5 μmol/μl), 1 μl PCR Reverse Primer (5 μmol/μl), 1 μl cDNA (10 ng), and 7 μl RNase-free water. The RT-qPCR reaction was performed as follows: initial denaturation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s, and a final extension at 72°C for 5 min. Each sample was analyzed in triplicate. The threshold cycle (CT) value was calculated as the mean of the triplicate values. Quantitative differential expression was calculated according to the 2–ΔΔCt method. Statistical analysis was performed using Student’s t-test with SPSS software (Version 16.0 SPSS Inc.). Statistical significance was set at p < 0.05.
Table II.
Primer sequences for UBE2CP3 and ACTB detection
| Gene | Primer | Strand |
|---|---|---|
| UBE2CP3 | H-UBE2CP3-FO-1 | AAAAGGCTACAGCAGGAGTTGG |
| H-UBE2CP3-RE-1 | GTAGCAGGGCGTGAGGAAC | |
| ACTB | hactB F primer | CCCTGGCACCCAGCAC |
| hactB R primer | GCCGATCCACACGGAGTAC |
Construction of stable cell lines with downregulated lncRNA UBE2CP3-001
The lentiviral vectors expressing short hairpin RNA (shRNA) were successfully constructed by GenePharma (Shanghai, China). Table III shows the designed sequence of the UBE2CP3 shRNAs. As shown in Table III, six short hairpin RNA were designed. However, shRNA No. 6 was the most effective and was used in the following experiments. U87 cells transfected with UBE2CP3 shRNAnc was used as a negative control group. Stable cell lines were also selected and cultured by GenePharma (Shanghai, China). Then cells were further used in the in vitro and in vivo experiments.
Table III.
Designed shRNA sequence for UBE2CP3
| No. | Gene name | Strand | |
|---|---|---|---|
| 1 | UBE2CP3-122 | Target | GCACAGCATATGAAGACCTGA |
| 2 | UBE2CP3-143 | Target | GGTATAAGCTCTCCCTAGAGC |
| 3 | UBE2CP3-328 | Target | GGCAAACCCAACATTGATAGC |
| 4 | UBE2CP3-329 | Target | GCAAACCCAACATTGATAGCT |
| 5 | UBE2CP3-28 | Target | GTGACCTTCACAATGCCTAGT |
| 6 | UBE2CP3-408 | Target | GCAAGAAACCTACTTAAAGCA |
Construction of overexpression vector of TRAF3IP2
The lentiviral vector of overexpression of TRAF3IP2 (Lenti-GFP-TRAF3IP2) was successfully constructed by GenePharma (Shanghai, China). It was then transfected into U87 cells, and its overexpression in U87 cells was confirmed by Western blot. After that, Lenti-GFP-TRAF3IP2 was applied to the following experiment.
Cell proliferation assay
Cell proliferation assays were performed using the Cell Counting kit-8 (Donjindo, Kumamoto, Japan). Cells after treatment were plated in 96-well plates in triplicate at approximately 3 × 1000 cells per well. Then, 100 μl of DMEM containing 10% fetal bovine serum complete medium was added. PBS was added to prevent evaporation, and the cells were then incubated in 5% CO2 at 37°C. A total of 100 μl serum free medium containing 10 μl CCK8 reagents was added to each well after 24, 48, 72, and 96 h. The OD value was measured using ELISA at a wavelength of 450 nm.
Cell wound healing assay
After treatment, U87 cells were plated into 6-well plates. A total of 10 μl of AXYGEN was used to remove cells from the bottom of the monolayer cells after the cells had grown to 90% confluence. Floating cells in each well were washed away twice using PBS. Free serum DMEM was added to the plate. Scratches were photographed at the beginning of the experiment and after 24 h.
Cell in vitro invasion assay
After treatment, U87 cells were cultured for 72 h and transferred onto the top of Matrigel-coated invasion chambers (24-well insert, 8-μm pore size; BD Biosciences) in serum-free DMEM. DMEM containing 10% FBS was then added to the lower chamber as a chemoattractant. After incubation for 48 h, non-invading cells were removed from the inner part of the insert using a cotton swab. Cells on the lower membrane surface were fixed with 4% formaldehyde and stained with 0.1% crystal violet. Invading cells were manually counted under a microscope in five randomly selected fields, and photographs were taken.
Flow cytometric analysis
After treatment, U87 cells were plated in free serum DMEM to induce cell apoptosis in a 6-well plate. Approximately 2 × 105 cells were plated per well and incubated at 37°C with 5% CO2 for 24 h. After digestion with no EDTA and trypsin, cells were collected into 10 ml test tubes to be centrifuged at 1000 rpm for 3–5 min. Cells were washed once in PBS. Then, 3 μl of Annexin-V with 3 μl of 7-ADD was added to the cell suspension. Cells were incubated for 15 min with 300 μl of buffer solution. Flow cytometry was used to determine the result with FlowJo 7.5 software, and data were analyzed.
Western blot analysis
Total protein was isolated from cells during exponential growth, separated by SDS-polyacrylamide gel electrophoresis, and then transferred to nitrocellulose membrane. After being blocked with 5% non-fat milk powder, the membranes were incubated with the following primary antibodies: anti-MMP-9 (Cell Signaling, USA; 1 : 1000 dilution) and anti-TRAF3IP2 (Abcam, USA; 1 : 1000 dilution); then, the membranes were incubated with horseradish peroxidase conjugated secondary antibody (Beyotime, China; 1 : 2,000 dilution). Bands were visualized using the super ECL Detection System (Beyotime). Actin was used as a loading control. The results of Western blot analysis represent an average of three individual experiments. Data were analyzed using ImageJ software.
In vivo experiments
We established a subcutaneous xenograft nude mouse model. Cells were plated with free serum DMEM in each well and incubated in CO2 conditions at 37.5°C. When cells had grown to 90% confluence, the cells were harvested and inoculated into 5 nude mice by subcutaneous injection. Fifteen days after implantation, the tumors were measured using calipers and weighed on a precision electronic balance.
Statistical analysis
All statistical analyses were carried out using SPSS 20 software. Experimental results were analyzed using the χ2 test (or Fisher’s exact test) and one-way ANOVA. All cell culture experiments were performed in triplicate. Data are presented as the mean ± standard deviation (SD). A level of p < 0.05 was considered statistically significant.
Results
Upregulated expression of lncRNA UBE2CP3-001 was observed in gliomas
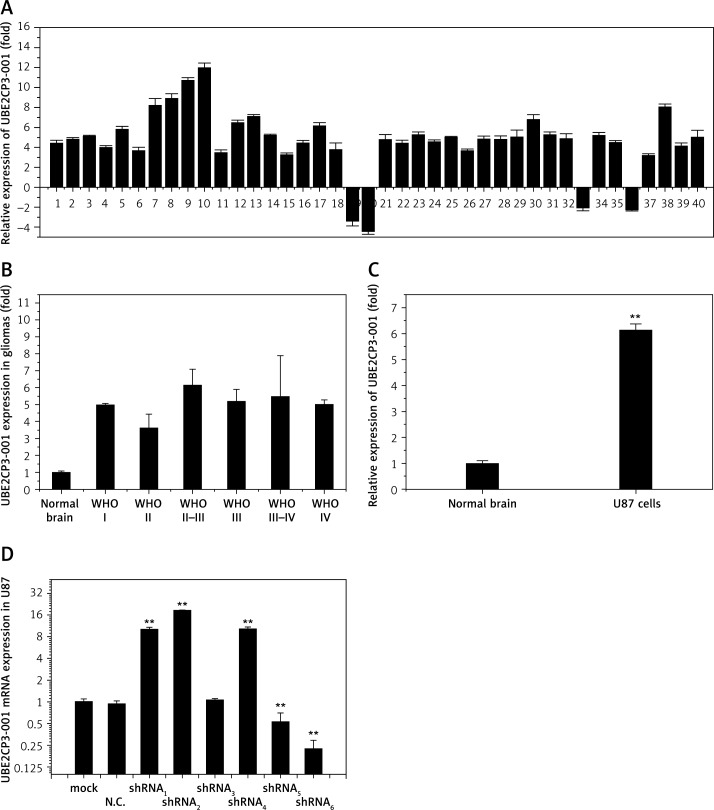
In our previous study, some lncRNAs were found differently expressed between tumor and adjacent normal tissues from glioma patients through lncRNA expression profiling microarrays. Then we examined the lncRNA UBE2CP3-001 expression level using quantitative real-time PCR. Figure 1 A shows that expression of UBE2CP3-001 increased remarkably in 90% of gliomas in comparison with the normal tissue. As shown in Figure 1 B, the expression of UBE2CP3-001 was not positively correlated with histologic grades of gliomas. Statistical analysis indicated that there was no difference in the expression of UBE2CP3-001 at different grades of gliomas (p > 0.05).
Figure 1.
Overexpression of the long non-coding RNA UBE2CP3 in gliomas. A – RNA was extracted with Trizol from human glioma and adjacent tissues. UBE2CP3 expression was assessed in human glioma tissues and adjacent normal tissues using real-time RT-PCR. Bars represent the ratio between expression in glioma and in adjacent normal tissues. B – Statistical analysis shows the expression of UBE2CP3-001 in different grades of gliomas. C – The expression of UBE2CP3-001 in U87 cells compared with normal astrocyte cells was detected using real-time RT-PCR. D – RT-PCR was used to detect lncRNA UBE2CP3-001 expression in U87 stable cells with transfected shRNA1-6
In order to further study the mechanism of UBE2CP3-001 in gliomas, we selected U87 cells as the research object in vitro. Although the commonly used version of U87MG was found to be non-identical to its patient of origin, U87 is a human primary glioblastoma cell line in cell biology [17, 18]. We detected the expression of UBE2CP3-001 in U87 cells, and found it was 6.3-fold higher compared with normal astrocyte cells (Figure 1 C). Then, we designed six kinds of UBE2CP3-001 shRNA (shRNA1-6) and packaged them with lentivirus. UBE2CP3-001 shRNA1-6 were transferred into U87 cells, and the expression of UBE2CP3-001 in the stable U87 cells was detected by RT-PCR. As shown in Figure 1 D, UBE2CP3-001 shRNA6 showed the strongest inhibitory effect, and we used it in the following in vitro and in vivo experiments.
Downregulation of lncRNA UBE2CP3-001 expression suppressed cell migration ability and reduced the invasiveness of glioma in vitro
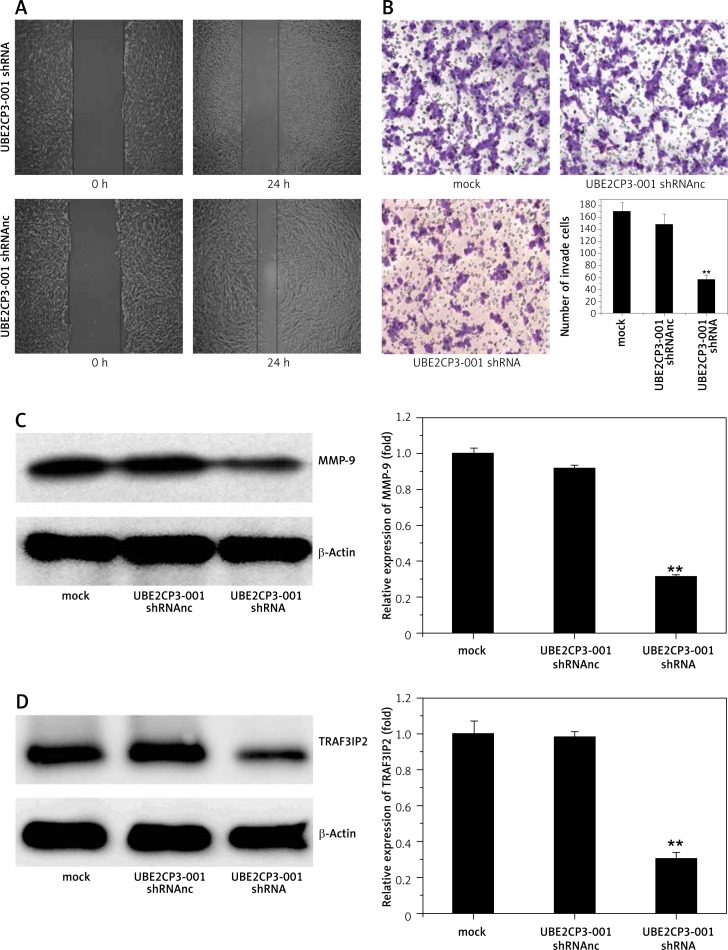
The wound healing assay showed that knockdown of lncRNA UBE2CP3-001 by transfecting UBE2CP3-001 shRNA could effectively suppress the cell migration ability of U87 cells compared with those treated with mock and UBE2CP3-001 shRNAnc (Figure 2 A). To assess the effect of the downregulation of UBE2CP3-001 on the invasiveness of glioma cells, we used a transwell invasion system assay. The number of invasive U87 cells transfected with UBE2CP3-001 shRNA was significantly lower compare to those treated with mock and UBE2CP3-001 shRNAnc (Figure 2 B). These data suggested that knockdown of lncRNA UBE2CP3-001 could effectively inhibit both the migration and invasiveness ability in U87 cells.
Figure 2.
UBE2CP3-001 downregulation inhibited the cell migration and invasiveness of glioma in vitro. A – Cell wound assay shows that downregulation of lncRNA UBE2CP3-001 inhibited cell migration. Cell migration rate of U87 which was transfected with lncRNA UBE2CP3-001 shRNA for 24 h was decreased compared to with U87 cells transfected with lncRNA UBE2CP3-001 negative control (short for UBE2CP3-001 shRNAnc). B – Cell invasive assay shows that downregulation of lncRNA UBE2CP3-001 inhibited cell invasiveness. The data show that the number of invasive cells transfected with lncRNA UBE2CP3-001 shRNA was less than the control cells. C – MMP-9 protein expression was downregulated in U87 transfected with lncRNA UBE2CP3-001 compared to mock cells and negative control cells. D – TRAF3IP2 mRNA expression was downregulated in U87 transfected with lncRNA UBE2CP3-001 shRNA compared to the mock cells and negative control cells
Downregulation of lncRNA UBE2CP3-001 led to downregulated expression of MMP-9 and TRAF3IP2
The wound healing assay and transwell invasion system assay showed that the downregulation of UBE2CP3-001 inhibited migration ability and invasiveness of glioma cells. To further explore the molecular associations between lncRNA UBE2CP3-001 and invasiveness in human glioma, we assayed the expression of MMP-9 after UBE2CP3-001 shRNA treatment using Western blot analysis. As shown in Figure 2 C, data showed that MMP-9 expression was effectively inhibited by downregulation of lncRNA UBE2CP3-001. Interestingly, the network regulation diagram based on the interaction of lncRNAs and target genes suggested that TRAF3IP2 mRNA levels showed a high degree of correlation with lncRNA UBE2CP3-001 expression. As shown in Figure 2 D, our data also showed that downregulation of lncRNA UBE2CP3-001 could inhibit the expression of TRAF3IP2 in U87 cells. Considering that TRAF3IP2 is an activator of the NF-ΚB/MMP9 signaling pathway, we deduced that downregulation of UBE2CP3-001 may suppress NF-ΚB/MMP9 signaling to inhibit proliferation in U87 cells and suppress their migration and invasive abilities.
Downregulation of lncRNA UBE2CP3-001 inhibited glioma cell proliferation and promoted cell apoptosis
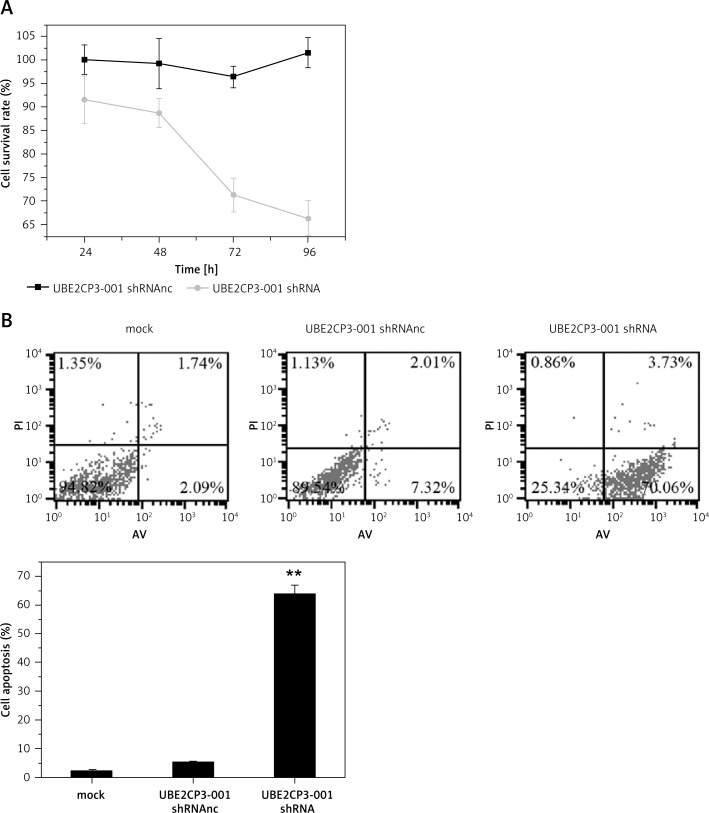
The CCK-8 assay was used to test the effects of UBE2CP3-001 knockdown on U87 cell proliferation. We found that downregulation of lncRNA UBE2CP3-001 in U87 glioma cells by UBE2CP3-001 shRNA transfection resulted in reduced cell proliferation compared to mock and UBE2CP3-001 shRNAnc transfection (Figure 3 A). Furthermore, flow cytometry analysis demonstrated that downregulation of lncRNA UBE2CP3-001 also increased the cell apoptosis of U87 cells after transfection with lncRNA UBE2CP3-001 shRNA compared to mock and UBE2CP3-001 shRNAnc (Figure 3 B).
Figure 3.
UBE2CP3-001 downregulation inhibited cells and induced cell apoptosis of glioma in vitro. A – The cell survival rates of U87 cells transfected with lncRNA UBE2CP3-001 shRNA, negative control or mock at different time points were analyzed by CCK8 assay. B – The cell apoptosis rates of U87 cells transfected with lncRNA UBE2CP3-001 shRNA, negative control or mock were analyzed by flow cytometry analysis
Downregulation of lncRNA UBE2CP3-001 reduced glioma growth in an in vivo mouse model
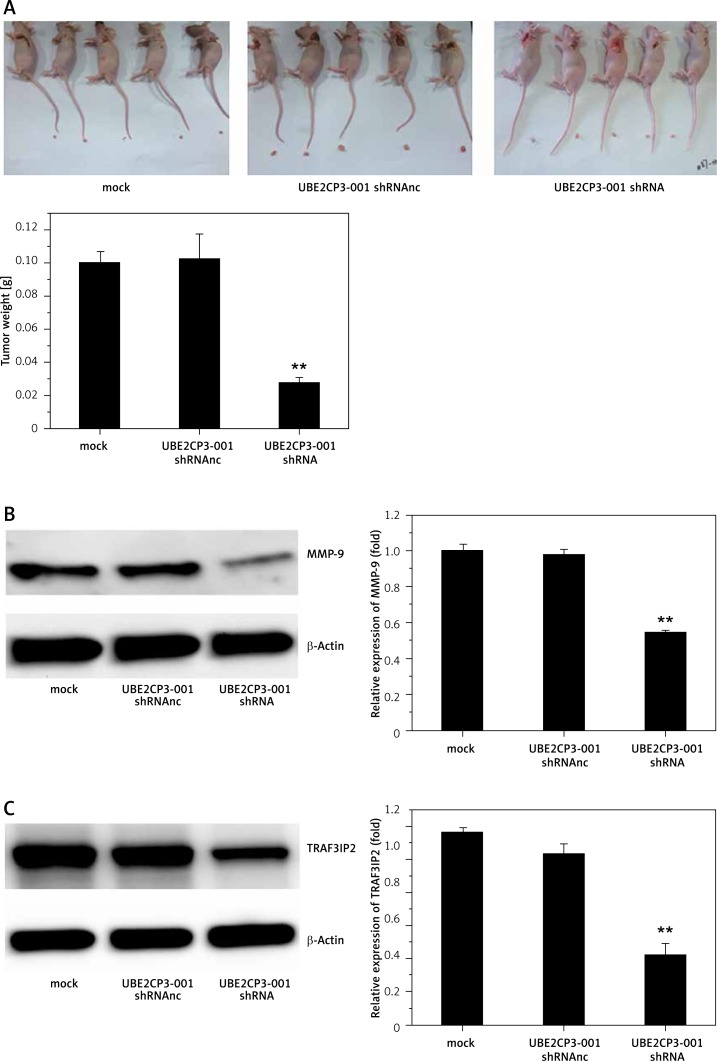
Finally, to test the effects of downregulation of lncRNA UBE2CP3-001 on glioma growth in vivo, we constructed a mouse model. All subcutaneous tumors were removed and weighed by a precision electronic balance. As shown in Figure 4 A, the tumors’ weights were efficiently decreased after UBE2CP3-001 shRNA transfection compared with mock and UBE2CP3-001 shRNAnc transfection. TRAF3IP2 and MMP-9 protein expression levels were also reduced in U87 tumor tissues transfected with lncRNA UBE2CP3-001 shRNA compared with other controls (Figure 4 B).
Figure 4.
UBE2CP3-001 downregulation reduced glioma growth in an in vivo mouse model. A – Tumor weight from xenograft tumors transfected with lncRNA UBE2CP3-001 shRNA decreased compared with tumors from mouse injected with cells transfected with U87 mock and U87 negative control cells. B, C – MMP9 protein and TRAF3IP2 mRNA expression was found to be downregulated in xenograft tumors transfected with lncRNA UBE2CP3-001 shRNA compared to xenograft tumors from transfected with U87 mock and U87 negative control cells
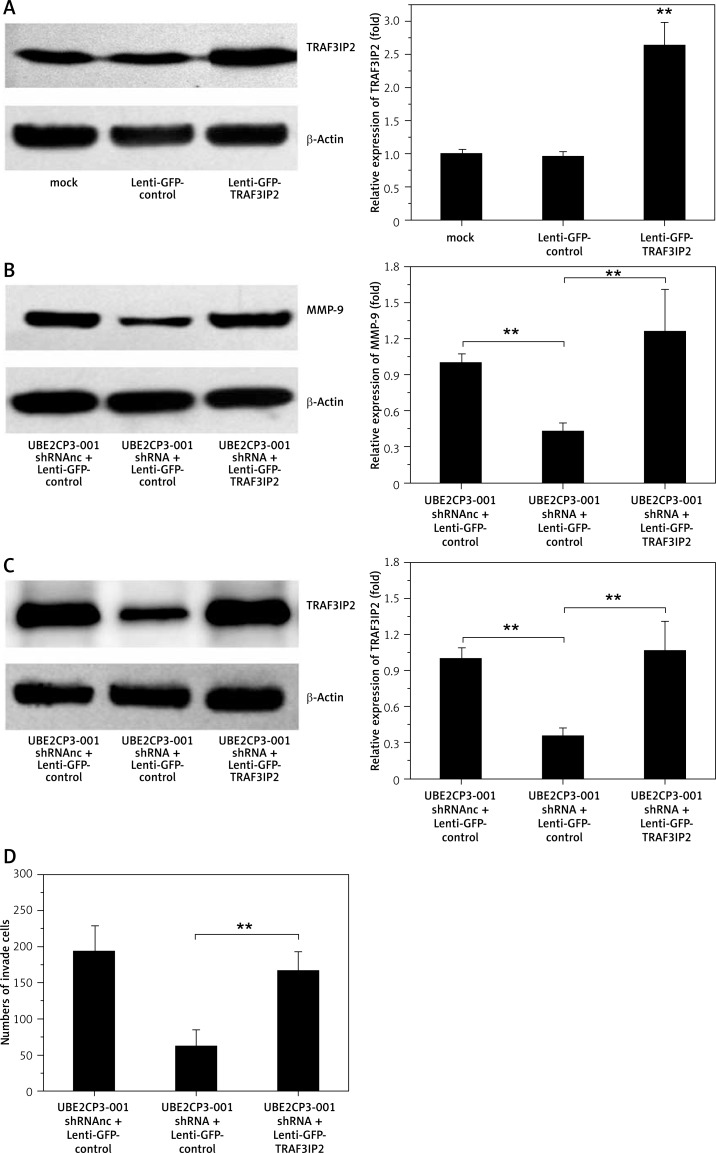
Overexpression of TRAF3IP2 reverses the anti-tumor effects of lncRNA UBE2CP3-001 shRNA in U87 glioma cells
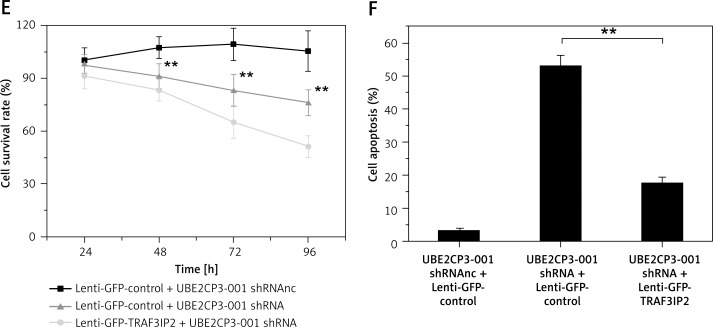
We further examined whether overexpression of TRAF3IP2 counteracts the anti-tumor effects of the lncRNA UBE2CP3-001 shRNA. U87 cells were transfected with the TRAF3IP2 overexpression vector (Lenti-GFP-TRAF3IP2). As shown in Figure 5 A, TRAF3IP2 expression was upregulated after Lenti-GFP-TRAF3IP2 transfection in U87 cells. The downstream of MMP-9 downregulated by UBE2CP3-001 shRNA was also increased after Lenti-GFP-TRAF3IP2 transfection (Figure 5 B), and the downregulation of TRAF3IP2 induced by UBE2CP3-001 shRNA was rescued by Lenti-GFP-TRAF3IP2 transfection (Figure 5 B). More importantly, the resulting overexpression of TRAF3IP2 levels could rescue the cell invasiveness, apoptosis and growth induced by UBE2CP3-001 shRNA (Figures 5 D–F).
Figure 5.
Overexpression of TRAF3IP2 reverses the anti-tumor effects of lncRNA UBE2CP3-001 shRNA. A – U87 cells were transfected with Lenti- GFP-TRAF3IP2, and the expression of TRAF3IP2 was analyzed by Western blot. B – The expression of MMP-9 was analyzed by Western blot after UBE2CP3-001 shRNA and Lenti-GFP-TRAF3IP2 transfection. C – The downregulation of TRAF3IP2 induced by UBE2CP3-001 shRNA rescued in the “UBE2CP3-001 shRNA + lenti-GFP-TRAF3IP2” cell line was measured by Western blot. D – The cell invasiveness of U87 cells upon transfection with UBE2CP3-001 shRNA and/or lenti-GFP-TRAF3IP2 was measured by the Transwell assay. E – The viability of U87 cells upon transfection with UBE2CP3-001 shRNA and/or lenti-GFPTRAF3IP2 was measured by the Cell Counting kit-8 assay. F – The cell apoptosis of U87 cells upon transfection with UBE2CP3-001 shRNA and/or Lenti-GFP-TRAF3IP2 was measured by flow cytometric analysis
Discussion
LncRNAs are non-protein-coding transcripts that contain more than 200 nucleotides [7]. In contrast with other smaller non-coding RNAs, the functions of most lncRNAs are not yet fully understood [19]. LncRNAs are pervasively transcribed and exhibit spatial and temporal regulation expression patterns [20]. With advances in transcriptome profiling, aberrant expression of long non-coding RNA (lncRNA) has been implicated in various diseases, such as cancer [2, 21]. Identification of lncRNAs that are important for disease progression could also result in new options for glioma prognosis and treatment. Recent research has shown that long non-coding RNA expression profiles can predict clinical phenotypes of gliomas [22]. The well-studied lncRNA HOTAIR was first found to be overexpressed in primary breast and metastatic breast cancer [23]. Some studies have shown that HOTAIR is a favorable factor for malignant progression and poor prognosis in glioma patients and exhibits pro-oncogenic activity by modulating the cell cycle [14, 24]. Previous studies suggested that HOTAIR can target PRC2, leading to H3K27me3 modification, and can also promote metastasis [25]. LncRNA MEG3, another imprinted gene, plays an important role in cell proliferation control by interacting with cyclic AMP and p53 [26]. Overexpression of lncRNA MEG3, a growth differentiation factor (GDF15) [26, 27], can impair in vitro glioma cell proliferation [28]. TSLCA-AS1, a novel lncRNA, is also overexpressed in glioma [29]. Through downregulating the expression of TSLCA-AS1, the proliferation abilities and invasiveness of glioma, which is mainly controlled by promoter hypermethylation, can be increased [30].
Taken together, our data show that aberrant expression of lncRNA could be an important element in molecular mechanisms of glioma. In our previous study, using expression profiling microarrays we found that some lncRNAs were differentially expressed between tumor and adjacent tissue. Included among these was lncRNA UBE2CP3-001, which we selected, and it was highly expressed in most gliomas relative to adjacent normal tissues. However, our literature search found no studies that reported the expression profiles or functional roles of lncRNA UBE2CP3-001.
In this study, our data firstly showed that UBE2CP3-001 was overexpressed in most glioma tissues. Downregulation of UBE2CP3-001 in U87 cells inhibited the proliferation and induced the cell apoptosis. Also it could result in reduced cell migration and invasiveness ability. The TRAF3IP2-NF-ΚB/MMP-9 pathway was considered by us to be the underlined anti-glioma mechanism of UBE2CP3-001 knockdown. TRAF3IP2 was reported to be an activator of the NF-ΚB/MMP-9 pathway. Our data showed that downregulation of UBE2CP3-001 could directly inhibit the expression of MMP-9 and TRAF3IP2 proteins. Thus, these results suggested that UBE2CP3-001 knockdown could inhibit activation of the TRAF3IP2-NF-ΚB/MMP-9 pathway.
Conventional therapies for treating glioma, including surgical resection and combined radiation and chemotherapy, have failed to improve the survival rate [3]. Molecular mechanisms mediating invasion may provide a foundation for decreasing glioma recurrence and for prolonging patient survival [31, 32]. Tumor cell invasion and metastatic spread are indicative of advanced tumor progression and depend on changes in cell-cell and cell-matrix adhesion. Thus, the NF-ΚB signaling pathway plays a crucial role in tumor development through the transcriptional regulation of genes associated with tumor growth, invasion, and metastasis [33]. Expression of MMPs, especially MMP2 and MMP9, is upregulated in glioma and correlates with progression, tumor aggressiveness, and poor prognosis [34–36]. Dimerization of the NF-ΚB transcription factor at the ΚB sequence in the MMP9 promoter initiates MMP9 transcription, and then upregulates MMP9 protein expression [37]. Therefore, blocking NF-ΚB transcriptional activity could inhibit glioma cell invasion. In our study, we found that downregulating lncRNA UBE2CP3-001 could suppress glioma growth by regulating MMP9 protein expression in vitro and in vivo. TRAF3IP2 is an upstream regulator of both IKK/NF-ΚB and JNK/AP-1 [38]. Some studies have demonstrated that the TRAF3IP2 gene deletion blunts Ang-II-induced cardiac fibrosis [39]. Thus, we hypothesized that lncRNA UBE2CP3 could affect the NF-ΚB/MMP9 signaling pathway by mediating TRAF3IP2, the activator of NF-ΚB/MMP9 pathways [40].
In conclusion, our results add mechanistic insight regarding the role of a novel lncRNA in the growth and aggressiveness of glioma. LncRNA UBE2CP3-001 can be identified as a positive regulator of glioma in vitro and in vivo, which can control cell growth, proliferation, apoptosis, migration, and invasion. As glioma presents a therapeutic challenge due to the lack of molecular targets, this study suggests the potential application of lncRNA UBE2CP3-001 in glioma therapy.
Acknowledgments
Zhengxiang Luo, Junchen Pan, Yi Ding contributed equally to this work.
This work was supported by the Starting Fund for Young Scientists of Nanjing Public Health Bureau (QYK11126).
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Yang H, Zhang Q, Pei D, Xu F, Li Y, Yu R. FK506-binding protein 5 inhibits proliferation and stimulates apoptosis of glioma cells. Arch Med Sci. 2015;11:1074–80. doi: 10.5114/aoms.2015.54864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang KC, Rao PH, Lau CC, et al. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol Cancer Ther. 2002;1:769–76. [PubMed] [Google Scholar]
- 3.Vlachostergios PJ, Papandreou CN. Efficacy of low dose temozolomide in combination with bortezomib in U87 glioma cells: a flow cytometric analysis. Arch Med Sci. 2015;11:307–10. doi: 10.5114/aoms.2013.36919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maher EA, Furnari FB, Bachoo RM, et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–33. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 5.Yang TQ, Lu XJ, Wu TF, et al. MicroRNA-16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor-kappaB1/MMP9 signaling pathway. Cancer Sci. 2014;105:265–71. doi: 10.1111/cas.12351. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Zhang X, Sun S, Pu JK, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. 2010;19:R162–8. doi: 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mariner PD, Walters RD, Espinoza CA, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Beltran M, Puig I, Peña C, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–69. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakaya HI, Amaral PP, Louro R, et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reis EM, Nakaya HI, Louro R, et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–92. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- 12.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–40. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JX, Han L, Bao ZS, et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol. 2013;15:1595–603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773–9. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, Wang Y, Luan W, et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rock K, McArdle O, Forde P, et al. A clinical review of treatment outcomes in glioblastoma multiforme: the validation in a non-trial population of the results of a randomised phase III clinical trial: has a more radical approach improved survival? Br J Radiol. 2012;85:e729–33. doi: 10.1259/bjr/83796755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark MJ, Homer N, O’Connor BD, et al. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin W, Shi Y, Zhao B, et al. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. doi: 10.1371/journal.pone.0009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–15. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farazi TA, Spitzer JI, Morozov P, Tuschl T. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Sun S, Pu JK, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Lu L, Zhu G, Zhang C, et al. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat. 2012;136:875–83. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 24.Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 25.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim EL, Wüstenberg R, Rübsam A, et al. Chloroquine activates the p53 pathway and induces apoptosis in human glioma cells. Neuro Oncol. 2010;12:389–400. doi: 10.1093/neuonc/nop046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–74. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 29.Houshmandi SS, Surace EI, Zhang HB, et al. Tumor suppressor in lung cancer-1 (TSLC1) functions as a glioma tumor suppressor. Neurology. 2006;67:1863–6. doi: 10.1212/01.wnl.0000244472.56198.84. [DOI] [PubMed] [Google Scholar]
- 30.Qin X, Yao J, Geng P, Fu X, Xue J, Zhang Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int J Clin Exp Pathol. 2014;7:3065–72. [PMC free article] [PubMed] [Google Scholar]
- 31.Nogueira L, Ruiz-Ontañon P, Vazquez-Barquero A, Moris F, Fernandez-Luna JL. The NFkappaB pathway: a therapeutic target in glioblastoma. Oncotarget. 2011;2:646–53. doi: 10.18632/oncotarget.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nogueira L, Ruiz-Ontañon P, Vazquez-Barquero A, et al. Blockade of the NFkappa B pathway drives differentiating glioblastoma-initiating cells into senescence both in vitro and in vivo. Oncogene. 2011;30:3537–48. doi: 10.1038/onc.2011.74. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Ling N, Bai Y, et al. MiR-16-1 plays a role in reducing migration and invasion of glioma cells. Anat Rec (Hoboken. 2013;296:427–32. doi: 10.1002/ar.22626. [DOI] [PubMed] [Google Scholar]
- 34.Wild-Bode C, Weller M, Wick W. Molecular determinants of glioma cell migration and invasion. J Neurosurg. 2001;94:978–84. doi: 10.3171/jns.2001.94.6.0978. [DOI] [PubMed] [Google Scholar]
- 35.Bjorklund M, Koivunen E. Gelatinase-mediated migration and invasion of cancer cells. Biochim Biophys Acta. 2005;1755:37–69. doi: 10.1016/j.bbcan.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Rao JS, Gondi C, Chetty C, Chittivelu S, Joseph PA, Lakka SS. Inhibition of invasion, angiogenesis, tumor growth, and metastasis by adenovirus-mediated transfer of antisense uPAR and MMP-9 in non-small cell lung cancer cells. Mol Cancer Ther. 2005;4:1399–408. doi: 10.1158/1535-7163.MCT-05-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato H, Seiki M. Regulatory mechanism of 92 kDa type IV collagenase gene expression which is associated with invasiveness of tumor cells. Oncogene. 1993;8:395–405. [PubMed] [Google Scholar]
- 38.Valente AJ, Clark RA, Siddesha JM, Siebenlist U, Chandrasekar B. CIKS (Act1 or TRAF3IP2) mediates angiotensin-II-induced interleukin-18 expression, and Nox2-dependent cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2012;53:113–24. doi: 10.1016/j.yjmcc.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valente AJ, Sakamuri SS, Siddesha JM, et al. TRAF3IP2 mediates interleukin-18-induced cardiac fibroblast migration and differentiation. Cell Signal. 2013;25:2176–84. doi: 10.1016/j.cellsig.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu L, Zepp J, Li X. Function of Act1 in IL-17 family signaling and autoimmunity. Adv Exp Med Biol. 2012;946:223–35. doi: 10.1007/978-1-4614-0106-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]