Abstract
Introduction
Genetic variants have been replicated for association with type 2 diabetes mellitus (T2D) and many of them with diabetes-related traits. Because T2D is highly prevalent in Mexico, this study aimed to test the association of CDKN2A/B, PPARGC1A, VEGFA, SIRT1 and UCP2 gene polymorphisms (rs10811661, rs8192678, rs2010963, rs7896005 and rs659366 respectively) with metabolic traits in 415 unrelated Mexican mestizos with T2D under three models of inheritance.
Material and methods
A total of 415 unrelated Mexican mestizos were genotyped by TaqMan assays. Triglycerides, cholesterol, glucose, high-density lipoprotein cholesterol (HDL-C), insulin and anthropometric measurements were determined and the HOMA-IR was calculated. Association studies were tested by the Kruskal-Wallis test, linear regression, statistical power analysis, Bonferroni correction, paired SNP analysis, and physical interaction by GeneMANIA.
Results
All polymorphisms were in Hardy-Weinberg equilibrium, and the association by genotype with T2D-related traits displayed nominal significance for rs8192678 with glucose (p = 0.023) and triglycerides (p = 0.013); rs2010963 with diastolic blood pressure (DBP) (p = 0.012) and cholesterol (p = 0.013); rs7896005 with DBP (p = 0.012) and insulin (p = 0.011); and rs659366 with cholesterol (p = 0.034), glucose (p = 0.031) and triglycerides (p = 0.028); and the association of rs2010963 with HDL-C (p = 0.0007) was significant. Linear regression performed with three models of inheritance, adjusted by age + sex + BMI and corrected with Bonferroni, showed a significant association of rs2010963 with HDL-C in an additive model (p = 0.007); and rs7896005 was significantly associated with DBP in the recessive model (p = 0.006).
Conclusions
Rigorous analysis evidenced the association of VEGFA rs2010963 and SIRT1 rs7896005 with HDL-C and DBP respectively; these traits are known predictors of cardiovascular complications, which increase the risk of cardiovascular diseases in this population.
Keywords: polymorphisms, high-density lipoproteins, diastolic pressure, Mexicans, type 2 diabetes
Introduction
Type 2 diabetes mellitus (T2D) is a complex disease that has a strong impact on populations with genetic predisposition [1, 2], suggesting that susceptibility genes are triggered by shifts in non-genetic factors [3], such as diet and sedentary lifestyles. The risk of developing T2D is 40% for individuals who have one parent with this disease and almost 70% if both parents are affected [4]. Different approaches to understanding the background of the genetic variability in this complex and polygenic disease have been conducted in the past, including the candidate gene approach, linkage analysis, and genome-wide association studies [5], identifying approximately 153 genetic loci associated with risk of T2D and 24 genetic loci related to glycaemic traits [6]. These loci, previously linked to complex traits, should be replicated by various groups around the world and different populations for validation and used as genetic markers in diagnosis, prevention and treatment of the disease. This approach is relevant for Mexican mestizos because of the relatively recent admixture approximately 500 years ago [7].
The polymorphism rs10811661 upstream of CDKN2A/B, which may confer increased risk for T2D by affecting β-cell function, has been associated with T2D in Asian and European subjects in a meta-analysis [8]. PPARGC1A (rs8192678) has been associated with T2D in populations from Denmark and northern China [9, 10], and the UCP2 (rs659366) gene has been associated with T2D and diabetic retinopathy in Shanghai, China [11]. These 2 variants have not been included in association studies with T2D in any Mexican mestizo population. Furthermore, the VEGFA (rs2010963) gene variant has been suggested to play a major role in proliferative diabetic retinopathy in subjects from India [12] and in a Chinese population [13], although is not associated with T2D in a United Kingdom Caucasian population [14]; and the SIRT1 (rs7896005) gene variant had a reduced acute insulin response nominally associated with T2D in a Pima population [15]. Consequently, all five variants have the possibility to be associated with T2D in the Mexican mestizo population.
The aim of this study was to assess the association of metabolic traits with five single nucleotide polymorphisms (SNPs) (rs10811661, rs8192678, rs2010963, rs7896005 and rs659366) located in different genes (CDKN2A/B, PPARGC1A, VEGFA, SIRT1 and UCP2 respectively) in 415 unrelated Mexican mestizos from Mexico City with T2D.
Material and methods
Subjects
Unrelated Mexican mestizos with T2D (301 female and 114 male, with a median age of 56 years) referred to the Metabolic Diseases Research Unit between 2010 and 2014 with signed informed consent were enrolled in the study, which was previously approved by the institutional ethical and research committees of Centro Medico Nacional Siglo XXI of IMSS in Mexico City. A complete medical history of each patient was taken, including anthropometric measures, glucose levels, cholesterol, triglycerides, insulin and glycated haemoglobin (HbA1c). The diagnosis was confirmed according to the clinical criteria for T2D from the American Diabetes Association: fasting glucose greater than 126 mg/dl on more than one occasion, random glucose level higher than 200 mg/dl on at least two occasions or HbA1c greater than 6.5%. The diagnosis of hypertension was made when systolic blood pressure (SBP) was ≥ 140 mm Hg and/or diastolic blood pressure (DBP) ≥ 90 mm Hg [16]. All subjects were considered Mexican mestizos if they were born in Mexico City, as traced back to the third generation [17]. In addition, Mexican mestizos are defined as Mexicans who have a relatively recent admixture (approximately 500 years ago) and exhibit strong genetic components of Native and European origin and, to a lesser extent, Asian and African descent [17]. Recent studies have shown no significant stratification effect in diabetes [18] and cardiovascular [19] studies in Mexican mestizos from Mexico City.
Exclusion criteria
The following exclusion criteria were included in this study: a) presence of clinically significant renal, respiratory, haematological, gastrointestinal, hepatic, neurological or other inherited disorder different to T2D capable of altering the glucose metabolism; b) volunteers born in other states of the country, with less than three generations living in Mexico City; c) self-administration of insulin or another medication to treat diabetes before the T2D diagnosis; d) familial relationship between participants; e) presence of substance abuse or alcoholism; f) pregnant women; g) volunteers with significant incomplete values in demographic or phenotypic data that could affect the statistical analysis.
Anthropometric variables and biochemical assays
Height and waist and hip circumference were measured to the nearest centimetre with a measuring tape. Patients were weighed before breakfast, at the same hour each day, without shoes and in light clothing. Body mass index (BMI) was calculated with the following formula: weight (kg)/height2 (m2). The homeostasis model assessment of insulin resistance index (HOMA-IR) was evaluated according to the HOMA calculator (version 2.2.3 HOMA, Oxford, England). Triglycerides, cholesterol, glucose and high-density lipoprotein cholesterol (HDL-C) were determined using the Biotecnica Automatic Analyser BT3000 (Biotecnica Instruments S.p.A, Rome, Italy). Insulin was determined using the Monobind Human Insulin ELISA (enzyme-linked immunosorbent assay) Kit (Monobind, Inc., Lake Forest, California, USA). Samples from individuals included in this study were obtained before the patient received any medical treatment and all T2D patients were newly diagnosed.
DNA isolation and genotyping
DNA was isolated with the Qiagen Gentra Puregene Blood kit (Qiagen, Hilden, Germany) from human nucleated cells from 5 ml of whole blood and quantified by a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA); five polymorphisms (rs10811661, rs8192678, rs2010963, rs7896005 and rs659366) located in five genes associated with diabetes or related traits in European and Asian populations (Table I) were selected and genotyped using 5′ exonuclease TaqMan assay on a StepOnePlus System (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Ten percent of DNA samples were genotyped twice to confirm concordance.
Table I.
Single nucleotide polymorphisms studied
| Gene | dbSNP | Type of polymorphism | Alleles | Location |
|---|---|---|---|---|
| CDKN2A/B | rs10811661 | Intergenic | T/C | 9:22134095 |
| PPARGC1A | rs8192678 | Missense | C/T | 4:23814039 |
| VEGFA | rs2010963 | 5 prime UTR | C/G | 6:43770613 |
| SIRT1 | rs7896005 | Intron | A/G | 10:67891367 |
| UCP2 | rs659366 | Upstream gene | C/T | 11:73983709 |
dbSNP – the single nucleotide polymorphism database.
Statistical analysis
Statistical analyses were performed with IBM SPSS Statistics 24.0 (SPSS, Inc., Chicago, Illinois, USA). Baseline results were expressed as mean ± standard deviation (SD), and median (25th–75th percentiles) and genotype frequencies were calculated by direct counting. The χ2 test for Hardy-Weinberg equilibrium was used to evaluate the deviation of equilibrium. Comparison of between-group differences in clinical and anthropometric variables was performed with ANOVA or the Kruskal-Wallis test, and categorical variables were analysed with the χ2 or Fisher’s exact test.
Using the software Quanto 1.2.4 (University of Southern California, Los Angeles, California, USA) the main genetic effect (βg) was calculated with a statistical power of 80% and α = 0.05, taking into account the global minor allelic frequency informed in the Single Nucleotide Polymorphism Database (dbSNP) of Nucleotide Sequence Variation (https://www.ncbi.nlm.nih.gov/snp/) and the mean and standard deviation for each trait after normalisation of the values, using the dominant, recessive and additive models of inheritance. Linear regression analysis was used to test the association of polymorphisms and diabetes traits under the three models of inheritance. Those variables with non-normal distribution were rank-based inverse normal transformed to avoid confounding factors due to the preponderance of one sex and obese patients, and the linear regression analysis was adjusted for age, sex, and BMI. Additionally, an association test among paired SNPs was performed, and an interaction analysis between candidate genes associated in the paired SNP analysis was performed with GeneMANIA software (www.genemania.org) [20]. Additionally, a power analysis was performed in the most unbalanced variable. A value of p between < 0.05 and > 0.01 was considered to have nominal significance, and in post-Bonferroni correction 0.05/5, p ≤ 0.01 was significant.
Results
Univariate analysis of risk factors for T2D in Mexican mestizos
The present analysis was performed to determine differences in the clinical, anthropometric and biochemical parameters of 415 (women = 301 and men = 114) unrelated Mexican mestizos with T2D. Table II shows the univariate analysis of the population divided by sex. Both groups (women and men) have a similar distribution in age, and there were no differences in SBP, DBP, glucose, triglycerides, cholesterol, insulin, HDL-C or HbA1c levels. Body mass index was significantly higher in women (29.13 ±7.19) than in men (26.95 ±3.85) with p < 0.001, and HOMA-IR was also substantially higher in women (2.65 (1.69–4.49)) than in men (2.26 (1.41–3.72)), with p < 0.001.
Table II.
Biochemical and anthropometric data of Mexicans with T2D
| Clinical data | All patients (N = 415) | Women (n = 301) | Men (n = 114) | P-value |
|---|---|---|---|---|
| Age [years] | 57.85 ±11.67 | 57.51 ±11.60 | 58.76 ±11.81 | 0.74 |
| BMI [kg/m2] | 28.52 ±6.54 | 29.13 ±7.19 | 26.95 ±3.85 | < 0.001* |
| Weight [kg] | 68 ±13 | 66 ±13 | 73 ±13 | < 0.001* |
| Height [cm] | 154 ±10 | 151 ±7 | 164 ±9 | 0.014 |
| WHR | 0.91 ±0.07 | 0.89 ±0.07 | 0.95 ±0.06 | 0.565 |
| HIP [cm] | 103 ±13 | 105 ±14 | 100 ±9 | < 0.001* |
| Waist [cm] | 97 ±11 | 97 ±11 | 96 ±11 | < 0.001* |
| SBP [mm Hg] | 133.91 ±22.02 | 134.13 ±22.84 | 133.35 ±19.71 | 0.474 |
| DBP [mm Hg] | 75.17 ±11.42 | 74.51 ±11.69 | 76.92 ±10.48 | 0.8314 |
| Glucose [mg/dl] | 129.08 ±63.52 | 128.92 ±60.90 | 129.52 ±29.96 | 0.8307 |
| Cholesterol [mg/dl] | 191.64 ±40.80 | 196.16 ±41.67 | 180.15 ±35.60 | 0.1274 |
| TRG [mmol/l] | 1.90 (1.42–2.77) | 1.93 (1.46–2.76) | 1.08 (1.19–2.84) | 0.1897 |
| HDL [mg/dl] | 39.39 ±10.89 | 41.20 ±11.16 | 34.96 ±8.39 | 0.0926 |
| Insulin [µIU/ml] | 8.15 ±9.75 | 8.59 ±10.68 | 7.12 ±6.57 | 0.1499 |
| HbA1c (%) | 8.26 ±2.58 | 8.23 ±2.60 | 8.35 ±2.53 | 0.4085 |
| HOMA-IR | 2.47 (1.56–4.33) | 2.65 (1.69–4.49) | 2.26 (1.41–3.72) | < 0.001* |
N – total sample size, n – sub-group size, BMI – body max index, WHR – waist to hip ratio, SBP – systolic blood pressure, DBP – diastolic blood pressure, TRG – triglycerides, HDL – high-density lipoprotein, HbA1c – glycated haemoglobin, HOMA-IR – homeostasis model assessment of insulin resistance. Variables are expressed as means ± SD and median (25th–75th percentile).
P-value < 0.05.
Comparison of allelic frequencies with HapMap and 1000 Genomes Project
The five analysed polymorphisms were in Hardy-Weinberg equilibrium. Allelic frequencies of the five polymorphisms are displayed in Table III and were compared to those reported in HapMap and the 1000 Genomes Project for Mexicans. The frequencies of rs10811661, rs8192678, rs659366, rs2010963 and rs7896005 agreed with these projects.
Table III.
Allelic frequencies for six SNPs in diverse studies and patients with T2D
| dbSNP | Actual project | HapMap MXL | 1000 GMXL | 1000 GCEU | 1000 GYRI | 1000 GCHB |
|---|---|---|---|---|---|---|
| rs10811661 | T = 0.9 (749) | T = 0.905 (105) | T = 0.891 (114) | T = 0.801 (181) | T = 0.976 (285) | T = 0.555 (152) |
| C = 0.1 (81) | C = 0.095 (11) | C = 0.109 (14) | C = 0.199 (45) | C = 0.024 (7) | C = 0.445 (122) | |
| rs8192678 | C = 0.8 (665) | C = 0.802 (93) | C = 0.773 (99) | C = 0.65 (147) | C = 0.949 (277) | C = 0.581 (158) |
| T = 0.2 (165) | T = 0.198 (23) | T = 0.227 (29) | T = 0.35 (79) | T = 0.051 (15) | T = 0.419 (114) | |
| rs7896005 | A = 0.68 (562) | A = 0.612 (71) | A = 0.617 (79) | A = 0.288 (65) | A = 0.976 (287) | A = 0.826 (223) |
| G = 0.32 (268) | G = 0.388 (45) | G = 0.383 (49) | G = 0.712 (161) | G = 0.024 (7) | G = 0.174 (47) | |
| rs659366 | C = 0.53 (435) | C = 0.561 (64) | C = 0.562 (72) | C = 0.633 (143) | C = 0.534 (157) | C = 0.544 (147) |
| T = 0.47 (391) | T = 0.439 (50) | T = 0.438 (56) | T = 0.367 (83) | T = 0.466 (137) | T = 0.456 (123) | |
| rs2010963 | C = 0.35 (292) | – | C = 0.336 (43) | C = 0.328 (65) | C = 0.306 (66) | C = 0.447 (92) |
| G = 0.65 (538) | – | G = 0.664 (85) | G = 0.672 (133) | G = 0.694 (150) | G = 0.553 (114) |
dbSNP – the single nucleotide polymorphism database, Actual Project – T2D patients of this study, Data obtained from: MXL – the HapMap project of USA residents with Mexican ancestry, 1000 GMXL – 1000 Genomes Project from USA residents with Mexican ancestry, 1000 CEU – Utah residents with Northern and Western European Ancestry, 1000 YRI – Yoruba from Ibadan, Nigeria, CHB – Han Chinese from Beijing, China.
SNP association with T2D traits
Comparison of genotype frequencies with traits is displayed in Table IV and concerning obesity in Table V. Genotyping frequencies of polymorphisms in patients with T2D are not associated with obesity. These frequencies will be useful as a reference for future studies. Table II displays the results of the association analysis of genotype frequencies and for quantitative traits; nominal significance was identified for rs8192678 (PPARGC1A) with glucose (p = 0.023) and triglycerides (p = 0.013); rs2010963 (VEGFA) with DBP (p = 0.012) and cholesterol (p = 0.013) and a significant association with HDL-C (p = 0.0007); rs7896005 (SIRT1) with DBP (p = 0.012) and insulin (p = 0.01); and rs659366 (UCP2) with cholesterol (p = 0.034), glucose (p = 0.031) and triglycerides (p = 0.028).
Table IV.
Metabolic traits of the samples stratified by genotype
| dbSNP Genotype | BMI [kg/m2] | SBP [mm Hg] | DBP [mm Hg] | CHOL [mg/dl] | Glucose [mg/dl] | TRG [mg/dl] | HDL [mg/dl] | Insulin [mU/ml] | HbA1c (%) | HOMA-IR | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10811661 | T/T | 28 (26–32) | 131 (120–148) | 76 (68.5–83) | 195 (169.5–218) | 119 (94–164.5) | 171 (127.5–245.5) | 39 (34–46) | 9 (6–13) | 8 (7–10) | 2.82 (1.81–4.43) |
| C/T | 28.5 (25–31.75) | 135 (121–151) | 77 (69–82) | 186 (165–214) | 122 (97–155) | 168 (111–214) | 38 (33–44) | 5 (4–9) | 8 (7–10) | 1.74 (0.96–3.88) | |
| C/C | 31 (26–33) | 137 (120–138) | 76 (69–77) | 211 (197–217) | 150 (141–312) | 168 (149–272) | 47 (42–51) | 4 (3–6) | 14 (7–15) | 2.22 (1.18–2.38) | |
| P | 0.966 | 0.096 | 0.988 | 0.089 | 0.093 | 0.08 | 0.086 | 0.111 | 0.133 | 0.089 | |
| rs8192678 | C/C | 28 (26–31) | 132 (120–148) | 76 (68–82) | 187 (168–216) | 121 (95.5–172.5) | 169 (125.5–245) | 38 (33–45) | 7 (4–11) | 8 (7–10) | 2.28 (140–3.85) |
| C/T | 29 (25.5–32) | 134 (123–148) | 76 (69–83) | 199 (171.5–220.5) | 114 (94–150.5) | 173 (131–254) | 42 (35–49) | 10 (7–14) | 8 (7–10) | 3.00 (1.86–4.68) | |
| T/T | 27 (25.5–30.5) | 136 (121.5–158.5) | 79 (70.5–85) | 199 (179.5–224.5) | 130 (97–192.5) | 156 (128–178) | 42 (33–45) | 12.5 (8–24.5) | 8 (7–10) | 4.60 (2.88–6.45) | |
| P | 0.651 | 0.078 | 0.073 | 0.063 | 0.023 | 0.013 | 0.087 | 0.092 | 0.314 | 0.065 | |
| rs2010963 | C/C | 29 (26–32) | 137 (123.5–148.75) | 77 (70–82.75) | 183 (164.75–214.75) | 128 (97–174.5) | 149.5 (109.75–248) | 38 (33.75–47.25) | 4 (3–9) | 8 (7–10) | 1.74 (0.97–3.79) |
| C/G | 29 (26–33) | 130 (118–147) | 74 (67–81) | 188 (169–214) | 119 (96–168) | 169 (127–231) | 38 (33–44) | 7 (5–11) | 8.5 (7–10.75) | 2.33 (1.47–3.64) | |
| G/G | 28 (25–31) | 134.5 (122–148) | 76 (69–83) | 199 (170.75–224.25) | 115.5 (93.75–157) | 174 (132.75–252.25) | 41 (34.75–49) | 10 (7–14) | 8 (7–10) | 3.05 (1.90–4.90) | |
| P | 0.175 | 0.06 | 0.012 | 0.013 | 0.14 | 0.081 | 0.0007* | 0.105 | 0.416 | 0.09 | |
| rs7896005 | A/A | 28 (25–31) | 132 (120–148) | 76 (67–80.25) | 187.5 (168–215.25) | 122 (95–172.25) | 168 (122.75–245.25) | 38 (33–45) | 6 (4–11) | 8 (7–11) | 2.19 (1.30–3.56) |
| A/G | 28 (26–32) | 135 (122.25–147.75) | 76 (69–83) | 195.5 (168–217.75) | 119.5 (96.5–157.75) | 174 (131.25–245.5) | 39 (34–48.75) | 9 (6–13) | 8 (7–10) | 2.65 (1.79–4.58) | |
| G/G | 28 (25–32) | 134 (118.5–154) | 79 (69.5–86) | 203 (182–231) | 110 (92.5–166.5) | 161 (127–248.5) | 42 (35–45) | 12 (10–19.5) | 8 (6.5–10) | 3.72 (2.88–6.33) | |
| P | 0.867 | 0.075 | 0.012 | 0.065 | 0.07 | 0.051 | 0.106 | 0.011 | 0.592 | 0.107 | |
| rs659366 | C/C | 29 (26–32) | 134.5 (120–148) | 76 (68.75–80) | 181 (164–216) | 117.5 (94.5–156.5) | 169.5 (109.75–242) | 32 (28–45) | 5 (4–10) | 8 (7–10) | 1.89 (1.14–3.54) |
| C/T | 28 (25–31) | 130 (120–147.25) | 76 (69–84) | 199 (170.5–217) | 121 (95.75–175.25) | 173 (130–248) | 39 (34–46) | 8 (5–12) | 8 (7–10) | 2.68 (1.82–4.17) | |
| T/T | 28 (26–32) | 136 (123–151) | 76 (69–82.5) | 199 (177–229) | 118 (94–155.5) | 165 (128.5–238) | 42 (35–49) | 11 (7–16) | 8 (7–10) | 3.11 (2.04–6.16) | |
| P | 0.401 | 0.06 | 0.634 | 0.034 | 0.031 | 0.028 | 0.06 | 0.17 | 0.286 | 0.104 | |
dbSNP – the single nucleotide polymorphism database, medians (25–75 percentile), BMI – body max index, SBP – systolic blood pressure, DBP – diastolic blood pressure, CHOL – cholesterol, TRG – triglycerides, HDL – high-density lipoprotein, HbA1c – glycated haemoglobin, HOMA-IR – homeostasis model assessment of insulin resistance. Nominal significance between 0.01 > and < 0.05
Significant p-value < 0.01, after Bonferroni correction.
Table V.
Single-nucleotide polymorphism genotype frequencies in T2D patients with respect to obesity
| Gene and dbSNP | Genotype | UDW | NW | OW | CIO | CIIO | CIII0 | Total |
|---|---|---|---|---|---|---|---|---|
| rs10811661a | T/T | 1 | 69 | 155 | 77 | 29 | 8 | 339 |
| C/T | 0 | 14 | 29 | 18 | 7 | 3 | 71 | |
| C/C | 0 | 2 | 2 | 0 | 1 | 0 | 5 | |
| rs8192678 | C/C | 1 | 49 | 127 | 61 | 23 | 8 | 269 |
| C/T | 0 | 32 | 50 | 2 | 13 | 3 | 100 | |
| T/T | 0 | 4 | 9 | 5 | 1 | 0 | 19 | |
| rs2010963 | C/C | 1 | 9 | 28 | 11 | 8 | 0 | 57 |
| C/G | 0 | 40 | 71 | 46 | 17 | 4 | 178 | |
| G/G | 0 | 36 | 87 | 37 | 12 | 7 | 179 | |
| rs7896005 | A/A | 0 | 38 | 89 | 46 | 18 | 3 | 194 |
| A/G | 0 | 35 | 78 | 20 | 15 | 7 | 155 | |
| G/G | 1 | 12 | 19 | 10 | 4 | 1 | 47 | |
| rs659366a | C/C | 1 | 23 | 63 | 23 | 8 | 3 | 121 |
| C/T | 0 | 41 | 78 | 16 | 22 | 5 | 162 | |
| T/T | 0 | 20 | 44 | 24 | 6 | 3 | 97 |
Polymorphisms associated with obesity in previous studies
UDW – underweight, NW – normal weight, OW – over weight, CIO – class I obesity, CIIO – class II obesity, CIIIO – class III obesity.
SNP association with T2D traits by models of inheritance
All five polymorphisms were studied for association with several traits in linear regression under three models of inheritance (dominant, recessive and additive). Table VI shows the nominal and significant polymorphisms associated with the best model of inheritance after the Bonferroni correction for multiple testing: VEGFA polymorphism rs2010963 was significantly associated with HDL-C (p = 0.007) in the additive model, and with nominal significance with HbA1c (p = 0.020) in the dominant model, and DBP (p = 0.032) in the recessive model. The UCP2 polymorphism rs659366 was nominally significantly associated with SBP (p = 0.025) in the additive model, and polymorphism rs7896005 located in SIRT1 was significantly associated with DBP (p = 0.006) in the recessive model. All models were adjusted for age + sex + BMI, and they were designated following the best Akaike and Bayesian criteria.
Table VI.
Nominal and significant polymorphisms by trait and model of inheritance
| dbSNP | Model | Coef. β (95% CI) | Trait | P-value |
|---|---|---|---|---|
| rs2010963 | Additive | –0.12 (–0.20 – –0.03) | HDL | 0.007* |
| rs2010963 | Dominant | –0.17 (–0.30 – –0.03) | HbA1C | 0.02 |
| rs2010963 | Recessive | 0.05 (0.00–0.09) | DBP | 0.032 |
| rs659366 | Additive | 0.02 (0.00–0.05) | SBP | 0.025 |
| rs7896005 | Recessive | –0.06 (–0.11– –0.02) | DBP | 0.006* |
dbSNP – the single nucleotide polymorphism database, models adjusted by age + sex + BMI, DBP – diastolic blood pressure, HbA1c – glycated haemoglobin, HDL – high-density lipoprotein, SBP – systolic blood pressure, the coefficient β is shown (95% CI) with log-transformed values, nominal significance between 0.01 > and < 0.05
significant p-value < 0.01, after Bonferroni correction.
SNP association with T2D traits by pairwise association test
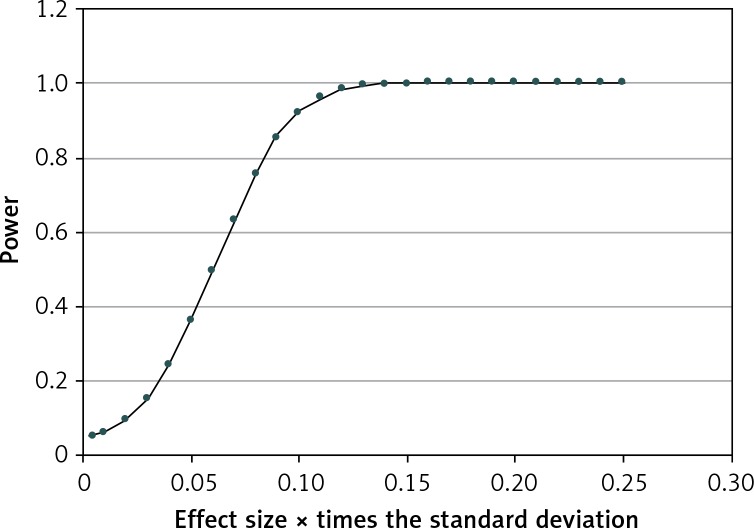
Results of the association test among paired SNPs (Table VII) showed that all SNPs are independent of one another. Further, if the analysis is performed with all the SNPs independently of HOMA-IR, the results are the same, as SNPs are independent of one another. However, a significant value was found for the pair rs8192678 and rs10811661 (exact Pr ≥ χ2 0.966). Finally, because arterial systemic hypertension displayed an unbalanced distribution with a high coefficient of variation, to determine its influence on the results, a power analysis was performed in the unbalanced distribution by dividing the variable of HBP (high blood pressure) into three subgroups (Table VIII and Figure 1). The results showed that a difference between means of around 0.10 times had an SD = 0.55, which can be detected as significant with a probability of 0.92. The genetic effect calculated (Tables IX and X) showed good power (0.80) with an accuracy of approximately 10% in most cases. We marked in bold the percentages greater than 10%.
Table VII.
Association test among paired SNPs
| dbSNP | Exact Pr ≥ χ2 | |
|---|---|---|
| rs8192678 | rs10811661 | 0.9666 |
| rs2010963 | rs10811661 | 0.9301 |
| rs659366 | rs2010963 | 0.693 |
| rs8192678 | rs7896005 | 0.3482 |
| rs659366 | rs8192678 | 0.3005 |
| rs659366 | rs10811661 | 0.287 |
| rs2010963 | rs7896005 | 0.287 |
| rs659366 | rs7896005 | 0.2568 |
| rs8192678 | rs2010963 | 0.1948 |
| rs7896005 | rs10811661 | 0.1005 |
dbSNP – single nucleotide polymorphism database.
Table VIII.
Power analysis for arterial systemic hypertension categorical variable
| Average N per group | Number of groupsa | Total N | α | β | Standard deviation between individuals | Effect size, X times Std. Dev. | Power |
|---|---|---|---|---|---|---|---|
| 138.3 | 3 | 415 | 0.05 | 0.9474 | 0.55 | 0.005 | 0.0526 |
| 138.3 | 3 | 415 | 0.05 | 0.9396 | 0.55 | 0.010 | 0.0604 |
| 138.3 | 3 | 415 | 0.05 | 0.9064 | 0.55 | 0.020 | 0.0933 |
| 138.3 | 3 | 415 | 0.05 | 0.8461 | 0.55 | 0.030 | 0.1531 |
| 138.3 | 3 | 415 | 0.05 | 0.7558 | 0.55 | 0.040 | 0.2427 |
| 138.3 | 3 | 415 | 0.05 | 0.6376 | 0.55 | 0.050 | 0.3600 |
| 138.3 | 3 | 415 | 0.05 | 0.5015 | 0.55 | 0.060 | 0.4954 |
| 138.3 | 3 | 415 | 0.05 | 0.3635 | 0.55 | 0.070 | 0.6332 |
| 138.3 | 3 | 415 | 0.05 | 0.2403 | 0.55 | 0.080 | 0.7565 |
| 138.3 | 3 | 415 | 0.05 | 0.1436 | 0.55 | 0.090 | 0.8537 |
| 138.3 | 3 | 415 | 0.05 | 0.0772 | 0.55 | 0.100 | 0.9209 |
| 138.3 | 3 | 415 | 0.05 | 0.0370 | 0.55 | 0.110 | 0.9618 |
| 138.3 | 3 | 415 | 0.05 | 0.0158 | 0.55 | 0.120 | 0.9835 |
| 138.3 | 3 | 415 | 0.05 | 0.0060 | 0.55 | 0.130 | 0.9937 |
| 138.3 | 3 | 415 | 0.05 | 0.0020 | 0.55 | 0.140 | 0.9979 |
| 138.3 | 3 | 415 | 0.05 | 0.0006 | 0.55 | 0.150 | 0.9994 |
| 138.3 | 3 | 415 | 0.05 | 0.0002 | 0.55 | 0.160 | 0.9998 |
| 138.3 | 3 | 415 | 0.05 | 0.0000 | 0.55 | 0.170 | 1.0000 |
| 138.3 | 3 | 415 | 0.05 | 0.0000 | 0.55 | 0.180 | 1.0000 |
| 138.3 | 3 | 415 | 0.05 | 0.0000 | 0.55 | 0.190 | 1.0000 |
| 138.3 | 3 | 415 | 0.05 | 0.0000 | 0.55 | 0.200 | 1.0000 |
| 138.3 | 3 | 415 | 0.05 | 0.0000 | 0.55 | 0.210 | 1.0000 |
| 138.3 | 3 | 415 | 0.05 | 0.0000 | 0.55 | 0.220 | 1.0000 |
| 138.3 | 3 | 415 | 0.05 | 0.0000 | 0.55 | 0.230 | 1.0000 |
| 138.3 | 3 | 415 | 0.05 | 0.0000 | 0.55 | 0.240 | 1.0000 |
| 138.3 | 3 | 415 | 0.05 | 0.0000 | 0.55 | 0.250 | 1.0000 |
The total number of individuals tested was divided into three groups and means were compared.
Figure 1.
These results are for the worst case that corresponds to the variable HBP and has a high variance coefficient and the largest unbalanced distribution with the gene rs10811661 in the three groups. A difference between means around 0.10 times the standard deviation can be detected as significant with a probability of 0.92
Table IX.
Effect βg calculated with Quanto software by regression coefficient according to three models of inheritance
| Variable | DBP [mm Hg] | Glucose [mg/dl] | HbA1c (%) | CHOL [mg/dl] | HDL [mg/dl] | SBP [mm Hg] | TRG [mg/dl] | HOMA-IR | BMI [kg/m2] | Insulin [mU/ml] | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dbSNP | MAF | N | 406 | 406 | 408 | 406 | 406 | 406 | 406 | 355 | 409 | 357 |
| Mean | 77.4 | 102 | 11.2 | 179 | 49.8 | 143 | 154 | 2.20 | 31.3 | 6.96 | ||
| SD | 11.41 | 63.56 | 2.582 | 40.78 | 10.84 | 22.04 | 165.6 | 3.960 | 5.071 | 9.687 | ||
| rs10811661 | 0.10 | Additive | 3.80 | 21.2 | 0.861 | 13.6 | 3.61 | 7.35 | 55.2 | 1.62 | 1.69 | 3.95 |
| Dominant | 16.22 | 90.3 | 3.670 | 58.0 | 15.41 | 31.33 | 235.4 | 6.89 | 7.21 | 16.86 | ||
| Recessive | 4.11 | 22.9 | 0.931 | 14.7 | 3.91 | 7.95 | 59.7 | 1.75 | 1.83 | 4.28 | ||
| rs2010963 | 0.35 | Additive | 2.39 | 13.3 | 0.541 | 8.5 | 2.27 | 4.62 | 34.7 | 1.02 | 1.06 | 2.49 |
| Dominant | 4.92 | 27.4 | 1.114 | 17.6 | 4.68 | 9.51 | 71.4 | 2.09 | 2.19 | 5.12 | ||
| Recessive | 3.27 | 18.2 | 0.739 | 11.7 | 3.10 | 6.31 | 47.4 | 1.39 | 1.45 | 3.40 | ||
| rs659366 | 0.47 | Additive | 2.29 | 12.7 | 0.517 | 8.2 | 2.17 | 4.42 | 33.2 | 0.97 | 1.02 | 2.38 |
| Dominant | 3.89 | 21.7 | 0.880 | 13.9 | 3.70 | 7.51 | 56.5 | 1.65 | 1.73 | 4.04 | ||
| Recessive | 3.59 | 20.0 | 0.813 | 12.8 | 3.41 | 6.94 | 52.1 | 1.53 | 1.60 | 3.73 | ||
| rs7896005 | 0.32 | Additive | 2.45 | 13.6 | 0.554 | 8.7 | 2.32 | 4.72 | 35.5 | 1.04 | 1.09 | 2.54 |
| Dominant | 5.32 | 29.6 | 1.204 | 19.0 | 5.06 | 10.28 | 77.2 | 2.26 | 2.37 | 5.53 | ||
| Recessive | 3.24 | 18.0 | 0.732 | 11.6 | 3.07 | 6.25 | 47.0 | 1.38 | 1.44 | 3.37 | ||
| rs8192678 | 0.20 | Additive | 2.85 | 15.9 | 0.646 | 10.2 | 2.71 | 5.51 | 41.4 | 1.21 | 1.27 | 2.97 |
| Dominant | 8.23 | 45.9 | 1.863 | 29.4 | 7.82 | 15.91 | 119.5 | 3.50 | 3.66 | 8.56 | ||
| Recessive | 3.36 | 18.7 | 0.761 | 12.0 | 3.19 | 6.49 | 48.8 | 1.43 | 1.49 | 3.50 | ||
dbSNP – the single nucleotide polymorphism database, MAF – minor allele frequency, N – number of individuals per variable, SD – standard deviation, DBP – diastolic blood pressure, HbA1c – glycated haemoglobin, CHOL – cholesterol, HDL – high-density lipoprotein, SBP – systolic blood pressure, TRG – triglycerides, HOMA-IR – homeostasis model assessment of insulin resistance, BMI – body max index. All the traits were rank-based inverse normal transformed. Accuracy greater than 10% is marked in bold numbers.
Table X.
Effect βg calculated with Quanto software as percentage of the mean according to three models of inheritance
| Variable | DBP [mmHg] | Glucose [mg/dl] | HbA1c (%) | CHOL [mg/dl] | HDL [mg/dl] | SBP [mm Hg] | TRG [mg/dl] | HOMA-IR | BMI [kg/m2] | Insulin [mU/ml] | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dbSNP | MAF | N | 406 | 406 | 408 | 406 | 406 | 406 | 406 | 355 | 409 | 357 |
| Mean | 77.4 | 102 | 11.2 | 179 | 49.8 | 143 | 154 | 2.20 | 31.3 | 6.96 | ||
| SD | 11.41 | 63.56 | 2.582 | 40.78 | 10.84 | 22.04 | 165.6 | 3.960 | 5.071 | 9.687 | ||
| rs10811661 | 0.10 | Additive | 4.9 | 20.9 | 7.7 | 7.6 | 7.3 | 5.1 | 35.7 | 73.6 | 5.4 | 56.8 |
| Dominant | 20.9 | 88.9 | 32.8 | 32.4 | 31.0 | 21.9 | 152.4 | 314.0 | 23.0 | 242.2 | ||
| Recessive | 5.3 | 22.6 | 8.3 | 8.2 | 7.9 | 5.6 | 38.7 | 79.6 | 5.8 | 61.4 | ||
| rs2010963 | 0.35 | Additive | 3.1 | 13.1 | 4.8 | 4.8 | 4.6 | 3.2 | 22.5 | 46.3 | 3.4 | 35.7 |
| Dominant | 6.4 | 27.0 | 9.9 | 9.8 | 9.4 | 6.7 | 46.3 | 95.3 | 7.0 | 73.5 | ||
| Recessive | 4.2 | 17.9 | 6.6 | 6.5 | 6.2 | 4.4 | 30.7 | 63.3 | 4.6 | 48.8 | ||
| rs659366 | 0.47 | Additive | 3.0 | 12.5 | 4.6 | 4.6 | 4.4 | 3.1 | 21.5 | 44.3 | 3.2 | 34.1 |
| Dominant | 5.0 | 21.3 | 7.9 | 7.8 | 7.4 | 5.3 | 36.6 | 75.3 | 5.5 | 58.1 | ||
| Recessive | 4.6 | 19.7 | 7.3 | 7.2 | 6.9 | 4.9 | 33.7 | 69.5 | 5.1 | 53.6 | ||
| rs7896005 | 0.32 | Additive | 3.2 | 13.4 | 4.9 | 4.9 | 4.7 | 3.3 | 23.0 | 47.4 | 3.5 | 36.5 |
| Dominant | 6.9 | 29.2 | 10.8 | 10.6 | 10.2 | 7.2 | 50.0 | 103.1 | 7.6 | 79.5 | ||
| Recessive | 4.2 | 17.7 | 6.5 | 6.5 | 6.2 | 4.4 | 30.4 | 62.7 | 4.6 | 48.3 | ||
| rs8192678 | 0.20 | Additive | 3.7 | 15.6 | 5.8 | 5.7 | 5.4 | 3.9 | 26.8 | 55.2 | 4.1 | 42.6 |
| Dominant | 10.6 | 45.1 | 16.6 | 16.4 | 15.7 | 11.1 | 77.4 | 159.4 | 11.7 | 123.0 | ||
| Recessive | 4.3 | 18.4 | 6.8 | 6.7 | 6.4 | 4.5 | 31.6 | 65.1 | 4.8 | 50.2 | ||
dbSNP – the single nucleotide polymorphism database, MAF – minor allele frequency, N – number of individuals per variable, SD – standard deviation, DBP – diastolic blood pressure, HbA1c – glycated haemoglobin, CHOL – cholesterol, HDL – high-density lipoprotein, SBP – systolic blood pressure, TRG – triglycerides, HOMA-IR – homeostasis model assessment of insulin resistance, BMI – body max index. All the traits were rank-based inverse normal transformed. Accuracy greater than 10% is marked in bold numbers.
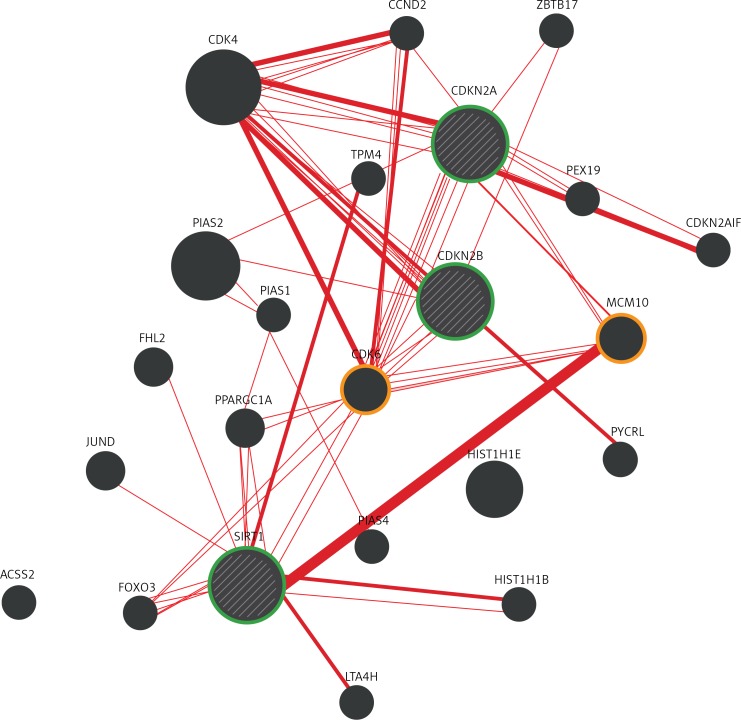
The GeneMANIA bioinformatics analysis for interaction among CDKN2A/CDKN2B and SIRT1 with CDK6 and MCM10 genes are displayed in Figure 2. The results showed that CDKN2A, CDKN2B and SIRT1 interact with CDK6; and CDKN2A and SIRT1 interact with the MCM10 homologue gene (Figure 2).
Figure 2.
Physical interaction analysis for CDKN2A, CDKN2B and SIRT1. This figure displays the analysis of the three genes (green circle) by the GeneMANIA bioinformatics, showing that CDKN2A, CDKN2B and SIRT1 interact with CDK6, and CDKN2A and SIRT1 interact with the homologous gene MCM10
Discussion
Diabetes is the third cause of death in Mexico. It is the leading cause of death in women and the second in men [21]. One of the efforts to counteract the impact of this disease has been to study the genetic influences in the Mexican mestizo population to establish primary prevention measures and improve treatment. Therefore, rs10811661, rs8192678, rs7896005, rs659366 and rs2010963, which have been associated with T2D in European, Chinese, Pima and Maya populations [8–15, 22–25], were selected to study their association with traits related to diabetes.
CDKN2A/B genes are expressed in adipocytes and pancreatic islets and play a role in β-cell function and regeneration. The polymorphism rs10811661 located 125 kb upstream of these genes has been identified as a risk allele in a contemporaneous Mayan population [25] containing lower rates of dispersion and a high predisposition to obesity, diabetes and consanguineous marriage [26]. Our results showed no statistically significant difference in allele frequency of this variant compared to HapMap and the 1000 Genomes Project, or with any of the traits related to T2D. Future analysis of rs10811661 in multi-ethnic Mexican cohorts to elucidate its implications and impact on T2D is suggested.
PPARGC1A is a coactivator of nuclear receptors and other transcription factors that regulate mitochondrial biogenesis, respiration, hepatic gluconeogenesis and muscle fibre type switching [27, 28]. The frequencies obtained for rs8192678 had a difference of 2.2% compared with the 1000 Genomes Project and a difference of 10% with Mexicans from West Mexico (Jalisco State) [29]. The distribution difference obtained between these populations could represent different ancestry and migration history between central and western Mexico. Stratification of genotype frequencies resulted in significant nominal values for glucose (p = 0.023) and triglyceride levels (p = 0.013). This polymorphism has been associated with T2D in populations from China [9], and Denmark [10]; however, it has not been studied widely in Mexican populations for T2D related traits [30]. Consequently, association studies for this gene must be replicated.
Polymorphism rs2010963 in VEGFA has been associated with diseases, such as amyotrophic lateral sclerosis [31]. Allelic frequencies of this polymorphism in Mexicans reported in a case-control study with preeclampsia showed no evidence of an association of VEGFA alleles or haplotype frequencies with this disease [32]. However, diabetes complications, such as retinopathy with vascular implications, have been associated with rs2010963 in case-control studies [12, 13, 33]. Furthermore, metabolic traits analysed in T2D patients of this study showed a significant p-value for low HDL-C levels (Coef β (95% CI); –0.12 (‒0.20 – ‒0.03), p = 0.007) in the additive model and after correcting for multiple comparisons. The impact of the risk allele in the additive model could predispose people with T2D to cardiovascular diseases. A Slovenian group of 143 subjects with T2D and myocardial infarction (MI) was compared with 228 diabetic subjects without the disease, and the polymorphism rs2010963 was associated with MI [34]. These results are in agreement with our results, suggesting an impact of this polymorphism on low HDL-C levels and in the development of MI in patients with T2D. The present findings strengthen the contribution of VEGFA to variation in blood lipid levels, mainly for HDL-C. Additionally, a nominal association with HbA1c (p = 0.020; dominant model) and DBP (p = 0.032; recessive model) was also observed in our study. To the authors’ best knowledge, however, no information has become available on the association of polymorphism rs2010963 with HDL-C levels. These results support the association of rs2010963 in VEGFA with MI in patients with T2D [35]. The identification of genes sharing this covariance can be a target for future functional or clinical-intervention studies [36].
SIRT1 is a stress-response and chromatin-silencing factor, and in Pima Native American populations, rs7896005 has been associated with reduced acute insulin in response to an intravenous glucose bolus (adjusted p = 0.045) [15, 37]. This polymorphism displayed a significant association with DBP (Coef β (95% CI): –0.06 (–0.11 – –0.02), p = 0.006) in the recessive model, suggesting a protective role. This is the first study testing the association between rs7896005 and DBP in Mexicans. In mice, SIRT1 overexpression in vascular smooth muscle cells has been reported to reduce SBP [38], but not DBP.
UCP2 separates oxidative phosphorylation from ATP synthesis with energy dissipated as heat, also referred to as mitochondrial proton leak [39]. Allele frequencies displayed a difference of 3% with the reports from the international projects for Mexicans from Los Angeles, without significance. A recent meta-analysis using three cohorts from Europeans (20,242 individuals) showed that rs659366 was associated with liver dysfunction because γ-glutamyl transferase levels varied by genotype with an interaction with waist-to-hip ratio and body mass index [40]. However, in the present study no association with any of the T2D-related traits was found.
Diabesity is used to refer to a form of diabetes that typically develops due to obesity; however, only a fraction of people with obesity develop diabetes (< 10%) [41]. Previous studies have shown that rs10811661, rs8192678 and rs659366 were associated with obesity and T2D [8–11, 40], but our results did not show an association between these SNPs and obesity in this Mexican mestizo population. Furthermore, when the group of patients with T2D was divided and analysed by the binary variable obesity in the logistic regression, no association with SNPs was found (results not included). Association of the polymorphism rs10811661 at CDKN2A/2B with obesity and diabetes has been replicated in low-medium size samples. Therefore, the lack of association in this study may be due to the sample size or the stratification in the analysis. Nevertheless, some studies have shown that most obese, insulin-resistant individuals do not develop hyperglycaemia, maintaining a healthy β-cell level despite obesity [42, 43]. Furthermore, only three loci have been found to share a strong association (FTO, MC4R and QPCTL/GIPR) and genetic correlation between T2D and obesity [44]; therefore, there remains a need to study traits as new sources of covariance for diabesity.
According to the results of the association test among paired SNPs, for rs8192678 and rs10811661 (0.9666 for exact Pr ≥ χ2), a bioinformatics analysis for genetic interaction was performed to support these results. However, a direct interaction between these SNPs was not found, but identification of intermediary genes, such as the rich human β-cell proliferation CDK6 [45] interaction with these two SNPs in SIRT1 and the CDKN2A/2B locus, was observed. CDK6 codes for a kinase regulated by cyclin D. A mutation in this kinase has been reported to reduce cell proliferation and impair cell motility and polarity (https://www.ncbi.nlm.nih.gov/gene/1021). A stronger interaction was observed between CDKN2A and CDK6 than between CDKN2B and CDK6 (Figure 2). A robust interaction was also observed between SIRT1 and CDKN2A with MCM10, which is required for both initiation and elongation during chromosomal DNA replication [46]. These interactions support a role of regulatory factors during β-cell-cycle progression in the pathogenesis of T2D, and as a result, this may influence insulin synthesis and secretion.
The non-coding regulating polymorphisms in the CDKN2A and SIRT1 genes could affect β-cell replication through the interaction with CDK6 and MCM10 and the regulation of the cell cycle in pancreatic cells. Consequently, additional bioinformatics analysis and functional studies in vitro and in vivo are needed to test this hypothesis.
In conclusion, our results indicated that VEGFA (rs2010963) and SIRT1 (rs7896005) gene polymorphisms are associated with the cardiometabolic traits HDL-C and DBP respectively in T2D patients. These findings may explain the susceptibility to develop cardiovascular complications and derived comorbidity as the main cause of death in Mexican patients with T2D. This association should be replicated in other Mexican populations to strengthen these findings.
Acknowledgments
We thank the study participants, CINVESTAV and CONACYT (scholarship number 382646, for their support to the Ph.D. studies of ATS) and CONACYT (grant number CB258103).
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Knowler WC, Saad MF, Pettitt DJ, Nelson RG, Bennett PH. Determinants of diabetes mellitus in the Pima Indians. Diabetes Care. 1993;16:216–27. doi: 10.2337/diacare.16.1.216. [DOI] [PubMed] [Google Scholar]
- 2.Ravussin E, Valencia ME, Esparza J, Bennett PH, Schulz LO. Effects of a traditional lifestyle on obesity in Pima Indians. Diabetes Care. 1994;17:1067–74. doi: 10.2337/diacare.17.9.1067. [DOI] [PubMed] [Google Scholar]
- 3.Pratley RE. Gene-environment interactions in the pathogenesis of type 2 diabetes mellitus: lessons learned from the Pima Indians. Proc Nutr Soc. 1998;57:175–81. doi: 10.1079/pns19980029. [DOI] [PubMed] [Google Scholar]
- 4.Franks PW, Pearson E, Florez JC. Gene-environment and gene-treatment interactions in type 2 diabetes: progress, pitfalls, and prospects. Diabetes Care. 2013;36:1413–21. doi: 10.2337/dc12-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Köbberling J, Tillil H. Empirical risk figures for first degree relatives of non-insulin dependent diabetics. In: Köbberling J, Tattersall R, editors. The genetics of diabetes mellitus. London: Academic Press; 1982. pp. 201–9. [Google Scholar]
- 6.Prasad RB, Groop L. Genetics of type 2 diabetes – pitfalls and possibilities. Genes (Basel) 2015;6:87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreno-Estrada A, Gignoux CR, Fernández-López JC, et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344:1280–5. doi: 10.1126/science.1251688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Tang X, Liu Q, Wang Y. Association between type 2 diabetes and rs10811661 polymorphism upstream of CDKN2A/B: a meta-analysis. Acta Diabetol. 2013;50:657–62. doi: 10.1007/s00592-012-0400-7. [DOI] [PubMed] [Google Scholar]
- 9.Sun L, Yang Z, Jin F, et al. The Gly482Ser variant of the PPARGC1 gene is associated with type 2 diabetes mellitus in northern Chinese, especially men. Diabet Med. 2006;23:1085–92. doi: 10.1111/j.1464-5491.2006.01949.x. [DOI] [PubMed] [Google Scholar]
- 10.Ek J, Andersen G, Urhammer SA, et al. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to type II diabetes mellitus. Diabetologia. 2001;44:2220–6. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y, Wen Z, Wang N, et al. Investigation of variants in UCP2 in Chinese type 2 diabetes and diabetic retinopathy. PLoS One. 2014;9:e112670. doi: 10.1371/journal.pone.0112670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhuri S, Chowdhury IH, Das S, et al. Role of NF-kappaB activation and VEGF gene polymorphisms in VEGF up regulation in non-proliferative and proliferative diabetic retinopathy. Mol Cell Biochem. 2015;405:265–79. doi: 10.1007/s11010-015-2417-z. [DOI] [PubMed] [Google Scholar]
- 13.Fan X, Wu Q, Li Y, et al. Association of polymorphisms in the vascular endothelial growth factor gene and its serum levels with diabetic retinopathy in Chinese patients with type 2 diabetes: a cross-sectional study. Chin Med J (Engl) 2014;127:651–7. [PubMed] [Google Scholar]
- 14.Freathy RM, Weedon MN, Shields B, et al. Functional variation in VEGF is not associated with type 2 diabetes in a United Kingdom Caucasian population. JOP. 2006;7:295–302. [PubMed] [Google Scholar]
- 15.Dong Y, Guo T, Traurig M, et al. SIRT1 is associated with a decrease in acute insulin secretion and a sex-specific increase in risk for type 2 diabetes in Pima Indians. Mol Genet Metab. 2011;104:661–5. doi: 10.1016/j.ymgme.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James PA, Oparil S, Carter BL, et al. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8) JAMA. 2014;311:507–20. doi: 10.1001/jama.2013.284427. [DOI] [PubMed] [Google Scholar]
- 17.López-Beltrán C, García Deister V. Scientific approaches to the Mexican mestizo. Hist Cienc Saude-Manguinhos. 2013;20:391–410. doi: 10.1590/s0104-597020130002000002. [DOI] [PubMed] [Google Scholar]
- 18.Gamboa-Meléndez MA, Huerta-Chagoya A, Moreno-Macías H, et al. Contribution of common genetic variation to the risk of type 2 diabetes in the Mexican Mestizo population. Diabetes. 2012;61:3314–21. doi: 10.2337/db11-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Posadas-Sánchez R, Pérez-Hernández N, Rodríguez-Pérez JM, et al. Interleukin-27 polymorphisms are associated with premature coronary artery disease and metabolic parameters in the Mexican population: the genetics of atherosclerotic disease (GEA) Mexican study. Oncotarget. 2017;8:64459–70. doi: 10.18632/oncotarget.16223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuberi K, Franz M, Rodriguez H, et al. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013;41:W115–22. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization . Global report on diabetes. Geneva, Switzerland: OMS; 2016. Available at http://apps.who.int/iris/bitstream/10665/204871/1/9789241565257_eng.pdf Accessed November 29, 2017. [Google Scholar]
- 22.Jing C, Xueyao H, Linong J. Meta-analysis of association studies between five candidate genes and type 2 diabetes in Chinese Han population. Endocrine. 2012;42:307–20. doi: 10.1007/s12020-012-9643-x. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Wei M, Wang Q, et al. Association of genetic variants of SIRT1 with type 2 diabetes mellitus. Gene Expr. 2015;16:177–85. doi: 10.3727/105221615X14399878166195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurylowicz A. In search of new therapeutic targets in obesity treatment: sirtuins. Int J Mol Sci. 2016;17:572. doi: 10.3390/ijms17040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lara-Riegos JC, Ortiz-López MG, Peña-Espinoza BI, et al. Diabetes susceptibility in Mayas: evidence for the involvement of polymorphisms in HHEX, HNF4alpha, KCNJ11, PPARgamma, CDKN2A/2B, SLC30A8, CDC123/CAMK1D, TCF7L2, ABCA1 and SLC16A11 genes. Gene. 2015;565:68–75. doi: 10.1016/j.gene.2015.03.065. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez-Escalante VM, Nava Gonzalez EJ, Voruganti VS, et al. Replication of obesity and diabetes-related SNP associations in individuals from Yucatán, México. Front Genet. 2014;5:380. doi: 10.3389/fgene.2014.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Deng X, Shi Y, Su Y, Wei J, Duan H. PGC-1alpha, glucose metabolism and type 2 diabetes mellitus. J Endocrinol. 2016;229:R99–115. doi: 10.1530/JOE-16-0021. [DOI] [PubMed] [Google Scholar]
- 28.Fanelli M, Filippi E, Sentinelli F, et al. The Gly482Ser missense mutation of the peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1 alpha) gene associated with reduced insulin sensitivity in normal and glucose-intolerant obese subjects. Dis Markers. 2005;21:175–80. doi: 10.1155/2005/576748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vázquez-Del Mercado M, Guzmán-Ornelas MO, Corona Meraz FI, et al. The 482Ser of PPARGC1A and 12Pro of PPARG2 alleles are associated with reduction of metabolic risk factors even obesity in a Mexican-Mestizo population. Biomed Res Int. 2015;2015:285491. doi: 10.1155/2015/285491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulloa-Martínez M, Burguete-García AI, Murugesan S, Hoyo-Vadillo C, Cruz-Lopez M, García-Mena J. Expression of candidate genes associated with obesity in peripheral white blood cells of Mexican children. Arch Med Sci. 2016;12:968–76. doi: 10.5114/aoms.2016.58126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambrechts D, Storkebaum E, Morimoto M, et al. VEGF is a modifier of amyotrophic lateral sclerosis in mice and humans and protects motoneurons against ischemic death. Nat Genet. 2003;34:383–94. doi: 10.1038/ng1211. [DOI] [PubMed] [Google Scholar]
- 32.Garza-Veloz I, Castruita-De la Rosa C, Cortes-Flores R, et al. No association between polymorphisms/haplotypes of the vascular endothelial growth factor gene and preeclampsia. BMC Pregnancy Childbirth. 2011;11:35. doi: 10.1186/1471-2393-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stathopoulou MG, Bonnefond A, Ndiaye NC, et al. A common variant highly associated with plasma VEGFA levels also contributes to the variation of both LDL-C and HDL-C. J Lipid Res. 2013;54:535–41. doi: 10.1194/jlr.P030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrovič D, Verhovec R, Globočnik-Petrovič M, Osredkar J, Peterlin B. Association of vascular endothelial growth factor gene polymorphism with myocardial infarction in patients with type 2 diabetes. Cardiology. 2007;107:291–5. doi: 10.1159/000099064. [DOI] [PubMed] [Google Scholar]
- 35.Barbarash O, Gruzdeva O, Uchasova E, Belik E, Dyleva Y, Karetnikova V. Biochemical markers of type 2 diabetes as a late complication of myocardial infarction: a case-control study. Arch Med Sci. 2017;13:311–20. doi: 10.5114/aoms.2017.65240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazaherioun M, Saedisomeolia A, Javanbakht MH, Koohdani F, Eshraghian MR, Djalali M. Beneficial effects of n-3 polyunsaturated fatty acids on adiponectin levels and AdipoR gene expression in patients with type 2 diabetes mellitus: a randomized, placebo-controlled, double-blind clinical trial. Arch Med Sci. 2017;13:716–24. doi: 10.5114/aoms.2016.62139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang F, Kume S, Koya D. SIRT1 and insulin resistance. Nat Rev Endocrinol. 2009;5:367–73. doi: 10.1038/nrendo.2009.101. [DOI] [PubMed] [Google Scholar]
- 38.Gao P, Xu TT, Lu J, et al. Overexpression of SIRT1 in vascular smooth muscle cells attenuates angiotensin II-induced vascular remodeling and hypertension in mice. J Mol Med (Berl) 2014;92:347–57. doi: 10.1007/s00109-013-1111-4. [DOI] [PubMed] [Google Scholar]
- 39.Jastroch M, Divakaruni AS, Mookerjee S, Treberg JR, Brand MD. Mitochondrial proton and electron leaks. Essays Biochem. 2010;47:53–67. doi: 10.1042/bse0470053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vimaleswaran KS, Cavadino A, Verweij N, Nolte IM, Mateo Leach I, Life Lines Cohort Study, et al. Interactions between uncoupling protein 2 gene polymorphisms, obesity and alcohol intake on liver function: a large meta-analysed population-based study. Eur J Endocrinol. 2015;173:863–72. doi: 10.1530/EJE-15-0839. [DOI] [PubMed] [Google Scholar]
- 41.Chadt A, Scherneck S, Joost HG, Al-Hasani H. Molecular Links between Obesity and Diabetes: “Diabesity”. South Dartmouth: MA: 2014. Available at https://www.ncbi.nlm.nih.gov/books/NBK279051/ Accessed November 29, 2017. [Google Scholar]
- 42.Kahn BB, Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106:473–81. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kahn SE. Clinical Review 135: The importance of β-cell failure in the development and progression of type 2 diabetes. J Clin Endocrinol Metab. 2001;86:4047–58. doi: 10.1210/jcem.86.9.7713. [DOI] [PubMed] [Google Scholar]
- 44.Grarup N, Sandholt CH, Hansen T, Hansen T, Pedersen O. Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia. 2014;57:1528–41. doi: 10.1007/s00125-014-3270-4. [DOI] [PubMed] [Google Scholar]
- 45.Fiaschi-Taesch NM, Salim F, Kleinberger J, et al. Induction of human beta-cell proliferation and engraftment using a single G1/S regulatory molecule, cdk6. Diabetes. 2010;59:1926–36. doi: 10.2337/db09-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izumi M, Mizuno T, Yanagi KI, et al. The Mcm2-7-interacting domain of human mini-chromosome maintenance 10 (Mcm10) protein is important for stable chromatin association and origin firing. J Biol Chem. 2017;292:13008–21. doi: 10.1074/jbc.M117.779371. [DOI] [PMC free article] [PubMed] [Google Scholar]